Abstract
Dehydratases catalyze the breakage of a carbon—oxygen bond leading to unsaturated products via the elimination of water. The 1.6-Å resolution crystal structure of 4-hydroxybutyryl-CoA dehydratase from the γ-aminobutyrate-fermenting Clostridium aminobutyricum represents a new class of dehydratases with an unprecedented active site architecture. A [4Fe–4S]2+ cluster, coordinated by three cysteine and one histidine residues, is located 7 Å from the Re-side of a flavin adenine dinucleotide (FAD) moiety. The structure provides insight into the function of these ubiquitous prosthetic groups in the chemically nonfacile, radical-mediated dehydration of 4-hydroxybutyryl-CoA. The substrate can be bound between the [4Fe–4S]2+ cluster and the FAD with both cofactors contributing to its radical activation and catalytic conversion. Our results raise interesting questions regarding the mechanism of acyl-CoA dehydrogenases, which are involved in fatty acid oxidation, and address the divergent evolution of the ancestral common gene.
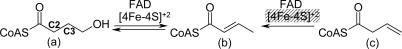
Dehydration is a very common reaction in biochemical pathways. More than 100 dehydratases (carbon–oxygen lyases, EC 4.2.-.-) are known [Expert Protein Analysis System (ExPASy), www.expasy.org]. Most of these enzymes catalyze the α,β-elimination of water, during which the α-hydrogen (C2 position) to be removed as a proton is activated by an adjacent electron-withdrawing carboxylate, carbonyl, or CoA-thiol ester group, and the hydroxyl group leaves from the β-position (C3 position) (1). In anaerobic microorganisms, the absence of the biradical dioxygen allows a rich chemistry of radical reactions involved in the removal of hydrogen atoms from nonactivated positions such as the β- or γ-carbons of 2-, 4-, or 5-hydroxyacyl-CoA derivatives. These atypical dehydratases contain one or more prosthetic groups such as Fe–S clusters or flavins (2). An example is 4-hydroxybutyryl-CoA dehydratase (4-BUDH) from Clostridium aminobutyricum, which catalyzes both the reversible oxygen-sensitive dehydration of 4-hydroxybutyryl-CoA (Scheme 1a) and the oxygen-insensitive isomerization of vinylacetyl-CoA (Scheme 1c) to crotonyl-CoA (Scheme 1b) (3). The dehydration reaction requires the removal of a hydrogen atom from the least activated C3 position of the butyryl chain (pKa ≈ 40) and is the mechanistically most demanding step in the fermentation of γ-aminobutyrate (GABA) to ammonia, acetate, and butyrate by C. aminobutyricum (4). 4-BUDH is active as a homotetramer with up to one [4Fe–4S]2+ cluster and one noncovalently bound flavin adenine dinucleotide (FAD) moiety per 54-kDa subunit. Despite the presence of a [4Fe–4S]2+ cluster and the need of an oxidized FAD for catalysis, the overall reaction occurs with no net redox change (4). These observations have led to the proposal of a radical-based mechanism for the dehydration, where the one-electron oxidation of the enolate of 4-hydroxybutyryl-CoA to the enoxy radical makes the C3-proS-hydrogen (5) acidic enough [pKa = 14 (6)] for deprotonation to a ketyl radical anion (Fig. 3). The now facile elimination of the hydroxyl group leads to the dienoxy radical, which upon reduction to the dienolate and protonation yields the product crotonyl-CoA (2). The addition of crotonyl-CoA to the enzyme raises the redox potentials of the flavin and the [4Fe–4S]2+ cluster by ≈200 mV (7). Under these conditions, the [4Fe–4S]2+ cluster can be readily reduced to [4Fe–4S]1+ by dithionite, and the enzyme is able to stabilize the neutral flavin semiquinone (FADH·) and an additional radical, presumably a substrate-derived radical interacting with the [4Fe–4S]1+ cluster (8). Although the functional role of the FAD appears to be established, the task of the [4Fe–4S]2+ cluster remains unresolved (3, 9).
Scheme 1.
The chemical reactions catalyzed by 4-BUDH. C2 and C3 indicate the α- and β-positions, respectively. The masked [4Fe-4S]2+ cluster indicates that the isomerization reaction does not require its participation.
