Abstract
Protein posttranslational modifications such as neddylation play crucial roles in regulating protein function. Only a few neddylated substrates have been validated to date, and the role of neddylation remains poorly understood. Here, using Trypanosoma brucei as the model organism, we investigated the function and substrates of TbNedd8. TbNedd8 is distributed throughout the cytosol but enriched in the nucleus and the flagellum. Depletion of TbNedd8 by RNAi abolished global protein ubiquitination, caused DNA re-replication in postmitotic cells, impaired spindle assembly, and compromised the flagellum attachment zone filament, leading to flagellum detachment. Through affinity purification and mass spectrometry, we identified 70 TbNedd8-conjugated and -associated proteins, including known Nedd8-conjugated and -associated proteins, putative TbNedd8 conjugation system enzymes, proteins of diverse biological functions, and proteins of unknown function. Finally, we validated six Cullins as bona fide TbNedd8 substrates and identified the TbNedd8 conjugation site in three Cullins. This work lays the foundation for understanding the roles of protein neddylation in this early divergent parasitic protozoan.
Keywords: DNA replication, mitosis, mitotic spindle, parasitology, Trypanosoma brucei, Cullin, Nedd8, flagellum, neddylation
Introduction
Posttranslational protein modifications play important roles in regulating protein function. Ubiquitination and ubiquitin-like protein modifications have emerged as important regulatory mechanisms in controlling various cellular processes in eukaryotes. Among the ubiquitin-like modifiers, Nedd8 is the closest relative of ubiquitin and is similarly conjugated to its substrates in a process known as neddylation. Analogous to ubiquitination, neddylation also involves the E1 activating enzyme, the E2 conjugating enzyme, and the E3 ligase (1). Nedd8 is synthesized as a precursor that is further processed at the well conserved Gly-76 residue in the C terminus by the Nedd8 carboxyl-terminal hydrolase (1, 2). The exposed C-terminal glycine in Nedd8 is adenylated by the Nedd8-activating enzyme E1 (NAE) composed of the NAE1 (APPBP1) and Uba3 heterodimer. Subsequently, the activated Nedd8 is transferred to the E2 Nedd8-conjugating enzyme Ubc12. Finally, an E3 Nedd8 ligase transfers Nedd8 to the lysine residues of Nedd8 substrates (1). The conjugated Nedd8 can be hydrolyzed by deneddylation enzymes, and the most studied deneddylation enzyme is the COP9 signalosome, which is mainly involved in deneddylation of the Cullin subunit in the Cullin-RING ubiquitin ligase (CRL)5 (3).
Many putative Nedd8-associated proteins in humans have been identified through proteomic approaches (4–7), but their candidacy as genuine Nedd8 substrates remains to be experimentally confirmed. Nevertheless, these studies all demonstrated that Cullins are the most abundant Nedd8 substrates. Other Nedd8 substrates include a number of oncogenic proteins, such as Von Hippel-Lindau (VHL) tumor suppressor, p53, MDM2, BCA3, EGF receptor, and some ribosomal proteins. Neddylation of these proteins regulates transactivation function and protein-protein interaction (p53 and BCA3), increases protein stability (MDM2 and ribosomal proteins), or stimulates ubiquitination (EGF receptor) (1, 8). Intriguingly, some neddylated proteins are substrates or components of the Skp1-Cullin-F box protein (SCF) complex, suggesting an intimate relationship between neddylation and ubiquitination (8).
Trypanosoma brucei, a parasitic protozoan causing human sleeping sickness, expresses a number of ubiquitin-like proteins, such as TbUrm1 (9), TbSUMO (10), TbATG8 (11, 12), and TbNedd8 (Tb927.4.2540). Nothing is known about the function of TbNedd8 in T. brucei or its conjugation system and substrates. Given the unusual biology of T. brucei, investigation of the cellular function of TbNedd8 and identification of its conjugated substrates may uncover novel regulatory pathways, which could provide hints about the evolution of the neddylation pathway.
In this report, we characterized the function of TbNedd8 and identified its conjugated and associated proteins by affinity purification and mass spectrometry. Our work showed that depletion of TbNedd8 caused DNA re-replication, disrupted the spindle, and impaired flagellum-cell body adhesion and identified 70 TbNedd8-conjugated and -associated proteins, including putative TbNedd8 conjugation system enzymes, Cullins, proteins of diverse cellular functions, and proteins of unknown function. This work lays the foundation for understanding the roles of protein neddylation in T. brucei.
Results
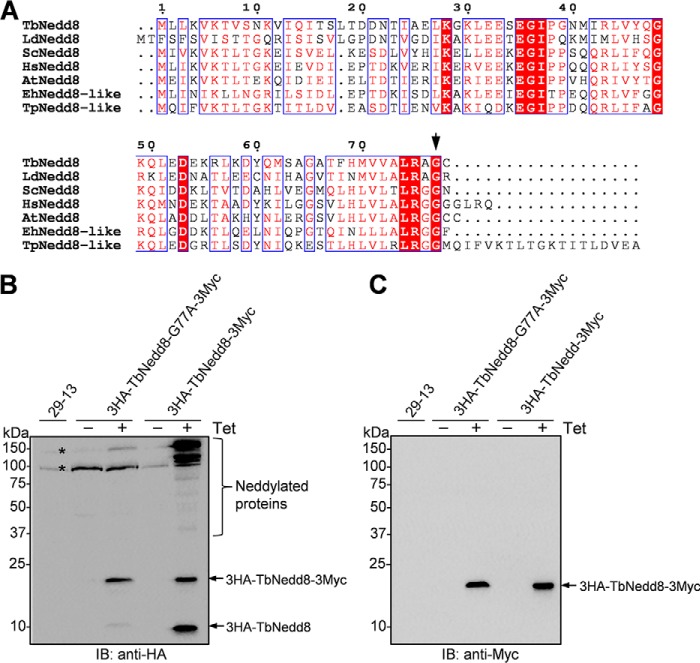
Requirement of the Sole Glycine Residue, Gly-77, for TbNedd8 Maturation
TbNedd8 exhibits a high sequence similarity to the Nedd8 homologs from diverse eukaryotes, but it contains only a single glycine residue, Gly-77, at the C terminus instead of the diglycine motif found in its counterparts from other non-kinetoplastid organisms (Fig. 1A). Gly-77 in TbNedd8 aligns with Gly-76 in other Nedd8 homologs (Fig. 1A), suggesting that the TbNedd8 precursor may be processed at Gly-77 for maturation. To test this possibility, we mutated Gly-77 to alanine and ectopically expressed the G77A mutant and wild-type TbNedd8 in T. brucei. The G77A mutant and wild-type TbNedd8 were each tagged with a triple HA epitope at the N terminus and a triple Myc epitope at the C terminus. In cells expressing the wild-type TbNedd8, Western blotting with anti-HA antibody detected numerous TbNedd8-conjugated proteins and two free TbNedd8 bands, one with a molecular mass of ∼20 kDa and the other with a molecular mass of ∼10 kDa. As expected, in cells expressing the wild-type TbNedd8 (Fig. 1B), Western blotting with anti-Myc antibody detected only the 20-kDa band (Fig. 1C). These results indicate that the 20-kDa band is the precursor containing both the N-terminal 3HA and the C-terminal 3Myc epitopes and that the 10-kDa band is the mature form containing only the N-terminal 3HA tag. In cells expressing the G77A mutant, however, anti-HA immunoblotting detected the 20-kDa band and a faint 10-kDa band (Fig. 1C), suggesting that G77A mutation impaired TbNedd8 processing and maturation.
FIGURE 1.
