Abstract
A novel alanine dehydrogenase (AlaDH) showing no significant amino acid sequence homology with previously known bacterial AlaDHs was purified to homogeneity from the soluble fraction of the hyperthermophilic archaeon Archaeoglobus fulgidus. AlaDH catalyzed the reversible, NAD+-dependent deamination of l-alanine to pyruvate and NH4+. NADP(H) did not serve as a coenzyme. The enzyme is a homodimer of 35 kDa per subunit. The Km values for l-alanine, NAD+, pyruvate, NADH, and NH4+ were estimated at 0.71, 0.60, 0.16, 0.02, and 17.3 mM, respectively. The A. fulgidus enzyme exhibited its highest activity at about 82°C (203 U/mg for reductive amination of pyruvate) yet still retained 30% of its maximum activity at 25°C. The thermostability of A. fulgidus AlaDH was increased by more than 10-fold by 1.5 M KCl to a half-life of 55 h at 90°C. At 25°C in the presence of this salt solution, the enzyme was ∼100% stable for more than 3 months. Closely related A. fulgidus AlaDH homologues were found in other archaea. On the basis of its amino acid sequence, A. fulgidus AlaDH is a member of the ornithine cyclodeaminase-μ-crystallin family of enzymes. Similar to the μ-crystallins, A. fulgidus AlaDH did not exhibit any ornithine cyclodeaminase activity. The recombinant human μ-crystallin was assayed for AlaDH activity, but no activity was detected. The novel A. fulgidus gene encoding AlaDH, AF1665, is designated ala.
Alanine dehydrogenase (AlaDH) catalyzes the reversible deamination of l-alanine to pyruvate (NAD+ + l-alanine + H2O ↔ NADH + H+ + pyruvate + NH4+).
The enzyme can serve both assimilatory and dissimilatory metabolic functions in various organisms. The characteristic NH4+ Km value for AlaDH generally has been used as an arbitrary predictor of the assimilatory versus dissimilatory role of AlaDHs. AlaDHs from Streptomyces fradiae and pea and soybean bacteroids with relatively high affinities for ammonia (Km, <10 mM) function primarily for NH4+ assimilation (3, 41, 49), while AlaDHs from other organisms have lower affinities for ammonia (Km, >10 mM) and are therefore expected to act effectively in an assimilatory fashion only when high concentrations of NH4+ are present or when other pathways are repressed (6, 14, 29, 30, 35). In Bacillus sp., AlaDH has been implicated as the critical dissimilatory enzyme for generating pyruvate, the energy source for sporulation (39).
AlaDHs have been purified from a variety of bacteria and consist of a single catalytic subunit that, depending on the organism, assumes a homohexameric (11, 14, 23, 29, 30, 32, 35, 47), homotetrameric (5, 37, 41, 48), or homooctameric (49) quaternary complex. A comparison of the bacterial AlaDH amino acid sequences indicates that the enzyme is highly conserved, with ∼50% sequence identity between individual AlaDHs (4). Significant sequence homology is also present between the bacterial AlaDHs and the alpha subunits and the N termini of bacterial and eukaryotic nicotinamide nucleotide transhydrogenase, respectively (4, 52). The AlaDH and pyridine nucleotide families of enzymes have glycine-rich regions in common that have been suggested to form a nucleotide-binding Rossman fold domain. Recently, the three-dimensional structure of the AlaDH from the cyanobacterium Phormidium lapideum was solved (4). Each AlaDH subunit of the hexameric enzyme consists of two compact domains that are separated by a cleft. The C terminus assumes a Rossman fold domain that binds NAD+, and the substrate-binding site is located deep within the cleft and close to the binding site of the nicotinamide ring. Given the amino acid sequence similarity among the AlaDHs characterized to date, it can be expected that they will assume a similar tertiary structure.
Very little is known about archaeal AlaDHs. Two AlaDHs were purified or partially enriched from the halophilic archaea Halobacterium salinarum and Halobacterium cutirubrum, respectively (15, 17). Because the purification of these enzymes was performed about 20 years ago, no nucleotide sequence information for their genes is available. In addition, the ald gene encoding the bacterial-type AlaDH appears to be absent in the archaeal genomes that have been published thus far. The goals of this work were to establish whether AlaDH is also present in other archaea and to elucidate how it differs from the bacterial AlaDHs. With a native polyacrylamide gel electrophoresis (PAGE) activity staining procedure, AlaDH activity was detected in the hyperthermophilic archaeon Archaeoglobus fulgidus. A. fulgidus grows optimally at 83°C and uses sulfate reduction for energy generation (43, 44, 54). We describe here the purification and characterization of a novel archaeal AlaDH with no homology to known bacterial AlaDHs. Homologues of the A. fulgidus enzyme are also present in other archaea, suggesting that the archaeal AlaDHs belong to a distinct evolutionary class of enzymes that is related to the μ-crystallin and ornithine cyclodeaminase (OCD) enzyme families.
MATERIALS AND METHODS
Organisms and growth conditions.
A. fulgidus VC-16 (DSM 4304) served as the source of wild-type AlaDH and of the genomic DNA used in the PCR procedure to obtain the gene AF1665 encoding AlaDH. A. fulgidus was grown anaerobically on 10 mM sodium lactate and 30 mM sodium sulfate at 83°C, essentially as described by Zellner et al. (54) and modified as described by Vadas et al. (45). A. fulgidus biomass was produced in a 70-liter culture grown in a custom-built 100-liter glass-lined steel fermentor (Pfaudler, Rochester, N.Y.). The cells were harvested in the late log phase (∼24 h after inoculation) at an optical density at 660 nm of 0.5 to 0.6 by concentration with an A/G Technology hollow-fiber unit (nominal molecular mass cutoff, 500 kDa) and subsequent centrifugation for 20 min at 16,000 × g. Cell yields were approximately 80 g (wet weight) per 70-liter culture. Escherichia coli strains DH5α (NEB) and BL21(λDE3) (Stratagene) were used for plasmid maintenance and gene overexpression, respectively. For the culture of the E. coli strains, Luria-Bertani liquid and solid media containing ampicillin at a 100-mg/liter concentration were used.
Detection of AlaDH activity in native gels.
The soluble fraction was prepared by a previously published high-pressure homogenization and centrifugation protocol (45) and separated by native PAGE with stacking and resolving gels of 4 and 8% polyacrylamide, respectively, with the Bio-Rad Mini-PROTEAN system. After electrophoresis, the gel was transferred to a 15-ml anaerobic culture tube sealed with a butyl rubber stopper to which the following anaerobic solutions were added: 5.0 ml of 100 mM Tricine buffer (pH 8.0), with 10 mM l-alanine, 0.05 mM phenazine methosulfate, 0.1 mg of methyl thiazolyl tetrazolium/ml, and 0.5 mM NAD+. After incubation for 5 min at 85°C, AlaDH activity was identified as a dark purple formazan band in the gel (24).
Enzyme and protein assays.