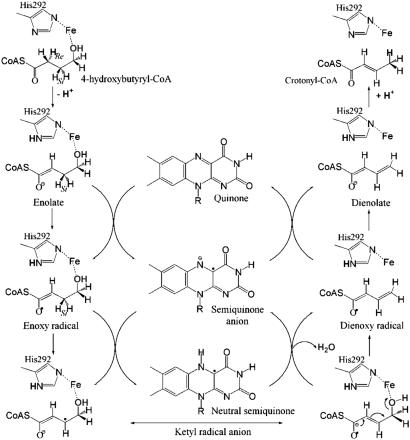
Fig. 3.
Proposed radical-mediated mechanism of 4-BUDH. The catalytic base His-292 abstracts the C2-proR-hydrogen and protonates the C4 position of the last reaction intermediate, dienolate. The C3-proS-hydrogen is abstracted by the semiquinone anion and, together with the hydroxyl group, forms the byproduct water molecule.
4-BUDH belongs to an increasing number of enzymes that use a radical-based mechanism. Well characterized biological systems include the tyrosyl radical-dependent aerobic ribonucleotide reductase and the large family of enzymes in which the 5′-deoxyadenosyl radical derived from coenzyme B12 or S-adenosylmethionine acts as a radical generator (1, 10, 11). In all these systems the substrate-derived radical is generated by hydrogen atom abstraction, whereas with 4-BUDH the enoxy radical seems to be formed by one-electron transfer from the enolate to the flavin. Interestingly, the activation of the unusual clostridial 2-hydroxyglutaryl-CoA dehydratase also requires one-electron transfer but is driven by ATP hydrolysis in the reverse direction (12). To understand the molecular basis for atypical dehydrations, we have determined the crystal structure of 4-BUDH. This work describes the mechanistic significance of the distinctive active site for radical-based dehydration and addresses the unpredicted evolutionary relationship between radical-mediated dehydratase and dehydrogenase activities. Thus, the elucidation of the structure of 4-BUDH sheds light on the mechanism of acyl-CoA dehydrogenases and challenges their generally accepted mechanism (13).
Materials and Methods
Anoxic Purification and Crystallization. Purification and crystallization of 4-BUDH were performed under strictly anoxic conditions (atmosphere of 95% N2 and 5% H2, with a palladium catalyst and oxygen sensor) by using an anaerobic chamber (Coy Laboratory Products, Ann Arbor, MI). Crystals were obtained by the sitting-drop vapor diffusion method at 22°C by mixing 2 μl of protein solution (20 mg/ml in 10 mM Bis-Tris-propane (1,3-bis[tris(hydroxymethyl)methylamino]propane), pH 7, supplemented with 8–10 mM crotonyl-CoA and 5 mM sodium dithionite) with or without 8–10 mM FAD plus 2 μl of precipitant containing 5 mM sodium dithionite and 0.3 μl of FAD (to have 8–10 mM in the drop). Dark brown crystals grown within 1–2 weeks were harvested in precipitant supplemented with 2–5% d(-)-2,3-butanediol and shock-frozen in liquid nitrogen.
Structure Determination and Refinement. The crystal structure was solved by multiple wavelength anomalous dispersion (MAD) methods at the Fe K-edge. Diffraction data were collected at the synchrotron beamline PX (Swiss Light Source at the Paul Scherrer Institute, Villigen, Switzerland) by using an orthorhombic crystal (space group P212121 with cell constants of a = 102.3 Å, b = 130.2 Å, and c = 175.5 Å, corresponding to one homotetramer per asymmetric unit and a solvent content of 45%). The data were processed and scaled with xds (14). The four [4Fe–4S] clusters were localized by using the Shake-and-Bake algorithm (bnp interface) (15), and phases were calculated with sharp (16).** Individual Fe atoms were manually positioned in one of the [4Fe–4S] clusters, and phases were recalculated with sharp. The consequently averaged electron density map [created with ave (17)] showed clear electron density for that [4Fe–4S] cluster, and noncrystallographic symmetry (NCS) operators [determined with ncs6d (17) and sharp] were used to generate the NCS-related Fe positions. Model building, averaging, and refinement were performed with main (18), ave, and cns (19), respectively. In the final cycles of refinement, the NCS restraints were lowered and finally were completely released. shelx (20) was used for final anisotropic refinement of the B factor values for the [4Fe–4S] clusters. The model was refined to 1.6-Å resolution with Rcryst of 0.163, Rfree of 0.212, and a good stereochemistry. (For details on the data collection and structure refinement statistics, see Table 1, which is published as supporting information on the PNAS web site.) The refined model contains the complete set of four molecules (Met-1 to Lys-490) with one [4Fe–4S]2+ cluster and one FAD moiety per monomer, and 2,338 water molecules. The B factor values for all of the Fe and S atoms of the four clusters are in the same range of the observed values for the neighboring atoms. The final electron density showed that residues Gly-167 and Asp-357 were incorrectly assigned in the protein sequence P55792 and were replaced by Asp and Gly, respectively. The side chains for the few residues at the surface of the molecules for which there was no clear electron density were modeled according to the more stable conformer. Gln-101, the only residue located in a disallowed region of the Ramachandran plot (21), displays well defined electron density and is located in the loop containing the [4Fe–4S]2+ cluster ligands Cys-99 and Cys-103.
