With an estimate of 350 million people chronically infected with the hepatitis B virus (HBV) worldwide, it is critically important to understand how persistent HBV infection is maintained and linked to chronic hepatitis, cirrhosis, and development of liver cancer (hepatocellular carcinoma [HCC]) (39). This review will focus on the HBV nonstructural X protein (HBx), a key regulatory protein of the virus that is at the intersection of HBV infection, replication, pathogenesis, and possibly carcinogenesis. The exact role of HBx in viral replication has yet to be established, and its link to the progression of HCC remains controversial. Moreover, it is still unclear whether development of HCC associated with chronic infection by HBV involves a viral protein, is solely the consequence of a continual inflammatory response to infection, or requires both. Understanding the role of HBx in HBV replication and its effect on hepatocyte biology may help resolve this issue. This review describes key studies and activities of HBx, often interpreted within the context of the viral life cycle. The reader is referred to several comprehensive reviews on the reported biological properties of HBx (3, 78, 137).
HBV
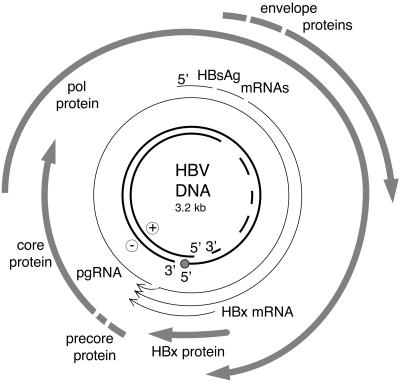
HBV is a member of the Hepadnaviridae family of viruses. The natural host for HBV is humans (39), although similar viruses have been isolated from apes, woodchucks (woodchuck hepatitis virus [WHV]), squirrels (ground squirrel hepatitis virus [GSHV]), herons (heron hepatitis B virus), ducks (duck hepatitis B virus), geese (goose hepatitis B virus), and cranes (crane hepatitis B virus). The hepadnavirus genome is a partially double-stranded circular DNA structure that is encapsidated within the enveloped viral particle (115). Upon infection of hepatocytes, viral DNA is transported to the nucleus, where it is converted to a covalently closed, circular, double-stranded DNA (cccDNA). The cccDNA is essentially an episome that does not replicate but functions solely as the template for all viral transcription (115). The HBV genome is highly compact, such that over 50% of the coding capacity lies in at least two open reading frames (Fig. 1). Genomic and subgenomic viral transcripts, all of which utilize the same polyadenylation signal, are all transcribed from cccDNA. Viral mRNAs encode the viral core (HBcAg), envelope (HBsAgs), polymerase/reverse-transcriptase (pol), and HBx polypeptides. The largest HBV transcript, the pregenomic RNA (pgRNA), is also the template for viral replication and is reverse transcribed by the viral pol, similar to the replication of retroviruses (114). However, in contrast to that of conventional retroviruses, reverse-transcribed HBV DNA is not integrated into the host cell genome as a requisite step in the viral life cycle. Instead, an intermediate in the replication reaction, a partially double-stranded viral DNA genome, is encapsidated within the mature viral particle (Fig. 1). Viral pgRNA is encapsidated in cytoplasmic viral particles comprised of core protein, known as core particles, within which reverse transcription and DNA replication occur. As the virus replicates, it buds into the endoplasmic reticulum by envelopment within the viral HBsAg proteins and is eventually secreted from the infected cell (39). While all hepadnaviruses can establish persistent infections in their respective hosts, only chronic infection by mammalian hepadnaviruses is associated with significant development of HCC (39). As avian viruses either lack the HBx gene or, at best, encode a highly divergent form, the potential role of HBx in the development of HCC in mammals has been an area of intense interest for research (21).
FIG. 1.
A diagrammatic representation of the HBV genome. The inner circle represents the virion genomic DNA that is packaged within viral particles in the cytoplasm of infected cells, and the dashes indicate the region of the genome which is incompletely synthesized. The thick arrows represent open reading frames corresponding to core, envelope (surface antigen), polymerase (pol), and HBx proteins. The thin lines represent HBV RNAs.
GENERAL FEATURES OF HBx
HBx is encoded by the smallest open reading frame of mammalian hepadnaviruses and translated from a small mRNA controlled by the HBx promoter (41, 113). The designation X gene/protein originally reflected its unknown function and lack of homology with known proteins. An HBx mRNA has been detected in the livers of WHV-infected woodchucks and in HBV-infected human liver tissue, and antibodies to HBx have been detected in some infected individuals (31, 49, 66). Studies suggest that the WHV HBx protein (also referred to as WHx) accumulates to 10,000 to 50,000 copies per cell (31).
HBx is 154 amino acids in size, with a molecular mass of approximately 17.5 kDa. Due to a lack of results from X-ray crystallography (HBx has defied high-resolution crystallization) and nuclear magnetic resonance, little is known of HBx three-dimensional structure. Comparative analyses of HBx gene sequences from mammalian hepadnaviruses of different species revealed areas of high conservation, including presumptive helical domains located in the amino- and carboxy-terminal regions, and a potential coiled-coil motif (28, 56). There is some evidence that disulfide bonds may link cysteine residues in the N terminus of HBx with cysteine residues in the C terminus (67, 127). Several groups have also reported that HBx can be phosphorylated when expressed both in insect cells and HepG2 cells, a human hepatoblastoma cell line (98, 127), although other studies have failed to detect phosphorylation (115). Acetylation of HBx was observed when it was expressed in insect cells (127). The significance of phosphorylation or acetylation of HBx for its reported activities is currently unknown. HBx amino acids 52 to 148 are essential for its various reported activities (reviewed in reference 137). Interestingly, deletion of the amino-terminal 1 to 50-amino-acid fragment up-regulates HBx transcriptional functions, suggesting that it may be a negative regulatory element (79). Higher-order structures of HBx are not well established, but there is some evidence that the protein may form dimers (reviewed in reference 39). HBx from both HBV and WHV displays a bimodal half-life that is influenced by the intracellular location of the protein; HBx associated with the cytoskeleton and nuclear framework has a longer half-life (∼3 h) than the cytosolic soluble protein (15 to 20 min) (30, 98). The significance of a differential half-life for HBx function is unclear but could indicate that HBx protein associated with the cytoskeleton has a more-sustained activity.
Most studies have found HBx to be located largely in the cytoplasm of cells but certainly detectable in the nucleus (30, 31, 34, 44, 45, 104, 133). Similar patterns of distribution have been observed in cells transfected with HBx expression vectors and during HBV and WHV infections. A few studies have found HBx largely in the nucleus in some cell lines (46, 102). More recently, it has been reported that HBx is predominantly nuclear when expressed in cells at very low levels but becomes largely cytoplasmic as its expression level increases (44, 52). Although these studies involved HBx expression in the absence of HBV, a dynamic distribution of HBx could be important, considering the multiple functions of HBx during the HBV life cycle, and could influence its effects on transcriptional activation in the nucleus and viral replication in the cytoplasm. Another group reported that HBx shuttles between the nucleus and the cytoplasm through a Crm-1-dependent nuclear export pathway. HBx shuttling may serve to sequester Crm-1 in the cytoplasm and enhance NF-κB localization to the nucleus, with subsequent activation of NF-κB-dependent transcription (38).
HBx IS IMPORTANT FOR THE MAMMALIAN HEPADNAVIRUS LIFE CYCLE
Two different laboratories have demonstrated that HBx is essential for WHV infection of woodchucks (22, 142). Wild-type WHV genomic DNA can be directly injected into the livers of young woodchucks, establishing a WHV infection and viremia. However, if WHx expression is abolished by mutating the WHx gene without altering the overlying polymerase reading frame, neither viral infection nor viremia is detectable. This finding is consistent with colocalization of WHx in active centers of WHV replication in the livers of chronically infected woodchucks (31). Using a similar approach, another group found that the WHx mutant WHV produced a low level of viremia after an extended period of time, indicative of infection or replication (139). However, all WHVs that emerged in these woodchucks were found to be wild-type WHx revertants. While these results were interpreted as indicating that HBx is not essential for viral infection or replication because only viral revertants were recovered, we believe that these data actually support the importance of HBx in viral infection rather than negate it.
Evidence for the importance of HBx in HBV replication has not been found in transgenic mice expressing the entire HBV genome as a liver transgene (96, 136). In these mice, HBx mutants replicate identically to wild-type HBV. It is possible that replication of HBV as a transgene inserted into the mouse chromosome or selection for mice that tolerate HBV and HBx expression (which may be cytotoxic) could select for viruses with HBx independence. Indeed, transcomplementation experiments carried out by crossing HBx-expressing mice with HBV-expressing mice suggest that HBx can augment HBV replication and persistence of infection in the hybrid animals (136). An important but nonessential role for HBx in human HBV replication has been demonstrated in HepG2 cells in tissue culture (17, 77). However, cell culture replication of HBV in the human hepatoma cell line Huh7 is not obviously influenced by HBx expression (13). Consequently, additional studies need to explore the requirement for HBx in HBV replication in primary human (and mouse) hepatocytes in culture. Collectively, studies indicate that HBx in mammalian hepadnaviruses performs several important but perhaps not absolute functions in viral infection and replication.