The protein concentration was measured by the Bradford assay (Bio-Rad) with bovine serum albumin as the standard. AlaDH activity was measured as oxidative deamination of alanine and as reductive amination of pyruvate by procedures adapted from previously described methods (28, 30). The assays were performed aerobically in stoppered quartz cuvettes at 82°C unless indicated otherwise. All assays were performed in triplicate, and the error range was within 6%. The oxidative deamination assay mixture contained 100 mM Tricine-KOH (pH 8.5) (titrated at 25°C), 5 mM l-alanine, and 2.5 mM NAD+. The reductive amination assay mixture consisted of 100 mM Tricine-KOH (pH 8.5), 700 mM NH4Cl, 5 mM pyruvic acid, and 0.08 mM NADH unless indicated otherwise. Rates were measured spectrophotometrically by following the reduction of NAD+ for the deamination reaction or the oxidation of NADH for the amination reaction at 340 nm. One unit of activity is defined as 1 μmol NAD(H) reduced (or oxidized) per min. OCD activity was measured by a modified version of a previously published assay (33, 34). The reaction mixture, which contained 100 mM Tricine-KOH (pH 8.0) (titrated at 25°C), 10 mM ornithine, 1.0 mM NAD+, and 0.3 μg of purified AlaDH, was incubated for 24 h at 37 or 80°C. The samples then were separated on cellulose K2F thin-layer chromatography plates (Whatman Ltd.) with 3:1 n-propanol-NH3OH as the mobile phase. The presence of primary and secondary amines was detected with 0.2% ninhydrin dissolved in ethanol (10, 33). l-Proline and ornithine were used as standards.
Purification of AlaDH from A. fulgidus.
All purification procedures were carried out aerobically at 25°C. The soluble fraction was prepared by a previously published high-pressure homogenization and centrifugation protocol (45). The soluble fraction (∼400 mg of protein) was applied to a 5-ml Q Sepharose column (Pharmacia) equilibrated with 20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)-NaOH (pH 7.0). After the column was washed with PIPES buffer containing 180 mM NaCl, AlaDH activity was eluted at 240 mM NaCl. The eluted protein was diluted with buffer to 100 mM NaCl and applied to a 5-ml Red Agarose 120 affinity column (Sigma) equilibrated with 20 mM PIPES-NaOH (pH 7.0)-100 mM NaCl. The column was washed with the equilibration buffer, and AlaDH was eluted with 350 mM NaCl in PIPES buffer. Ammonium sulfate was added to the eluted protein to a final concentration of 1.0 M. The solution was centrifuged and loaded onto a 1-ml Butyl Sepharose hydrophobic-interaction column (Pharmacia) equilibrated with PIPES buffer containing 1.0 M (NH4)2SO4. The column was washed with 0.75 M (NH4)2SO4, and AlaDH was eluted with 0.55 M (NH4)2SO4 in PIPES buffer. The eluted protein was passed through a Centricon filter with a 100-kDa molecular mass cutoff (Amicon) and subsequently concentrated with a Centricon filter with a 30-kDa molecular mass cutoff. The concentrated protein was stored at −80°C.
SDS-gel electrophoresis.
Proteins were separated on precast 20% homogeneous polyacrylamide gels under denaturing conditions with the Pharmacia PhastSystem. The protein samples were heated for 30 min at 100°C in 1% sodium dodecyl sulfate (SDS) prior to loading (24). The apparent molecular masses of the subunits of the purified AlaDH were estimated from the electrophoretic motility of the denatured protein. The molecular mass standards for SDS-PAGE were bovine albumin (66 kDa), chicken egg albumin (45 kDa), glyceraldehyde-3-posphate dehydrogenase (36 kDa), bovine carbonic anhydrase (29 kDa), bovine pancreas trypsinogen (24 kDa), soybean trypsin inhibitor (20 kDa), and bovine milk α-lactalbumin (14 kDa) (Sigma).
Native molecular weight determination.
The native molecular weight of the AlaDH was determined with a Superose 6 (Pharmacia) gel filtration column. RNase A (13.7 kDa), chymotrypsinogen A (25 kDa), ovalbumin (43 kDa), and bovine serum albumin (67 kDa) were used as standards. The column was run with 20 mM PIPES buffer-300 mM NaCl (pH 7.0).
AlaDH pH and temperature optima.
The pH optima for the AlaDH-catalyzed deamination and amination reactions were determined with the following buffers and pH values at 82°C: 100 mM morpholineethanesulfonic acid (MES) at pHs 6.1 and 6.15; 100 mM Tricine at pHs 6.5, 6.75, 6.8, 7.3, and 7.4; and 100 mM glycine at pHs 7.5, 7.9, 8.0, and 8.5. Temperature optima were determined at the indicated temperatures.
pH stability.
The concentrated enzyme preparation was diluted 30-fold in the following buffers and pH values at 25°C: 100 mM glycine at pH 9.5, 100 mM Bicine at pH 9.0, 100 mM Tricine at pHs 8.5 and 8.0, 100 mM potassium phosphate at pHs 7.5 and 7.0, and 100 mM MES at pH 6.5. The samples were stored at 25°C in sterile microcentrifuge tubes, and the activity was monitored with the reductive amination assay over a period of 2 weeks.
Effect of salt on stability and activity.
The desalted enzyme was concentrated with a centrifugal filter with a 30-kDa molecular mass cutoff (Amicon). Various salts [NaCl, KCl, (NH4)2SO4, or K2HPO4] were added to the concentrated enzyme to give a final concentration of 1.0 M in PIPES buffer (pH 7.0). The samples were sealed in serum vials with butyl rubber stoppers and incubated at 90°C for 68 h. Periodically, samples were removed from the vials and AlaDH activity was measured with the reductive amination assay. The effect of the KCl concentration on enzyme activity was determined by adding 0.0, 0.5, 1.0, 1.5, and 2.0 M KCl to the reductive amination assay mixture.
Temperature-dependent stability.
The AlaDH in 20 mM PIPES buffer (pH 7.0) (titrated at 25°C)-1.5 M KCl was incubated in stoppered serum vials at 25, 60, 90, and 100°C. The activity was monitored with the reductive amination assay on samples immediately after their removal from the vials.
N-terminal amino acid sequence analysis.
The purified protein was run on an SDS-20% polyacrylamide gel and transferred to a Sequi-Blot polyvinylidene difluoride membrane (Bio-Rad). N-terminal amino acid sequence analysis was performed by the Protein Microsequencing Facility of the University of California, Los Angeles.
Phylogenetic analysis.
Amino acid sequences homologous to the A. fulgidus AlaDH encoded by the AF1665 gene were identified with FASTA and BLASTp databases. The homologous amino acid sequences were aligned with Multialign (9). The amino acid alignment of selected proteins was displayed with GeneDoc (27). For database accession numbers, see the legend to Fig. 6. A phylogenetic tree was constructed with the neighbor-joining algorithm of TREECON (50). Evolutionary distances were calculated by Poisson correction with 1,000 bootstrap replicates.
FIG. 6.
Comparison of the amino acid sequence of A. fulgidus AlaDH to those of selected homologues. For accession numbers, see the legend to Fig. 7. The thick line denotes the proposed NAD-binding site. The aligned C-terminal 23 amino acid residues of A. fulgidus AlaDH are not shown since the alignment did not reveal any significant conservation.
Cloning and overexpression of ala and purification of recombinant AlaDH.
Cloning and overexpression of ala and purification of recombinant AlaDH were done in parallel and in accordance with the protocol described by Smith et al. (42).
RESULTS
Detection of AlaDH activity in A. fulgidus.
To examine the properties of an archaeal AlaDH, two archaeal representatives, A. fulgidus and Pyrobaculum aerophilum, were assayed spectrophotometrically for the reductive amination of pyruvate and for the oxidative deamination of alanine; however, no activity was detectable in either organism. Since both archaea had been cultured in the presence of yeast extract and peptone, it was possible that feedback inhibition by external amino acids resulted in very low AlaDH activity. To detect very low enzyme activity, a high-temperature, native PAGE activity assay was used. The advantage of such an approach is that low-abundance proteins are separated and assayed in situ, resulting in sensitive enzyme detection. Following electrophoreses, the gels were stained for oxidative deamination of alanine and concomitant reduction of NAD+. The purple formazan band indicating AlaDH activity in A. fulgidus formed within 5 min at 85°C and was dependent on the presence of l-alanine in the assay buffer (data not shown). In contrast, no AlaDH activity could be detected in the soluble fraction of P. aerophilum (data not shown). AlaDH activity in the A. fulgidus soluble fraction was below the detection limit of the spectrophotometric assay used and could be measured only after enrichment of the enzyme.