Results and Discussion
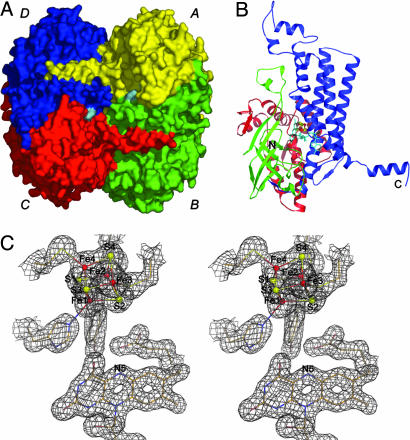
Overall Structure. The 4-BUDH crystal structure reveals the presence of a homotetramer with local D2-symmetry and a size of 92 × 91 × 85 Å3 (Fig. 1A). Within the homotetramer, the four monomers clearly assemble into two catalytically functional dimers (AB and CD in Fig. 1 A) with two active sites per dimer. Each active site contains one [4Fe–4S]2+ cluster and one FAD moiety. The two dimers associate by docking their extruding C-terminal α-helices at the relevant parts of the partner dimer. The monomer consists of three domains. The N- and C-terminal domains (residues Met-1 to Leu-143 and residues Glu-277 to Lys-490, respectively) are mainly α-helical, whereas the middle domain (residues Ile-144 to Gln-276) is predominantly β-structured (Fig. 1B). A similar fold is found in FAD-containing medium chain acyl-CoA dehydrogenase (MCAD) from pig (Sus scrofa) liver (22), which catalyzes the reversible oxidation of an acyl-CoA derivative to form the α,β-double bond in the corresponding enoyl-CoA (23). This is surprising, because the two enzymes show only 16% amino acid sequence identity (Fig. 4, which is published as supporting information on the PNAS web site), and 4-BUDH contains the unprecedented His-coordinated [4Fe–4S]2+ cluster. Interestingly, the catalytic reactions of both 4-BUDH and MCAD require the rupture of nonactivated β-C—H bonds.
Fig. 1.
Crystal structure. (A) Surface representation of the 4-BUDH homotetramer. Each monomer's surface is individually colored, and the FAD's surface is shown in cyan. (B) Secondary structure topology of the monomer with the N-terminal domain (residues Met-1 to Leu-143) shown in red, the middle domain (residues Ile-144 to Gln-276) shown in green, and the C-terminal domain (residues Glu-277 to Lys-490) shown in blue. The [4Fe–4S]2+ cluster is shown as ball-and-sticks with Fe and S atoms shown in red and yellow, respectively. The FAD is displayed in sticks and shown in cyan. The N and C termini of the polypeptide chain are marked. (C) Example of the well defined electron density for the refined model (2 Fo - Fc electron density map contoured at 1.0 σ and shown in gray). Atom color code for protein residues and FAD is cream for carbon, red for oxygen, blue for nitrogen, and yellow for sulfur. Color code for the [4Fe–4S]2+ cluster is as in B. This figure and Fig. 2 were prepared with pymol (44).