HBx IS A MODEST TRANSCRIPTIONAL ACTIVATOR
Work from many laboratories has demonstrated that the X proteins of HBV, WHV, and GSHV can moderately stimulate transcription of many different viral and cellular transcription elements (137). Promoters and enhancers stimulated by HBx typically contain DNA binding sites for NF-κB, AP-1, AP-2, c-EBP, ATF/CREB, or the calcium-activated factor NF-AT (36, 59, 65, 70, 74, 75, 135). Biochemical evidence for activation of the DNA binding activity of NF-κB, AP-1, ATF/CREB, and NF-AT by HBx has also been demonstrated (6, 10, 24, 59, 74, 83, 108, 133, 135). HBx can also stimulate RNA pol I- and pol III-dependent promoters, demonstrating pleitropy in its effects (4, 58, 128-130).
HBx does not directly bind DNA, and its ability to activate transcription likely involves several different properties, including direct interaction with nuclear transcription components and activation of cytosolic signal transduction pathways. Through interaction and cloning approaches, it was shown that HBx associates with several components of the basal transcriptional machinery, including TFIIB, TFIIH, the RPB5 subunit of RNA polymerases, and the TATA-binding protein (TBP) (23, 43, 68, 80, 92). HBx also directly interacts in vitro and in vivo with CREB, a member of the basic leucine zipper (b-Zip) family of transcription factors. Interaction likely occurs via the b-Zip region of CREB, which is involved in its DNA binding activity, and a predicted coiled-coil motif in HBx. This interaction increases CREB affinity for its DNA binding site by an order of magnitude, enhances in vitro occupation of the HBV enhancer I ATF/CREB binding site, and correlates with in vivo CREB-dependent transcriptional activation by HBx (74, 135). A similar mechanism of action was described for HBx interaction with c/EBPα, another b-Zip transcription factor (27). However, all studies demonstrating in vivo interaction between HBx and transcriptional components were performed with significant overexpression of both proteins and in the absence of viral infection. To date there has been no demonstration of an in vivo interaction between HBx expressed in the context of replicating HBV and members of the endogenous transcription machinery. Nevertheless, support for the importance of nuclear transcription functions of HBx and its interaction with the transcriptional machinery is also derived from studies in which HBx protein was engineered to contain a nuclear localization signal (NLS), resulting in extensive nuclear accumulation. Results from these studies demonstrate that an NLS-HBx protein more strongly stimulates transcription from HBV enhancer I and other elements than wild-type HBx protein but no longer activates transcription dependent upon NF-κB and AP-1 (34, 119).
HBx STIMULATES CYTOPLASMIC SIGNAL TRANSDUCTION PATHWAYS
Many groups have reported that the predominant location of HBx is in the cytoplasm of cells (30, 31, 34, 44, 45, 59, 104, 111). Moreover, the inability of HBx to activate NF-κB- or AP-1-dependent transcription when targeted to the nucleus and the wide variety of transcription factors and/or elements stimulated by HBx led many investigators to analyze its potential to stimulate cytoplasmic signaling pathways. Numerous HBx studies have probed pathways by using dominant-inhibiting mutant signaling proteins, and some have used biochemical analyses. It has been established that HBx can stimulate various cytoplasmic signal transduction pathways (Fig. 2). HBx was shown to activate the extracellular signal-regulated kinases (ERKs), the stress-activated protein kinases/NH2-terminal-Jun kinases (SAPK/JNKs), and the p38 kinase (10, 118-120). This activity has been demonstrated in a variety of cell lines, including those of liver origin, and requires HBx to be located in the cytoplasm, although the extent of activation and its longevity can vary in different cells (34, 118-120). Moreover, HBx activation of mitogen-activated protein kinases (MAPKs) and JNKs was demonstrated in intact mouse liver (85). No direct interaction has been found between HBx and any of these protein kinases, directing attention to the upstream activators. In this regard, HBx-dependent activation of transcription factor AP-1, as well as RNA pol I- and pol III-dependent transcription, is blocked by dominant-inhibiting forms of Ras and the kinase Raf, an effector of the Ras-Raf-MAP kinase signal transduction pathway (8, 29, 83, 84, 128). Moreover, HBx stimulates Ras-Raf-MAP kinase signal transduction when expressed in the context of replicating HBV or WHV in hepatoma cells and when expressed in primary mouse hepatocytes in culture (54, 55, 85).
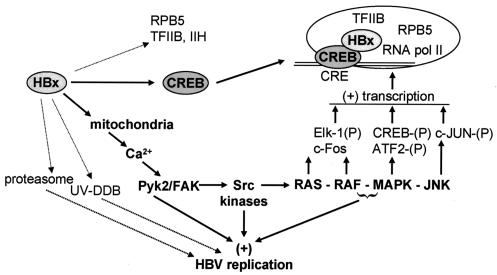
FIG. 2.
Targets of HBx activity. Arrows indicate pathways and targets of HBx activity. Filled arrows represent more-certain primary targets or activities of HBx, whereas dotted arrows are likely secondary targets or have not been widely validated. HBx likely binds directly to the transcription factor CREB and possibly to transcription factors TFIIB, TFIIH, and the RNA polymerase II-associated protein RPB5, but the latter might interact with HBx via secondary interactions with CREB. HBx-CREB transcription complexes have been shown to stimulate transcription. HBx is thought to act on the mitochondria, causing calcium release, in turn activating the Pyk2/FAK and Src kinase families, leading to stimulation of a variety of cytoplasmic signal transduction pathways. HBx activation of signaling pathways can stimulate phosphorylation and activation of transcription factors, as indicated, as well as viral replication. Interaction of HBx with the UV-DDB proteins (1 and/or 2) is implicated in stimulation of viral replication. HBx interaction with the proteasome is also reported to stimulate HBV replication.
Activation of Ras by HBx is indirect, as HBx does not associate with Ras, Ras-GAP, the GTP exchange factor Sos, or Ras adapter proteins Grb2 or Shc (10, 55). It has now been established that HBx activates Ras, JNK, and the JAK-STAT signaling pathway by activating nonreceptor tyrosine kinases of the Src family, which are upstream activators of Ras GTPases (55, 64, 120). HBx constitutively activates Src kinases when expressed from WHV or HBV genomes (replicons) during viral replication in cultured hepatoma cells and when expressed independently of the viral genome (54, 55). Inhibition of Src kinases prevents HBx activation of transcription by AP-1 and STAT3, as well as viral replication (as described later) (54, 55, 120). HBx activation of Src family kinases also promotes alteration of cellular adherens junctions, another activity associated with HBx expression in liver cell lines (60). No direct interaction between Src kinases and HBx has been observed (54).
Clues to how HBx activates Src kinases and the Ras-Raf-MAPK pathway came from work by Lopez-Cabrera's group, when they demonstrated that HBx activates the nuclear factor of activated T cells, NF-AT (59). Activation of this protein is strongly regulated by cytosolic calcium signaling, another known regulator of Src kinases (reviewed in reference 95). This finding led to studies which showed that HBx activates another nonreceptor tyrosine kinase family, consisting of focal adhesion kinase (FAK) and the proline-rich tyrosine kinase (Pyk2) (17). Moreover, the ability of HBx to activate FAK/Pyk2 kinases is dependent on cytosolic calcium modulation and is linked to downstream activation of Src family kinases. HBx activation of Pyk2 and Src kinases is blocked by a dominant-inhibiting mutant of Pyk2, the intracytosolic calcium chelator BAPTA-AM, as well as by inhibitors of the mitochondrial transition pore, such as cyclosporine A and its nonimmunosuppressive derivative, SDZ NIM 811 (16, 17). Notably, a retrospective evaluation of many (but certainly not all) of the activities ascribed to the expression of HBx can be explained by invoking pathways known to be regulated by cytosolic calcium modulation. Recent work from the Andrisani group has confirmed the importance of calcium signaling in HBx activation of STAT3 and MAPK p38, an HBx activity that this group also directly linked to stimulation of Src kinases (120), and two groups have directly measured modulation of cellular calcium by HBx (20, 86). Further investigation is required to determine the exact mechanism by which HBx influences cytosolic calcium levels.
A number of key issues regarding HBx activation of NF-κB remain unresolved or mired in claims and counterclaims. The extent to which activation of cytoplasmic signal transduction pathways is important for HBx activation of transcription factor NF-κB and the mechanism for HBx activation of NF-κB are unresolved. It is also unclear whether HBx activation of NF-κB involves requisite activation of protein kinase C or Ras (24, 34, 70, 80, 108). Whether HBx acts directly on NF-κB itself, its IκB regulators, or predominantly on the upstream signal transduction pathways of NF-κB is also in dispute (24, 108, 133). These discrepancies may result from the fact that NF-κB consists of multiple related proteins that may be activated to different extents in different ways in different cells and therefore could be significantly impacted by the experimental systems in which HBx studies were conducted.
MOLECULAR FUNCTIONS OF HBx IN THE VIRAL LIFE CYCLE
Apart from studies demonstrating an essential requirement for HBx in WHV infection in woodchucks, the molecular function of HBx in the viral life cycle in animals and humans has not been well investigated. However, studies have examined the function of HBx in WHV and HBV replication in cultured hepatocytic cells. While it has been unequivocally established that HBx stimulates transcription, the effect of HBx on HBV, WHV, and GSHV transcription is modest in most experimental systems, increasing viral transcription from three- to eightfold (17, 28, 54, 142). It is possible during whole animal infection that HBx might stimulate viral transcription much more strongly than in cultured cells, but presently, little evidence supports this contention. Transgenic mice that specifically express HBx in the liver, when crossed with mice that express a reporter gene controlled by the human immunodeficiency virus long-terminal repeat, increase its expression by only three- to sixfold (5). However, one study reported a strong increase in HBV core promoter activity by HBx when HBV and HBx transgenic mice were crossed (97). It is therefore entirely possible that the importance of HBx for stimulation of HBV transcription has simply not been adequately recapitulated in existing experimental systems. The importance of HBx activation of transcription for establishment of virus infection and replication therefore remains to be fully investigated in more representative models, including replicon delivery to primary rodent and human hepatocytes in culture. As it stands, very few activities of HBx, including its role in the viral life cycle, have been examined in primary hepatocyte systems which are amenable to many of the types of studies that have been conducted in hepatoma cell lines.