Requirement of the C-terminal glycine residue for TbNedd8 maturation and protein neddylation. A, Nedd8 sequences from the five eukaryotic supergroups, Excavata (T. brucei and Leishmania donovani), Archaeplastida (A. thaliana), Chromalveolata (Tetrahymena pyriformis), Amoebozoa (Entamoeba histolytica), and Opisthokonta (budding yeast and humans), were aligned. TbNedd8, T. brucei Nedd8 (Q584D3); LdNedd8, L. donovani Nedd8 (E9BQY7); HsNedd8, Homo sapiens Nedd8 (Q15843); ScNedd8, Saccharomyces cerevisiae Nedd8 (A6ZY99); AtNedd8, A. thaliana Nedd8 (O65381); EhNedd8-like, E. histolytica, nedd8-like (N9TFZ1); TpNedd8-like, T. pyriformis Nedd8-like (Q6LDR3). Note that only a single glycine residue (Gly-77, arrow) is present in TbNedd8 and LdNedd8, which was mutated to alanine to test its requirement for TbNedd8 maturation. B and C, requirement of Gly-77 for TbNedd8 maturation and substrate neddylation. Wild-type TbNedd8 and the TbNedd8-G77A mutant were each tagged with an N-terminal triple HA epitope and a C-terminal triple Myc epitope and ectopically expressed in T. brucei in a tetracycline-inducible manner. The crude cell lysate was immunoblotted (IB) with anti-HA mAb (B) and anti-Myc mAb (C), respectively. The asterisks indicate nonspecific bands detected by anti-HA antibody.
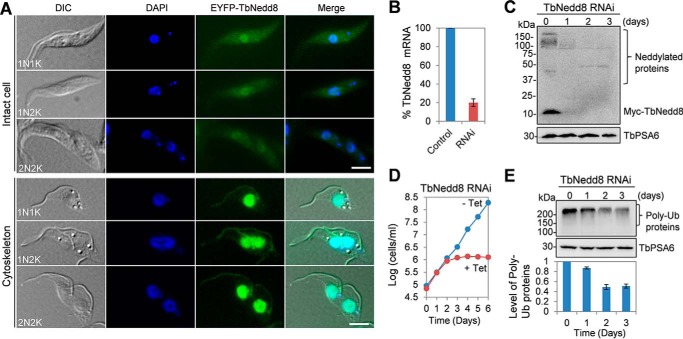
TbNedd8 Localizes to the Nucleus, Flagellum, and Cytosol
To determine the subcellular localization of TbNedd8, we tagged TbNedd8 at the N terminus at one of its endogenous loci in T. brucei and then examined the cells under a fluorescence microscope. EYFP-tagged TbNedd8 appeared to spread throughout the cell and was enriched in the nucleus at all cell cycle stages (Fig. 2A). On detergent-extracted cytoskeletons, TbNedd8 was localized to the flagellum and the nucleus (Fig. 2A). Given that TbNedd8 was detected as both free and conjugated forms (Fig. 1B), these results suggest that either free TbNedd8 or TbNedd8-conjugated proteins or both are present in the cytoplasm, the nucleus, and the flagellum.
FIGURE 2.
Subcellular localization and knockdown of TbNedd8 in the procyclic form. A, subcellular localization of TbNedd8 in procyclic trypanosomes. TbNedd8 was endogenously tagged at the N terminus with EYFP, and the localization of EYFP-TbNedd8 was examined in paraformaldehyde-fixed intact cells and in detergent-extracted cytoskeletons. DIC, differential interference contrast. Scale bars = 5 μm. B, quantitative RT-PCR to monitor the mRNA level of TbNedd8 before and after RNAi induction for 24 h. C, Western blotting to detect the level of TbNedd8 and its conjugated proteins. TbNedd8 was tagged at the N terminus with a Myc epitope at one of the two endogenous loci in cells harboring the pSL-TbNedd8 RNAi construct. The crude cell lysate was immunoblotted with anti-Myc mAb to detect Myc-TbNedd8 and Myc-TbNedd8-conjugated proteins. The same blot was reprobed with anti-TbPSA6 pAb as the loading control. D, effect of TbNedd8 RNAi on cell proliferation. E, effect of TbNedd8 RNAi on global protein ubiquitination (Ub). Control and TbNedd8 RNAi cells were treated with MG-132 for 8 h and then analyzed by Western blotting with anti-ubiquitin mAb. The level of TbPSA6, T. brucei proteasome subunit α-6, served as the loading control. The histogram below the Western blots shows the quantitative data of the level of poly-ubiquitinated proteins, which was normalized against the level of TbPSA6. Error bars indicate standard deviation calculated from three independent experiments.
RNAi of TbNedd8 Abolishes Global Protein Ubiquitination and Causes Mitotic Defects and DNA Re-replication
To investigate the function of TbNedd8, inducible RNAi was carried out in the procyclic form of T. brucei. Quantitative RT-PCR showed that, upon RNAi induction for 1 day, the TbNedd8 mRNA level was decreased ∼80% (Fig. 2B). To further assess the efficiency of TbNedd8 RNAi, TbNedd8 was tagged with a Myc epitope at the N terminus in TbNedd8 RNAi cells, and Western blotting with anti-Myc antibody showed that the level of Myc-TbNedd8 as well as the levels of Myc-TbNedd8-conjugated proteins were significantly decreased on day 1 of RNAi and depleted from day 2 of RNAi (Fig. 2C). This depletion of TbNedd8 resulted in a severe growth defect and eventual cell death after 4 days (Fig. 2D), suggesting that protein neddylation is essential for cell viability in T. brucei.
Given that the main function of Nedd8 is to regulate the Cullin subunit of the CRL-type ubiquitin ligase in diverse eukaryotes, including budding yeast, Arabidopsis thaliana, and humans (13), we examined whether depletion of TbNedd8 affected global protein ubiquitination. Western blotting with anti-ubiquitin antibody showed that the level of polyubiquitinated proteins was gradually decreased to ∼50% of the control level after TbNedd8 RNAi induction for 3 days (Fig. 1E), suggesting that TbNedd8 is required for protein ubiquitination in trypanosomes.
To determine whether the growth defect was attributed to any cell cycle defect, we carried out flow cytometry analysis, which showed that cells with 2C DNA content decreased from ∼55% to ∼16% and cells with 4C DNA content increased from ∼25% to ∼50% after TbNedd8 RNAi induction for 4 days (Fig. 3, A and B), suggesting defective mitosis. Intriguingly, cells with the DNA content greater than 4C but less than 8C also increased from ∼1% to ∼25% after 4 days of RNAi (Fig. 3, A and B), implying that some mitotic cells may have undergone DNA re-replication. To test this possibility, we carried out a BrdU incorporation assay. Non-induced control cells and TbNedd8 RNAi-induced cells were incubated with BrdU for 2 h and then immunostained with anti-BrdU antibody. Within this short incubation time (about of the doubling time of the 29-13 cell line), only cells that are undergoing DNA replication will incorporate BrdU and, therefore, can be detected by anti-BrdU immunostaining. In non-induced control cells, ∼30% of the 1N1K (one nucleus and one kinetoplast) cells, but none of the 2N cells (2N2K), were BrdU-positive (Fig. 3, C and D). In TbNedd8 RNAi cells, ∼32% of 1N1K cells and ∼21% of 2N cells (2N2K and 2N1K) were BrdU-positive (Fig. 3, C and D). These results, together with the flow cytometry data (Fig. 3, A and B), suggest DNA re-replication in mitotic and postmitotic cells upon TbNedd8 RNAi.
FIGURE 3.