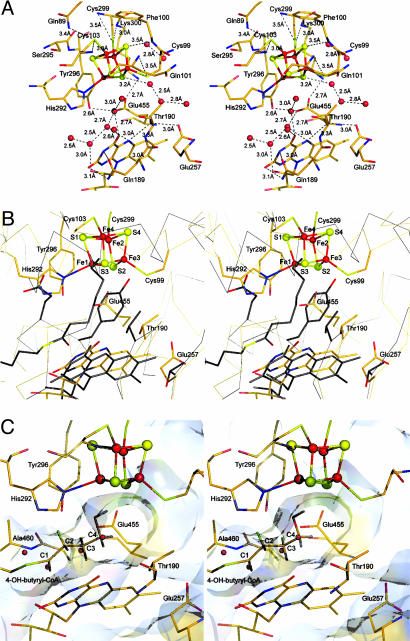
Active Site Architecture. The active site, embedded in the center of the protein, is accessed by a narrow substrate binding channel leading from the surface of the molecule to the [4Fe–4S]2+ cluster and FAD. The walls of this channel are created by residues from both monomers and the elongated FAD molecule (Ade and isoalloxazine rings distal to each other). The isoalloxazine ring is located at the crevice between the three domains of the monomer, and the Ade moiety lies at the dimer interface (Fig. 1 A and B). The binding of the FAD is accomplished by residues from both middle and C-terminal domains from one monomer and the C-terminal domain of the neighboring monomer (Scheme 2, which is published as supporting information on the PNAS web site). The [4Fe–4S]2+ cluster forms the bottom of the channel and is ≈7 Å from the Re-side of the isoalloxazine ring (distance between Fe1 and N5 of FAD) (Fig. 1C). The Fe atoms of the [4Fe–4S]2+ cluster are covalently bound to the protein by three Cys residues and one His residue (Fig. 2A), which originate from the N-terminal (Cys-99 and Cys-103) and C-terminal (His-292 and Cys-299) domains. This noncanonical [4Fe–4S]2+ cluster coordination motif, CX3CX179–183HisX6C, is also found in homologous amino acid sequences from Clostridium difficile, Porphyromonas gingivalis, Fusobacterium nucleatum, Archaeoglobus fulgidus, Pyrobaculum aerophilum, Sulfolobus solfataricus, and Sulfolobus tokodaii (Fig. 4B). Additionally, the presence of a FAD moiety and the absence of sequence similarity to other Fe–S cluster-containing dehydratases suggest that 4-BUDH and counterparts constitute a new class within the Fe–S cluster-dependent dehydratase protein family.
Fig. 2.
Active site. (A) Stereoview of the 4-BUDH active site environment with color code as in Fig. 1. The labels for the [4Fe–4S]2+ cluster atoms were omitted for simplicity (see B). Water molecules are represented as red spheres. Dashed lines indicate interactions in hydrogen bonding distance (values of 3.4 and 3.5 Å suggest a weak character for the respective interactions). Note the asymmetry in the distribution of these interactions, which are concentrated on the [4Fe–4S]2+ cluster side opposite to His-292. Glu-455 interacts with Tyr-296 and Lys-300, residues belonging to the helix coordinating the [4Fe–4S]2+ cluster. These residues are well or strictly conserved among all 4-BUDHs (Fig. 4B, which is published as supporting information on the PNAS web site). (B) Stereoview of the superposition of the active sites of 4-BUDH and MCAD from pig liver (in gray except for the carboxylate group of the active base Glu-376, which is in red, and for the sulfur atom and thiol ester carbonyl group of the ligand octanoyl-CoA, which are in yellow and red, respectively). (C) Stereoview of the putative binding mode for 4-hydroxybutyryl-CoA in the 4-BUDH active site. The C2-proR- and C3-proR-hydrogens are shown in green, whereas the C2-proS- and C3-proS-hydrogens are shown in gray.
Environment of the [4Fe–4S]2+ Cluster and Its Catalytic Role. Protein preparation-dependent variations on cofactor content (and consequent disparities in specific activity) made it difficult to determine whether the [4Fe–4S]2+ cluster has a solely structural function or plays an active role in the catalysis, as Lewis acid, in the elimination of the hydroxyl group (3, 9). Aconitases as well as bacterial l-serine dehydratases and fumarases contain [4Fe–4S]2+ clusters, which are not involved in electron transfer but act as Lewis acids facilitating the leaving of the substrate's hydroxyl group (24). Crystallographic studies of aconitase, the prototype of the dehydratase protein family, have shown that one Fe atom of the [4Fe–4S]2+ cluster is not coordinated by the protein scaffold but interacts with the substrate (25). Although present in the crystallization solution, the crystal structure of 4-BUDH displays no extra electron density that could account for the product crotonyl-CoA, which could have been hydrolyzed during crystal growth. The Fe atoms of the [4Fe–4S]2+ cluster are tetrahedrally coordinated with Fe1 coordinated by Nε2 of His-292. The distances between the Fe atoms and their coordinating sulfides and thiolates, which range from 2.3 to 2.4 Å (Table 2, which is published as supporting information on the PNAS web site), are similar to those usually found in [2Fe–2S], [3Fe–4S], and [4Fe–4S] clusters. In contrast to Rieske proteins (26), which have a [2Fe–2S] cluster with two Cys and two His ligands, and the Fe–Ni hydrogenase, where the distal [4Fe–4S] cluster is coordinated by three Cys and one His residues (27), the Fe1 atom is not coordinated by the Nδ1 atom but by the Nε2 atom of His-292. A similar coordination by the Nε2 atom has been found only in the iron-only hydrogenase of Clostridium pasteurianum (28), the hybrid-cluster protein (29), and the nickel-containing carbon monoxide dehydrogenases (30–33). A common feature for such coordination in [4Fe–4S] clusters seems to be the modulation of the redox properties of the coordinated Fe atom. Interestingly, although in all these metalloproteins the value observed for the Fe—Nε2 bond length ranges from 1.9 to 2.1 Å, in 4-BUDH the average Fe1—Nε2 bond length is 2.4 Å (Table 2). It is tempting to propose that this long bond contributes to the tuning of the electronic and bonding properties of Fe1, which, like the labile Fe atom in aconitase (25), is ideally positioned for interaction with the hydroxyl group of the substrate. The redox potential of Fe–S clusters is affected by several environmental factors, such as hydrogen bonding, net charges of the ligands, peptide dipoles and specific charged residues, and solvent exposure (ref. 34 and references therein). Hydrogen bonding to the Fe–S cluster ligands reduces their charge donation to the metals and, thus, decreases the metal–ligand covalence (for similar bond lengths, the strength of the hydrogen bond increases from a thiolate to a sulfide ligand). This results in the stabilization of the electron density on the Fe–S cluster and a higher effective nuclear charge on the metal ion. The larger the effective nuclear charge, the easier the metal can be reduced, resulting in an increase of its redox potential (ref. 35 and references therein). In 4-BUDH, the [4Fe–4S]2+ cluster is solvent-exposed and has few NH/OH···S interactions involving only two sulfide ligands (Fig. 2 A). Thus, the overall negative charge of the [4Fe–4S]2+ cluster is not completely stabilized, resulting in the increase of the total iron–sulfide covalence, which explains its low redox potential [approximately -600 mV (7)]. By modulating such a low redox potential of the [4Fe–4S]2+ cluster, the enzyme assures that the transient one-electron oxidation of the substrate results only in the reduction of FAD. The redox potential of the [4Fe–4S]2+ cluster increases by ≈200 mV in the presence of substrate (7), which indicates a drastic alteration of the [4Fe–4S]2+ cluster environment. Both the consequent solvent protection (displacement of solvent molecules by substrate) and protonation of the His-292 ligand [imidazolate state (36)] would explain this shift.
Structural Basis for Radical-Mediated Catalysis. The structural similarity of 4-BUDH with MCAD (37) (Figs. 2B and 4A) suggests a similar binding mode for the respective substrates. In 4-BUDH, the dimensions of the channel leading to the active site indicate that it can accommodate the substrate 4-hydroxybutyryl-CoA with its hydroxyacyl part sandwiched between the two prosthetic groups. Additionally, and as observed for MCAD with bound ligand (37), the substrate could be positioned by hydrogen bonds between its CoA-thiol ester carbonyl group and both the 2′-OH of FAD and the polypeptide backbone. In the active site there are three well defined solvent molecules involved in a network of hydrogen-bond interactions (Fig. 2 A and C). Thus, substrate binding would displace the water molecules from the active site [as observed in MCAD upon substrate binding (37)]. Fitting of 4-hydroxybutyryl-CoA into the active site of 4-BUDH, with the CoA-thiol ester carbonyl group in hydrogen-bonding distance to the 2′-OH of FAD and the backbone amide of Ala-460, positions the hydroxyl group at interaction distance from the Fe1 atom (modeled distance of ≈2.7 Å) (Fig. 2C). It has been shown for MCAD that this arrangement lowers the pKa of the α-hydrogen (C2 position) of 4-hydroxybutyryl-CoA from 21 (38) to ≈8 (13). Thus, the first step of our postulated ketyl radical mechanism (Fig. 3) is the deprotonation of the C2 position by a base. Apart from His-292, three other well conserved residues among 4-BUDHs line the active site cavity, namely, Tyr-296, Glu-257, and Glu-455 (Fig. 2). Both Glu-455 and His-292 at 3 Å (modeled distance) from the C2 position are