Purification of AlaDH.
The enzyme was purified to apparent homogeneity by Q Sepharose ion-exchange chromatography, Red Agarose 120 affinity chromatography, Butyl Sepharose hydrophobic interaction chromatography, and centrifugal filtration through a membrane with a 100-kDa cutoff (Table 1). After the final purification step, the enzyme was enriched 572-fold relative to the preparation obtained from the first chromatography column. Since AlaDH activity could not be quantified in the soluble fraction, recovery and fold purification were based on the AlaDH activity recovered from the Q Sepharose column. The purified AlaDH protein has an Mr of 35,000 as determined by SDS-PAGE. By size exclusion chromatography, the apparent molecular weight of the native protein was determined to be 76,000, suggesting that the enzyme is a homodimer (data not shown). The purified enzyme did not exhibit any significant absorption in the visible range, indicating that the enzyme does not contain chromophores such as flavins, Fe-S centers, or cytochromes.
TABLE 1.
Purification of AlaDH from A. fulgidus
| Prepn | Protein (mg) | Activity (Ua) | Recoveryc (%) | Sp act (U/mg) | Purification (fold) |
|---|---|---|---|---|---|
| Soluble fraction | 399 | NDb | ND | ND | ND |
| Q Sepharose | 48.3 | 17.2 | 100 | 0.3 | 1 |
| Red agarose | 0.8 | 16.7 | 97 | 22 | 62 |
| Butyl Sepharose | 0.4 | 10.4 | 60 | 25 | 70 |
| 100-kDa filter | 0.04 | 7.3 | 43 | 203 | 572 |
One unit is defined as the oxidation of 1 μmol of NADH per min in the reductive amination of pyruvate to l-alanine.
ND, not detected.
Recovery and fold purification assume that all of the AlaDH was recovered from the Q Sepharose step.
Kinetic properties of AlaDH.
After the final purification step, AlaDH catalyzed the oxidative deamination of alanine with a specific activity of 10.6 U/mg (kcat, 6.1 s−1) and the reductive amination of pyruvate with a specific activity of 203 U/mg (kcat, 118 s−1) (Table 2). The apparent affinity for pyruvate was fourfold higher than that for l-alanine (Table 2). The enzyme has an affinity for NH4+ that is in the same range as bacterial dissimilatory-type AlaDHs; however, the low abundance of AlaDH in A. fulgidus grown under rich-medium conditions argues against a dissimilatory function for this enzyme. An assimilatory function for A. fulgidus AlaDH is also supported by its low Km for NADH. Its Km for NAD+ was more than an order of magnitude higher than that for NADH. A. fulgidus AlaDH is specific for NAD+; NADP+ was not used as a substrate. As is characteristic of many bacterial AlaDHs, the A. fulgidus enzyme exhibits substrate inhibition by pyruvate and NADH, but not by NAD+ (Table 2) (2, 13, 29, 41).
TABLE 2.
Kinetic properties of A. fulgidus AlaDH
| Assay and substrate | kcat (s−1) | Km (mM) | kcat/Km (M−1 s−1, 103) | Ki (mM) |
|---|---|---|---|---|
| Oxidative deaminationa | ||||
| l-Alanine | 6.1 | 0.71 | 9 | 16.8 |
| l-2-Aminobutyrate | 9.6 | 0.085 | 113 | 4.7 |
| NAD+ | 6.1 | 0.60 | 10 | NIb |
| Reductive aminationa | ||||
| Pyruvate | 118 | 0.16 | 739 | 27.8 |
| Ketobutyrate | 143 | 0.48 | 298 | 4.8 |
| Oxaloacetate | 113 | 0.97 | 116 | NI |
| NADHd | 118 | 0.02 | 5,900 | 0.12 |
| NH4+ | 118 | 17.3 | 7 | NDc |
The assay was performed at pH 8.5 (titrated at 25°C) and 82°C.
NI, no inhibition.
ND, not determined.
With pyruvate as the substrate.
Substrate specificity of AlaDH.
A variety of l-amino acids and 2-oxo acids were tested as substrates for A. fulgidus AlaDH. In the deamination direction, only l-2-aminobutyrate is deaminated efficiently, at a rate about 1.5-fold higher than that determined for alanine (Tables 2 and 3). A ninefold lower Km for l-2-aminobutyrate versus l-alanine suggests that l-2-aminobutyrate is the preferred substrate in the deamination reaction (Table 2). l-Valine, l-serine, l-threonine, l-aspartate, and l-isoleucine also served as substrates but at rates ≤12% of that for l-alanine (Table 3). Activity with several other natural amino acids was not detected. In the reductive amination direction, the enzyme exhibited high activity with 2-ketobutyrate and oxaloacetate compared to pyruvate (Tables 2 and 4). The Michaelis constants for 2-ketobutyrate and oxaloacetate were three- and sixfold larger, respectively, compared to the Km for pyruvate (Table 2). Larger 2-keto acids of five and six carbons such as 2-ketovalerate and 2-ketocaproate were not suitable substrates (Table 4). The pyruvate derivatives 3-fluoropyruvate and 3-hydroxypyruvate resulted in lower activity compared to that obtained with pyruvate. The more bulky compound phenylpyruvate was not aminated reductively at a measurable rate.
TABLE 3.
Substrate specificity for the oxidative deamination reaction of AlaDH
| Substratea | Sp actc (U/mg) |
|---|---|
| l-Alanine | 10.6 ± 0.6 |
| l-2-Aminobutyrateb | 16.4 ± 1.0 |
| l-Valine | 1.30 ± 0.05 |
| l-Serine | 1.06 ± 0.05 |
| l-Threonine | 0.85 ± 0.05 |
| l-Isoleucine | 0.42 ± 0.02 |
| l-Aspartate | 0.42 ± 0.02 |
| l-Lysine | <0.01 |
| l-Tyrosine | <0.01 |
| l-Proline | <0.01 |
| l-Tryptophan | <0.01 |
| l-Phenylalanine | <0.01 |
Substrate concentration, 5 mM.
l-2-Aminobutyrate concentration, 1 mM.
The assays were performed at pH 8.5 (titrated at 25°C) and 82°C.
TABLE 4.
Substrate specificity of the reductive amination reaction of AlaDH
| Substrate (concn, mM) | Sp acta (U/mg) |
|---|---|
| Pyruvate (5) | 203 ± 12 |
| 3-Fluoropyruvate (5) | 40.6 ± 2.4 |
| Hydroxypyruvate (5) | 20.3 ± 1.2 |
| Oxaloacetate (1) | 195 ± 12 |
| Ketobutyrate (1) | 246 ± 15 |
| Ketovalerate (10) | 12.2 ± 0.7 |
| Ketocaproate (10) | 4.1 ± 0.2 |
| Phenylpyruvate (10) | <0.1 |
The assays were performed at pH 8.5 (titrated at 25°C) and 82°C.
pH and temperature optima of AlaDH.