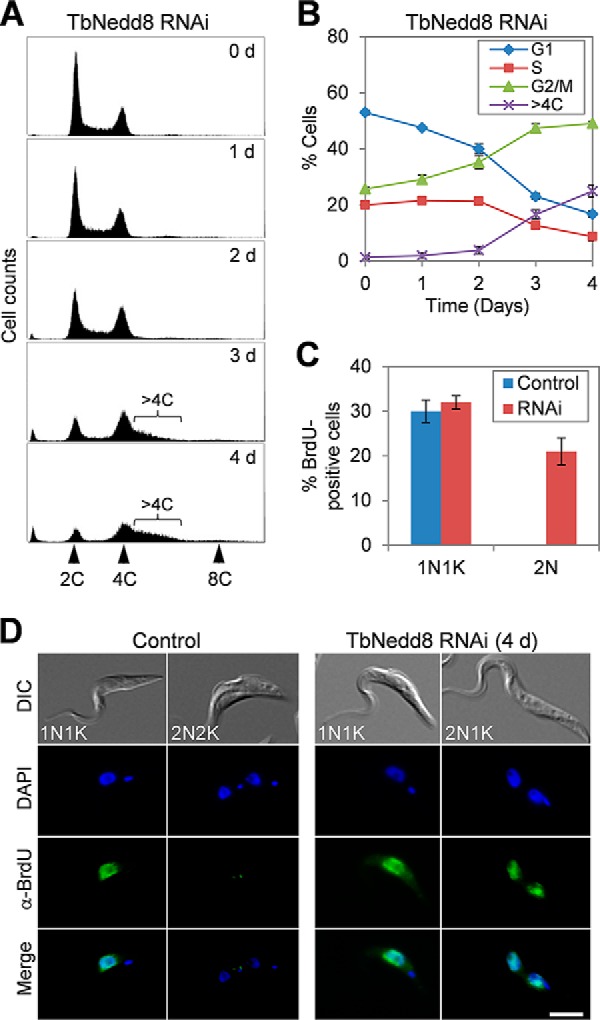
RNAi of TbNedd8 results in DNA re-replication. A, flow cytometry analysis of TbNedd8 RNAi cells. Cells before and after RNAi induction were analyzed by flow cytometry, and shown are the flow cytometry histograms. d, day. B, quantification of flow cytometry assays. The percentage of cells at G1, S, and G2/M phases and the percentage of cells with more than 4C DNA content but less than 8C DNA content were plotted against time (days of RNAi induction). Error bars represent standard deviation calculated from three independent experiments. C, percentage of BrdU-positive cells in control and TbNedd8 RNAi cells (day 3). Data represent the average of three independent experiments, and the error bars represent standard deviation. D, BrdU incorporation in control and TbNedd8 RNAi cells (day 3). Cells were incubated with BrdU for 2 h and immunostained with anti-BrdU mAb. DIC, differential interference contrast. Scale bar = 5 μm.
Depletion of TbNedd8 Impairs Spindle Assembly and Chromosome Segregation
To ascertain which mitotic stage was defective upon TbNedd8 RNAi, we stained the cells with DAPI and counted the number of cells with different numbers of nuclei and kinetoplasts. The results showed that TbNedd8 RNAi led to an increase in 1N2K cells from ∼13% to ∼37%, which was accompanied by a gradual decrease in 1N1K cells from ∼79% to ∼34% after 4 days (Fig. 4A), suggesting that chromosome segregation was defective. To investigate whether this defect in chromosome segregation was attributed to any defect in the spindle structure, we immunostained the cells with KMX-1, which labels β-tubulin in the spindle of mitotic trypanosome cells (14). In non-induced control cells, ∼22% of 1N2K cells contained a metaphase spindle (Fig. 4, B and C), whereas, in TbNedd8 RNAi cells, only ∼3% of 1N2K cells contained a metaphase spindle (Fig. 4, B and C). These results suggest that TbNedd8 RNAi disrupted spindle assembly in 1N2K cells, leading to the increase of 1N2K cells (Fig. 4A). In contrast, there was only a slight decrease in 2N2K cells containing the anaphase spindle after TbNedd8 RNAi (Fig. 4, B and C), suggesting that TbNedd8 depletion did not affect the spindle structure in 2N2K cells, likely because the spindle has already formed in the 2N2K cells when RNAi was induced.
FIGURE 4.
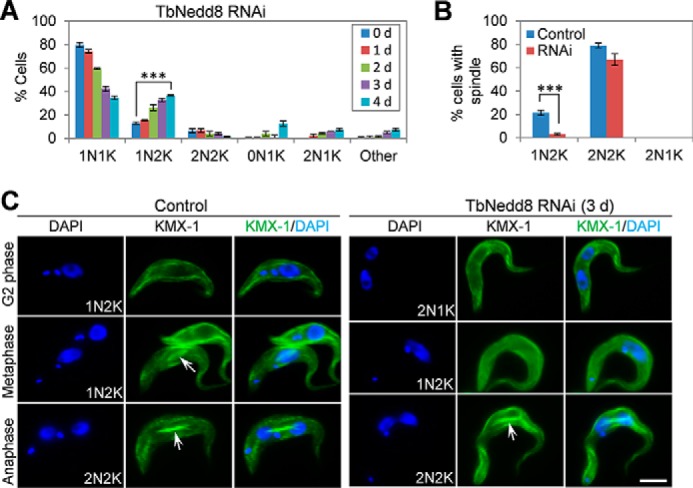
The effect of TbNedd8 RNAi on spindle assembly and chromosome segregation. A, RNAi of TbNedd8 resulted in a significant increase in 1N2K cells. A total of 300 cells from each time point were counted for the numbers of nuclei and kinetoplasts. Error bars indicate standard deviation from three independent experiments. ***, p < 0.001. d, day. B, percentage of cells with visible mitotic spindle structure in control and TbNedd8 RNAi cells. Cells were stained with KMX-1, and 300 cells from control and TbNedd8 RNAi cells (day 3) were counted. Error bars represent standard deviation from three independent experiments. ***, p < 0.001. C, KMX-1 immunostaining to detect the spindle (arrows) in control and TbNedd8 RNAi cells. Scale bar = 5 μm.
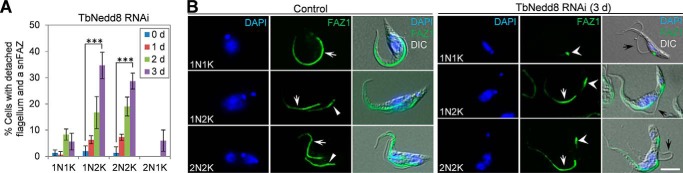
Deficiency in TbNedd8 Disrupts the Elongation of the FAZ Filament, Causing Flagellum Detachment
TbNedd8 RNAi also caused flagellum detachment, which was first observed after day 1 of RNAi but became prominent after RNAi induction for 3 days in 1N2K and 2N2K cells, with ∼35% of the former and ∼29% of the latter possessing a detached flagellum (Fig. 5A). Flagellum detachment in 1N1K and 2N1K cells was also observed but was much less prominent (Fig. 5A). In most cases, the detached flagellum formed a loop, with the anterior tip of the flagellum tethered to the old flagellum (Fig. 5B, 1N2K). In some other cases, the flagellum was fully detached (Fig. 5B, 2N2K). Because the flagellum is attached to the cell body via a specialized cytoskeletal structure known as the FAZ filament (15), and defects in the FAZ filament cause flagellum detachment (16), we investigated the integrity of the FAZ filament in TbNedd8 RNAi cells by immunostaining the cells with L3B2, which detects the FAZ1 protein in the FAZ filament (17). Unlike the control 1N2K and 2N2K cells that all contained two FAZ filaments, the TbNedd8-deficient 1N2K and 2N2K cells contained a full-length, old FAZ filament associated with the attached, old flagellum and a short new FAZ (snFAZ) filament located at the proximal base of the detached, new flagellum (Fig. 5B), indicating defective assembly of the new FAZ filament. This result suggests the involvement of TbNedd8 in maintaining flagellum-cell body adhesion in trypanosomes.
FIGURE 5.
The effect of TbNedd8 RNAi on FAZ filament assembly and flagellum attachment. A, percentage of cells with a detached flagellum and an snFAZ in control and TbNedd8 RNAi cells. Cells were immunostained with anti-FAZ1 antibody, and a total of 300 cells from each time point were counted. Error bars represent standard deviation from three independent experiments. ***, p < 0.001. d, day. B, assembly of the FAZ filament in control and TbNedd8 RNAi cells. Cells were immunostained with anti-FAZ1 antibody and counterstained with DAPI. The white arrows indicate the old FAZ filament, the solid white arrowheads indicate the new FAZ filament, and the open white arrowheads indicate the snFAZ. Black arrows show the detached flagellum. DIC, differential interference contrast. Scale bar = 5 μm.
Identification of TbNedd8-conjugated and -associated Proteins by Affinity Purification and Mass Spectrometry
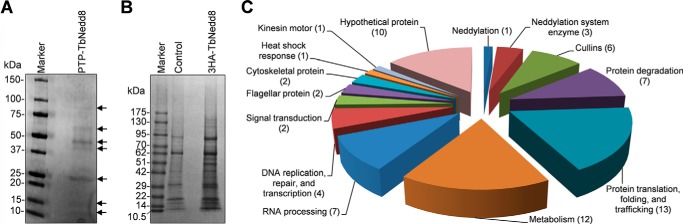
We attempted to identify TbNedd8-conjugated and -associated proteins by proteomic analysis of TbNedd8 co-precipitates. To this end, we first carried out tandem affinity purification using procyclic cells expressing an endogenously PTP-tagged TbNedd8. After two-step purification through the IgG column, tobacco etch virus protease cleavage, and then the protein C column, the final eluate was separated by SDS-PAGE and stained with Coomassie Blue (Fig. 6A). Several protein bands were detected (Fig. 6A, arrows), which were excised from the gel and analyzed by LC-MS/MS. This analysis allowed the identification of 16 proteins, including three Cullins, putative neddylation system enzymes, and a few hypothetical proteins (supplemental Table S1, highlighted in yellow and green). Given that the T. brucei genome encodes at least six Cullin-like proteins and only three of them were identified by tandem affinity purification, it suggests that this method was inefficient to identify TbNedd8-conjugated and -associated proteins.