Using cell culture systems and an HBV replicon, the activation of Src kinases by HBx protein was found to stimulate HBV and WHV replication in established liver cell lines by 5- to 20-fold in an Src-dependent/Pyk2-dependent, but Ras-independent, manner (17, 54). In this system, abolishment of HBx expression impaired HBV and WHV replication by 5- to 20-fold. Inhibition of Src kinases impaired HBV and WHV replication by 10- to 15-fold, and a similar effect on HBV replication was observed when Pyk2 kinases were inhibited (17, 54). Other studies have confirmed the lack of requirement for Ras activation for HBV replication (140). Analysis of viral RNAs and the activity of viral polymerase in cell extracts suggested that loss of HBx expression or inhibition of Src/Pyk2 kinases negatively regulates HBV replication at the level of either pgRNA encapsidation, DNA replication, or both, as RNA levels were only slightly decreased in this system. However, it is not known whether HBx stimulates Pyk2 or Src kinases during infection in animals or whether Pyk2 or Src kinases are involved in viral replication in animals. Importantly, activation of Pyk2 or Src kinases cannot account for the entire function of HBx in viral replication, even in cultured cells, since a constitutively activated form of Src only partially replaces HBx function (54).
MOLECULAR TARGETS AND MECHANISM OF HBx ACTION
The biochemical and molecular bases for HBx activity and its direct cellular targets have been only partially explored. There have been many attempts to identify HBx binding partners, using yeast two-hybrid studies or systems in which HBx and putative interacting proteins were overexpressed. These studies have contributed to the large number of cellular proteins reported to interact with HBx. Unfortunately, none of these studies have identified cellular proteins that interact with HBx under conditions that recapitulate HBV infection in hepatocytes; evaluation of these putative interactions in biologically relevant systems is still needed. Cellular factors that were reported to interact with HBx include a number of transcriptional components, such as transcription factors (ATF/CREB, ATF3, ICERIIγ, c/EBP, NF-IL-6, Egr1, Ets, Oct1, RXR receptor, SMAD4, and IκBα, among others) and components of the basal transcription machinery (RPB5, TFIIB, TBP, and TFIIH, among others) (see reference 39 for a complete list and more references). Other reported interaction partners include the proteosome subunit protein C7, cell cycle regulatory protein p53, the UV light-damaged DNA binding protein complex UVDDB, and the mitochondrial voltage-dependent anion channel-3 protein (HVDAC3), a component of the mitochondrial transition pore that regulates calcium flux (37, 47, 94) (for other reported proteins that interact with HBx, see reviews [references 3, 39, 78, and 137]).
Of the myriad components reported to interact with HBx, investigation of just a few have suggested that they are primary molecular targets. Certain observations, such as the direct interaction of HBx with p53 (37, 125, 132), have not been substantiated in more physiologically relevant systems (91, 112). Moreover, while some studies suggest that p53 and HBx can colocalize and that HBx may sequester p53 in the cytoplasm of cells and potentially inhibit its activity (35, 37, 52, 116), others have reported that HBx does not colocalize with p53 and does not sequester it in the cytoplasm (112). Since HBx-associated activities that are potentially linked to p53-dependent pathways can still be observed in the absence of p53, further analyses are required to determine whether there is a functional consequence to putative interactions of HBx and p53 in the context of viral replication in hepatocytes (40, 123).
A compelling group of molecular targets of HBx are the b-ZIP transcription factors, such as CREB (3, 6, 74, 78, 135). The b-ZIP factors are the only components involved in transcription that have been demonstrated to interact with HBx in vitro and for which the interaction is functionally important for activity (3, 6, 27, 74). HBx likely interacts with b-ZIP proteins via a leucine-rich putative coiled-coil domain, which promotes binding to and activation of the HBV enhancer II/pgRNA promoter (6, 27). Notably, activated CREB or other b-ZIP factors (ATF3, ICERIIγ, or NF-IL-6) interact with the basal transcription machinery through TFIID/hTAFII130, which in turn can involve the RPB5 subunit of RNA polymerase II or TFIIB (3). The reported interaction of HBx with components of the basal transcriptional machinery might therefore reflect weak secondary interactions arising from primary strong interactions with b-ZIP factors, which promote complex assembly with basal transcription components (6). Nevertheless, this scenario cannot explain some of the other reported HBx interactions with transcription components or factors, which might occur in vitro only or with strong overexpression in vivo.
Several studies concur that a fraction of HBx interacts in vitro and in vivo with the DNA repair proteins DDB1 (127 kDa) and DDB2 (48 kDa) (7, 12, 105). Together, DDB1 and DDB2 form a complex that binds tightly to DNA and transcription factor E2F1, implicating roles in both DNA repair and cell cycle control (reviewed in reference 18). Several groups have studied the interaction of HBx with DDB1. While all agree that HBx and WHx proteins interact with DDB1, there are conflicting results regarding the functional consequences of this interaction. Using essentially similar assay systems to measure excision repair fidelity and frequency, some researchers observed a slight to moderate decrease upon expression of HBx, whereas others did not (7, 11, 40, 48). Perhaps most important, transgenic mice that express HBx in the liver do not demonstrate an altered frequency of spontaneous mutations (72). However, HBx does appear to slightly but reproducibly increase the sensitivity of hepatocytes in animals to effects of low levels of chemical mutagens (71, 141). Thus, it is possible that HBx may subtly but importantly contribute to mutagenesis during chronic HBV infection but is not itself a primary mutagen. Among other reported functions of the HBx-DDB interaction, one report found that HBx interaction with DDB1 is important for WHV replication in woodchucks (105). The interaction of HBx with DDB1, while correlative, is likely important for WHV infection, as HBx mutants that abolish this interaction impair viral replication. The mechanism by which HBx-DDB1 interaction promotes viral replication is not known. Moreover, it is not known whether HBx interacts exclusively with DDB1 or DDB2 or what the consequence of this for formation of the DDB1/2 complex may be, nor is it known how this interaction impacts on HBx-associated activation of cellular RNA-transcription pathways (14, 81, 134). One study also indicated that the nuclear interaction of HBx with DDB1 and DDB2 is tightly regulated in hepatocytes, probably occurring predominantly during the late G1 phase of the cell cycle (81). Consequently, HBx interaction with DDB1 and DDB2 might be involved in delay or inhibition of cell cycle progression beyond the G1/S phase border. This finding takes on particular significance, since HBV replicates very poorly in S phase cells but replicates well in nondividing cells (87). However, as the same interaction of HBx with DDB1 was reported by other groups to promote cell death, a role in promoting viral replication is in conflict with a role in cell death (11, 14, 69). Studies need to determine at a molecular level the functional importance of HBx-DDB interaction and whether cell death mediated by HBx interaction with DDB1 and DDB2 is an artifact of cell culture systems.
HBx AND APOPTOSIS
Few areas of HBV biology have been as mired in opposing results as the association of HBx expression with promotion or inhibition of apoptosis. Indeed, HBx has been reported to strongly mediate apoptosis, sensitize cells to proapoptotic stimuli, prevent apoptosis, or have no impact on apoptosis (reviewed in reference 39). Some studies found enhanced apoptotic turnover of hepatocytes in mice expressing HBx (122, 123), whereas others did not (72). HBx also failed to promote cell death when expressed in mouse mammary epithelium (53). Equally perplexing are the opposing reports of HBx apoptotic activity in cell lines in culture. HBx has been reported to block cell death mediated by TNF, Fas, p53, or TGFβ (35, 88, 99). One report also found HBx in association with a complex containing the antiapoptotic protein survivin, which suppressed caspase action and apoptosis (76). In contrast, many other reports indicate that HBx promotes apoptosis in a variety of cells when expressed independently of virus replication (25, 51, 52, 100, 101, 109, 110, 123). The recognition that at least some fraction of HBx likely interacts with or acts on the mitochondrial transition pore suggests a possible mechanism for HBx-induced modulation of apoptotic pathways, although this association in itself does not imply either a pro- or antiapoptotic effect (94, 101). HBx has been shown to induce mitochondrial aggregation and cytochrome c release, which is indicative of induction of apoptosis through mitochondrial dysfunction (101, 117). However, aggregation of mitochondria by HBx also suggests that the protein was strongly overexpressed in some of these studies, as aggregation of mitochondria is not observed during HBV replication in cultured cells or during natural infection. It is important that few studies have been performed in the context of HBV replication. However, HBx expressed from a replicating HBV genome in cells in culture provides hypersensitivity to killing by tumor necrosis factor (TNF); this hypersensitivity likely requires a particular set of conditions involving HBx activation of c-Jun N-terminal kinase and Myc (109, 110). In these studies, HBx expressed during HBV replication did not directly promote apoptosis in the absence of proapoptotic stimuli but it did promote hypersensitivity to TNF-mediated cell death at levels well below that at which cells are normally sensitive to TNF (109). This observation has recently been confirmed by another group (52). It is possible that the dueling influences of HBx on cell death or survival are a result of its overexpression in different studies or the inherently different cell systems that were used or that these studies were conducted in the absence of HBV replication.