possible candidates. The occurrence of His-292 as imidazolate ligand of the [4Fe–4S]2+ cluster makes this residue ideally positioned to act as a catalytic base. Furthermore, His-292 has a similar structural position as the active base in MCAD (Fig. 2B). The next step is the one-electron oxidation of the enolate of 4-hydroxybutyryl-CoA by FAD to form the enoxy radical and the flavin semiquinone anion. The latter acts as a base and abstracts the C3-proS-hydrogen (the modeled distance between N5 of FAD and the C3 position is ≈3 Å) (5), forming the ketyl radical anion and the neutral semiquinone FADH· (39). It has been calculated that the C3-proton of the enoxy radical has a pKa of 14 (6), which could be further lowered to ≈7, because the corresponding base, the ketyl radical anion, could be stabilized by hydrogen bonds as suggested for the enolate. The ketyl radical anion then eliminates the hydroxyl group, which is polarized by the [4Fe–4S]2+ cluster (acting as Lewis acid). The released hydroxyl ion can abstract the proton at N5 of FADH· to form the byproduct water molecule. Next comes the reduction of the dienoxy radical to dienolate by the FAD semiquinone anion's restoration of the oxidized FAD (quinone state). Final protonation at the C4 position of the dienolate by His-292 yields the product crotonyl-CoA. The produced water molecule may exit the active site through a second channel leading from the bottom of the active site to the protein surface. A number of well ordered water molecules present in the channel indicates the solvent accessibility of the active site (Fig. 2A). The extreme oxygen sensitivity of the dehydratase activity compared with the more stable isomerization reaction (3) can be explained by the requirement of a functional [4Fe–4S]2+ cluster for only the former reaction. The protective effect observed in the presence of substrate results from the displacement of the solvent molecules upon substrate binding.
Evolutionary Link with the MCAD Family and Mechanistic Implications. The generally accepted mechanism for MCAD involves the deprotonation at the C2 position of acyl-CoA concomitant with a hydride transfer from the C3 position to the FAD molecule (13). This view does not take into account the ability of flavins to stabilize a semiquinone radical, which provides the basis of an alternative radical pair-dependent mechanism. This 45-year-old proposal starts with a one-electron transfer from the enolate of acyl-CoA to FAD, yielding the enoxy radical and the flavin semiquinone anion, which acts as a base [pKa = 8.3 (40)] and removes the C3 proton. The resulting ketyl radical anion transfers a second electron to the flavin, whereby FAD hydroquinone anion and the product enoyl-CoA are formed (41). This proton–electron–proton–electron mechanism explains the experimental observations with the radical clock (methylenecyclopropyl)acetyl-CoA that rearranges at the level of the enoxy radical before the second electron transfer (42). However, it has been argued that with normal substrates the reaction appears to be concerted without accumulation of an enolate intermediate able to be oxidized by one-electron transfers (43). Probably, rapid, successive single electron and proton transfers cannot be distinguished from a hydride transfer by the currently available methods. 4-BUDH, which exhibits no butyryl-CoA dehydrogenase activity, catalyzes the exchange of a proton of butyryl-CoA and the elimination of the C3-proS-hydrogen of 4-hydroxylbutyryl-CoA, which corresponds to the C3-proR-hydrogen removed by butyryl-CoA dehydrogenase (5). This similarity suggests that the introduction of a [4Fe–4S]2+ cluster converted the acyl-CoA dehydrogenase into a 4-BUDH, a process called “evolution by selection of (small) modifications of existing catalytic systems” (J. W. Cornforth, personal communication).
Outlook. The advantage of using radical intermediates in enzymatic transformations is their high reactivity and special properties, thus allowing their possible application in new reactions for the metabolism of otherwise refractory compounds (1). Besides 4-BUDH and acyl-CoA dehydrogenases, the 2-hydroxyacyl-CoA dehydratases (12) form a third family of enzymes that may make use of the facile deprotonation of an enoxy radical. The results presented here provide a basis for further structural and computational investigations of radical-mediated dehydrations in biological systems.