FIGURE 6.
Identification of TbNedd8-conjugated and -associated proteins by affinity purification and mass spectrometry. A, procyclic cells expressing PTP-tagged TbNedd8 were lysed by sonication, and the cell lysate was used for tandem affinity purification. The final EGTA/EDTA eluate was loaded onto SDS-PAGE and stained with Coomassie Blue. Arrows indicate the protein bands that were analyzed by LC-MS/MS. B, immunoprecipitation of 3HA-TbNedd8-conjugated and -associated proteins for LC-MS/MS. Control cells and cells expressing 3HA-TbNedd8 were lysed and then incubated with anti-HA-agarose beads. C, putative TbNedd8-conjugated and -associated proteins identified by affinity purification were grouped according to their molecular functions. The numbers in the parenthesis indicate the numbers of proteins in each function group.
We next carried out immunoprecipitation with HA-tagged TbNedd8. To this end, TbNedd8 was tagged with a triple HA epitope at the N terminus, and immunoprecipitation was performed with anti-HA-agarose beads. The immunoprecipitates from the control cells and 3HA-TbNedd8-expressing cells were separated on SDS-PAGE and stained with Coomassie Blue (Fig. 6B). Gel slices from both the control and 3HA-TbNedd8 immunoprecipitates were then digested with trypsin and analyzed by LC-MS/MS. Three independent immunoprecipitations and mass spectrometry analyses were performed, and only proteins that were detected in all three repeats and not in the control immunoprecipitation were considered putative TbNedd8-conjugated and -associated proteins.
Together, these proteomic approaches identified a total of 70 putative TbNedd8-conjugated and -associated proteins (Fig. 6C and supplemental Table S1). The affinity purification appeared to be very efficient, as it identified six Cullin homologs, which are the best known Nedd8 substrates in humans, and the enzymes involved in neddylation (TbUba3, TbAPPBP1, and TbUbc12), suggesting conservation of the neddylation pathway in T. brucei and humans. Additionally, three proteins, DNA damage binding protein 1 or DDB1, translation elongation factor α1, and a chaperone protein, DnaJ, which have been reported previously to co-precipitate with human Nedd8 (4), were also co-precipitated with TbNedd8. It should be noted that DDB1 has been identified previously as a component of the CUL4-containing CRL ubiquitin ligase complex, which is involved in the degradation of Cdt1 in animals (13) and was co-purified with Nedd8 in humans (4). Therefore, the DDB1 homolog in T. brucei is probably not a substrate of TbNedd8 but a component of the CUL4-containing complex and, therefore, was co-precipitated with TbNedd8-conjugated TbCUL4.
Subunits of the 19S proteasome regulatory complex were also co-precipitated with TbNedd8 (Fig. 6C and supplemental Table S1). It is likely that the 19S proteasome subunits make transient physical contact with the CRL-type ubiquitin ligase, which ubiquitinates substrate proteins for degradation by the proteasome. The rest of the TbNedd8 co-precipitated proteins are either involved in diverse cellular processes, including protein translation, folding, and trafficking, DNA replication and repair, and heat shock response, etc., or are hypothetical proteins of unknown function (Fig. 6C and supplemental Table S1).
Sec13 and Sec23, two essential subunits of the coat protein complex II, were also co-precipitated with TbNedd8, suggesting that they may be neddylated. Alternatively, they could be co-precipitated with a CRL ubiquitin ligase complex because the human CRL complex CUL3-KLHL12 was found to interact with the coat protein complex II (18).
The Six Cullin-like Proteins, TbCUL1-TbCUL6, Are Bona Fide Substrates of TbNedd8
Cullins are known substrates of Nedd8 in yeast and humans and are conjugated by Nedd8 at a conserved lysine residue at the C terminus (4–7). Our affinity purification and mass spectrometry analysis of TbNedd8-conjugated and -associated proteins identified six Cullin-like proteins, named TbCUL1-TbCUL6, that all contain the conserved Nedd8-conjugating lysine residue at the C terminus (Fig. 7A), suggesting that these Cullins are likely to be conjugated by TbNedd8. To validate the candidacy of the six Cullins as TbNedd8 substrates, they were each tagged with a triple HA epitope at the N terminus and ectopically expressed in T. brucei. The 3HA-tagged Cullins were then immunoprecipitated with anti-HA antibody and detected by Western blotting with anti-HA antibody and anti-TbNedd8 antibody. The results showed that anti-HA Western blotting detected at least two bands for all six Cullins, with the slowest migrating band labeled by the anti-TbNedd8 antibody (Fig. 7B, arrowheads), indicating that the slowest migrating band is the neddylated form of these Cullins.
FIGURE 7.
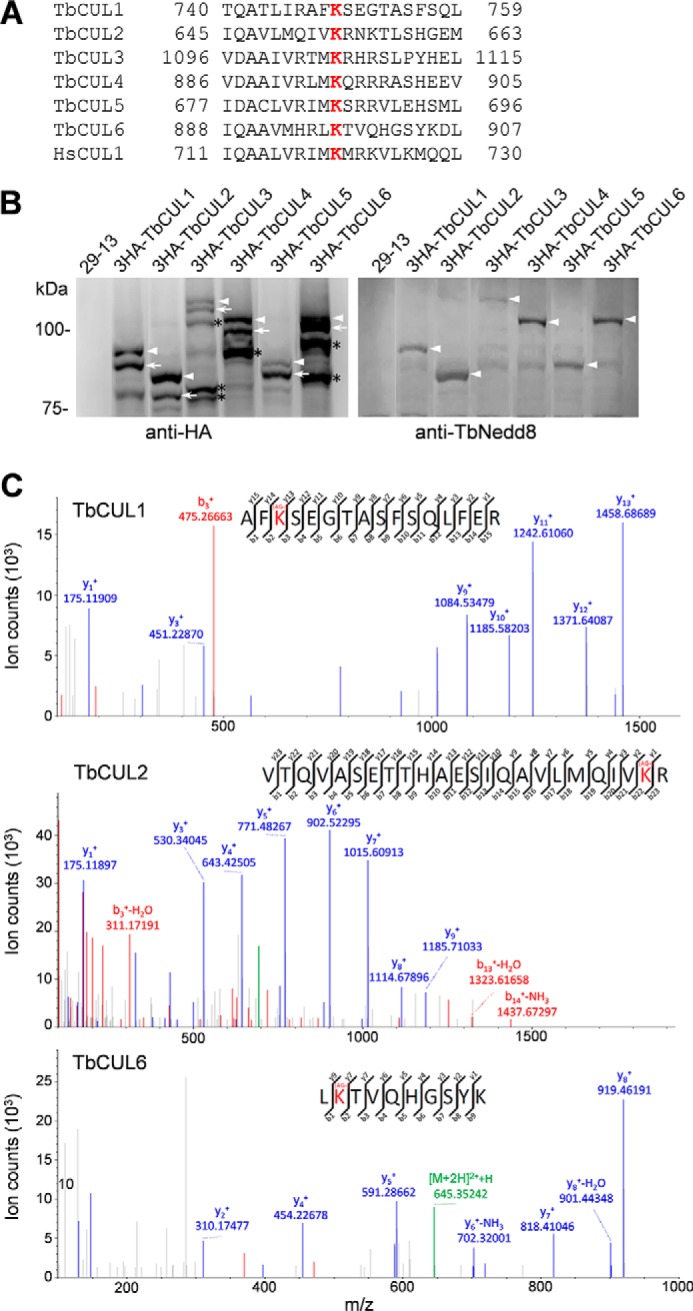
Confirmation of Cullins as TbNedd8 substrates and identification of TbNedd8-conjugated residue in Cullins. A, alignment of the conserved TbNedd8-conjugating lysine residue (red) of the six T. brucei Cullin homologs (TbCUL1-TbCUL6) with that of the human CUL1 (HsCUL1). B, Cullin homologs in T. brucei are neddylated in vivo in trypanosomes. The six Cullin homologs, TbCUL1-TbCUL6, were each tagged with an N-terminal triple HA epitope and ectopically expressed in T. brucei. 3HA-tagged proteins were immunoprecipitated from crude cell lysate and then immunoblotted with anti-HA monoclonal antibody and anti-TbNedd8 polyclonal antibody. Arrows indicate the non-conjugated form of Cullins, and arrowheads indicate the TbNedd8-conjugated form of Cullins. The asterisks indicate the truncated forms of TbCUL3, TbCUL4, and TbCUL6. C, MS/MS spectra of tryptic peptides. The sequences were determined as AFK*SEGTASFSQLFER, VTQVASETTHAESIQAVLMQIVK*R, and LK*TVQHGSYK, corresponding to TbCUL1, TbCUL2, and TbCUL6, respectively. K*, lysine residue modified with arginine-glycine.
