Abstract
Immediate-early viral gene products of human cytomegalovirus (HCMV) are derived from several genomic loci and largely serve to establish a cellular environment conducive to viral replication. We have further examined an unusual immediate-early transcript known as the 5-kb RNA, concluding that it is a stable intron encoded by HCMV. The 5-kb RNA is highly AT rich in sequence and lacks open reading frames likely to be translated into protein. We confirmed the absence of polyadenylation of the transcript and showed that it is primarily nuclear localized during viral infection. We mapped the 5′ end of the 5-kb RNA to a consensus splice donor site and localized the 3′ end in the vicinity of a splice acceptor site. In transfection studies, we showed that the 5-kb RNA can be spliced from a heterologous primary transcript. Using bacterial artificial chromosome technology, we constructed a viral recombinant containing a mutation in the 5′ splice donor site that defines the 5′ end of the RNA and found that this mutation eliminates expression of the 5-kb RNA during viral infection. This mutant grows in human fibroblasts without complementation. Taken together, these data support the conclusion that the 5-kb RNA is a stable intron expressed by HCMV.
Human cytomegalovirus (HCMV) is a member of the betaherpesvirus family (reviewed in reference 24). Infection is widespread and generally asymptomatic in healthy individuals. The virus infects a wide variety of cell types such as endothelial cells, fibroblasts, smooth muscle cells, macrophages, and cells of the bone marrow. Following primary exposure, HCMV establishes a lifelong latent infection in the host and latent virus is found in cells of the myeloid lineage (23, 36). Primary or reactivated HCMV infections can cause severe disease in AIDS patients and transplant patients, including retinitis and pneumonia, and are associated with acute organ rejection (28). Congenital HCMV infections are the leading cause of virally induced birth defects, and neonates can suffer life-threatening complications following HCMV infection.
During productive infection, HCMV genes are expressed in an organized cascade of immediate-early, early, and late transcription (reviewed in reference 24). The immediate-early gene products serve to establish a cellular environment conducive to viral replication, functioning largely in transcriptional regulation and immune evasion. The expression of early and late genes is dependent upon the activities of the immediate-early gene products. Immediate-early transcripts originate from several loci in the HCMV genome. The most abundant transcripts are derived from the UL123 and UL122 genes, encoding the family of IE1 and IE2 proteins, respectively. The UL36-38, UL115-119, UL69, TRS1, IRS1, and US3 genes encode the remaining immediate-early proteins. Together, the immediate-early genes of HCMV encode a variety of proteins with diverse regulatory activities, serving essential and dispensable functions in cultured cells during HCMV replication.
An additional immediate-early transcript of HCMV has been identified: the so-called 5-kb RNA accumulates during the immediate-early and late phases of infection, and its level drops during the early phase (12, 20, 30, 32). Plachter et al. (30) reported that the 5-kb RNA was equally abundant in cytoplasmic and nuclear compartments and evenly distributed between the polyadenylated and nonpolyadenylated fractions of total RNA from infected cells. They also mapped the 5′ end of the transcript to a G residue at position 159627 in the AD169 genome. The 3′ end of the 5-kb RNA was mapped downstream of a consensus polyadenylation sequence at position 154829. The full-size 5-kb RNA was reported to be ∼4,800 nucleotides (nt) long, and high-resolution Northern blot analysis detected two bands that differ by ∼400 nt. No evidence was found for splicing within the 5-kb RNA, and Plachter et al. (30) attributed the size heterogeneity to potential differences in 3′ termination and processing events.
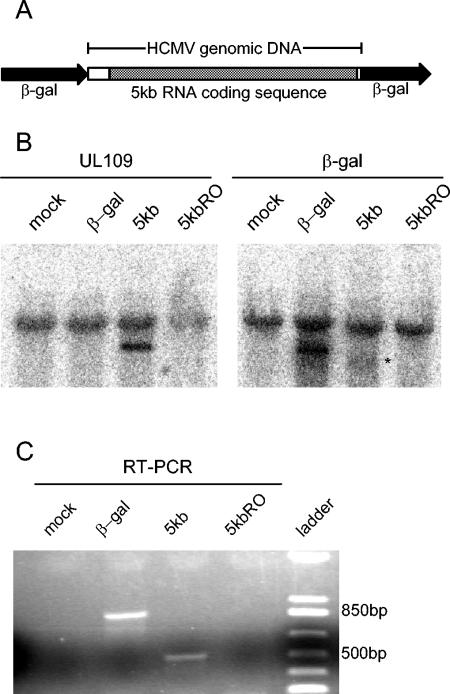
The genomic sequence from which the 5-kb RNA is expressed contains a number of unusual characteristics. The 5-kb RNA has an AT content of ∼60%, considerably higher than that of the overall HCMV genome, and includes numerous stretches of homopolymeric A or T residues (ranging from 5 to 11 nt). Previous analysis revealed no open reading frames (ORFs) longer than 99 amino acids, with none having significant homology to sequences in the databases existing at the time (30). Subsequent annotation of the HCMV AD169 genome identified six ORFs ranging in length from 93 to 153 amino acids in the region spanned by the transcript (UL106 through UL111; Fig. 1) (8). Only UL109 and UL106 have AUG initiator codons, neither of which are located within Kozak consensus sites. Frequent start and stop codons define additional small ORFs in all three reading frames within the region. Consequently, there is no compelling evidence that the 5-kb RNA encodes a protein, although it remains possible that spliced RNAs encode proteins in this region.
FIG. 1.
Schematic diagram of the 5-kb-RNA locus of HCMV strain AD169. Tick marks on the black bar mark every 1,000 nt. ORFs annotated by Chee et al. (8) are indicated with shaded arrows. The two exons of the spliced 1.1-kb late RNA are indicated with open rectangles (31). The solid black arrow above the diagram indicates the position of the 5-kb RNA as determined in this study.