Supplementary Material
Acknowledgments
We thank J. W. Cornforth (University of Sussex, Sussex, U.K.) and B. T. Golding (University of Newcastle upon Tyne, Newcastle upon Tyne, U.K.) for helpful discussions; N. Mühlhäuser (Philipps-Universität, Marburg, Germany) for technical assistance; G. Bourenkov (BW6, Deutsches Elektronen Synchrotron, Hamburg, Germany), M. Walsh (BM14, European Synchrotron Radiation Facility, Grenoble, France), and C. Schultz-Briese (PX, Swiss Light Source at the Paul Scherrer Institute, Villigen, Switzerland) for help during synchrotron data collection; R. Huber (Max-Planck-Institut fur Biochemie, Martinsried, Germany) for continuous support; and S. Macedo-Ribeiro (Centro de Neurociências e Biologia Celular, Coimbra, Portugal) for help at the initial stage of the project. This work was supported by the Priority Program “Radicals in Enzymatic Catalysis” of the Deutsche Forschungsgemeinschaft (Bonn). B.M.M. was supported in part by PRAXIS XXI (fellowship BD/9656/96 of the Fundação para a Ciência e a Tecnologia, Lisbon).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: 4-BUDH, 4-hydroxybutyryl-CoA dehydratase; FAD, flavin adenine dinucleotide; MCAD, medium chain acyl-CoA dehydrogenase.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1U8V).
Footnotes
De La Fortelle, E., Irwin, J. J. & Bricogne, G., Crystallographic Computing 7: Proceedings from the Macromolecular Crystallography Computing School, eds. Bourne, P. E. & Watenpaugh, K., Aug. 17–22, 1996, Seattle, WA, pp. 1–9.
References
- 1.Buckel, W. & Golding, B. T. (1999) FEMS Microbiol. Rev. 22, 523-541. [Google Scholar]
- 2.Buckel, W. (1996) FEBS Lett. 389, 20-24. [DOI] [PubMed] [Google Scholar]
- 3.Müh, U., Cinkaya, I., Albracht, S. P. & Buckel, W. (1996) Biochemistry 35, 11710-11718. [DOI] [PubMed] [Google Scholar]
- 4.Gerhardt, A., Cinkaya, I., Linder, D., Huisman, G. & Buckel, W. (2000) Arch. Microbiol. 174, 189-199. [DOI] [PubMed] [Google Scholar]
- 5.Scott, R., Näser, U., Friedrich, P., Cinkaya, I., Buckel, W. & Golding, B. T. (2004) Chem. Commun. (Cambridge, U.K.) 10, 1210-1211. [DOI] [PubMed] [Google Scholar]
- 6.Smith, D. M., Buckel, W. & Zipse, H. (2003) Angew. Chem. Int. Ed. Engl. 42, 1867-1870. [DOI] [PubMed] [Google Scholar]
- 7.Çinkaya, I. (2003) Ph.D. thesis (Philipps-Universität, Marburg, Germany).
- 8.Näser, U., Pierik, A. J., Scott, R., Çinkaya, I., Buckel, W. & Golding, B. T. (2004) Bioorg. Chem., in press. [DOI] [PubMed]
- 9.Müh, U., Buckel, W. & Bill, E. (1997) Eur. J. Biochem. 248, 380-384. [DOI] [PubMed] [Google Scholar]
- 10.Stubbe, J. & van Der Donk, W. A. (1998) Chem. Rev. 98, 705-762. [DOI] [PubMed] [Google Scholar]
- 11.Frey, P.A. (2001) Annu. Rev. Biochem. 70, 121-148. [DOI] [PubMed] [Google Scholar]
- 12.Hans, M., Bill, E., Cirpus, I., Pierik, A. J., Hetzel, M., Alber, D. & Buckel, W. (2002) Biochemistry 41, 5873-5882. [DOI] [PubMed] [Google Scholar]
- 13.Ghisla, S. & Thorpe, C. (2004) Eur. J. Biochem. 271, 494-508. [DOI] [PubMed] [Google Scholar]
- 14.Kabsch, W. (1993) J. Appl. Crystallogr. 26, 795-800. [Google Scholar]
- 15.Weeks, C.M. & Miller, R. (1997) J. Appl. Crystallogr. 27, 613-621. [Google Scholar]
- 16.De La Fortelle, E., Irwin, J. J. & Bricogne, G. (1997) sharp, A Program for Statistical Heavy-Atom Refinement and Phasing (Global Phasing Ltd., Cambridge, U.K.).
- 17.Jones, T. A. (1992) in Molecular Replacement, eds. Dodson, E. J., Gover, S. & Wolf, W. (SERC Daresbury Laboratory, Warrington), pp. 91-105.
- 18.Turk, D. (1992) Ph.D. thesis (Technische Universität München, Munich).
- 19.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta. Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 20.Sheldrick, G. M. & Schneider, T. R. (1997) Methods Enzymol. 277, 319-343. [PubMed] [Google Scholar]
- 21.Laskowski, R. A., McArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 22.Kim, J.-J-P. & Wu, J. (1988) Proc. Natl. Acad. Sci. USA 85, 6677-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beinert, H. (1963) in Enzymes, eds. Pyer, P. D., Lardy, H. & Myrback, K. (Academic, New York), pp. 447-466.