The neddylated substrates that have been digested with trypsin are characterized by the remnant of two glycine residues (GG) attached to an internal lysine residue of the substrate, which can be determined by MS/MS, similar to that for identifying the ubiquitin-conjugated residues (19). Given that TbNedd8 contains a sole glycine residue immediately following an alanine residue (Fig. 1A), we searched for the Cullin peptide(s) that contain(s) the AG-modified lysine residue and identified such a peptide for TbCUL1, TbCUL2, and TbCUL6 (Fig. 7C). The TbNedd8-conjugated lysine residue in these three Cullins appeared to be well conserved in all six T. brucei Cullins and aligned well with the neddylation sequence of the human CUL1 (Fig. 7A), suggesting that Nedd8-conjugation of Cullins is evolutionarily conserved from the early-branching T. brucei to humans.
Identification of Putative TbNedd8 Conjugation Enzymes
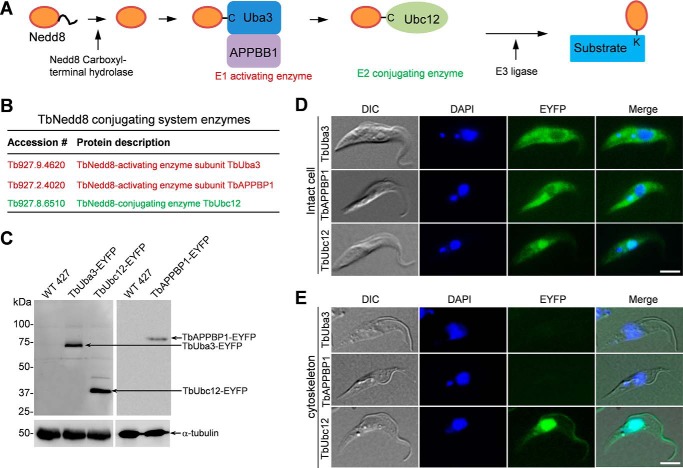
The maturation of Nedd8 and subsequent conjugation of mature Nedd8 to its substrates involve four enzymes, the Nedd8 carboxyl-terminal hydrolase, the Nedd8-activating E1 enzyme complex composed of Uba3 and APPBP1, the Nedd8 conjugating E2 enzyme Ubc12, and the Nedd8 E3 ligase (Fig. 8A) (8). The trypanosome genome encodes 16 putative ubiquitin C-terminal hydrolases, 9 putative E1 ubiquitin-activating enzymes, 21 putative E2 ubiquitin-conjugating enzymes, 11 HECT-containing ubiquitin ligases, and 69 RING finger- and U box-containing proteins that are putative E3 ligases, but it is unclear which enzyme is involved in neddylation. To examine whether TbNedd8 conjugation system enzymes were precipitated by TbNedd8 affinity purification, we searched the list of TbNedd8-conjugated and -associated proteins (supplemental Table S1) for putative conjugation enzymes and identified two subunits of ubiquitin-activating enzyme (Tb927.9.4620 and Tb927.2.4020) that are homologous to Uba3 (supplemental Fig. S1) and APPBP1 (supplemental Fig. S2), respectively, and an ubiquitin-conjugating enzyme (Tb927.8.6510) that is homologous to Ubc12 (supplemental Fig. S3). We named them TbUba3, TbAPPBP1, and TbUbc12, respectively (Fig. 8B).
FIGURE 8.
Subcellular localizations of the neddylation enzymes. A, enzymes involved in Nedd8 maturation and conjugation to substrates. B, TbNedd8 conjugating enzymes identified by affinity purification and mass spectrometry. C, Western blotting to monitor the expression of EYFP-tagged TbUba3, TbUbc12, and TbAPPBP1. EYFP-tagged proteins were detected by anti-GFP antibody. Levels of α-tubulin served as the loading control. D and E, subcellular localization of TbAPPBP1, TbUba3, and TbUbc12 in intact cells (D) and on the cytoskeleton (E). TbAPPBP1, TbUba3, and TbUbc12 were each tagged with a C-terminal EYFP from the endogenous locus and visualized under the fluorescence microscope. DIC, differential interference contrast. Scale bars = 5 μm.
We next investigated the subcellular localization of TbUba3, TbAPPBP1, and TbUbc12 by C-terminal EYFP tagging in their respective endogenous loci. Expression of EYFP-tagged proteins was confirmed by Western blotting (Fig. 8C). In intact cells, TbAPPBP1 and TbUba3 appeared to spread throughout the cell body but were less abundant in the nucleus (Fig. 8D). In contrast, TbUbc12 was highly enriched in the nucleus but less abundant in the cytosol (Fig. 8D), which resembles the localization of TbNedd8 (Fig. 2A). On detergent-extracted cytoskeletons, EYFP-tagged TbAPPBP1 and TbUba3 were not detectable (Fig. 8E), indicating that the two E1 components do not associate with the cytoskeleton. In contrast, EYFP-tagged TbUbc12 was detected in the flagellum and the nucleus (Fig. 8E), similar to the localization of EYFP-tagged TbNedd8, which further confirms that protein neddylation occurs in the flagellum.
Discussion
In this work, we report the functional characterization of the Nedd8 homolog in T. brucei and demonstrate the involvement of TbNedd8 in DNA replication, spindle assembly, and FAZ filament assembly (Figs. 3–5). Among these, the requirement of Nedd8 for preventing DNA re-replication has been well understood in other systems (20, 21). In animals, degradation of the DNA licensing factor Cdt1, which is mediated by the CRL-type ubiquitin ligase complexes SCF-Skp2 and CUL4-DDB1, prevents DNA re-replication (22, 23). Because neddylation activates Cullin in the SCF complex, inhibition of Cullin neddylation results in stabilization of Cdt1 and, hence, promotes DNA re-replication (20). In T. brucei, however, the Cdt1 homolog has not been identified, and the mechanisms underlying DNA replication licensing remain largely elusive (24). It is possible that T. brucei expresses either a highly divergent Cdt1 ortholog that has escaped a homology-based search or a trypanosome-specific functional, but not structural, homolog of Cdt1, as are the cases of the chromosomal passenger complex (25) and the origin recognition complex (26, 27), both of which are composed of some trypanosome-specific proteins.
The finding that TbNedd8 RNAi caused defective spindle assembly and chromosome segregation (Fig. 4) is unexpected because similar defects have not been reported in any other organisms. Although the underlying mechanisms remain elusive, this interesting observation prompted us to postulate that TbNedd8 or neddylation of a regulator of spindle assembly is required for spindle formation. Alternatively, the result may suggest the involvement of CRL-type ubiquitin ligase activity in spindle assembly. Indeed, components of the CRL-type ubiquitin ligase complex have been detected previously in centrosomes in animal and human cells and are required for centrosome duplication (28–30). Unlike animals, however, trypanosomes undergo a closed mitosis and do not possess centrosomes. Therefore, assembly of the mitotic spindle is probably achieved through mechanisms distinct from that in animals. On the other hand, although it is well accepted that Nedd8 plays essential roles in cell cycle control in fungi and animals, the primary function of Nedd8 is to promote S phase entry by activating the CRL-type ubiquitin ligase for degradation of the S phase CDK inhibitor Sic1p in yeast and p27kip1 in animals (31, 32). Strikingly, trypanosomes appear to lack S phase CDK and cyclin (24), and the currently available experimental evidence suggests that an S phase CDK inhibitor is probably not present in trypanosomes because inhibition of proteasome activity does not compromise G1/S transition but, instead, arrests cells at the G2/M boundary, likely because of the incapability of degrading the anaphase inhibitor securin ortholog (33, 34).