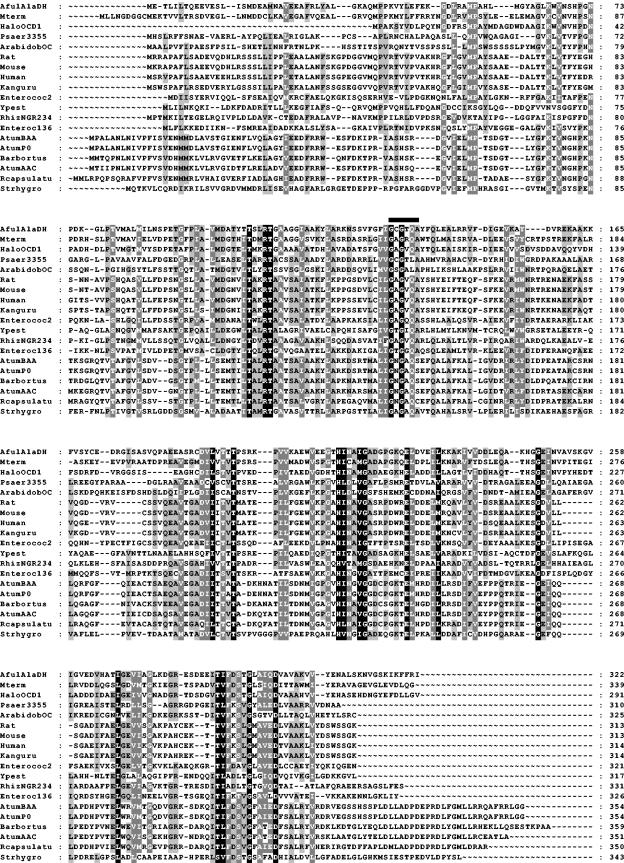
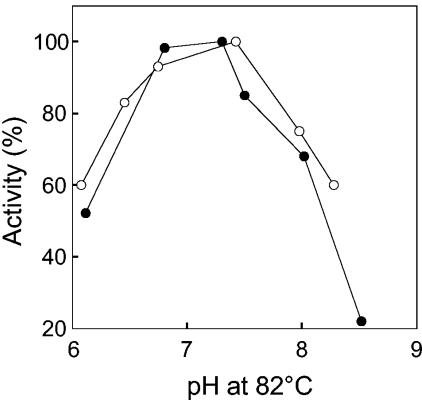
The pH optimum of AlaDH was ∼7.0 for both the deamination and amination reactions (Fig. 1). AlaDH was not affected by prolonged incubation in buffers at the various pH values, indicating that the observed pH optimum is a reflection of the protonation and deprotonation of active site amino acid residues. The temperature optimum was approximately 82°C as measured for the reductive amination of pyruvate assay (Fig. 2). This temperature optimum correlates well with the optimum growth temperature of A. fulgidus (83°C) (54). It is noteworthy that 30% of the activity remained at 25°C.
FIG. 1.
pH dependence of A. fulgidus AlaDH activity for oxidative deamination (filled circles) and reductive amination (open circles). One hundred percent activity corresponds to 203 U/mg.
FIG. 2.
Temperature dependence of A. fulgidus AlaDH activity. One hundred percent activity corresponds to 203 U/mg.
Effects of salt on AlaDH temperature stability.
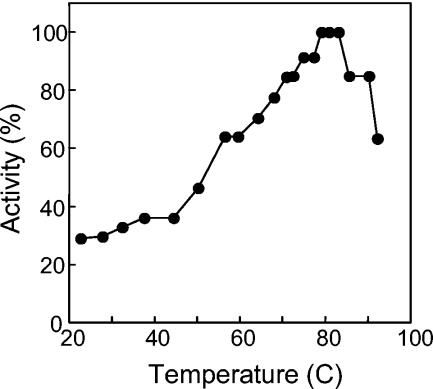
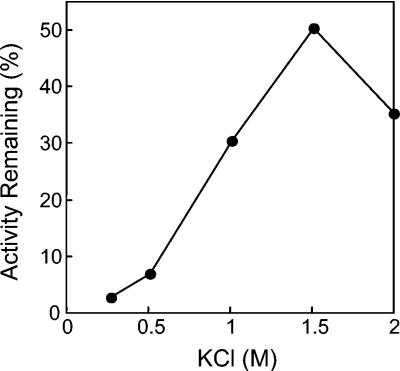
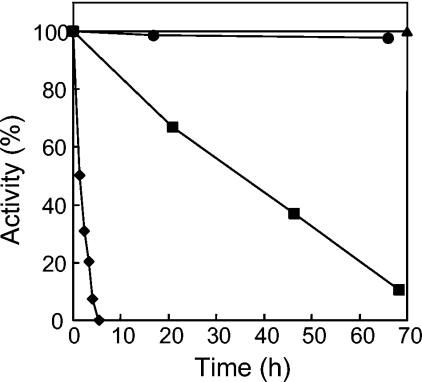
When incubated at 90°C for 2 h, AlaDH lost its activity completely (Fig. 3). However, the addition of various salts at a 1.0 M concentration increased stability at 90°C by more than an order of magnitude (Fig. 3). Of these, KCl proved to be optimal for stabilizing enzyme activity. The optimum KCl concentration for stability was determined to be 1.5 M (Fig. 4). In 1.5 M KCl, the enzyme half-life at 90°C was extended to 55 h (Fig. 5). At 25°C, no loss of activity could be detected for more than 3 months in the presence of 1.5 M KCl. AlaDH activity was not measurably affected by the addition of up to 2.0 M KCl to the enzyme assay.
FIG. 3.
Effect of salt on the temperature stability of A. fulgidus AlaDH. The enzyme was incubated at 90°C in the presence of no added salt (filled diamonds), 1.0 M KCl (filled triangles), 1.0 M NaCl (filled circles), 1.0 M (NH4)2SO4 (filled squares), or 1.0 M K2HPO4 (open circles). One hundred percent activity corresponds to 203 U/mg.
FIG. 4.
Activity of A. fulgidus AlaDH after 55 h at 90°C as a function of KCl concentration. One hundred percent activity corresponds to 203 U/mg.
FIG. 5.
Temperature stability of A. fulgidus AlaDH. The enzyme was incubated with 1.5 M KCl at ambient temperature (triangles), 60°C (circles), 90°C (squares), or 100°C (diamonds). One hundred percent activity corresponds to 203 U/mg.
N-terminal amino acid sequence analysis of AlaDH.
The N-terminal sequence of A. fulgidus AlaDH was determined to be Met-Glu-Thr-Leu-Ile-Leu-Thr-Gln-Glu-Glu-Val-Glu-Ser-Leu. This sequence corresponds to the translated AF1665 gene locus in the A. fulgidus genome (19). The protein predicted by AF1665 consists of 323 amino acids and has a calculated molecular weight of 34,828, which is consistent with the value estimated for the purified AlaDH protein by SDS-PAGE. The theoretical pI of the protein is 5.18.
OCD activity.
The gene locus AF1665 had been annotated to encode an OCD. OCD catalyzes the conversion of ornithine to proline in the catabolism of opines, which are synthesized in crown gall tumors of plants infected by Agrobacterium tumefaciens (7, 33, 36). To test whether AlaDH also exhibited OCD activity, the purified enzyme was incubated with ornithine at 80 and 37°C; however, no proline was detected (data not shown). This suggests that A. fulgidus AlaDH is not an OCD. The detection limit of the method was approximately 1% of the activity measured for l-alanine deamination.
Expression of ala and characterization of the recombinant enzyme.