- 24.Flint, D. H. & Allen, R. M. (1996) Chem. Rev. 96, 2315-2334. [DOI] [PubMed] [Google Scholar]
- 25.Beinert, H., Kennedy, M. C. & Stout, C. D. (1996) Chem. Rev. 96, 2335-2374. [DOI] [PubMed] [Google Scholar]
- 26.Link, T. A. (1997) FEBS Lett. 412, 257-264. [DOI] [PubMed] [Google Scholar]
- 27.Volbeda, A., Charon, M. H., Piras, C., Hatchikian, E. C., Frey, M. & Fontecilla-Camps, J. C. (1995) Nature 373, 580-587. [DOI] [PubMed] [Google Scholar]
- 28.Peters, J. W., Lanzilotta., W. N., Lemon, B. J. & Seefeldt, L. C. (1998) Science 282, 1853-1858. [DOI] [PubMed] [Google Scholar]
- 29.Cooper, S. J., Garner, C. D., Hagen, W. R., Lindley, P. F. & Bailey, S. (2000) Biochemistry 33, 15044-15054. [DOI] [PubMed] [Google Scholar]
- 30.Dobbek, H., Svetlitchnyi, V., Gremer, L., Huber, R. & Meyer, O. (2001) Science 293, 1281-1285. [DOI] [PubMed] [Google Scholar]
- 31.Drennan, C. L., Heo, J., Sintchak, M. D., Schreiter, E. & Ludden, P. W. (2001) Proc. Natl. Acad. Sci. USA 98, 11973-11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doukov, T. I., Iverson, T. M., Seravalli, J., Ragsdale, S. W. & Drennan, C.L. (2002) Science 298, 567-572. [DOI] [PubMed] [Google Scholar]
- 33.Darnault, C., Volbeda, A., Kim, E. J., Legrand, P., Vernede, X., Lindahl, P. A. & Fontecilla-Camps, J. C. (2003) Nat. Struct. Biol. 10, 271-279. [DOI] [PubMed] [Google Scholar]
- 34.Hunsicker-Wang, L. M., Heine, A., Chen, Y., Luna, E. P., Todaro, T., Zhang, Y. M., Williams, P. A., McRee, D. E., Hirst, J., Stout, C. D. & Fee, J. A. (2003) Biochemistry 42, 7303-7317. [DOI] [PubMed] [Google Scholar]
- 35.Anxolabéhère-Mallart, E., Glaser, T., Frank. P., Aliverti, A., Zanetti, G., Hedman, B., Hodgson, K. O. & Solomon, E. I. (2001) J. Am. Chem. Soc. 123, 5444-5452. [DOI] [PubMed] [Google Scholar]
- 36.Holm, R. H., Kennepohl, P. & Solomon, E. I. (1996) Chem. Rev. 96, 2239-2314. [DOI] [PubMed] [Google Scholar]
- 37.Kim, J.-J. P., Wang, M. & Paschke, R. (1993) Proc. Natl. Acad. Sci. USA 90, 7523-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amyes, T. L. & Richard, J. P. (1992) J. Am. Chem. Soc. 114, 10297-10302. [Google Scholar]
- 39.Çinkaya, I., Buckel, W., Medina, M., Gomez-Moreno, C. & Cammack, R. (1997) Biol. Chem. Hoppe-Seyler 378, 843-849. [DOI] [PubMed] [Google Scholar]
- 40.Müller, F., Ghisla, S. & Bacher, A. (1988) in Vitamine II: Wasserlösliche Vitamine, eds. Isler, O., Brubacher, G., Ghisla, S. & Kräutler, B. (Thieme, Stuttgart), pp. 51-159.
- 41.Cornforth, J. W. (1959) J. Lipid Res. 1, 3-28. [Google Scholar]
- 42.Lenn, N. D., Shih, Y., Stankovich, M. T. & Liu, H.-W. (1989) J. Am. Chem. Soc. 111, 3065-3067. [Google Scholar]
- 43.Wang, W., Fu, Z., Zhou, J. Z., Kim, J.-J. P. & Thorpe, C. (2001) Biochemistry 40, 12266-12275. [DOI] [PubMed] [Google Scholar]
- 44.DeLano, W. L. (2003) The PyMol Molecular Graphics System (DeLano Scientific LLC, San Carlos, CA).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.