Another relevant feature of the region encoding the 5-kb RNA is the expression of a l.1-kb spliced late RNA. The presence of this spliced transcript was predicted by a computer analysis that searched for consensus splice donor and acceptor sites; confirmation was then achieved by sequencing cDNA clones from an HCMV library (31). The two exons of this spliced RNA flank the region spanned by the 5-kb RNA (Fig. 1). The intron of this mRNA is calculated to be 4,528 nt in length, very close to the length of the 5-kb RNA. Interestingly, the 3′ end of the first exon of the spliced RNA also falls close to, but not directly adjacent to, the 5′ end of the 5-kb RNA as defined by Plachter et al. (30). The spliced 1.1-kb RNA is detected only at late times during infection and is predicted to encode a protein of 31 amino acids in length that bears no similarity to other known proteins (31). Nothing is known about the function of this spliced transcript or the protein it putatively encodes.
Given the incongruous features of the 5-kb RNA and its uncertain relationship to the 1.1-kb late spliced RNA, we sought to further characterize the transcript in an effort to determine a function for such a large and abundant immediate-early RNA. We reexamined the subcellular localization and polyadenylation status of the transcript, and we determined that the 5-kb RNA is a transcription product of RNA polymerase II. We also remapped the termini of the transcript and found subtle differences from previously published results. Based on these observations, we hypothesized that the 5-kb RNA is a stable intron derived from processing of a precursor RNA. We observed that the 5-kb RNA could be spliced out of a heterologous reporter gene in transfection experiments, indicating that the generation of the 5-kb RNA could result from a splicing event and was independent of viral infection. Lastly, we constructed a viral mutant lacking the splice donor site we believed to function in the generation of the 5-kb RNA. While this viral mutant replicates efficiently in cultured fibroblasts, it failed to express the 5-kb transcript, supporting our hypothesis that expression of the 5-kb RNA was the result of a splicing event.
MATERIALS AND METHODS
Cell culture and virus.
Primary human foreskin fibroblast cells and H1299 lung carcinoma cells were propagated in Dulbecco's modified Eagle medium with 10% fetal calf serum or newborn calf serum at 37°C in a 5% CO2 atmosphere. HCMV strain AD169 was used in this study (33). Lipofectamine was used for transfection of H1299 cells, as recommended by the manufacturer (Invitrogen, Carlsbad, Calif.). Virus titers were determined in human foreskin fibroblasts by plaque assay.
RNA analysis.
Total RNA was prepared from cells, using Trizol as recommended by the manufacturer (Invitrogen). Immediate-early RNA was prepared from fibroblasts infected with AD169 at a multiplicity of 2 PFU/cell in the presence of 100 μg of cycloheximide/ml at 18 h postinfection. Late RNA was prepared from fibroblasts infected with the same multiplicity at 48 h postinfection. Polyadenylated RNA was isolated by using an Oligotex mRNA Midi kit (QIAGEN, Valencia, Calif.). Nuclear and cytoplasmic RNA fractions were derived from infected cells collected by trypsinization at the appropriate time postinfection using previously described methods (43). Briefly, the cell pellet was resuspended in RSB (10 mM NaCl, 3 mM MgCl2, 10 mM Tris-HCl [pH 7.4]) and 20× detergent (10% sodium deoxycholate and 20% Tween 40 in RSB) was added to a 1× final concentration. Cells were vortexed and incubated on ice for 5 min. Nuclei were pelleted by centrifugation at 1,000 × g for 3 min at 4°C. The supernatant was processed for total cytoplasmic RNA, and the pellet was processed for total nuclear RNA, using Trizol.
For Northern blot analysis, RNA was glyoxalated (2), subjected to 0.5% agarose gel electrophoresis, and transferred to a GeneScreen Plus nylon membrane (Perkin-Elmer Life Sciences, Boston, Mass.). Riboprobes were radiolabeled with [α-32P]UTP, using the SP6/T7 Riboprobe Combination system (Promega, Madison, Wis.). PCR-amplified DNA sequence from IE1 (UL123), UL106, UL107, UL109, or the polyadenylation sequence between UL105 and UL106 was cloned into pSP72 (Promega) to produce strand-specific antisense riboprobes. A KpnI/EcoRV-generated restriction fragment from the β-galactosidase gene in pCH110 (Amersham, Piscataway, N.J.) was cloned into pBluescript KS(+) (Stratagene, LaJolla, Calif.) to generate a strand-specific antisense riboprobe for β-galactosidase. Northern hybridization of riboprobes was performed as described elsewhere (2) and detected by phosphorimager analysis.
Nuclear run-on assays were performed as described previously (3) with the following modifications. Nuclei were prepared from mock- or HCMV-infected fibroblasts at 6 h postinfection. Confluent monolayers on 15-cm tissue culture dishes were rinsed and scraped off in ice-cold phosphate-buffered saline. The cell pellet was resuspended in ice-cold ID buffer (0.32 M sucrose, 3 mM CaCl2, 2 mM magnesium acetate, 0.1 mM EDTA, 10 mM Tris HCl [pH 8.0]). Cells were then disrupted in a Dounce homogenizer. Resulting nuclei were collected by centrifugation and resuspended in ice-cold GSB (5 mM MgCl2, 40% glycerol, 0.1 mM EDTA, 50 mM Tris [pH 8.3]). RNA purified from nuclear run-on assays performed in the presence or absence of α-amanitin was hybridized to filters in Church buffer (9). Plasmids containing the 28S rRNA 5′ external transcribed spacer (ETS) and U6 sequences were kindly provided by J. Flint (Princeton University). Plasmids containing IE1 or UL106 were the same constructs as those used for generation of riboprobes. Plasmid DNA was denatured and immobilized on a GeneScreen Plus nylon membrane following standard protocols (3). Hybridized run-on RNAs were detected by phosphorimager analysis.