To confirm that the AF1665 gene locus encodes AlaDH, the AF1665 gene was cloned from the A. fulgidus chromosome into an E. coli overexpression vector (42). After induction of gene expression in E. coli, a 35-kDa protein was produced. The activity of the recombinant protein in the E. coli soluble cell fraction was barely detectable, i.e., <0.05 U/mg, but increased to 113 U/mg after incubation for 20 min at 80°C, suggesting that the enzyme requires an elevated temperature to assume an active conformation. Recombinant A. fulgidus AlaDH was purified to homogeneity, and the measured kinetic and stability properties of the recombinant AlaDH protein were indistinguishable from those of the native enzyme (data not shown).
Analysis of primary structure and phylogenetic relationships.
FASTA and BLASTP searches with the A. fulgidus AlaDH amino acid sequence revealed homologous proteins, which were aligned with ClustalW (Fig. 6). No apparent homology to any bacterial AlaDHs was identified, suggesting that A. fulgidus AlaDH constitutes an independent class of enzymes. The AF1665 gene is now designated ala to distinguish it from the ald genes encoding bacterial AlaDHs.
A. fulgidus AlaDH is 31 to 34% identical to the family of OCD enzymes in A. tumefaciens. It is also about 33 to 35% identical to the mammalian μ-crystallin family that includes the human thyroid hormone-binding protein (THBP) (51). μ-Crystallins are structural proteins present in the eye lenses of marsupials and are found in various tissues in other organisms, including humans, where their function is uncertain (18, 38, 53). Other homologous proteins (about 27% identity) include the lysine cyclodeaminase from Streptomyces hygroscopicus (16), an enzyme that is more closely related to the OCD family, and several hypothetical proteins from various bacteria and archaea. The proteins most closely related to A. fulgidus AlaDH are encoded by open reading frames from other archaea, the hyperthermophile Methanothermobacter thermoautotrophicum and the mesophile Halobacterium sp. strain NCR-1, with 49 and 46% sequence identity, respectively, which supports a prediction that these proteins will prove to be AlaDHs.
Visual inspection of the aligned amino acid sequences revealed highly conserved residues (Fig. 6). Among these, a GxGxxA/S motif (Gly132, Gly134, and Ala137 in the A. fulgidus sequence), suggesting a NAD+ binding site, is located in the central part of the proteins. While the glycine residues are conserved in the entire family of enzymes, Gly/Ala is not conserved completely.
On the basis of the sequence alignment, a phylogenetic tree was constructed (Fig. 7). The tree branches into 10 major groups with long lineages, indicating the functional diversity of the proteins. Because of the long lineages, it was difficult to assign an unambiguous relationship of each group to another. Therefore, the tree is shown in a collapsed form, in which the relationship of the groups to one another is not taken into consideration. Three of the groups contain proteins with a known function. They include group I with the subgroup of μ-crystallin proteins, group V with OCDs and lysine cyclodeaminases (RapL of S. hygroscopicus), and group X containing A. fulgidus AlaDH. While nine groups appear to consist of proteins with functions that are predicted to be identical or very similar, group I contains proteins from very diverse organisms such as mammals, insects, yeast, plants, bacteria, and a strikingly large number of hyperthermophilic archaea. Whether proteins in this group have the same or diverse activities has yet to be established.
FIG. 7.
Condensed phylogenetic tree of the archaeal AlaDH-OCD-μ-crystallin family. The values at the internodes are bootstrap values. The Roman numerals indicate the 10 subfamilies. The letter coding behind each genus name indicates the gene locus or protein, where available. In some cases the beginning of the accession number was used to differentiate paralogues. The organisms, in alphabetical order with the accession numbers in parentheses, are Aeropyrum pernix APE0916 (Q9YDJ7), A. tumefaciens AGR_L_247 (AAK88692), A. tumefaciens AAF (AAF77139), A. tumefaciens AAL44 (AAL44751), A. tumefaciens AAL45 (AAL45552), A. tumefaciens AAL46 (AAL46252), A. tumefaciens Q44 (Q44332), A. tumefaciens Q9W (Q9WWA2), A. tumefaciens AGR_L_1807 (AAK89477), A. tumefaciens AGR_PTI_54 (AAK90974), A. tumefaciens AGRT4 (Q59701), A. tumefaciens AGRT5 (P09773), A. tumefaciens TIORF188 (Q9R693), A. tumefaciens TORF21 (Q9R468), Arabidopsis thaliana (Q9FLY0), A. fulgidus AlaDH AF1665 (O28608), Brucella abortus (Q59175), Brucella melitensis BMEII0397 (Q8YCY1), Corynebacterium glutamicum (Q9Z468), Drosophila melanogaster CG4872 (AAL68180), Halobacterium sp. strain NRC-1 OCD1 (Q9HN71), Halobacterium sp. strain NRC-1 OCD2 (Q9HQ24), Homo sapiens Mu (AAH18061), H. sapiens THBP (Q14894), Macropus fuliginosus Mu (Q28488), Mesorhizobium loti MLL6776 (Q988E2), M. loti MLR1729 (Q98JY2), M. loti MLR2282 (Q98IR6R), M. loti MLR3204 (Q98GR8), M. loti MLR7139 (Q987A2), Methanothermobacter thermautotrophicum MTH1495 (027539), Mus musculus Mu (O54983), Pseudomonas aeruginosa PA4908 (Q9HUQ5), P. aerophilum PAE2445 (Q8ZV60), Ralstonia solanacearum RSP0418 (Q8XSP9), R. solanacearum RSP0483 (Q8XSJ1), Rattus norvegicus Mu (Q9QYU4), Rhizobium meliloti P33 (P33728), R. meliloti Q92 (Q92U20), R. meliloti OCD1 (P58338), R. meliloti OCD2 (P58339), R. meliloti RA0256 (Q930E2), Rhizobium sp. strain NGR234 Y4TK (P55665), R. capsulatus (O68052), Schizosaccharomyces pombe SPAP11E10.01 (Q9HDZ0), Staphylococcus aureus SA0113 (Q99X98), Streptomyces avermitilis (Q93H28), S. hygroscopicus var. ascomyceticus FKBL (Q9KIE2), S. hygroscopicus RAPL lysine cyclodeaminase (Q54304), Sulfolobus solfataricus SSO0378 (Q980D6), S. solfataricus SSO3243 (Q97TY8), S. tokodaii ST2179 (Q96YJ3), S. tokodaii ST2334 (Q96Y33), Thermoplasma acidophilum TA0480 (Q9HKW3), T. volcanium TVG0859714 (Q97AH2), and Yersinia pestis YPO4090 (Q8Z9V2).
Assay of human μ-crystallin for AlaDH activity.
Many eye lens proteins exhibit enzyme activity, such as ɛ-crystallin, which was shown to have lactate dehydrogenase activity (53). To establish whether the human μ-crystallin exhibits AlaDH activity, purified recombinant human μ-crystallin was generously provided by G. Wistow (National Institutes of Health) and assayed for reversible AlaDH activity at 37°C; however, no activity was detected.
DISCUSSION
We report here the characterization of a novel, extremely thermostable AlaDH protein that shows no significant sequence homology to previously known AlaDHs. The A. fulgidus enzyme exhibits kinetics similar to those of AlaDHs from bacteria, albeit at a far higher temperature of 82°C, which is consistent with the optimal growth temperature for A. fulgidus (54). Previously described AlaDHs from thermophilic bacteria show remarkable thermal stability, yet the A. fulgidus enzyme is extraordinary by comparison. For example, the AlaDHs of Bacillus sphaericus and Thermus thermophilus exhibit half-lives of ∼70 and ∼10 min at 80 and 85°C, respectively (29, 47), whereas the A. fulgidus enzyme has a half-life of 50 min at 90°C at low ionic strength and ∼55 h at 90°C in 1.5 M KCl (Fig. 3 and 5). KCl is one of the known osmolytes in marine archaea and has been suggested to serve as a general stabilizer for proteins (21, 31). It is particularly noteworthy that this enzyme retains 30% of its maximal activity at 25°C. A. fulgidus AlaDH therefore represents a striking counterexample to the widely held generalization that extremely thermostable enzymes from hyperthermophiles should be expected to show very little or no activity at or near 25°C (20). Because of its high stability and activity at 25°C, A. fulgidus AlaDH lends itself to the in vitro synthesis of l-alanine (46).