Another intriguing phenotype caused by TbNedd8 RNAi is defective FAZ filament assembly and flagellum detachment (Fig. 5). Assembly of the FAZ filament is known to require many FAZ filament proteins, including FAZ1 (17), FAZ2 (35), CC2D (16), FLA1 (36), FLA3 (37), and KMP-11 (35) and the flagellar protein FLAM3, which appears to constitute a component of the flagellum-FAZ connector linking the flagellum to the FAZ filament (38). The defective flagellum adhesion caused by TbNedd8 RNAi (Fig. 5) and the localization of TbNedd8 and TbUbc12 to the flagellum (Figs. 2A and 8E) suggest an involvement of the neddylation pathway in maintaining flagellum-cell body adhesion, likely by regulating certain flagellar protein(s) that is/are required for flagellum attachment. Indeed, two flagellar proteins were co-precipitated with TbNedd8 (Fig. 6C and supplemental Table S1), which may be TbNedd8 substrates or may associate with TbNedd8 or with neddylated proteins.
The subcellular localizations of TbNedd8 are correlated with its functions. The enrichment of TbNedd8 in the nucleus (Fig. 2A) suggests its involvement in certain nuclear events, such as DNA replication and mitosis, which was further corroborated by RNAi studies (Figs. 3 and 4). Localization of TbNedd8 to the flagellum (Fig. 2A) indicates its role in flagellum-related function(s), and, indeed, RNAi of TbNedd8 caused flagellum detachment (Fig. 5). Because TbNedd8 was detected as free molecule and protein conjugates (Figs. 1B and 5), the observed subcellular distribution of TbNedd8 may represent the localization of TbNedd8-conjugated protein(s) or the localization of both free TbNedd8 and TbNedd8-conjugated protein(s). For example, TbUbc12 and TbNedd8 display almost identical subcellular localization patterns (Figs. 2A and 8E). Therefore, we cannot rule out the possibility that the EYFP-TbNedd8 fluorescence signal detected in the flagellum and the nucleus may represent the fluorescent signal of TbNedd8-conjugated TbUbc12. Additionally, because several flagellum-associated proteins precipitated by TbNedd8 affinity purification are potentially conjugated with TbNedd8, the EYFP-TbNedd8 fluorescence signal in the flagellum may be attributed to the localization of the TbNedd8-conjugated flagellar protein(s). Nevertheless, no matter what form(s) of TbNedd8 localize(s) to the flagellum, these results provide another line of evidence to support the involvement of protein neddylation in flagellum-cell body adhesion.
A total of 70 putative TbNedd8-conjugated and -associated proteins were identified, among which are neddylation enzymes, proteasome components, the cullin subunits of the E3 ubiquitin ligase complexes, and many proteins of diverse cellular functions and of unknown function (Fig. 6C and supplemental Table S1). The functional groups of the TbNedd8-conjugated and -associated proteins matched very well with that of the human Nedd8-conjugated and -associated proteins identified by GST affinity purification (4), suggesting that Nedd8 may regulate similar cellular processes in T. brucei and humans. However, the individual proteins in these functional groups appear to differ between T. brucei and humans, indicating that Nedd8 may regulate distinct proteins of the same functional group in the two organisms.
Cullins are the best characterized Nedd8 substrates (39–41), and although a few non-Cullin proteins were also conjugated by Nedd8 (8), Cullins are likely the primary targets of Nedd8. In fungi and humans, Cullins are neddylated on a conserved lysine residue at the C terminus (4, 39, 40, 42, 43). Our results showed that six Cullin homologs in T. brucei, TbCUL1-TbCUL6, are also modified by TbNedd8, and we further identified the TbNedd8-modified lysine residue in three cullins, TbCUL1, TbCUL2, and TbCUL6 (Fig. 7). The other three cullins, TbCUL3, TbCUL4, and TbCUL5, also contain the conserved lysine residue at the C terminus (Fig. 7A). Although our mass spectrometry failed to detect the TbNedd8-conjugated peptide of these three Cullins, Western blotting with anti-TbNedd8 antibody confirmed that they are indeed neddylated (Fig. 7B). The failure to identify the TbNedd8-conjugated peptide for these Cullins could be due to the relatively low abundance of them in the total immunoprecipitates used for mass spectrometry.
Materials and Methods
Purification of Recombinant TbNedd8 and Antibody Production
The full-length coding sequence of TbNedd8 was cloned into pET22b (Novagen), and the resulting construct was transformed into the Escherichia coli BL21 strain. Recombinant TbNedd8-His6 was purified using His-nickel matrix (GE Healthcare), dialyzed to PBS, and used to immunize rabbits for polyclonal antibody production. Crude antisera from two rabbits were collected and used for Western blotting.
Trypanosome Cell Culture and RNAi
The procyclic 29-13 cell line (44), which was derived from the procyclic Lister 427 strain, was cultivated at 27 °C in SDM-79 medium containing 10% fetal bovine serum (Atlanta Biologicals, Inc.), 15 μg/ml G418, and 50 μg/ml hygromycin. The procyclic Lister 427 strain was cultured at 27 °C in SDM-79 medium supplemented with 10% fetal bovine serum.
RNAi of TbNedd8 was carried out using the stem-loop RNAi vector pSL (45). The full-length coding sequence of TbNedd8 was cloned into the HindIII/XhoI sites of the pSL vector, and the same DNA fragment was cloned into the AflII/BamHI sites in an opposite direction in the pSL vector. The resulting plasmid (pSL-TbNedd8) was linearized by NotI digestion and electroporated into the 29-13 cell line. Transfectants were selected with 2.5 μg/ml phleomycin and cloned by limiting dilution in a 96-well plate.
In Situ Epitope Tagging of Endogenous Proteins
The full-length coding sequence of TbNedd8 was cloned into the pN-EYFP-PAC and pN-PTP-PAC vector, and the resulting constructs were electroporated into the 427 cell line. TbNedd8 was also cloned into the pN-3Myc-PAC vector, and the resulting construct was transfected into the pSL-TbNedd8 RNAi cell line according to our published procedures (26, 46). Stable transfectants were selected under 1 μg/ml puromycin and cloned by limiting dilution in 96-well plates. Correct in situ tagging of one of the two TbNedd8 alleles was confirmed by PCR and subsequent sequencing of the PCR fragment as well as by Western blotting with anti-GFP antibody (JL-8 clone, Clontech) to detect EYFP-TbNedd8, anti-protein C antibody (Roche) to detect PTP-TbNedd8, or anti-Myc mAb (Sigma-Aldrich) to detect 3Myc-TbNedd8.
Endogenous tagging of TbAPPBP1, TbUba3, and TbUbc12 with a C-terminal EYFP was performed using the one-step PCR-based method described in a previous publication (47). Correct in situ tagging of TbAPPBP1, TbUba3, and TbUbc12 was confirmed by PCR and subsequent sequencing of the PCR fragment and by Western blotting with anti-GFP antibody (JL-8 clone, Clontech).
Ectopic Expression of Wild-type and Mutant TbNedd8 and Wild-type Cullins
The full-length coding sequence of TbNedd8 was fused with the sequences encoding a triple HA epitope and a triple Myc epitope and cloned into the pLew100 vector for expressing TbNedd8 with an N-terminal triple HA epitope and a C-terminal triple Myc epitope in T. brucei. Mutation of Gly-77 to alanine was performed by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Invitrogen). The full-length coding sequences of TbCUL1 to TbCUL6 were each cloned into the N′-3HA-pLew100 vector. The resulting constructs were transfected into the 29-13 cell line. Transfectants were selected under 2.5 μg/ml phleomycin and cloned by limiting dilution in a 96-well plate. Cells were induced with 1 μg/ml tetracycline to induce the expression of TbNedd8 and Cullins.