Primer extension assays were performed as described previously (3) using primer pcak15: 5′-TGG TTC CTA CAG AAA CTA TTC TAA CCG CGG AAG AAA GAA ATT-3′. Radiolabeled primer pcak15 was hybridized to 40 μg of total RNA prepared from mock-infected or AD169-infected fibroblasts at immediate-early or late time points after infection. The primer was extended by using SuperscriptII reverse transcriptase (Invitrogen), and extension products were analyzed by denaturing 10% urea-polyacrylamide gel electrophoresis followed by phosphorimager analysis.
Reverse transcription-PCR (RT-PCR) was performed on total RNA prepared from H1299 cells transfected with constructs for analysis of splicing from the 5-kb RNA locus. Total RNA was extensively treated with DNase I prior to RT-PCR amplification of 500 ng of RNA with β-galactosidase-specific primers (pcak41, 5′-ATG AAT CGT CTG ACC GAT GAT CC-3′; and pcak42, 5′-TGA TGT GCC CGG CTT CTG ACC-3′) using the Titan One Tube RT-PCR system (Roche, Indianapolis, Ind.). DNase I was removed from RNA samples by using the DNase Inactivation Reagent according to the product recommendations (Ambion, Austin, Tex.).
Construction of 5-kb-mutant virus.
The recombinant virus carrying a mutation in the splice donor site at the 5′ end of the region encoding the 5-kb RNA was constructed by an allelic exchange protocol (41). The mutation was created by mutagenizing the splice donor site on a small fragment of HCMV DNA cloned into pSP72. The mutagenesis was accomplished using the QuikChange XL Site-Directed Mutagenesis kit (Stratagene) as specified by the manufacturer. Two complementary primers containing the desired mutation were synthesized (pcak47A, 5′-TAA CAT AAA GGA CCA CCT CGA GGG GAC GCG CAG TTG GGC G-3′; and pcak47B, 5′-CGC CCA ACT GCG CGT CCC CTC GAG GTG GTC CTT TAT GTT A-3′). Successful mutation of the splice donor site resulted in the introduction of an XhoI restriction site and allowed for screening of clones for the desired mutation. Once the splice donor mutation was confirmed, the sequence was subcloned into the shuttle vector pGS284 along with additional flanking sequence to perform allelic exchange with bacterial artificial chromosome (BAC) TS149. This BAC contains a transposon insertion in the UL111 ORF, approximately 100 bp away from the splice donor site (40). The transposon contains two selectable markers: a kanamycin resistance gene and the lacZ gene, permitting blue-white selection of recombinants. Recombination of the target sequence containing the splice donor mutation removes the transposon mutation, reverting the BAC to a white phenotype, and introduces the splice donor site mutation into the viral genome. Recombination was performed with a recA+ strain of Escherichia coli (34). Recombinants were again screened for inclusion of the XhoI site by Southern blotting and virus, termed BADpm5kbSD, was recovered by electroporation of the BAC clone into fibroblasts.
Plasmid construction.
pCH110-5kb was constructed by inserting a BglII/BamHI restriction fragment spanning the 5-kb RNA locus into the unique BclI site within the β-galactosidase gene of pCH110 (Amersham). To remove sequence 3′ to the second exon of the 1.1-kb late RNA, including the polyadenylation sequence, this intermediate construct was digested with RsrII and SacI, treated with T4 DNA polymerase to blunt the ends, and closed by ligation with T4 DNA ligase. The resulting construct, pCH110-5kb, therefore lacks a portion of the β-galactosidase sequence as well as sequences 3′ to the second exon of the 1.1-kb late RNA.
RESULTS
The 5-kb RNA is not polyadenylated, and it is localized to the nucleus.
Previous analysis of the 5-kb RNA showed that it was expressed most abundantly during the immediate-early and late phases of viral replication (30). This initial work concluded that the transcript was equally abundant in cytoplasmic and nuclear fractions of total RNA and evenly distributed between the polyadenylated and nonpolyadenylated fractions of total RNA.
We reexamined the polyadenylation status of the 5-kb RNA, taking into account the vast differences between the absolute amounts of poly(A)+ and poly(A)− RNA in cells. The relative amounts of 5-kb RNA in these two fractions were determined by Northern blot analysis of oligo(dT)-selected RNA prepared from HCMV-infected cells. The majority of the 5-kb RNA was in the nonpolyadenylated fraction at both immediate-early and late times after infection (Fig. 2A). A small quantity of 5-kb RNA was detected in the polyadenylated fraction. The small amount of 5-kb RNA in this fraction may result from the binding of 5-kb RNA to the oligo(dT) matrix via the multiple homopolymeric stretches of A residues found throughout the 5-kb coding sequence. Northern blot analysis of the same samples for the IE1 (UL123) RNA showed that it was found exclusively in the polyadenylated fraction, as expected, demonstrating that our selection efficiently captured polyadenylated RNA.
FIG. 2.
Polyadenylation status and cellular localization of the 5-kb RNA. (A) Polyadenylation status of the 5-kb RNA. Total RNA was prepared from AD169-infected fibroblasts at immediate-early or late time points. Poly(A)+ or poly(A)− fractions were prepared from total RNA by oligo(dT) selection. Northern blot analysis of total (T), polyadenylated (A+), or nonpolyadenylated (A−) RNA was performed with a 5-kb RNA- or IE1-specific probe as indicated. (B) Cellular localization of the 5-kb RNA. Nuclear (N) or cytoplasmic (C) fractions were prepared from AD169-infected fibroblasts at immediate-early (IE) or late (L) time points. Northern blot analysis was performed on these fractions and on total RNA (T) from infected cells or mock-infected cells (m) with a 5-kb RNA- or IE1-specific probe as indicated.