Equally problematic is discerning why there would be a viral advantage for HBx promotion of pathogenesis and hepatocyte death during the viral life cycle, assuming that HBx sensitizes cells to proapoptotic stimuli. HBV has been generally thought of as a noncytopathic virus, capable of replicating without killing the infected hepatocyte in the absence of an inflammatory response (reviewed in reference 39). The proapoptotic activity of HBx is reported to act through association with and inactivation of c-FLIP, a protein which protects against TNF and Fas-mediated apoptosis, and this association might produce yet another potential molecular explanation for HBx sensitization of hepatocytes to TNF apoptotic activity (52). Of particular interest is the possibility that there is a physiological significance to hypersensitization of hepatocytes to TNF; TNF promotes growth and regeneration of hepatocytes during liver injury (33). Hypersensitization of hepatocytes to TNF-mediated apoptosis could promote hepatocyte regeneration, providing a larger reservoir of cells for new HBV infection. The important questions that now need to be addressed concern whether HBx indeed sensitizes, blocks, or has no impact on the effects of proinflammatory cytokines in the context of an authentic, in vivo HBV infection. It needs to be determined whether HBx sensitizes cells to proapoptotic stimuli, whether there is a distinct advantage for the virus, or whether apoptosis merely represents an unavoidable outcome linked to other HBx activities that are necessary for HBV replication but which predispose cells to apoptotic stimuli.
HBx AND THE CELL CYCLE
A number of researchers have focused on the impact of HBx expression on the cell cycle (reviewed in reference 73). Studies have been performed with established cell lines, primary mouse liver cells, and HBx transgenic mice (9, 15, 57, 73, 107). Some studies have shown that HBx can induce arrested cells to exit G0 and reenter the cell cycle, acting through activation of signal transduction factors such as Src kinases, MAPK, and JNK (9, 15, 57). In one system, inactivation of Src kinases by its negative regulator, Csk (c-terminal Src-kinase), prevented HBx from inducing cell cycle progression (15). Whether HBx causes cells to progress entirely through the cell cycle or to stall at the border of G1 and S phases is dependent on the presence of other factors. When growth-arrested cells transfected with HBx were provided medium supplemented with serum, cells progressed more rapidly through the cell cycle than cells without HBx (9). However, in the absence of serum, HBx-expressing cells exited G0 but stalled at the G1/S border (15). Importantly, it is not known whether cell cycle progression is induced during HBV replication or represents effects related to overexpression of HBx. Studies performed with HBx transgenic mouse models have revealed a potentially more complicated influence of HBx on cell cycle progression (71, 73). Results have ranged from only a slight influence on cellular proliferation observed in the transgenic mouse models that express HBx in the liver and do not develop spontaneous tumors to a more dramatic effect in some HBx models in which its expression in liver or mammary epithelium results in spontaneous development of tumors (53, 57, 71, 73).
In some established cell lines and in mouse primary hepatocyte cultures, HBx alters expression of p21 and p27 cell cycle regulation genes, influencing cell cycle progression profiles in these cells (1, 42, 61, 89, 93). However, there are no clear answers as to whether HBx induces or inhibits expression of p21 and p27, as conflicting reports have been published; these differences may reflect influences of the experimental systems or levels of HBx expression (61, 93). To some extent, different interpretations of essentially similar data are responsible for seeming discrepancies. For example, HBx expression that causes cells to exit G0 and stall at the G1/S border has been interpreted by some as inducing cells to progress into the cell cycle from a state of quiescence but by others as inhibition of cell cycle progression beyond this point, simply reflecting the interest of a particular study (15, 103). In addition, different cell lines can show different cycling profiles when HBx is expressed, as established by Andrisani and coworkers (62). Their studies demonstrated that HBx differentially regulates cell cycle progression in differentiated and dedifferentiated hepatocytic cell lines derived from the same parental liver cell line. These cells stably express HBx under an inducible tetracycline promoter. The differentiated hepatocytes display HBx-dependent G1, S, and G2/M progression, induction of cyclin D1, A, and B1, and induction of Cdc2. The dedifferentiated cells display HBx-dependent G1 and S phase entry only and pause early in S phase. Furthermore, the dedifferentiated cells show an HBx-induced expression of cyclin-dependent kinase inhibitor p21. HBx promotion of cell cycle progression in immortalized cells raises a possible connection to a role for HBx in HBV oncogenesis. By inducing cell cycle progression of normally quiescent hepatocytes, HBx may render hepatocytes more sensitive to other potentially carcinogenic signals which, when combined with both an immune response to viral infection and the interaction of HBx with other cellular proteins, could substantially impact hepatocytic transformation (15, 62, 71, 73). While this may ultimately be deleterious to the host, the induction of cell cycle progression may be necessary to induce cellular expansion of available deoxynucleoside triphosphates that are required for viral replication (15).
WHAT IS THE ROLE OF HBx IN HBV-MEDIATED CARCINOGENESIS?
The study with the first transgenic mouse model to express HBx in the liver reported development of liver tumors directly related to HBx expression (50). These mice were generated with an HBx expression vector that included the HBV X coding sequence as well as the HBV enhancer I region, the X gene promoter, and the HBV polyadenylation signal (Fig. 1). Using an identical expression construct, Yu et al. subsequently confirmed this observation in another lineage of HBx transgenic mice (138). However, several other groups derived HBx transgenic mice in which the expression of HBx was controlled by the human alpha-1-antitrypsin regulatory element or the human antithrombin gene regulatory sequence; none of these studies supported a strong and direct connection between HBx and tumorigenesis (31, 63, 106, 121). The reasons for these discrepant results are unclear and may not simply reflect the use of different expression vectors. Indeed, oncogenesis has not been observed in HBV transgenic mice that contain the entire HBV genome as a transgene and therefore presumably express HBx that is controlled by authentic HBV elements. A strong, direct role for HBx in liver carcinoma also does not reflect the biology of HCC development, which involves decades of chronic HBV infection, often (although not always) accompanied by a necro-inflammatory response resulting in years of liver destruction and renewal which can be highly mutagenic in its own right (reviewed in reference 26). However, subtle HBx-induced changes in cellular physiology may eventually have detrimental consequences for hepatocytes. Thus, if HBx plays a role in HBV oncogenesis, it would most reasonably be as an assisting, weakly proto-oncogenic factor. In this regard, several studies described such an effect in HBx transgenic mice treated with low levels of carcinogenic agents or in HBx transgenic mice coexpressing a constitutively activated Myc protein. The HBx mice display higher levels of early neoplastic lesions and somewhat higher levels of tumor development (71, 106, 121). Exactly what HBx-associated activities are involved in its cofactor role are unclear and will require a better understanding of its in vivo role during authentic infections and subsequent characterization of the potential cofactor role of these activities.
Integration of HBV genomic DNA into cellular chromosomes occurs during the viral life cycle and is observed in approximately 85% of HBV-associated tumors (reviewed in reference 39). HBV-integrated DNA is usually highly rearranged and, for human HBV, is more or less without specificity (82, 124). However, while rare, some integration events may place HBV enhancer or promoter sequences in the vicinity of cellular genes and induce inappropriate expression of cellular proteins. Conversely, integration may disrupt cellular genes. Such human HBV integration events have been described as rare, but interesting, and include examples such as fusions with the retinoic acid receptor and cyclin A (32, 131). A recent reinvestigation of HBV integration sites in cellular sequences of 22 unselected HCC tumors suggests that integration-mediated mutagenesis that induces the expression of specific classes of proteins may be more common than previously appreciated (90). Of 22 tumors analyzed, more than half contained HBV DNA integration sites in genes involved in cell cycle control, signal transduction, and cell survival. However, these results do not necessarily confirm preferential integration sites for HBV; integration may be random, but it is possible that only integration events that confer a survival advantage will be identified in tumors from HBV-associated HCC. It is not yet known whether any of the integration events from the 22 analyzed tumors are productive. Of particular interest to the role of HBV integration in associated HCC is the observation that, while most HBV genes are not expressed in these tumors, recent studies have suggested that HBx, or truncated versions of HBx, may continue to be expressed (103, 126). Interestingly, the truncated HBx proteins derived from integrated HBV sequences were incapable of promoting apoptosis but promoted Myc plus Ras transformation in cell assays (126). It is therefore possible that HBx has another role, other than that of sensitizing cells to mutagenic stimuli, in the development of HCC or subsequent survival and proliferation of tumor cells. However, as the truncated versions of HBx identified in these liver tumors have lost many of the activities often associated with wild-type HBx expression, it is unclear how HBx may be influencing hepatocyte transformation. Taken together, these findings suggest that HBx likely plays a role in the development of hepatocellular carcinoma but is probably not directly oncogenic when expressed in the context of an authentic infection.