Consistent with most of the characterized bacterial AlaDHs, the reductive amination of pyruvate to l-alanine is catalyzed by the archaeal enzyme at a higher maximum rate in vitro than is the deamination of l-alanine (14, 28-30, 32, 35, 37). However, enzymes with Km values above 10 mM may not be very effective for assimilation since intracellular ammonium concentrations usually do not exceed 10 mM. Experimentally determined kcat/Km values provide for a better comparison of AlaDH efficiency in the oxidative and reductive directions, yet the kcat/Km values compiled to date do not show a strong correlation with the physiological function of bacterial AlaDHs deduced from gene regulation experiments. A. fulgidus AlaDH exhibits similar catalytic efficiencies in both directions (kcat/Km,Ala = 9 ×103 M−1 s−1; kcat/Km,NH4+ = 7 ×103 M−1 s−1; Table 2). It is therefore not possible to discern the physiological role of AlaDH on the basis of its kinetic properties. A. fulgidus also contains a very abundant NADP+-dependent glutamate dehydrogenase with an ammonium Km of 4 mM that could serve as the primary nitrogen assimilatory enzyme (1). It is not known whether the ala gene is regulated in A. fulgidus; however, studies to elucidate the physiological role of this enzyme are under way.
The AlaDH protein from A. fulgidus is selective for l-alanine and l-2-aminobutyrate in the oxidative direction, although 12% or lower activity is also obtained with other l-amino acids such as valine, serine, threonine, isoleucine, and aspartate. Minor deamination activities of other l-amino acids (about 5% of the activity with l-alanine) are common features of many bacterial AlaDHs, although enzymes with high substrate specificity also exist (8, 23, 28, 30, 47, 49). However, A. fulgidus AlaDH distinguishes itself by its unusually high l-2-aminobutyrate deamination activity. Substrate specificity for the other known archaeal AlaDHs from H. salinarum and H. cutirubrum were not determined (15, 17).
Recently, an l-2-aminobutyrate dehydrogenase was purified from the archaeon Halobacterium saccharovorum (25). The H. saccharovorum l-2-aminobutyrate dehydrogenase also deaminates l-alanine, but at a twofold slower rate. The Km values for l-2-aminobutyrate and l-alanine are similar to those of A. fulgidus AlaDH. The subunit molecular weight of the H. saccharovorum l-2-aminobutyrate dehydrogenase (Mr, 54,000) is larger than that of A. fulgidus AlaDH, and it assumes a tetrameric quaternary structure while A. fulgidus AlaDH constitutes a dimer. Although the amino acid sequence of the H. saccharovorum l-2-aminobutyrate dehydrogenase is not known, it is possible that this enzyme is related to A. fulgidus AlaDH. As is typical for enzymes from halophiles, the H. saccharovorum l-2-aminobutyrate dehydrogenase is only stable in the presence of high salt concentrations.
It is possible that archaeal AlaDHs function as an AlaDH and as an l-2-aminobutyrate dehydrogenase. While 2-ketobutyrate is a common pathway intermediate, l-2-aminobutyrate is a so-called “unnatural” amino acid that can be formed when 2-ketobutyrate accumulates. High levels of 2-ketobutyrate have been shown to be toxic to Salmonella enterica serovar Typhimurium (22). Therefore, amination of 2-ketobutyrate to the less inhibitory compound l-2-aminobutyrate may serve as a detoxification mechanism.
In the reductive amination direction, A. fulgidus AlaDH exhibits a broader substrate range, which appears to be a feature common to many AlaDHs (14, 17, 28, 30, 49). 2-Ketobutyrate, oxaloacetate, and pyruvate are aminated at similar rates (Tables 2 and 4). However, a comparison of the catalytic efficiencies indicates that pyruvate is the preferred substrate for reductive amination. It is noteworthy that A. fulgidus AlaDH also acts as a reversible serine dehydrogenase, since it both aminates hydroxypyruvate to l-serine and deaminates l-serine, but at 10% of the rates obtained with pyruvate and alanine (Tables 3 and 4).
Strikingly, A. fulgidus AlaDH has no significant amino acid sequence similarity to previously described bacterial AlaDHs and thus belongs to an independent class of enzymes. Instead, A. fulgidus AlaDH exhibits homology to bacterial OCD and the mammalian μ-crystallin family of proteins (Fig. 6 and 7). The highest sequence similarity, however, exists to proteins from two archaea, M. thermoautotrophicum and Halobacterium sp. strain NRC-1, suggesting that these proteins may function as AlaDHs and not as OCDs as they have been annotated (26, 40). There is no amino acid sequence information available for the H. salinarum and H. cutirubrum AlaDH enzymes (15, 17). Interestingly, Graupner and White detected OCD activity in cell extracts of the hyperthermophilic methanogen Methanococcus jannaschii (12). This archaeon lacks any AlaDH or OCD homologue, suggesting that proline formation from ornithine is catalyzed by a novel, not yet characterized, enzyme.
Recently, the three-dimensional structure of the AlaDH enzyme from the cyanobacterium P. lapideum was solved (4). On the basis of the analysis of this structure, P. lapideum AlaDH belongs to a family of d-2-hydroxy acid dehydrogenases that also includes d-lactate dehydrogenases, d-glycerate dehydrogenase, d-phosphoglycerate dehydrogenase, NAD+-dependent formate dehydrogenase, and d-2-hydroxyisocaproate dehydrogenase (4). Despite the structural similarity, there is no significant sequence similarity in this enzyme family, with the exception of an NAD+ binding site, i.e., the common dinucleotide-binding fingerprint, GxGxxA/G (4). This motif is present as GxGxxA/S in A. fulgidus AlaDH and homologues, with the notable exception of the putative OCD from Rhodobacter capsulatus, where the motif is GxGxxC (Fig. 6).
Smith et al. recently solved the three-dimensional structure of A. fulgidus AlaDH, and a detailed description of this structure will follow (42). The crystal structure confirmed the dimeric quaternary structure of A. fulgidus AlaDH, demonstrating that each monomer consists of two domains, a Rossman fold domain for NAD+ binding and a catalytic domain that is, however, structurally unrelated to the catalytic domain of P. lapideum AlaDH (11a).
The different primary and tertiary structures of bacterial and archaeal AlaDHs suggest that these enzymes did not evolve from a common ancestor but that evolution occurred along two separate pathways. Since A. fulgidus AlaDH homologues are present in all three domains of life, archaea, bacteria, and eucarya, it is likely that this type of enzyme was present in the last common ancestor. Alternatively, an early horizontal gene transfer event among the three domains could have occurred. After the divergence of the bacteria, archaea, and eucarya, the predecessor protein appears to have evolved into a diverse group of proteins with specialized functions. Thus, A. fulgidus AlaDH does not exhibit OCD activity and the human μ-crystallin protein lacks both OCD and AlaDH activities. Whether any of the OCDs have AlaDH activity is not known.