The subcellular localization of the 5-kb RNA was then determined by Northern blot analysis of RNA prepared from nuclear or cytoplasmic fractions of cells infected with HCMV at immediate-early or late times after infection (Fig. 2B). The 5-kb RNA is almost exclusively nuclear localized during the immediate-early phase and predominantly nuclear localized during the late phase of viral replication. A small fraction of the 5-kb RNA was detected in the cytoplasmic fraction of the late RNA. This might result from nuclear leakage, due in part to the substantial cytopathic effect observed at this point during infection. Northern blot analysis of the same RNA preparations for the IE1 RNA showed that substantial amounts of this RNA were present in the cytoplasmic fraction, consistent with an RNA that was exported from the nucleus to the cytoplasm for translation into protein. In contrast to a previous report (30), we conclude that the 5-kb RNA is not polyadenylated to any significant degree and is localized primarily to the nucleus throughout viral infection.
The 5-kb RNA is transcribed by RNA polymerase II.
Given the absence of polyadenylation and the nuclear localization of the 5-kb RNA, we considered that this transcript might not be a standard mRNA transcribed by RNA polymerase II. Protein-coding mRNAs transcribed by RNA polymerase II are rapidly processed for export to the cytoplasm and translated into protein. It was possible that the 5-kb RNA is transcribed by RNA polymerase I or III, although it lacks features consistent with RNAs transcribed by these polymerases. Nevertheless, to test this possibility, we performed nuclear run-on analysis of 5-kb transcription. Nuclei were prepared from mock- or HCMV-infected fibroblasts, and run-on assays were performed in the presence or absence of α-amanitin, a potent RNA polymerase II inhibitor at low concentrations. Run-on RNAs were purified and hybridized to filters carrying RNA polymerase I-, II-, or III-specific genes. As shown in Fig. 3, α-amanitin inhibited the run-on transcription of the 5-kb transcript, as well as that of IE1 RNA, an HCMV mRNA that is capped, polyadenylated, and processed for translation into protein. RNA polymerase I and III transcription was not inhibited under these conditions in HCMV-infected cells. We conclude that the 5-kb RNA is transcribed by RNA polymerase II.
FIG. 3.
Nuclear run-on analysis of 5-kb RNA transcription. Nuclei were prepared from mock- or AD169-infected fibroblasts during the immediate-early phase of infection. Run-on transcription was completed in the presence or absence of 0.1 μg of α-amanitin/ml. At this concentration, α-amanitin is a potent inhibitor of RNA polymerase II but has little effect on RNA polymerase I or III. Run-on transcripts were purified and hybridized to filters carrying RNA polymerase I-specific sequence (28S rRNA 5′ ETS), RNA polymerase II-specific sequence (HCMV IE1), and RNA polymerase III-specific sequence (U6 snRNA) in addition to two sequences specific to the 5-kb RNA (UL106 and UL109).
The 5-kb RNA is a stable intron.
Since the 5-kb RNA is transcribed by RNA polymerase II and is nuclear localized and nonpolyadenylated, we more carefully examined the structure of this RNA, including the precise location of the transcript termini relative to the splice junctions of the 1.1-kb RNA, whose exons roughly flank the 5-kb-RNA coding region.
We employed a primer extension assay to locate the 5′ end of the 5-kb RNA. Primer extension reactions were performed by hybridizing a radiolabeled oligonucleotide probe to total RNA from mock-infected or HCMV-infected fibroblasts. Following extension of the probe by reverse transcriptase, the primer extension products were analyzed by denaturing polyacrylamide electrophoresis (Fig. 4). We observed a single extension product derived from immediate-early and late RNA preparations that was 88 nt in length. The size of this product is consistent with the 5′ end of the 5-kb RNA being located at a G residue in the repeat sequence CAGGTAGGT (underlined G, nt 159631). No extension product was produced when the primer extension assay was carried out using RNA from mock-infected cells. The 5′ end identified in this experiment is displaced 4 bp from the end proposed earlier by Plachter et al. (30), which placed the 5′ end in the second copy of the AGGT repeat at nt 159627. Our work indicates that the 5′ end of the 5-kb RNA is adjacent to the 3′ end of the first exon of the 1.1-kb RNA.
FIG. 4.
Location of the 5′ end of the 5-kb RNA. Primer extension analysis was performed on total RNA prepared from mock-infected (M) or AD169-infected fibroblasts at immediate-early (IE) or late (L) time points using primer 15. The left lane contains a radiolabeled 10-bp ladder (la). The 100-bp band in the ladder is twice as intense as the other bands for orientation purposes, and the sizes (in nucleotides) of the ladder bands are indicated to the left.
We used Northern blot analysis to roughly localize the position of the 3′ end of the 5-kb RNA. The 5-kb RNA has been reported to terminate at a CA dinucleotide just downstream of a putative polyadenylation sequence located between UL106 and UL105 (Fig. 5) (30). This polyadenylation site has also been reported to be the poly(A) signal for the spliced 1.1-kb late RNA (31). Northern blot analysis was performed on total RNA prepared from mock-infected or HCMV-infected fibroblasts, using antisense riboprobes to UL106 or the polyadenylation site downstream of UL106 (Fig. 5). As shown in Fig. 6, the UL106 probe specifically detected the 5-kb RNA in both immediate-early and late RNA preparations. However, the polyadenylation site probe does not hybridize to the 5-kb RNA, whereas this probe does hybridize to the 1.1-kb RNA in late RNA preparations. This suggests that the 5-kb RNA is unlikely to extend beyond the UL106 coding sequence to the polyadenylation site as previously suggested (30); rather, the 3′ end of the 5-kb RNA lies within the UL106 ORF. Interestingly, the splice acceptor site that defines the 5′ end of the second exon of the 1.1-kb RNA also falls within the UL106 ORF, consistent with the possibility that the 3′ end of the 5-kb RNA coincides with this junction.