SUMMARY
The exact function of HBx during HBV replication and its influence in HBV-associated hepatocellular carcinoma are still undefined, but common themes are emerging. Most studies agree that, if not absolutely essential for HBV infections, HBx at the very least plays a critical role in viral replication. HBx functions in the cytoplasm to activate various signaling pathways, many of which are controlled by modulation of cytosolic calcium. A number of groups have now demonstrated that HBx activities somehow involve modulation of cytosolic calcium, although the exact molecular mechanism is undefined. In the nucleus, HBx can regulate transcription through a direct interaction with different transcription factors and, in some cases, enhance their binding to specific transcription elements. Clearly, HBx can influence apoptotic and cell cycle regulatory pathways, but the exact consequences of these activities of HBx are likely affected by the cell types in which the studies have been conducted. In this regard, it is interesting to note that much of the seeming confusion or controversy regarding HBx activities is likely the result of the different assays performed, and the findings of different studies may ultimately be more compatible than expected. In most experiments, HBx activities have been analyzed based on the end product of what are long and complex pathways. For example, transcriptional reporter readout, apoptotic assays, cell cycle regulation, and mutagenic studies are judged based on an analysis of the final product of a given pathway without considering common effectors. HBx may initiate overlapping and very similar effectors in all the studies conducted, but the consequence of this will likely vary depending on the specific cellular phenotype and the assay involved. Moreover, the frequency or longevity of this stimulus may vary depending on the amount of HBx present, which in turn influences the results. Fluctuations in the frequency or longevity of a HBx-induced stimulus, such as calcium signaling, can have a dramatic influence on the final effect on a cell (reviewed in reference 19). Equally interesting is the possibility that HBx has different consequences for hepatocyte physiology as HBV-infected cells are targeted by the immune system or as hepatocytes in which HBx is expressed undergo transformation and progression to HCC. Both these processes are dynamic, and it is likely that cellular physiology is consequently altered. The demonstration that different signaling pathways can be activated based on the differentiation state of hepatocytes derived from the same parental cell line and suggestions that the cellular status of p53, p21, and p27 can affect HBx-dependent regulation of cell cycle control supports the notion that the consequence of HBx expression may change as hepatocytes become transformed (2, 62, 93). It remains to be determined whether similar results will be observed in the context of an authentic HBV infection. Such observations will add to the complexity of identifying and understanding the precise molecular mechanism of HBx activities during viral replication. However, there is the exciting possibility that the end product of HBx activities may change as the cell responds to the infection. Finally, studies of HBx activities during HBV replication should help clarify the many properties that have been ascribed to HBx expression, including those that are required for viral replication and those that may influence cellular transformation.
REFERENCES
- 1.Ahn, J. Y., E. Y. Chung, H. J. Kwun, and K. L. Jang. 2001. Transcriptional repression of p21waf1 promoter by hepatitis B virus X protein via a p53-independent pathway. Gene 275:163-168. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, J. Y., E. Y. Jung, H. J. Kwun, C. W. Lee, Y. C. Sung, and K. L. Jang. 2002. Dual effects of hepatitis B virus X protein on the regulation of cell-cycle control depending on the status of cellular p53. J. Gen. Virol. 83:2765-2772. [DOI] [PubMed] [Google Scholar]
- 3.Andrisani, O., and S. Barnabas. 1999. The transcriptional function of the hepatitis B virus X protein and its role in hepatocarcinogenesis. Int. J. Oncol. 15:1-7. [DOI] [PubMed] [Google Scholar]
- 4.Aufiero, B., and R. J. Schneider. 1990. The hepatitis B virus X-gene product trans-activates both RNA polymerase II and III promoters. EMBO J. 9:497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balsano, C., O. Billet, M. Bennoun, C. Cavard, A. Zider, G. Grimber, G. Natoli, P. Briand, and M. Levero. 1994. Hepatitis B virus X gene product acts as a transactivator in vivo. J. Hepatol. 21:103-109. [DOI] [PubMed] [Google Scholar]
- 6.Barnabas, S., T. Hai, and O. M. Andrisani. 1997. The hepatitis B virus X protein enhances the DNA binding potential and transcription efficacy of bZip transcription factors. J. Biol. Chem. 272:20684-20690. [DOI] [PubMed] [Google Scholar]
- 7.Becker, S. A., T.-H. Lee, J. S. Butel, and B. L. Slagle. 1998. Hepatitis B virus X protein interferes with cellular DNA repair. J. Virol. 72:266-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benn, J., and R. J. Schneider. 1994. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc. Natl. Acad. Sci. USA 91:10350-10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benn, J., and R. J. Schneider. 1995. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc. Natl. Acad. Sci. USA 92:11215-11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benn, J., F. Su, M. Doria, and R. J. Schneider. 1996. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J. Virol. 70:4978-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergametti, F., S. Prigent, B. Luber, A. Benoit, P. Tiollais, A. Sarasin, and C. Transy. 1999. The proapoptotic effect of hepatitis B virus HBx protein correlates with its transactivation activity in stably transfected cell lines. Oncogene 18:2860-2871. [DOI] [PubMed] [Google Scholar]
- 12.Bergametti, F., D. Sitterlin, and C. Transy. 2002. Turnover of hepatitis B virus X protein is regulated by damaged DNA-binding complex. J. Virol. 76:6495-6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blum, H. E., Z. Zhang, E. Galun, F. Weisacker, B. Garner, T. J. Liang, and J. R. Wands. 1992. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J. Virol. 66:1223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bontron, S., N. Lin-Marq, and M. Strubin. 2002. Hepatitis B virus X protein associated with UV-DDB1 induces cell death in the nucleus and is functionally antagonized by UV-DDB2. J. Biol. Chem. 277:38847-38854. [DOI] [PubMed] [Google Scholar]
- 15.Bouchard, M., S. Giannakopoulos, E. Wang, N. Tanese, and R. J. Schneider. 2001. Hepatitis B virus HBx protein activation of cyclin A-cyclin-dependent kinase 2 complexes and G1 transit via a Src kinase pathway. J. Virol. 75:4247-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouchard, M., R. Puro, L.-H. Wang, and R. J. Schneider. 2003. Activation and inhibition of cellular calcium and tyrosine kinase signaling pathways identify targets of the HBx protein involved in hepatitis B virus replication. J. Virol. 77:7713-7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376-2378. [DOI] [PubMed] [Google Scholar]
- 18.Butel, J. S., T.-H. Lee, and B. L. Slagle. 1996. Is the DNA repair system involved in hepatitis B virus mediated hepatocellular carcinogenesis? Trends Microbiol. 4:119-124. [DOI] [PubMed] [Google Scholar]
- 19.Carafoli, E., L. Santella, D. Branca, and M. Brini. 2001. Generation, control, and processing of cellular calcium signals. Crit. Rev. Biochem. Mol. Biol. 36:107-260. [DOI] [PubMed] [Google Scholar]
- 20.Chami, M., D. Ferrari, P. Nicotera, P. Paterlini-Brechot, and R. Rizzuto. 2003. Caspase-dependent alterations of Ca2+ signaling in the induction of apoptosis by hepatitis B virus X protein. J. Biol. Chem. 278:31745-31755. [DOI] [PubMed] [Google Scholar]
- 21.Chang, S. F., H. J. Netter, E. Hildt, R. Schuster, S. Schaefer, Y. C. Hsu, A. Rang, and H. Will. 2001. Duck hepatitis B virus expresses a regulatory HBx-like protein from a hidden open reading frame. J. Virol. 75:161-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, H., S. Kaneko, R. Girones, R. W. Anderson, W. E. Hornbuckle, B. C. Tennant, P. J. Cote, J. L. Gerin, R. H. Purcell, and R. H. Miller. 1993. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J. Virol. 67:1218-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheong, J.-H., M.-K. Yi, Y. Lin, and S. Murakami. 1995. Human RPB5, a subunit shared by eukaryotic nuclear polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 14:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chirillo, P., M. Falco, P. L. Puri, M. Artini, C. Balsano, M. Levrero, and G. Natoli. 1996. Hepatitis B virus pX activates NF-κB-dependent transcription through a Raf-independent pathway. J. Virol. 70:641-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chirillo, P., S. Pagano, G. Natoli, P. L. Puri, V. L. Burgio, C. Balsano, and M. Levrero. 1997. The hepatitis B virus X gene induces p53-mediated programmed cell death. Proc. Natl. Acad. Sci. USA 94:8162-8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chisari, F. 2000. Rous-Whipple Award Lecture: Viruses, immunity, and cancer: lessons from hepatitis B. Am. J. Pathol. 156:1118-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi, B. H., G. T. Park, and H. M. Rho. 1999. Interaction of hepatitis B viral X protein and CCAAT/enhancer-binding protein alpha synergistically activates the hepatitis B viral enhancer II/pregenomic promoter. J. Biol. Chem. 274:2858-2865. [DOI] [PubMed] [Google Scholar]