Interestingly, the μ-crystallin group of this diverse enzyme family includes several closely related archaeal homologues (group I in Fig. 7). The μ-crystallin proteins and the related protein THBP are found in human and animal eye lens retinas and in diverse tissues that respond to thyroid hormone regulation, including human, kangaroo, and rat brain, heart, skeletal muscle, and kidney tissues, respectively (18, 38). Neither the human μ-crystallin nor the P. aerophilum μ-crystallin homologue exhibits any detectable AlaDH activity, assuming that the latter protein is expressed. Elucidation of the function of the archaeal μ-crystallin homologues may aid in understanding the physiology of the human and animal μ-crystallins.
In conclusion, A. fulgidus AlaDH is a representative of a new archaeal line of AlaDHs that is distinct from the bacterial AlaDHs. We predict that once more archaeal genomes become available, the number of archaeal AlaDHs in group X will increase (Fig. 7).
Acknowledgments
We thank Graeme Wistow (National Institutes of Health) for the generous gift of recombinant human μ-crystallin. Funding for H.G.M. was provided by U.S. Department of Commerce-National Institute of Standards and Technology cooperative agreement 70NANB7H0009. I.S. was supported by the National Science Foundation (MCB-0091351). E.J. was supported by U.S. Public Health Service National Research Service award GM07185.
REFERENCES
- 1.Aalen, N., I. Steen, N. Birkeland, and T. Lien. 1997. Purification and properties of an extremely thermostable NADP+-specific glutamate dehydrogenase from Archaeoglobus fulgidus. Arch. Microbiol. 168:536-539. [DOI] [PubMed] [Google Scholar]
- 2.Aharonowitz, Y., and C. G. Friedrich. 1980. Alanine dehydrogenase of the β-lactam producer Streptomyces clavuligerus. Arch. Microbiol. 125:137-142. [DOI] [PubMed] [Google Scholar]
- 3.Allaway, D., E. M. Lodwig, L. A. Crompton, M. Wood, R. Parsons, T. R. Wheeler, and P. S. Poole. 2000. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 36:508-515. [DOI] [PubMed] [Google Scholar]
- 4.Baker, P. J., Y. Sawa, H. Shibata, S. E. Sedelnikova, and D. W. Rice. 1998. Analysis of the structure and substrate binding of Phormidium lapideum alanine dehydrogenase. Nat. Struct. Biol. 5:561-567. [DOI] [PubMed] [Google Scholar]
- 5.Bellion, E., and F. Tan. 1987. An NAD+-dependent alanine dehydrogenase from a methylotrophic bacterium. Biochem. J. 244:565-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caballero, F. J., J. Cardenas, and F. Castillo. 1989. Purification and properties of l-alanine dehydrogenase of the phototrophic bacterium Rhodobacter capsulatus E1F1. J. Bacteriol. 171:3205-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, K. Y., C. Fuqua, B. S. Martin, and S. C. Winans. 1996. Identification of Agrobacterium tumefaciens genes that direct the complete catabolism of octopine. J. Bacteriol. 178:1872-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury, E. K., T. Saitoh, S. Nagata, M. Ashiuchi, and H. Misono. 1998. Alanine dehydrogenase from Enterobacter aerogenes: purification, characterization, and primary structure. Biosci. Biotechnol. Biochem. 62:2357-2363. [DOI] [PubMed] [Google Scholar]
- 9.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dessaux, Y., A. Petit, J. Tempé, M. Demarez, C. Legrain, and J.-M. Wiame. 1986. Arginine catabolism in Agrobacterium strains: role of the Ti plasmid. J. Bacteriol. 166:44-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galkin, A., L. Kulakova, H. Ashida, Y. Sawa, and N. Esaki. 1999. Cold-adapted alanine dehydrogenases from two Antarctic bacterial strains: gene cloning, protein characterization, and comparison with mesophilic and thermophilic counterparts. Appl. Environ. Microbiol. 65:4014-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Gallagher, D. T., H. G. Monbouquette, I. Schröder, H. Robinson, M. J. Holden, and N. N. Smith. 2004. Structure of archaeal alanine dehydrogenase and relation to bacterial and human proteins. J. Mol. Biol. 342:119-130. [DOI] [PubMed] [Google Scholar]
- 12.Graupner, M., and R. H. White. 2001. Methanococcus jannaschii generates l-proline by cyclization of l-ornithine. J. Bacteriol. 183:5203-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimshaw, C. E., and W. W. Cleland. 1981. Kinetic mechanism of Bacillus subtilis l-alanine dehydrogenase. Biochemistry 20:5650-5655. [DOI] [PubMed] [Google Scholar]
- 14.Hutter, B., and M. Singh. 1999. Properties of the 40 kDa antigen of Mycobacterium tuberculosis, a functional l-alanine dehydrogenase. Biochem. J. 343:669-672. [PMC free article] [PubMed] [Google Scholar]
- 15.Keradjopoulos, D., and A. W. Holldorf. 1979. Purification and properties of alanine dehydrogenase from Halobacterium salinarium. Biochim. Biophys. Acta 570:1-10. [DOI] [PubMed] [Google Scholar]
- 16.Khaw, L. E., G. A. Bohm, S. Metcalfe, J. Staunton, and P. F. Leadlay. 1998. Mutational biosynthesis of novel rapamycins by a strain of Streptomyces hygroscopicus NRRL 5491 disrupted in rapL, encoding a putative lysine cyclodeaminase. J. Bacteriol. 180:809-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, E. K. and P. S. Fitt. 1977. Partial purification and properties of Halobacterium cutirubrum l-alanine dehydrogenase. Biochem. J. 161:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, R. Y., R. Gasser, and G. J. Wistow. 1992. Mu-crystallin is a mammalian homologue of Agrobacterium-ornithine cyclodeaminase and is expressed in human retina. Proc. Natl. Acad. Sci. USA 89:9292-9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, S. Peterson, C. I. Reich, L. K. McNeil, J. H. Badger, A. Glodek, L. Zhou, R. Overbeek, J. D. Gocayne, J. F. Weidman, L. McDonald, T. Utterback, M. D. Cotton, T. Spriggs, P. Artiach, B. P. Kaine, S. M. Sykes, P. W. Sadow, K. P. D'Andrea, C. Bowman, C. Fujii, S. A. Garland, T. M Mason, G. J. Olsen, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 20.Kohen, A., R. Cannio, S. Bartolucci, and J. P. Klinman. 1999. Enzyme dynamics and hydrogen tunneling in a thermophilic alcohol dehydrogenase. Nature 399:496-499. [DOI] [PubMed] [Google Scholar]
- 21.Lamosa, P., D. L. Turner, R. Ventura, C. Maycock, and H. Santos. 2003. Protein stabilization by compatible solutes. Effect of diglycerol phosphate on the dynamics of Desulfovibrio gigas rubredoxin studied by NMR. Eur. J. Biochem. 270:4606-4614. [DOI] [PubMed] [Google Scholar]
- 22.LaRossa, R. A., T. K. Van Dyk, and D. R. Smulski. 1987. Toxic accumulation of α-ketobutyrate caused by inhibition of the branched-chain amino acid biosynthetic enzyme acetolactate synthase in Salmonella typhimurium. J. Bacteriol. 169:1372-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laue, H., and A. M. Cook. 2000. Purification, properties and primary structure of alanine dehydrogenase involved in taurine metabolism in the anaerobe Bilophila wadsworthia. Arch. Microbiol. 174:162-167. [DOI] [PubMed] [Google Scholar]
- 24.Manchenko, G. 1994. A handbook of detection of enzymes on electrophoretic gels. CRC Press, Inc., Boca Raton, Fla.