FIG. 5.
Location of the 3′ end of the 5-kb RNA. Schematic diagram of the 3′ end of the 5-kb genomic locus. ORFs annotated by Chee et al. (8) are indicated with arrows. Sequences used to generate strand-specific riboprobes are indicated with stippled bars. Exon 2 of the 1.1-kb late RNA is indicated with an open bar. The consensus polyadenylation site (AATAAA) reported to be used for both the 5-kb RNA and the 1.1-kb RNA is also indicated (30, 31).
FIG. 6.
Location of the 3′ end of the 5-kb RNA. Northern blot analysis of the 3′ end of the 5-kb RNA. Total RNA from mock-infected (m) or AD169-infected HFF cells at immediate-early (IE) or late (L) times was hybridized with UL106- or polyadenylation site-specific riboprobes. The small arrow indicates the 5-kb RNA, and the asterisks indicate the 1.1-kb RNA.
Since the 5′ and 3′ termini of the 5-kb RNA are located at or near the splice donor and splice acceptor sites of the 1.1-kb late RNA, we hypothesized that the 5-kb RNA is a stable intron. As an initial test of this hypothesis, we asked if the 5-kb RNA could be stably expressed when spliced from a heterologous reporter gene transfected into tissue culture cells. We employed a strategy similar to that previously used to demonstrate that the herpes simplex virus type 1 (HSV-1) latency-associated transcript (LAT) RNA is an intron (18). These studies demonstrated that the LAT RNA could be stably expressed when spliced from a heterologous reporter gene transfected into HeLa cells.
A 5.7-kb HCMV DNA fragment spanning the entire 5-kb-RNA coding region and flanking sequence was cloned into the β-galactosidase gene of pCH110, creating pCH110-5kb (Fig. 7A). H1299 cells were transfected with pCH110-5kb or a control plasmid (pCH110 or pCH110-5kbRO containing the HCMV sequence inserted in the reverse orientation). Total RNA was isolated and analyzed by Northern blot analysis with probes specific for the 5-kb RNA or β-galactosidase sequence. As shown in Fig. 7B, the 5-kb RNA-specific probe hybridized to a large RNA from cells transfected with pCH110-5kb but not from control transfections. This RNA comigrates with the 5-kb RNA expressed in HCMV-infected fibroblasts (data not shown). A β-galactosidase-specific probe hybridized to an RNA of the expected size in cells transfected with pCH110. This probe also weakly detected an RNA in cells transfected with pCH110-5kb. The size of this RNA is consistent with it being the spliced exons from the HCMV sequence flanked by β-galactosidase sequence. This result also suggests that the 5-kb RNA is processed independently of other viral proteins and its stability does not require viral infection.
FIG. 7.
The 5-kb RNA is an intron. (A) Schematic diagram of the pCH110-5kb construct. Exons 1 and 2 of the 1.1-kb late RNA are indicated by open boxes. In pCH110, β-galactosidase is under the control of the simian virus 40 early promoter. (B) Northern blot analysis of total RNA prepared from H1299 cells transfected with the indicated construct. Duplicate blots were hybridized with 5-kb RNA-specific (UL109) or β-galactosidase-specific probes as indicated. The asterisk indicates the weakly hybridizing RNA that likely represents the spliced 1.1-kb RNA exons flanked by β-galactosidase sequence. (C) Ethidium bromide-stained agarose gel analysis of RT-PCR amplification products from total RNA prepared from H1299 cells transfected with the indicated constructs. RT-PCR was performed with primers specific for β-galactosidase sequence flanking the HCMV sequence insertion point.
To confirm the structures of the spliced RNAs, RT-PCR analysis was performed on total RNA from transfected H1299 cells, using primers specific for the β-galactosidase sequence flanking the point of insertion of HCMV DNA into pCH110-5kb. The predicted size of the RT-PCR product from normal β-galactosidase RNA is 875 bp. If the 5-kb RNA is correctly spliced from the β-galactosidase-5-kb RNA hybrid sequence, the resulting spliced RNA is predicted to yield a 509-bp RT-PCR product. The predicted products were observed with β-galactosidase-specific primers in RT-PCRs on total RNA (Fig. 7C). No RT-PCR products were obtained from control samples or in the absence of reverse transcriptase (data not shown).
Construction and analysis of a splice donor site mutant virus.