Flow Cytometry
Flow cytometry analysis of propidium iodide-stained trypanosome cells was carried out as described previously (48). Ethanol-fixed T. brucei cells were resuspended in PBS containing 10 μg/ml DNase-free RNase and 20 μg/ml propidium iodide and analyzed with a FACScan analytical flow cytometer (BD Biosciences). The percentage of cells at different cell cycle stages was determined by ModFit LT 3.1 software (BD Bioscience).
BrdU Incorporation and Detection
Non-induced control and TbNedd8 RNAi-induced cells (day 3) were incubated with 20 mm BrdU for 2 h and fixed with paraformaldehyde. Immunostaining with anti-BrdU antibody (Sigma-Aldrich, 1:40 dilution) was performed according to procedures described previously (49).
Affinity Purification of TbNedd8-conjugated and -associated Proteins
Tandem affinity purification was carried out according to our previous procedures (25, 50). Briefly, 500 ml (∼2 × 109) of cells expressing PTP-TbNedd8 from one of its endogenous loci was lysed by sonication, and the crude lysate was incubated with 200 μl of IgG-Sepharose 6 fast flow beads (Invitrogen) at 4 °C for 2 h. After overnight incubation with 200 units of tobacco etch virus protease (Promega), the eluate was incubated with 200 μl of anti-protein C beads at 4 °C for 2 h. Proteins were then eluted with EGTA/EDTA elution buffer. Proteins were further precipitated with 10 μl of StrataClean resin (Stratagene), separated on SDS-PAGE, and stained with silver solution (Pierce). Gel slices were excised from the gel and analyzed by LC-MS/MS for protein identification. As a control for non-specifically bound proteins, a parallel purification was performed using the wild-type 427 cell line.
Affinity purification with the anti-HA matrix was performed according to the instructions of the manufacturer (Pierce). Briefly, 1 liter (∼1 × 1010) of cells expressing 3HA-TbNedd8 from one of its endogenous loci or control cells was harvested and lysed by sonication. Crude cell lysate was centrifuged at 10,000 × g for 10 min at 4 °C, and the supernatant was incubated with 100 μl of anti-HA matrix (Pierce) at 4 °C for 2 h. The anti-HA matrix was then washed three times with 0.1% Triton X-100 in PBS. Proteins were eluted with 500 μl of 0.1 m glycine (pH 2.5) three times and then concentrated to a volume of 40 μl. Eluted proteins were separated by SDS-PAGE, stained with Coomassie Brilliant Blue, and excised from the gel for LC-MS/MS analysis.
In-gel Trypsin Digestion, LC-MS/MS, and Data Analysis
In-gel digestion of proteins was carried out according to our published procedures (50). Peptides extracted from gel slices were injected onto a Thermo LTQ Orbitrap XL and analyzed on a LTQ Orbitrap XL (Thermo Fisher Scientific, Bremen, Germany) interfaced with an Eksigent nano-LC 2D plus ChipLC system (Eksigent Technologies, Dublin, CA) at the Proteomics Core Facility of the University of Texas Health Science Center at Houston. Raw data files were searched using the Mascot search engine against the T. brucei database version 4, and the search conditions were set as follows: peptide tolerance of 10 ppm and MS/MS tolerance of 0.8 Da with the enzyme trypsin and two missed cleavages.
Immunofluorescence Microscopy
Cells were adhered to coverslips and then fixed in cold (−20 °C) methanol. The following antibodies were used: L3B2 (anti-FAZ1) mAb for the FAZ filament (1:50 dilution) (51) and KMX-1 mAb (Millipore) for the spindle (1:400 dilution). Cells were incubated with the primary antibody at room temperature for 1 h and washed three times with PBS containing 0.1% Triton X-100. Cells were then incubated with FITC-conjugated anti-mouse IgG (Sigma-Aldrich) at room temperature for 1 h. Slides were mounted in VectaShield mounting medium (Vector Labs) containing DAPI and examined using an inverted microscope (model IX71, Olympus) equipped with a cooled charge-coupled device camera (model Orca-ER, Hamamatsu) and a PlanApo N ×60 1.42 numerical aperture differential interference contrast objective. Images were acquired and processed using Slidebook5 software (Intelligent Imaging Innovations, Inc.).
Statistical Analysis
Statistical analysis was performed using the t test provided in the Microsoft Excel software. For the immunofluorescence microscopy experiments, images were taken randomly, and all cells in each image were counted.
Author Contributions
S. L., H. H., T. W., X. T., and Z. L. conceived and designed the experiments. S. L., H. H., and T. W. performed the experiments. S. L., X. T., and Z. L. analyzed the data. S. L., X. T., and Z. L. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. George A. M. Cross of Rockefeller University for providing the procyclic 29-13 cell line and the pLew100 vector. We also thank Dr. Arthur Günzl of the University of Connecticut Health Center for providing the pN-PTP-NEO vector and Dr. Keith Gull of the University of Oxford for providing the anti-FAZ1 antibody (L3B2).
This work was supported by National Natural Science Foundation of China Grants 31270780, U1332137, and 21272090 (to X. T.) and 31500601 (to S. L.), Fundamental Research Funds for the Central Universities Grant WK2070000020 (to X. T.), and National Institutes of Health Grants R01AI101437 and R01AI118736 (to Z. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1—S3 and Table S1.
- CRL
- Cullin-RING ligase
- SCF
- Skp1-Cullin-F box protein
- EYFP
- enhanced yellow fluorescence protein
- FAZ
- flagellum attachment zone
- snFAZ
- short new FAZ
- PTP
- protein A-tobacco etch virus-protein C
- CDK
- cyclin-dependent kinase.
References
- 1. Watson I. R., Irwin M. S., and Ohh M. (2011) NEDD8 pathways in cancer: sine quibus non. Cancer Cell 19, 168–176 [DOI] [PubMed] [Google Scholar]
- 2. Kamitani T., Kito K., Nguyen H. P., and Yeh E. T. (1997) Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem. 272, 28557–28562 [DOI] [PubMed] [Google Scholar]
- 3. Wei N., and Deng X. W. (2003) The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19, 261–286 [DOI] [PubMed] [Google Scholar]
- 4. Jones J., Wu K., Yang Y., Guerrero C., Nillegoda N., Pan Z. Q., and Huang L. (2008) A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J. Proteome Res. 7, 1274–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li T., Santockyte R., Shen R. F., Tekle E., Wang G., Yang D. C., and Chock P. B. (2006) A general approach for investigating enzymatic pathways and substrates for ubiquitin-like modifiers. Arch. Biochem. Biophys. 453, 70–74 [DOI] [PubMed] [Google Scholar]
- 6. Norman J. A., and Shiekhattar R. (2006) Analysis of Nedd8-associated polypeptides: a model for deciphering the pathway for ubiquitin-like modifications. Biochemistry 45, 3014–3019 [DOI] [PubMed] [Google Scholar]
- 7. Xirodimas D. P., Sundqvist A., Nakamura A., Shen L., Botting C., and Hay R. T. (2008) Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 9, 280–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rabut G., and Peter M. (2008) Function and regulation of protein neddylation: protein modifications: beyond the usual suspects review series. EMBO Rep. 9, 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang W., Zhang J., Xu C., Wang T., Zhang X., and Tu X. (2009) Solution structure of Urm1 from Trypanosoma brucei. Proteins 75, 781–785 [DOI] [PubMed] [Google Scholar]
- 10. Liao S., Wang T., Fan K., and Tu X. (2010) The small ubiquitin-like modifier (SUMO) is essential in cell cycle regulation in Trypanosoma brucei. Exp. Cell Res. 316, 704–715 [DOI] [PubMed] [Google Scholar]
- 11. Koopmann R., Muhammad K., Perbandt M., Betzel C., and Duszenko M. (2009) Trypanosoma brucei ATG8: structural insights into autophagic-like mechanisms in protozoa. Autophagy 5, 1085–1091 [DOI] [PubMed] [Google Scholar]
- 12. Li F. J., Shen Q., Wang C., Sun Y., Yuan A. Y., and He C. Y. (2012) A role of autophagy in Trypanosoma brucei cell death. Cell Microbiol. 14, 1242–1256 [DOI] [PubMed] [Google Scholar]
- 13. Pan Z. Q., Kentsis A., Dias D. C., Yamoah K., and Wu K. (2004) Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23, 1985–1997 [DOI] [PubMed] [Google Scholar]
- 14. Sasse R., and Gull K. (1988) Tubulin post-translational modifications and the construction of microtubular organelles in Trypanosoma brucei. J. Cell Sci. 90, 577–589 [DOI] [PubMed] [Google Scholar]