- 25.Nagata, S., Y. Kobayashi, S. Shinkawa, R. Katoh, T. Ohshima, and H. Misono. 2003. Novel halophilic 2-aminobutyrate dehydrogenase from Halobacterium saccharovorum DSM 1137. J. Mol. Catal. B Enzym. 23:223-230. [Google Scholar]
- 26.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. H. Jung, M. Alam, T. Freitas, S. B. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, R. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14. [Google Scholar]
- 28.Ohshima, T., and K. Soda. 1979. Purification and properties of alanine dehydrogenase from Bacillus sphaericus. Eur. J. Biochem. 100:29-39. [DOI] [PubMed] [Google Scholar]
- 29.Ohshima, T., M. Sakane, T. Yamazaki, and K. Soda. 1990. Thermostable alanine dehydrogenase from thermophilic Bacillus sphaericus DSM 462—purification, characterization and kinetic mechanism. Eur. J. Biochem. 191:715-720. [DOI] [PubMed] [Google Scholar]
- 30.Porumb, H., D. Vancea, L. Muresan, E. Presecan, I. Lascu, I. Petrescu, T. Porumb, R. Pop, and O. Barzu. 1987. Structural and catalytical properties of l-alanine dehydrogenase from Bacillus cereus. J. Biol. Chem. 262:4610-4615. [PubMed] [Google Scholar]
- 31.Roberts, M. F. 2000. Osmoadaptation and osmoregulation in archaea. Front. Biosci. 5:796-812. [DOI] [PubMed] [Google Scholar]
- 32.Rowell, P., and W. D. P. Stewart. 1976. Alanine dehydrogenase of the N2-fixing blue-green alga, Anabaena cylindrica. Arch. Microbiol. 107:115-124. [DOI] [PubMed] [Google Scholar]
- 33.Sans, N., U. Schindler, and J. Schröder. 1988. Ornithine cyclodeaminase from Ti plasmid C58: DNA sequence, enzyme properties and regulation of activity by arginine. Eur. J. Biochem. 173:123-130. [DOI] [PubMed] [Google Scholar]
- 34.Sans, N., G. Schröder, and J. Schröder. 1987. The Noc region of Ti plasmid C58 codes for arginine and ornithine cyclodeaminase. Eur. J. Biochem. 167:81-87. [DOI] [PubMed] [Google Scholar]
- 35.Sawa, Y., M. Tani, K. Murata, H. Shibata, and H. Ochiai. 1994. Purification and characterization of alanine dehydrogenase from a cyanobacterium, Phormidium lapideum. J. Biochem. 116:995-1000. [DOI] [PubMed] [Google Scholar]
- 36.Schindler, U., N. Sans, and J. Schröder. 1989. Ornithine cyclodeaminase from octopine Ti plasmid Ach5: identification, DNA sequence, enzyme properties, and comparison with gene and enzyme from nopaline Ti plasmid C58. J. Bacteriol. 171:847-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuffenhauer, G., T. Schrader, and J. R. Andreesen. 1999. Morpholine-induced formation of l-alanine dehydrogenase activity in Mycobacterium strain HE5. Arch. Microbiol. 171:417-423. [DOI] [PubMed] [Google Scholar]
- 38.Segovia, L., J. Horwitz, R. Gasser, and G. Wistow. 1997. Two roles for mu-crystallin: a lens structural protein in diurnal marsupials and a possible enzyme in mammalian retinas. Mol. Vis. 3:9. [PubMed] [Google Scholar]
- 39.Siranosian, K. J., K. Ireton, and A. D. Grossman. 1993. Alanine dehydrogenase (Ald) is required for normal sporulation in Bacillus subtilis. J. Bacteriol. 175:6789-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. M. Church, C. J. Daniels, J. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, M. T., and D. W. Emerich. 1993. Alanine dehydrogenase from soybean nodule bacteroids—kinetic mechanism and pH studies. J. Biol. Chem. 268:10746-10753. [PubMed] [Google Scholar]
- 42.Smith, N., M. Mayhew, H. Robinson, A. Heroux, D. Charlton, M. J. Holden, and D. T. Gallagher. 2003. Crystallization and phasing of alanine dehydrogenase from Archaeoglobus fulgidus. Acta Crystallogr. D Biol. Crystallogr. 59:2328-2331. [DOI] [PubMed] [Google Scholar]
- 43.Stetter, K. O. 1988. Archaeoglobus fulgidus gen. nov., sp. nov.: a novel taxon of extremely thermophilic archaebacteria. Syst. Appl. Microbiol. 10:172-173. [Google Scholar]
- 44.Stetter, K. O., G. Lauerer, M. Thomm, and A. Neuner. 1987. Isolation of extremely thermophilic sulfate reducers: evidence of a novel branch of archaebacteria. Science 236:822-824. [DOI] [PubMed] [Google Scholar]
- 45.Vadas, A., H. G. Monbouquette, and I. Schröder. 1999. Identification and characterization of a novel ferric reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus. J. Biol. Chem. 274:36715-36721. [DOI] [PubMed] [Google Scholar]
- 46.Vadas, A. J., I. Schröder, and H. G. Monbouquette. 2002. Room-temperature synthesis of l-alanine using the alanine dehydrogenase of the hyperthermophilic archaeon Archaeoglobus fulgidus. Biotechnol. Prog. 18:909-911. [DOI] [PubMed] [Google Scholar]
- 47.Váli, Z., F. Kilár, S. Lakatos, S. A. Venyaminov, and P. Závodszky. 1980. l-Alanine dehydrogenase from Thermus thermophilus. Biochim. Biophys. Acta 615:34-47. [DOI] [PubMed] [Google Scholar]
- 48.Vancura, A., I. Vancurová, J. Volc, S. K. T. Jones, M. Flieger, G. Basarová, and V. Behal. 1989. Alanine dehydrogenase from Streptomyces fradiae. Eur. J. Biochem. 179:221-227. [DOI] [PubMed] [Google Scholar]
- 49.Vancurová, I., A. Vancura, J. Volc, J. Neuzil, M. Flieger, G. Basarová, and V. Behal. 1988. Purification and partial characterization of alanine dehydrogenase from Streptomyces aureofaciens. Arch. Microbiol. 150:438-440. [Google Scholar]
- 50.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 51.Vie, M. P., C. Evrard, J. Osty, A. BretonGilet, P. Blanchet, M. Pomerance, P. Rouget, J. Francon, and J. P. Blondeau. 1997. Purification, molecular cloning, and functional expression of the human nicodinamide-adenine dinucleotide phosphate-regulated thyroid hormone-binding protein. Mol. Endocrinol. 11:1728-1736. [DOI] [PubMed] [Google Scholar]
- 52.Williams, R., N. P. J. Cotton, C. M. Thomas, and J. B. Jackson. 1994. Cloning and sequencing of the genes for the proton-translocating nicotinamide nucleotide transhydrogenase from Rhodospirillum rubrum and the implications for the domain structure of the enzyme. Microbiology 140:1595-1604. [DOI] [PubMed] [Google Scholar]
- 53.Wistow, G., and H. Kim. 1991. Lens protein expression in mammals—taxon-specificity and the recruitment of crystallins. J. Mol. Evol. 32:262-269. [DOI] [PubMed] [Google Scholar]
- 54.Zellner, G., E. Stackebrandt, H. Kneifel, P. Messner, U. B. Sleytr, E. C. De Marcario, H.-P. Zabel, K. O. Stetter, and J. Winter. 1989. Isolation and characterization of a thermophilic, sulfate reducing Archaebacterium, Archaeoglobus fulgidus strain Z. Syst. Appl. Microbiol. 11:151-160. [Google Scholar]