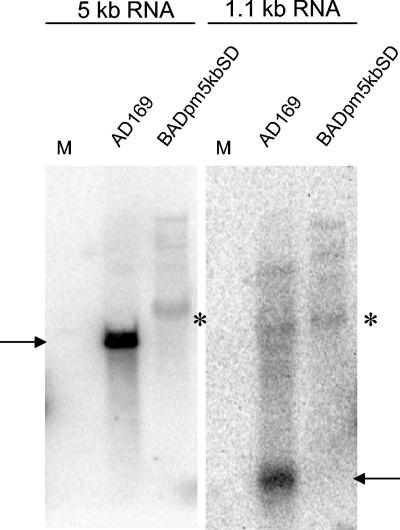
To confirm that the 5-kb RNA of HCMV is derived from a splicing event, we mutated the consensus splice donor site sequence at the 5′ end of the transcript in the context of the viral genome. We mutated 4 bp (in italics) in the splice donor sequence containing the 5′ end of the 5-kb RNA from CAGGTAGGT to CCTCGAGGT. The mutation was introduced into the viral genome by allelic exchange as described in Materials and Methods, generating the BAC clone BADpm5kbSD. Virus derived from BADpm5kbSD grew in fibroblasts without the need for complementation (data not shown). To determine if the splice donor mutant virus expressed the 5-kb transcript, Northern blot analysis was performed on total RNA from fibroblasts infected with wild-type AD169 or BADpm5kbSD prepared at 48 h postinfection. As shown in Fig. 8, while the 5-kb RNA was detected in AD169-infected cells, it was not detected in total RNA prepared from cells infected with BADpm5kbSD. Mutation of 4 bp within the splice donor site is sufficient to eliminate expression of the 5-kb RNA. Furthermore, Northern blot analysis of the same RNA samples with a riboprobe specific for the 1.1-kb late RNA detects this spliced RNA in AD169-infected cells but does not detect it in cells infected with BADpm5kbSD (Fig. 8). Again, this is consistent with a failure to splice the precursor RNA, abolishing expression of both the 5- and 1.1-kb RNAs. Interestingly, we do detect a larger RNA with both 5- and 1.1-kb RNA-specific probes in cells infected with BADpm5kbSD. The migration of this weakly hybridizing RNA suggests it could be the unspliced precursor RNA.
FIG. 8.
Northern blot analysis of 5-kb RNA expression from the recombinant splice donor mutant virus BADpm5kbSD. Total RNA was prepared from mock- (M), AD169-, or BADpm5kbSD-infected fibroblasts at 48 h postinfection and hybridized with a 5-kb-RNA-specific or 1.1-kb-RNA-specific riboprobe as indicated. The arrow indicates the position of the 5-kb or 1.1-kb RNA in each respective blot. The asterisks indicate the weakly hybridizing RNA that is likely to be the unspliced precursor RNA expressed in cells infected with the splice donor mutant virus.
DISCUSSION
We show here that the 5-kb RNA is a stable intron encoded by HCMV. The intronic origin of this transcript is consistent with its lack of polyadenylation and nuclear localization (Fig. 2), properties of the RNA first explored by Plachter et al. (30). The 5′ end of the RNA is located at the splice donor site of the 1.1-kb late mRNA (Fig. 4), and our 3′ end mapping places that end in the vicinity of the splice acceptor site of the 1.1-kb mRNA (Fig. 6). We also show that the 5-kb RNA can be spliced from a heterologous primary transcript (Fig. 7), and with the construction of a viral recombinant containing a mutation in the 5′ splice donor site (Fig. 8), we have confirmed that the transcript is generated from a splicing event, rather than from de novo initiation of transcription at the 5′ end of the RNA.
What is the function of the 5-kb RNA? We compared DNA sequences from the 5-kb RNA loci of laboratory-adapted and clinical strains of HCMV and determined that the overall sequence of the region is well conserved among strains. However, the specific ORFs annotated by Chee et al. (8) are not conserved (data not shown). These observations are consistent with published reports that indicate that this region lacks protein-coding ORFs (11, 25, 26). However, we have detected the expression of a 5-kb RNA by Northern blot analysis of total RNA prepared from fibroblasts infected with at least two clinical isolates of HCMV (data not shown), suggesting that the expression of the 5-kb RNA is not limited to laboratory-adapted strains of HCMV. Comparison of the 5-kb RNA locus of HCMV with the analogous region of CMVs that infect other species indicates that a noncoding transcript is likely to exist in many and perhaps all other CMVs (data not shown). In HCMV, the 5-kb RNA locus is flanked on either side by conserved genes that have been shown to play a role in regulating DNA replication: UL112/113 and UL105 (27). As noted by us and others, these conserved genes flank substantial gaps of sequence lacking predicted ORFs with strong potential to encode proteins in chimpanzee, rhesus, and mouse CMVs (Fig. 9) (16). This genomic organization extends to other betaherpesvirus family members: HHV6A, HHV6B, and HHV7. We have confirmed the expression of a large transcript in this region of the rhesus and mouse CMVs (C. A. Kulesza and T. Shenk, unpublished data).
FIG. 9.
Diagram of the genomic region bracketed by UL105 and UL112/113 in HCMV, chimpanzee CMV (ChCMV), rhesus CMV (RhCMV), and murine CMV (MCMV).
Given the conservation of this large noncoding RNA among clinical isolates of HCMV and in CMVs that infect other species, we anticipate that the 5-kb RNA plays an important role in viral replication or spread in the host, even though it is not required for efficient replication in cultured fibroblasts. The RNA might serve a critical function in other cell types such as endothelial or smooth muscle cells. Alternatively, it could serve a function, such as immune evasion, that is important for infection of the host organism but not in cultured cells.
Non-protein-coding RNAs, including stable introns, have been described for a number of biological systems, including bacteria, plants, mammals, and viruses (17, 35). These include rRNAs, tRNAs, and small nuclear RNAs (snRNAs), as well as catalytic RNAs such as ribozymes and gene- and protein-expression-regulating small interfering RNAs and microRNAs. These noncoding RNAs play diverse roles in cellular processes ranging from transcriptional activation, gene silencing, and RNA processing to protein translation and translocation. Other eukaryotic noncoding transcripts have been shown to function in genomic imprinting or regulation of gene expression in developmental events. Interestingly, a subset of the snRNAs, the small nucleolar RNAs, are almost entirely derived from introns (17). It is conceivable that the 5-kb RNA functions similarly to one of these cellular noncoding RNAs.