- 28.Colgrove, R., G. Simon, and D. Ganem. 1989. Transcriptional activation of homologous and heterologous genes by the hepatitis B virus X gene product in cells permissive for viral replication. J. Virol. 63:4019-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cross, J. C., P. Wen, and W. J. Rutter. 1993. Transactivation by hepatitis B virus X protein is promiscuous and dependent on mitogen activated cellular serine/threonine kinases. Proc. Natl. Acad. Sci. USA 90:8078-8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dandri, M., J. Petersen, R. J. Stockert, T. M. Harris, and C. E. Rogler. 1998. Metabolic labeling of woodchuck hepatitis B virus X protein in naturally infected hepatocytes reveals a bimodal half-life and association with the nuclear framework. J. Virol. 72:9359-9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dandri, M., P. Schirmacher, and C. E. Rogler. 1996. Woodchuck hepatitis virus X protein is present in chronically infected woodchuck liver and woodchuck hepatocellular carcinomas which are permissive for viral replication. J. Virol. 70:5246-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dejean, A., L. Bougueleret, K. H. Grzeschik, and P. Tiollais. 1986. Hepatitis B virus DNA integration in a sequence homologous to v-erb-A and steroid receptor genes in a hepatocellular carcinoma. Nature 322:70-72. [DOI] [PubMed] [Google Scholar]
- 33.Diehl, A., M. Yin, J. Fleckenstein, S. Yang, H. Lin, D. Brenner, J. Westwick, G. Bagby, and S. Nelson. 1994. Tumor necrosis factor-alpha induces c-jun during the regenerative response to liver injury. Am. J. Physiol. 267:G552-G561. [DOI] [PubMed] [Google Scholar]
- 34.Doria, M., N. Klein, R. Lucito, and R. J. Schneider. 1995. Hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 14:4747-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elmore, L. W., A. R. Hancock, S. F. Chang, X. W. Wang, S. Chang, C. P. Callahan, D. A. Geller, H. Will, and C. C. Harris. 1997. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc. Natl. Acad. Sci. USA 94:14707-14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faktor, O., and Y. Shaul. 1990. The identification of hepatitis B virus X gene responsive elements reveals functional similarity of X and HTLV-I tax. Oncogene 5:867-872. [PubMed] [Google Scholar]
- 37.Feitelson, M., M. Zhu, L. X. Duan, and W. T. London. 1993. Hepatitis B x antigen and p53 are associated in vitro and in liver tissues from patients with primary hepatocellular carcinoma. Oncogene 8:1109-1117. [PubMed] [Google Scholar]
- 38.Forgues, M., A. Marrogi, E. Spillare, C.-G. Wu, Q. Yang, M. Yoshida, and X. Wang. 2001. Interaction of the hepatitis B virus X protein with the Crm1-dependent nuclear export pathway. J. Biol. Chem. 276:22797-22803. [DOI] [PubMed] [Google Scholar]
- 39.Ganem, D., and R. J. Schneider. 2001. The molecular biology of the hepatitis B viruses, p. 2923-2970. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus, Fields virology, 4th ed, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 40.Groisman, I. J., R. Koshy, F. Henkler, J. D. Groopman, and M. A. Alaoui-Jamali. 1999. Downregulation of DNA excision repair by the hepatitis B virus-x protein occurs in p53-proficient and p53-deficient cells. Carcinogenesis 20:479-483. [DOI] [PubMed] [Google Scholar]
- 41.Guo, W. T., J. Wang, G. Tam, T. S. Yen, and J. S. Ou. 1991. Leaky transcription termination produces larger and smaller than genome size hepatitis B virus X gene transcripts. Virology 181:630-636. [DOI] [PubMed] [Google Scholar]
- 42.Han, H. J., E. Y. Jung, W. J. Lee, and K. L. Jang. 2002. Cooperative repression of cyclin-dependent kinase inhibitor p21 gene expression by hepatitis B virus X protein and hepatitis C virus core protein. FEBS Lett. 518:169-172. [DOI] [PubMed] [Google Scholar]
- 43.Haviv, I., M. Shamay, G. Doitsch, and Y. Shaul. 1998. Hepatitis B virus pX targets TFIIB in transcription coactivation. Mol. Cell. Biol. 18:1562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henkler, F., J. Hoare, N. Waseem, R. D. Goldin, M. J. McGarvey, R. Koshy, and I. A. King. 2001. Intracellular localization of the hepatitis B virus HBx protein. J. Gen. Virol. 82:871-882. [DOI] [PubMed] [Google Scholar]
- 45.Hoare, J., F. Henkler, J. Dowling, W. Errington, R. Goldin, D. Fish, and M. McGarvery. 2001. Subcellular localisation of the X protein of HBV infected hepatocytes. J. Med. Virol. 64:419-426. [DOI] [PubMed] [Google Scholar]
- 46.Hohne, M., S. Schaefer, M. Seifer, M. A. Feitelson, D. Paul, and W. H. Gerlich. 1990. Malignant transformation of immortalized transgenic hepatocytes after transfection with hepatitis B virus DNA. EMBO J. 9:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang, J., J. Kwong, E. C.-Y. Sun, and T. J. Liang. 1996. Proteosome complex as a potential cellular target of hepatitis B virus X protein. J. Virol. 70:5582-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia, L., X. W. Wang, and C. C. Harris. 1999. Hepatitis B virus X protein inhibits nucleotide excision repair. Int. J. Cancer 80:875-879. [DOI] [PubMed] [Google Scholar]
- 49.Kay, A., E. Mandart, C. Trepo, and F. Galibert. 1985. The HBV HBx gene expressed in E. coli is recognized by sera from hepatitis patients. EMBO J. 4:1287-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim, C.-M., K. Koike, I. Saito, T. Miyamura, and G. Jay. 1991. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature 353:317-320. [DOI] [PubMed] [Google Scholar]
- 51.Kim, H., H. Lee, and Y. Yun. 1998. X-gene product of hepatitis B virus induces apoptosis in liver cells. J. Biol. Chem. 273:381-385. [DOI] [PubMed] [Google Scholar]
- 52.Kim, K. H., and B. L. Seong. 2003. Pro-apoptotic function of HBV X protein is mediated by interaction with c-FLIP and enhancement of death-inducing signal. EMBO J. 22:2104-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klein, A., E. Guhl, Y. J. Tzeng, J. Fuhrhop, M. Levrero, M. Graessmann, and A. Graessmann. 2003. HBx causes cyclin D1 overexpression and development of breast cancer in transgenic animals that are heterozygous for p53. Oncogene 22:2910-2919. [DOI] [PubMed] [Google Scholar]
- 54.Klein, N., M. Bouchard, L.-H. Wang, C. Kobarg, and R. J. Schneider. 1999. Src kinases involved in hepatitis B virus replication. EMBO J. 18:5019-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klein, N., and R. J. Schneider. 1997. Activation of Src family kinases by hepatitis B virus HBx protein and coupled signaling to Ras. Mol. Cell. Biol. 17:6427-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kodama, K., N. Ogasawara, H. Yoshikawa, and S. Murakami. 1985. Nucleotide sequence of a cloned woodchuck hepatitis virus genome: evolutional relationship between hepadnaviruses. J. Virol. 56:978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koike, K., K. Moriya, H. Yotsuyanagi, Y. Shintani, H. Fujie, T. Tsutsumi, and S. Kimura. 1998. Compensatory apoptosis in preneoplastic liver of a transgenic mouse model for viral hepatocarcinogenesis. Cancer Lett. 134:181-186. [DOI] [PubMed] [Google Scholar]
- 58.Kwee, L., R. Lucito, B. Aufiero, and R. J. Schneider. 1992. Alternate translation initiation on hepatitis B virus X mRNA produces multiple polypeptides that differentially transactivate class II and III promoters. J. Virol. 66:4382-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lara-Pezzi, E., A. Armessila, P. Majano, J. Redondo, and M. Lopez-Cabrera. 1999. The hepatitis B virus X protein activates nuclear factor of activated T cells (NF-AT) by a cyclosporin A-sensitive pathway. EMBO J. 17:7066-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lara-Pezzi, E., S. Roche, O. Andrisani, F. Sanchez-Madrid, and M. Lopez-Cabrera. 2001. The hepatitis B virus HBx protein induces adherens junction disruption in a src-dependent manner. Oncogene 20:3323-3331. [DOI] [PubMed] [Google Scholar]
- 61.Leach, J. K., L. Qiao, Y. Fang, S. L. Han, D. Gilfor, P. B. Fisher, S. Grant, P. B. Hylemon, D. Peterson, and P. Dent. 2003. Regulation of p21 and p27 expression by the hepatitis B virus X protein and the alternate initiation site X proteins, AUG2 and AUG3. J. Gastroenterol. Hepatol. 18:376-385. [DOI] [PubMed] [Google Scholar]
- 62.Lee, S., C. Tarn, W. H. Wang, S. Chen, R. L. Hullinger, and O. M. Andrisani. 2002. Hepatitis B virus X protein differentially regulates cell cycle progression in X-transforming versus nontransforming hepatocyte (AML12) cell lines. J. Biol. Chem. 277:8730-8740. [DOI] [PubMed] [Google Scholar]
- 63.Lee, T.-H., M. J. Finegold, R. F. Shen, J. L. DeMayo, S. L. C. Woo, and J. S. Butel. 1990. Hepatitis B virus transactivator X protein is not tumorigenic in transgenic mice. J. Virol. 64:5939-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee, Y. H., and Y. Yun. 1998. HBx protein of hepatitis B virus activates Jak1-STAT signaling. J. Biol. Chem. 273:25510-25515. [DOI] [PubMed] [Google Scholar]