- 15. Sherwin T., and Gull K. (1989) Visualization of detyrosination along single microtubules reveals novel mechanisms of assembly during cytoskeletal duplication in trypanosomes. Cell 57, 211–221 [DOI] [PubMed] [Google Scholar]
- 16. Zhou Q., Liu B., Sun Y., and He C. Y. (2011) A coiled-coil- and C2-domain-containing protein is required for FAZ assembly and cell morphology in Trypanosoma brucei. J. Cell Sci. 124, 3848–3858 [DOI] [PubMed] [Google Scholar]
- 17. Vaughan S., Kohl L., Ngai I., Wheeler R. J., and Gull K. (2008) A repetitive protein essential for the flagellum attachment zone filament structure and function in Trypanosoma brucei. Protist 159, 127–136 [DOI] [PubMed] [Google Scholar]
- 18. Jin L., Pahuja K. B., Wickliffe K. E., Gorur A., Baumgärtel C., Schekman R., and Rape M. (2012) Ubiquitin-dependent regulation of COPII coat size and function. Nature 482, 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., and Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 20. Lin J. J., Milhollen M. A., Smith P. G., Narayanan U., and Dutta A. (2010) NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 70, 10310–10320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Milhollen M. A., Narayanan U., Soucy T. A., Veiby P. O., Smith P. G., and Amidon B. (2011) Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res. 71, 3042–3051 [DOI] [PubMed] [Google Scholar]
- 22. Nishitani H., Sugimoto N., Roukos V., Nakanishi Y., Saijo M., Obuse C., Tsurimoto T., Nakayama K. I., Nakayama K., Fujita M., Lygerou Z., and Nishimoto T. (2006) Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 25, 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li X., Zhao Q., Liao R., Sun P., and Wu X. (2003) The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J. Biol. Chem. 278, 30854–30858 [DOI] [PubMed] [Google Scholar]
- 24. Li Z. (2012) Regulation of the cell division cycle in Trypanosoma brucei. Eukaryot. Cell 11, 1180–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Z., Lee J. H., Chu F., Burlingame A. L., Günzl A., and Wang C. C. (2008) Identification of a novel chromosomal passenger complex and its unique localization during cytokinesis in Trypanosoma brucei. PLoS ONE 3, e2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dang H. Q., and Li Z. (2011) The Cdc45.Mcm2–7.GINS protein complex in trypanosomes regulates DNA replication and interacts with two Orc1-like proteins in the origin recognition complex. J. Biol. Chem. 286, 32424–32435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tiengwe C., Marcello L., Farr H., Gadelha C., Burchmore R., Barry J. D., Bell S. D., and McCulloch R. (2012) Identification of ORC1/CDC6-interacting factors in Trypanosoma brucei reveals critical features of origin recognition complex architecture. PLoS ONE 7, e32674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wojcik E. J., Glover D. M., and Hays T. S. (2000) The SCF ubiquitin ligase protein slimb regulates centrosome duplication in Drosophila. Curr. Biol. 10, 1131–1134 [DOI] [PubMed] [Google Scholar]
- 29. Freed E., Lacey K. R., Huie P., Lyapina S. A., Deshaies R. J., Stearns T., and Jackson P. K. (1999) Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev. 13, 2242–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Korzeniewski N., Zheng L., Cuevas R., Parry J., Chatterjee P., Anderton B., Duensing A., Münger K., and Duensing S. (2009) Cullin 1 functions as a centrosomal suppressor of centriole multiplication by regulating polo-like kinase 4 protein levels. Cancer Res. 69, 6668–6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feldman R. M., Correll C. C., Kaplan K. B., and Deshaies R. J. (1997) A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91, 221–230 [DOI] [PubMed] [Google Scholar]
- 32. Podust V. N., Brownell J. E., Gladysheva T. B., Luo R. S., Wang C., Coggins M. B., Pierce J. W., Lightcap E. S., and Chau V. (2000) A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 97, 4579–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mutomba M. C., To W. Y., Hyun W. C., and Wang C. C. (1997) Inhibition of proteasome activity blocks cell cycle progression at specific phase boundaries in African trypanosomes. Mol. Biochem. Parasitol. 90, 491–504 [DOI] [PubMed] [Google Scholar]
- 34. Li Z., and Wang C. C. (2002) Functional characterization of the 11 non-ATPase subunit proteins in the trypanosome 19 S proteasomal regulatory complex. J. Biol. Chem. 277, 42686–42693 [DOI] [PubMed] [Google Scholar]
- 35. Zhou Q., Hu H., He C. Y., and Li Z. (2015) Assembly and maintenance of the flagellum attachment zone filament in Trypanosoma brucei. J. Cell Sci. 128, 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. LaCount D. J., Barrett B., and Donelson J. E. (2002) Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 277, 17580–17588 [DOI] [PubMed] [Google Scholar]
- 37. Woods K., Nic a'Bhaird N., Dooley C., Perez-Morga D., and Nolan D. P. (2013) Identification and characterization of a stage specific membrane protein involved in flagellar attachment in Trypanosoma brucei. PLoS ONE 8, e52846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rotureau B., Blisnick T., Subota I., Julkowska D., Cayet N., Perrot S., and Bastin P. (2014) Flagellar adhesion in Trypanosoma brucei relies on interactions between different skeletal structures in the flagellum and cell body. J. Cell Sci. 127, 204–215 [DOI] [PubMed] [Google Scholar]
- 39. Lammer D., Mathias N., Laplaza J. M., Jiang W., Liu Y., Callis J., Goebl M., and Estelle M. (1998) Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12, 914–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liakopoulos D., Doenges G., Matuschewski K., and Jentsch S. (1998) A novel protein modification pathway related to the ubiquitin system. EMBO J. 17, 2208–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Osaka F., Kawasaki H., Aida N., Saeki M., Chiba T., Kawashima S., Tanaka K., and Kato S. (1998) A new NEDD8-ligating system for cullin-4A. Genes Dev. 12, 2263–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Laplaza J. M., Bostick M., Scholes D. T., Curcio M. J., and Callis J. (2004) Saccharomyces cerevisiae ubiquitin-like protein Rub1 conjugates to cullin proteins Rtt101 and Cul3 in vivo. Biochem. J. 377, 459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Osaka F., Saeki M., Katayama S., Aida N., Toh-E A., Kominami K., Toda T., Suzuki T., Chiba T., Tanaka K., and Kato S. (2000) Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 19, 3475–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wirtz E., Leal S., Ochatt C., and Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 45. Wei Y., Hu H., Lun Z. R., and Li Z. (2014) Centrin3 in trypanosomes maintains the stability of a flagellar inner-arm dynein for cell motility. Nat. Commun. 5, 4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu L., Hu H., and Li Z. (2012) A kinetoplastid-specific kinesin is required for cytokinesis and for maintenance of cell morphology in Trypanosoma brucei. Mol. Microbiol. 83, 565–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shen S., Arhin G. K., Ullu E., and Tschudi C. (2001) In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol. Biochem. Parasitol. 113, 171–173 [DOI] [PubMed] [Google Scholar]
- 48. Li Z., Tu X., and Wang C. C. (2006) Okadaic acid overcomes the blocked cell cycle caused by depleting Cdc2-related kinases in Trypanosoma brucei. Exp. Cell Res. 312, 3504–3516 [DOI] [PubMed] [Google Scholar]
- 49. Liu Y., Hu H., and Li Z. (2013) The cooperative roles of PHO80-like cyclins in regulating the G1/S transition and posterior cytoskeletal morphogenesis in Trypanosoma brucei. Mol. Microbiol. 90, 130–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hu H., Hu L., Yu Z., Chasse A. E., Chu F., and Li Z. (2012) An orphan kinesin in trypanosomes cooperates with a kinetoplastid-specific kinesin to maintain cell morphology by regulating subpellicular microtubules. J. Cell Sci. 125, 4126–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kohl L., Sherwin T., and Gull K. (1999) Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 46, 105–109 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.