Several viruses have evolved RNA-based immune-evasion mechanisms by targeting PKR and the interferon response pathway. The adenovirus VA RNAs and Epstein-Barr virus EBER RNAs are small noncoding RNAs transcribed by RNA polymerase III that inhibit activation of PKR, thereby preventing activation of the cellular antiviral response (7, 10). It is possible that the 5-kb transcript performs a similar function.
In comparison to most other cellular and viral RNA polymerase II-transcribed, noncoding RNAs, the 5-kb transcript is unusual in its large size and intronic origin. Examples of stable introns in other biological systems are not numerous, but one of the most well-studied stable introns is the LAT of HSV-1. LAT is the only viral transcript abundantly expressed in latently infected neurons and is a stable intron derived from the processing of an 8.3-kb precursor RNA (15). LAT is transcribed from the repeat regions of HSV-1 and is, therefore, diploid. In contrast, the 5-kb RNA is derived from the unique long region of HCMV and exists in only a single copy. LAT exists as a 2.0-kb major form expressed in both lytic and latent infections and a less-abundant 1.5-kb form, observed only in latent infections (38). Expression of 5-kb RNA is abundant during immediate-early and late phases of HCMV replication, and it will be interesting to determine the expression pattern of the transcript in latently infected cells. Although a function for LAT in latency has not been conclusively established, LAT has been implicated in the efficiency of establishment of latent infection and reactivation to productive infection in model systems (4, 5, 13, 14, 19, 22). It has been proposed that LAT promotes latency by protecting neuronal cells from apoptosis (1, 29, 37). It is conceivable that the 5-kb RNA plays a role in HCMV latency. LAT is a branched lariat, and the use of a nonconsensus branch point during the splicing process is thought to contribute to the stability of the LAT intron (21, 39, 42). We have not yet analyzed the structure of the 5-kb RNA to determine if it too is a branched lariat, nor have we determined the location of a putative branch point. However, in high-resolution Northern blot assays, we have observed two closely migrating forms of the 5-kb RNA (data not shown). The two forms might be branched and linear forms of the 5-kb transcript, but we have not yet experimentally determined the basis for the difference in migration. It is possible that a branched lariat structure combined with secondary and tertiary structure of the transcript confer considerable stability to the 5-kb RNA.
Our data lead to the conclusion that the 5-kb RNA, which is detected during the immediate-early and late phases of infection, is derived from splicing of a primary transcript that produces the 1.1-kb spliced RNA. This small, spliced transcript is detected only during the late phase of infection. If the two RNAs are produced from the same primary transcript, why is the 5-kb RNA, but not the 1.1-kb RNA, detected during the immediate-early phase of infection? A portion of the 5-kb RNA detected during the immediate-early phase of infection is likely delivered to cells by infecting virions, because the transcript is packaged in virions (6). Nevertheless, we have been able to detect the spliced exons of the 1.1-kb RNA by RT-PCR amplification of immediate-early RNA (data not shown), indicating that it is indeed present in small quantities during the immediate-early phase of the viral replication cycle. It appears likely that the 5-kb intron is more stable than the 1.1-kb RNA, and as a consequence it accumulates to a greater extent than the 1.1-kb RNA. The consequences of this differential stability between RNAs and phase of viral infection are unknown. The 1.1-kb RNA is predicted to encode a protein of 31 amino acids (31). It bears no similarity to proteins in the current databases, and the protein has not yet been identified in HCMV-infected cells, although it certainly has the potential to be translated. It is conceivable that the production of the 1.1-kb spliced RNA serves only to generate the stable 5-kb RNA; i.e., the 1.1-kb RNA itself might have no function.
Acknowledgments
This work was supported by Public Health Service grant CA085786 from the National Cancer Institute (T.S.) and National Research Service Award F32AI054034-01 from the National Institute of Allergy and Infectious Diseases (C.A.K.).
We thank members of the Shenk laboratory for critical reading of the manuscript.
REFERENCES
- 1.Ahmed, M., M. Lock, C. G. Miller, and N. W. Fraser. 2002. Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neuronal cells in vivo. J. Virol. 76:717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. L. M. 1999. Nucleic acid hybridization. Bios Scientific Publishers, Springer, Oxford, United Kingdom.
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2004. Current protocols in molecular biology, p. 4.0.1-4.10.11. John Wiley and Sons, Inc., New York, N.Y.