- 65.Levrero, M., C. Balsano, G. Natoli, M. L. Avantaggiati, and E. Elfassi. 1990. Hepatitis B virus X protein transactivates the long terminal repeats of human immunodeficiency virus types 1 and 2. J. Virol. 64:3082-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levrero, M., O. Jean-Jean, C. Balsano, H. Wills, and M. Perricaudet. 1990. Hepatitis B virus (HBV) X gene expression in human cells and anti-HBx antibodies detection in chronic HBV infection. Virology 174:299-304. [DOI] [PubMed] [Google Scholar]
- 67.Lin, M.-H., and S. Lo. 1989. Dimerization of hepatitis B viral X protein synthesized in a cell-free system. Biochem. Biophys. Res. Commun. 164:14-21. [DOI] [PubMed] [Google Scholar]
- 68.Lin, Y., T. Nomura, J. Cheong, D. Dorjsuren, K. Iida, and S. Murakami. 1997. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcription factor IIB and RNA polymerase II subunit 5. J. Biol. Chem. 272:7132-7139. [DOI] [PubMed] [Google Scholar]
- 69.Lin-Marq, N., S. Bontron, O. Leupin, and M. Strubin. 2001. Hepatitis B virus protein interferes with cell viability through interaction with the p127-kDa UV-damaged DNA-binding protein. Virology 287:266-274. [DOI] [PubMed] [Google Scholar]
- 70.Lucito, R., and R. J. Schneider. 1992. Hepatitis B virus X protein activates transcription factor NF-κB without a requirement for protein kinase C. J. Virol. 66:983-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Madden, C., M. Finegold, and B. Slagle. 2001. Hepatitis B virus X protein acts a a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J. Virol. 75:3851-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Madden, C. R., M. J. Finegold, and B. L. Slagle. 2000. Expression of hepatitis B virus X protein does not alter the accumulation of spontaneous mutations in transgenic mice. J. Virol. 74:5266-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Madden, C. R., and B. L. Slagle. 2001. Stimulation of cellular proliferation by hepatitis B virus X protein. Dis. Markers 17:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maguire, H. F., J. P. Hoeffler, and A. Siddiqui. 1991. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science 252:842-844. [DOI] [PubMed] [Google Scholar]
- 75.Mahe, Y., N. Mukaida, K. Kuno, M. Akiyama, N. Ikeda, K. Matshushima, and S. Murakami. 1991. Hepatitis B virus X protein transactivates human interleukin-8 gene through acting on nuclear factor κB and CCAAT/enhancer -binding protein-like cis elements. J. Biol. Chem. 266:13759-13763. [PubMed] [Google Scholar]
- 76.Marusawa, H., S. Matsuzawa, K. Welsh, H. Zou, R. Armstrong, I. Tamm, and J. C. Reed. 2003. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 22:2729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Melegari, M., P. P. Scaglioni, and J. R. Wands. 1998. Cloning and characterization of a novel hepatitis B virus x binding protein that inhibits viral replication. J. Virol. 72:1737-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murakami, S. 2001. Hepatitis B virus X protein: a multifunctional viral regulator. J. Gastroenterol. 36:651-660. [DOI] [PubMed] [Google Scholar]
- 79.Murakami, S., J. Cheong, and S. Kaneko. 1994. Human hepatitis virus X gene encodes a regulatory domain that represses transactivation of X protein. J. Biol. Chem. 269:15118-15123. [PubMed] [Google Scholar]
- 80.Murakami, S., J.-H. Cheong, S. Ohno, K. Matsushima, and S. Kaneko. 1994. Transactivation of human hepatitis B virus X protein, HBx, operates through a mechanism distinct from protein kinase C and okadaic acid activation pathways. Virology 199:243-246. [DOI] [PubMed] [Google Scholar]
- 81.Nag, A., A. Datta, K. Yoo, D. Bhattacharyya, A. Chakrobortty, X. Wang, B. Slagle, R. Costa, and P. Raychaudhuri. 2001. DDB2 induces nuclear accumulation of the hepatitis B virus X protein independently of binding to DDB1. J. Virol. 75:10383-10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nagaya, T., T. Nakamura, T. Tokino, T. Tsurimoto, M. Imai, T. Mayumi, K. Kamino, K. Yamamura, and K. Matsubara. 1987. The mode of hepatitis B virus DNA integration in chromosomes of human hepatocellular carcinoma. Genes Dev. 1:773-782. [DOI] [PubMed] [Google Scholar]
- 83.Natoli, G., M. L. Avantaggiati, P. Chirillo, A. Costanzo, M. Artini, C. Balsano, and M. Levrero. 1994. Induction of the DNA-binding activity of c-Jun/c-Fos heterodimers by the hepatitis B virus transactivator pX. Mol. Cell. Biol. 14:989-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Natoli, G., M. L. Avantaggiati, P. Chirillo, P. L. Puri, A. Ianni, C. Balsano, and M. Levrero. 1994. Ras- and Raf-dependent activation of c-jun transcriptional activity by the hepatitis B virus transactivator pX. Oncogene 9:2837-2843. [PubMed] [Google Scholar]
- 85.Nijhara, R., S. Jana, S. Goswami, A. Rana, S. Majumdar, V. Kumar, and D. Sarkar. 2001. Sustained activation of mitogen-activated protein kinases and activator protein 1 by the hepatitis B virus X protein in mouse hepatocytes in vivo. J. Virol. 75:10348-10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oh, J. C., D. L. Jeong, I. K. Kim, and S. H. Oh. 2003. Activation of calcium signaling by hepatitis B virus-X protein in liver cells. Exp. Mol. Med. 35:301-309. [DOI] [PubMed] [Google Scholar]
- 87.Ozer, A., V. I. Khaoustov, M. Mearns, D. E. Lewis, R. M. Genta, G. J. Darlington, and B. Yoffee. 1996. Effect of hepatocyte proliferation and cellular DNA synthesis on hepatitis B virus replication. Gastroenterology 110:1519-1528. [DOI] [PubMed] [Google Scholar]
- 88.Pan, J., L. X. Duan, B. S. Sun, and M. A. Feitelson. 2001. Hepatitis B virus X protein protects against anti-Fas-mediated apoptosis in human liver cells by inducing NF-κB. J. Gen. Virol. 82:171-182. [DOI] [PubMed] [Google Scholar]
- 89.Park, U. S., S. K. Park, Y. I. Lee, and J. G. Park. 2000. Hepatitis B virus-X protein upregulates the expression of p21waf1/cip1 and prolongs G1→S transition via a p53-independent pathway in human hepatoma cells. Oncogene 19:3384-3394. [DOI] [PubMed] [Google Scholar]
- 90.Paterlini-Brechot, P., K. Saigo, Y. Murakami, M. Chami, D. Gozuacik, C. Mugnier, D. Lagorce, and C. Brechot. 2003. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene 22:3911-3916. [DOI] [PubMed] [Google Scholar]
- 91.Puisieux, A., J. Ji, C. Guillot, Y. Legros, T. Soussi, K. Isselbacher, and M. Ozturk. 1995. p53-mediated cellular response to DNA damage in cells with replicative hepatitis B virus. Proc. Natl. Acad. Sci. USA 92:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qadri, I., J. W. Conaway, R. C. Conaway, J. Schaack, and A. Siddiqui. 1996. Hepatitis B virus transactivator protein, HBx, associates with the components of TFIIH and stimulates the DNA helicase activity of TFIIH. Proc. Natl. Acad. Sci. USA 93:10578-10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qiao, L., K. Leach, R. McKinstry, D. Gilfor, A. Yacoub, J.-S. Park, S. Grant, P. Hylemon, P. Fisher, and P. Dent. 2001. Hepatitis B virus X protein increases expression of p21Cip-1/WAF1/MDA6 and p27Kip-1 in primary mouse hepatocytes, leading to a reduced cell cycle progression. Hepatology 34:906-917. [DOI] [PubMed] [Google Scholar]
- 94.Rahmani, Z., K. W. Huh, R. Lasher, and A. Siddiqui. 2000. Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3, and alters its transmembrane potential. J. Virol. 74:2840-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rao, A., C. Luo, and P. Hogan. 1997. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15:707-747. [DOI] [PubMed] [Google Scholar]
- 96.Reifenberg, K., P. Nusser, J. Lohler, G. Spindler, C. Kuhn, F. von Weizsacker, and J. Kock. 2002. Virus replication and virion export in X-deficient hepatitis B virus transgenic mice. J. Gen. Virol. 83:991-996. [DOI] [PubMed] [Google Scholar]
- 97.Reifenberg, K., H. Wilts, J. Lohler, P. Nusser, R. Hanano, L. G. Guidotti, F. V. Chisari, and H. J. Schlicht. 1999. The hepatitis B virus X protein transactivates viral core gene expression in vivo. J. Virol. 73:10399-10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schek, N., R. Bartenschlager, C. Kuhn, and H. Schaller. 1991. Phosphorylation and rapid turnover of hepatitis B virus X protein expressed in HepG2 cells from a recombinant vaccinia virus. Oncogene 6:1735-1744. [PubMed] [Google Scholar]
- 99.Shih, W.-L., M.-L. Kuo, S.-E. Chuang, A.-L. Cheng, and S.-L. Doong. 2000. Hepatitis B virus X protein inhibits transforming growth factor-β-induced apoptosis through the activation of phosphatidylinositol 3-kinase pathway. J. Biol. Chem. 275:25858-25864. [DOI] [PubMed] [Google Scholar]
- 100.Shintani, Y., H. Yotsuyanagi, K. Moriya, H. Fujie, T. Tsutsumi, Y. Kanegae, S. Kimura, I. Saito, and K. Koike. 1999. Induction of apoptosis after switch-on of the hepatitis B virus X gene mediated by the Cre/loxP recombination system. J. Gen. Virol. 80:3257-3265. [DOI] [PubMed] [Google Scholar]
- 101.Shirakata, Y., and K. Koike. 2003. Hepatitis B virus X protein induces cell death by causing loss of mitochondrial membrane potential. J. Biol. Chem. 278:22071-22078. [DOI] [PubMed] [Google Scholar]
- 102.Siddiqui, A., S. Jameel, and J. Mapoles. 1987. Expression of the hepatitis B virus X gene in mammalian cells. Proc. Natl. Acad. Sci. USA 84:2513-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sirma, H., C. Giannini, K. Poussin, P. Paterlini, D. Kremsdorf, and C. Brechot. 1999. Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene 18:4848-4859. [DOI] [PubMed] [Google Scholar]
- 104.Sirma, H., R. Weil, O. Rosmorduc, S. Urban, A. Israel, D. Kremsdorf, and C. Brechot. 1998. Cytosol is the prime compartment of hepatitis B virus X protein where it colocalizes with the proteasome. Oncogene 16:2051-2063. [DOI] [PubMed] [Google Scholar]
- 105.Sitterlin, D., F. Bergametti, P. Tiollais, B. C. Tennant, and C. Transy. 2000. Correct binding of viral X protein to UVDDB-p127 cellular protein is critical for efficient infection by hepatitis B viruses. Oncogene 19:4427-4431. [DOI] [PubMed] [Google Scholar]
- 106.Slagle, B. L., T. H. Lee, D. Medina., M. J. Finegold, and J. S. Butel. 1996. Increased sensitivity to the hepatocarcinogen diethylnitrosamine in transgenic mice carrying the hepatitis B virus X gene. Mol. Carcinog. 15:261-269. [DOI] [PubMed] [Google Scholar]
- 107.Sohn, S., I. Jaitovitch-Groisman, N. Benlimame, J. Galipeau, G. Batist, and M. Alaoui-Jamali. 2000. Retroviral expression of the hepatitis B virus x gene promotes liver cell susceptibility to carcinogen-induced site specific mutagenesis. Mutat. Res. 460:17-28. [DOI] [PubMed] [Google Scholar]
- 108.Su, F., and R. J. Schneider. 1996. Hepatitis B virus HBx protein activates transcription factor NF-κB by acting on multiple cytoplasmic inhibitors of rel-related proteins. J. Virol. 70:4558-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Su, F., and R. J. Schneider. 1997. Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by TNFα. Proc. Natl. Acad. Sci. USA 94:8744-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Su, F., C. N. Theodosis, and R. J. Schneider. 2001. Role of NF-κB and Myc proteins in apoptosis induced by hepatitis B virus HBx protein. J. Virol. 75:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Su, Q., C. H. Schroder, W. J. Hofman, G. Otto, R. Pichlmayr, and P. Bannasch. 1998. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology 27:1109-1120. [DOI] [PubMed] [Google Scholar]
- 112.Su, Q., C. H. Schroder, G. Otto, and P. Bannasch. 2000. Overexpression of p53 protein is not directly related to hepatitis B x protein expression and is associated with neoplastic progression in hepatocellular carcinomas rather than hepatic preneoplasia. Mutat. Res. 462:365-380. [DOI] [PubMed] [Google Scholar]
- 113.Sugata, F., H. S. Chen, S. Kaneko, R. H. Purcell, and R. H. Miller. 1994. Analysis of the X gene promoter of woodchuck hepatitis virus. Virology 205:314-320. [DOI] [PubMed] [Google Scholar]
- 114.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 115.Summers, J., A. O'Connell, and I. Millman. 1975. Genome of hepatitis B virus: restriction enzyme cleavage and structure of DNA extracted from Dane particles. Proc. Natl. Acad. Sci. USA 72:4597-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Takada, S., N. Kaneniwa, N. Tsuchida, and K. Koike. 1997. Cytoplasmic retention of the p53 tumor suppressor gene product is observed in the hepatitis B virus X gene-transfected cells. Oncogene 15:1895-1901. [DOI] [PubMed] [Google Scholar]
- 117.Takada, S., Y. Shirakata, N. Kaneniwa, and K. Koike. 1999. Association of hepatitis B virus X protein with mitochondria causes mitochondrial aggregation at the nuclear periphery, leading to cell death. Oncogene 18:6965-6973. [DOI] [PubMed] [Google Scholar]
- 118.Tarn, C., M. L. Bilodeau, R. L. Hullinger, and O. M. Andrisani. 1999. Differential immediate early gene expression in conditional hepatitis B virus pX-transforming versus nontransforming hepatocyte cell lines. J. Biol. Chem. 274:2327-2336. [DOI] [PubMed] [Google Scholar]
- 119.Tarn, C., S. Lee, Y. Hu, C. Ashendel, and O. M. Andrisani. 2001. Hepatitis B virus X protein differentially activates RAS-RAF-MAPK and JNK pathways in X-transforming versus non-transforming AML12 hepatocytes. J. Biol. Chem. 276:34671-34680. [DOI] [PubMed] [Google Scholar]
- 120.Tarn, C., L. Zou, R. Hullinger, and O. Andrisani. 2002. Hepatitis B virus X protein activates the p38 mitogen-activated protein kinase pathway in dedifferentiated hepatocytes. J. Virol. 76:9763-9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Terradillos, O., O. Billet, C.-A. Rnard, R. Levy, T. Molina, P. Briand, and M. A. Buendia. 1997. The hepatitis B virus X gene potentiates c-myc-induced liver oncognesis in transgenic mice. Oncogene 14:395-404. [DOI] [PubMed] [Google Scholar]
- 122.Terradillos, O., A. de La Coste, T. Pollicino, C. Neuveut, D. Sitterlin, H. Lecoeur, M. L. Gougeon, A. Kahn, and M. A. Buendia. 2002. The hepatitis B virus X protein abrogates Bcl-2-mediated protection against Fas apoptosis in the liver. Oncogene 21:377-386. [DOI] [PubMed] [Google Scholar]
- 123.Terradillos, O., T. Pollicino, H. Lecoeur, M. Tripodi, M. L. Gougeon, P. Tiollais, and M. A. Buendia. 1998. p53-Independent apoptotic effects of the hepatitis B virus HBx protein in vivo and in vitro. Oncogene 17:2115-2123. [DOI] [PubMed] [Google Scholar]
- 124.Tokino, T., and K. Matsubara. 1991. Chromosomal sites for hepatitis B virus integration in human hepatocellular carcinoma. J. Virol. 65:6761-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Truant, R., J. Antunovic, J. Greenblatt, C. Prives, and J. A. Cromlish. 1995. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. J. Virol. 69:1851-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tu, H., C. Bonura, C. Giannini, H. Mouly, P. Soussan, M. Kew, P. Paterlini-Brechot, C. Brechot, and D. Kremsdorf. 2001. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 61:7803-7810. [PubMed] [Google Scholar]
- 127.Urban, S., E. Hildt, C. Eckerskorn, H. Sirma, A. Kekule, and P. H. Hofschneider. 1997. Isolation and molecular characterization of hepatitis B virus X-protein from a baculovirus expression system. Hepatology 26:1045-1053. [DOI] [PubMed] [Google Scholar]
- 128.Wang, H.-D., A. Trivedi, and D. L. Johnson. 1997. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol. Cell. Biol. 17:6838-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang, H.-D., C.-H. Yuh, C. V. Dang, and D. L. Johnson. 1995. The hepatitis B virus X protein increases the cellular level of TATA-binding protein which mediates transactivation of RNA polymerase III genes. Mol. Cell. Biol. 15:6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang, H. D., A. Trivedi, and D. L. Johnson. 1998. Regulation of RNA polymerase I-dependent promoters by the hepatitis B virus X protein via activated Ras and TATA-binding protein. Mol. Cell. Biol. 18:7086-7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang, J., X. Chenivesse, B. Henglein, and C. Brechot. 1990. Hepatitis B virus integration in a cyclin A gene in human hepatocellular carcinoma. Nature 343:555-557. [DOI] [PubMed] [Google Scholar]
- 132.Wang, X. W., K. Forrester, H. Yeh, M. A. Feitelson, J. Gu, and C. C. Harris. 1994. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc. Natl. Acad. Sci. USA 91:2230-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Weil, R., H. Sirma, C. Giannini, D. Kremsdorf, C. Bessia, C. Dargemont, C. Brechot, and A. Israel. 1999. Direct association and nuclear import of the hepatitis B virus X protein with the NF-κB inhibitor IκBα. Mol. Cell. Biol. 19:6345-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wentz, M., S. Becker, and B. Slagle. 2000. Dissociation of DDB1-binding and transactivation properties of the hepatitis B virus X protein. Virus Res. 68:87-92. [DOI] [PubMed] [Google Scholar]
- 135.Williams, J. S., and O. M. Andrisani. 1995. The hepatitis B virus X protein targets the basic region-leucine zipper domain of CREB. Proc. Natl. Acad. Sci. USA 92:3819-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xu, Z., T. S. Yen, L. Wu, C. R. Madden, W. Tan, B. L. Slagle, and J. H. Ou. 2002. Enhancement of hepatitis B virus replication by its X protein in transgenic mice. J. Virol. 76:2579-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yen, T. S. B. 1996. Hepadnaviral X protein: review of recent progress. J. Biomed. Sci. 3:20-30. [DOI] [PubMed] [Google Scholar]
- 138.Yu, D. Y., H. B. Moon, J. K. Son, S. Jeong, S. L. Yu, H. Yoon, Y. M. Han, C. S. Lee, J. S. Park, C. H. Lee, B. H. Hyun, S. Murakami, and K. K. Lee. 1999. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J. Hepatol. 31:123-132. [DOI] [PubMed] [Google Scholar]
- 139.Zhang, Z., N. Torii, Z. Hu, J. Jacob, and T. Liang. 2001. X-deficient woodchuck hepatitis virus mutants behave like attenuated viruses and induce protective immunity in vivo. J. Clin. Investig. 108:1523-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zheng, Y., J. Li, D. L. Johnson, and J. H. Ou. 2003. Regulation of hepatitis B virus replication by the Ras-mitogen-activated protein kinase signaling pathway. J. Virol. 77:7707-7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhu, H., Y. Wang, J. S. Chen, G. Cheng, and J. Xue. 2004. Transgenic mice expressing hepatitis B virus X protein are more susceptible to carcinogen induced hepatocarcinogenesis. Exp. Mol. Pathol. 76:44-50. [DOI] [PubMed] [Google Scholar]
- 142.Zoulim, F., J. Saputelli, and C. Seeger. 1994. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 68:2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]