- 4.Block, T. M., S. Deshmane, J. Masonis, J. Maggioncalda, T. Valyi-Nagi, and N. W. Fraser. 1993. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology 192:618-630. [DOI] [PubMed] [Google Scholar]
- 5.Bloom, D. C., G. B. Devi-Rao, J. M. Hill, J. G. Stevens, and E. K. Wagner. 1994. Molecular analysis of herpes simplex virus type 1 during epinephrine-induced reactivation of latently infected rabbits in vivo. J. Virol. 68:1283-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnahan, W. A., and T. E. Shenk. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 97:14506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgert, H. G., Z. Ruzsics, S. Obermeier, A. Hilgendorf, M. Windheim, and A. Elsing. 2002. Subversion of host defense mechanisms by adenoviruses. Curr. Top. Microbiol. Immunol. 269:273-318. [DOI] [PubMed] [Google Scholar]
- 8.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 9.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemens, M. J., K. G. Laing, I. W. Jeffrey, A. Schofield, T. V. Sharp, A. Elia, V. Matys, M. C. James, and V. J. Tilleray. 1994. Regulation of the interferon-inducible eIF-2 alpha protein kinase by small RNAs. Biochimie 76:770-778. [DOI] [PubMed] [Google Scholar]
- 11.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 12.DeMarchi, J. M. 1983. Post-transcriptional control of human cytomegalovirus gene expression. Virology 124:390-402. [DOI] [PubMed] [Google Scholar]
- 13.Devi-Rao, G. B., J. S. Aguilar, M. K. Rice, H. H. Garza, Jr., D. C. Bloom, J. M. Hill, and E. K. Wagner. 1997. Herpes simplex virus genome replication and transcription during induced reactivation in the rabbit eye. J. Virol. 71:7039-7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devi-Rao, G. B., D. C. Bloom, J. G. Stevens, and E. K. Wagner. 1994. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. J. Virol. 68:1271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobson, A. T., F. Sederati, G. Devi-Rao, W. M. Flanagan, M. J. Farrell, J. G. Stevens, E. K. Wagner, and L. T. Feldman. 1989. Identification of the latency-associated transcript promoter by expression of rabbit beta-globin mRNA in mouse sensory nerve ganglia latently infected with a recombinant herpes simplex virus. J. Virol. 63:3844-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolan, A., C. Cunningham, R. D. Hector, A. F. Hassan-Walker, L. Lee, C. Addison, D. J. Dargan, D. J. McGeoch, D. Gatherer, V. C. Emery, P. D. Griffiths, C. Sinzger, B. P. McSharry, G. W. Wilkinson, and A. J. Davison. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301-1312. [DOI] [PubMed] [Google Scholar]
- 17.Eddy, S. R. 1999. Noncoding RNA genes. Curr. Opin. Genet. Dev. 9:695-699. [DOI] [PubMed] [Google Scholar]
- 18.Farrell, M. J., A. T. Dobson, and L. T. Feldman. 1991. Herpes simplex virus latency-associated transcript is a stable intron. Proc. Natl. Acad. Sci. USA 88:790-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill, J. M., F. Sedarati, R. T. Javier, E. K. Wagner, and J. G. Stevens. 1990. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology 174:117-125. [DOI] [PubMed] [Google Scholar]
- 20.Jahn, G., E. Knust, H. Schmolla, T. Sarre, J. A. Nelson, J. K. McDougall, and B. Fleckenstein. 1984. Predominant immediate-early transcripts of human cytomegalovirus AD 169. J. Virol. 49:363-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krummenacher, C., J. M. Zabolotny, and N. W. Fraser. 1997. Selection of a nonconsensus branch point is influenced by an RNA stem-loop structure and is important to confer stability to the herpes simplex virus 2-kilobase latency-associated transcript. J. Virol. 71:5849-5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leib, D. A., K. C. Nadeau, S. A. Rundle, and P. A. Schaffer. 1991. The promoter of the latency-associated transcripts of herpes simplex virus type 1 contains a functional cAMP-response element: role of the latency-associated transcripts and cAMP in reactivation of viral latency. Proc. Natl. Acad. Sci. USA 88:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendelson, M., S. Monard, P. Sissons, and J. Sinclair. 1996. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J. Gen. Virol. 77:3099-3102. [DOI] [PubMed] [Google Scholar]
- 24.Mocarski, E. S. 1996. Cytomegaloviruses and their replication, p. 2447-2492. In B. N. Fields, D. M. Knipe, P. M. Howley et al. (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 25.Murphy, E., I. Rigoutsos, T. Shibuya, and T. E. Shenk. 2003. Reevaluation of human cytomegalovirus coding potential. Proc. Natl. Acad. Sci. USA 100:13585-13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2728. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott, Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 29.Perng, G. C., C. Jones, J. Ciacci-Zanella, M. Stone, G. Henderson, A. Yukht, S. M. Slanina, F. M. Hofman, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2000. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 287:1500-1503. [DOI] [PubMed] [Google Scholar]
- 30.Plachter, B., B. Traupe, J. Albrecht, and G. Jahn. 1988. Abundant 5 kb RNA of human cytomegalovirus without a major translational reading frame. J. Gen. Virol. 69:2251-2266. [DOI] [PubMed] [Google Scholar]
- 31.Rawlinson, W. D., and B. G. Barrell. 1993. Spliced transcripts of human cytomegalovirus. J. Virol. 67:5502-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romanowski, M., and T. Shenk. Unpublished results.
- 33.Rowe, W. P., J. W. Hartley, S. Waterman, H. C. Turner, and R. J. Huebner. 1956. Cytopathogenic agent resembling human salivary gland virus recovered from tissue cultures of human adenoids. Proc. Soc. Exp. Biol. Med. 92:418-424. [PubMed] [Google Scholar]
- 34.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storz, G. 2002. An expanding universe of noncoding RNAs. Science 296:1260-1263. [DOI] [PubMed] [Google Scholar]
- 36.Taylor-Wiedeman, J., J. G. Sissons, L. K. Borysiewicz, and J. H. Sinclair. 1991. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J. Gen. Virol. 72:2059-2064. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, R. L., and N. M. Sawtell. 2001. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J. Virol. 75:6660-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagner, E. K., W. M. Flanagan, G. Devi-Rao, Y. F. Zhang, J. M. Hill, K. P. Anderson, and J. G. Stevens. 1988. The herpes simplex virus latency-associated transcript is spliced during the latent phase of infection. J. Virol. 62:4577-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, T. T., Y. H. Su, T. M. Block, and J. M. Taylor. 1998. Atypical splicing of the latency-associated transcripts of herpes simplex type 1. Virology 243:140-149. [DOI] [PubMed] [Google Scholar]
- 40.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zabolotny, J. M., C. Krummenacher, and N. W. Fraser. 1997. The herpes simplex virus type 1 2.0-kilobase latency-associated transcript is a stable intron which branches at a guanosine. J. Virol. 71:4199-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zylber, E. A., and S. Penman. 1970. The effect of high ionic strength on monomers, polyribosomes, and puromycin-treated polyribosomes. Biochim. Biophys. Acta 204:221-229. [DOI] [PubMed] [Google Scholar]