Abstract
Mouse adenovirus type 1 (MAV-1) early region 1A (E1A) encodes a virulence gene in viral infection of mice. To broaden our understanding of the functions of E1A in MAV-1 pathogenesis, an unbiased experimental approach, glutathione S-transferase (GST) pulldown, was used to screen for cellular proteins that interact with E1A protein. We identified mouse Sur2, a subunit of Mediator complex, as a protein that binds to MAV-1 E1A. The interaction between Sur2 and MAV-1 E1A was confirmed in virus-infected cells. Conserved region 3 (CR3) of MAV-1 E1A was mapped as the region required for Sur2-E1A interaction, as is the case for human adenovirus E1A. Although it has been proposed that human adenovirus E1A recruits the Mediator complex to transactivate transcription of viral early genes, Sur2 function in adenovirus replication has not been directly tested previously. Studies on the functions of Sur2 with mouse embryonic fibroblasts (MEFs) showed that there was a multiplicity-dependent growth defect of MAV-1 in Sur2−/− MEFs compared to Sur2+/+ MEFs. Comparison of the viral DNA and viral mRNA levels in Sur2+/+ and Sur2−/− MEFs confirmed that Sur2 was important for efficient viral replication. The viral replication defects in Sur2−/− MEFs appeared to be due at least in part to a defect in viral early gene transcription.
Human adenovirus early region 1A (E1A) encodes two closely related proteins, 289R and 243R. The human adenovirus E1A 289R protein is one of the most extensively studied viral transcriptional regulators. E1A activates transcription of other viral early genes (9, 33) and is important for modulating the cell cycle to facilitate viral replication (27, 51, 54, 60). The E1A 289R and 243R proteins do this by various strategies involving both direct and indirect binding to cellular proteins. The identification of a set of cellular proteins that can be coimmunoprecipitated with human adenovirus E1A proteins, such as retinoblastoma protein (pRb), the Rb family proteins p107 and p130, and p300/CBP, was critical to understanding how E1A proteins manipulate cell cycle regulation (27, 60). CtBP is a transcriptional corepressor interacting with the conserved PXDLS motif near the C terminus of human adenovirus E1A proteins (12, 48). Additional newly identified cellular interacting proteins, such as Sur2 (13), p400 (25), and TRRAP/GCN5 (21, 38), continue to deepen our appreciation of the complexity of the molecular basis of virus-host interactions.
Mouse adenovirus type 1 (MAV-1) specifically targets endothelial cells and cells of the monocyte-macrophage lineage (34, 52), whereas epithelial cells are the main cell type for human adenovirus infection (31). Unlike human adenovirus, MAV-1 only produces a single 200-amino-acid E1A protein (4, 53). Although MAV-1 E1A is not an essential gene for viral replication in cell culture (61), it is a virulence gene in both outbred and inbred mice (52, 55), emphasizing the importance of MAV-1 E1A for viral pathogenesis in vivo. Since human adenovirus E1A itself does not bind to DNA (51), direct and/or indirect association with cellular proteins is essential for E1A to carry out its multiple functions during viral infection.
The function of MAV-1 E1A in viral pathogenesis is undoubtedly related to binding to cellular proteins. MAV-1 E1A contains all three conserved regions (CR1, CR2, and CR3) (4, 6) found in the human adenovirus 289R E1A protein (42). However, the C terminus of MAV-1 E1A, lacking the conserved motif PXDLS, is completely different from that of human adenovirus E1A (3), and thus it is unlikely to bind to a mouse homologue of CtBP. Like human adenovirus E1A, MAV-1 E1A interacts with pRb and p107, mainly through the CR1 domain (53). MAV-1 E1A is also similar to human adenovirus E1A in that it plays a major role in antagonizing the antiviral effects of interferons (35). However, all three conserved regions (CR1, CR2, and CR3) are required for MAV-1 resistance to the interferon responses, whereas only CR1 of human adenovirus E1A is needed for such resistance (1).
Because of these differences between MAV-1 and human adenovirus E1A and to broaden our understanding of the roles of E1A in viral pathogenesis, we looked for cellular proteins that interact with MAV-1 E1A. We used an unbiased experimental approach, glutathione S-transferase (GST) pulldown coupled with mass spectrometry analysis. In the work presented here, we identified mouse Sur2 as a protein that interacts with MAV-1 E1A.
Human Sur2, a subunit of Mediator, binds to CR3 of human adenovirus E1A (13). Mediator complexes function as molecular bridges to link the transcriptional regulators with RNA polymerase II to regulate transcription (11). The interaction between Sur2 and human adenovirus E1A-CR3 is required for binding Mediator and for transcriptional activation by human adenovirus E1A (13, 56), suggesting that recruitment of the Mediator complex transactivates the transcription of other viral early genes. Even though human Sur2 interacts with human adenovirus E1A in virus-infected cells (57), the importance of Sur2 in adenovirus replication has not been directly tested because Sur2−/− human cells have not been available. In the work reported here, we demonstrated that mouse Sur2 is a critical factor for MAV-1 replication in mouse embryonic fibroblasts (MEFs). Our findings indicate that Sur2 is important for efficient viral replication, providing a new molecular basis to investigate viral pathogenesis in mice.
MATERIALS AND METHODS
Cells and viruses.
Mouse NIH 3T6 fibroblast cells were maintained in Dulbecco's modified Eagle's medium supplemented with 5% heat-inactivated calf serum. MAV-1 E1A-expressing 37.1 cells (53) were maintained in Dulbecco's modified Eagle's medium containing 5% heat-inactivated calf serum and 200 μg of G418 per ml. Mouse NIH 3T3 fibroblast cells were maintained in Dulbecco's modified Eagle's medium with 10% heat-inactivated calf serum. Mouse brain microvascular endothelial cells (MBMECs) were maintained in complete MBMEC medium, purchased from Cell Applications, Inc.
Sur2−/− and Sur2+/+ MEFs were obtained as follows. Sur2 knockout mice were generated by injecting Sur2+/− embryonic stem (ES) cell clones (56) into C57BL/6 blastocysts. Sur2 heterozygous mice (129SvEV × C57BL/6) were intercrossed to generate Sur2−/− embryos. Generation and characterization of Sur2 knockout mice will be described in greater detail elsewhere (J. Stevens and A. Berk, unpublished data). Sur2+/+ and Sur2−/− MEFs were generated from individual embryonic day 9.5 (E9.5) littermate embryos. Embryos were dissociated by trypsin digestion and plated in single wells of a 24-well plate containing ES growth medium (15% fetal bovine serum [HyClone], 2 mM l-glutamine [Invitrogen], 50 μg of penicillin-streptomycin [Invitrogen], 0.1 mM nonessential amino acids [Cellgro], 10−4 M β-mercaptoethanol, 1,000 U of leukemia inhibitory factor [Chemicon] per ml). MEFs were passaged by a 3T3-like protocol until the cells escaped replicative senescence. They were then maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum.
Wild-type MAV-1 was the standard MAV-1 stock originally obtained from S. Larson (5). pmE109 is an MAV-1 E1A null mutant virus; the dlE105, dlE102, and dlE106 viruses are MAV-1 E1A CR1, CR2, and CR3 deletion mutants, respectively (53). Mouse gammaherpesvirus 68 was obtained from Jason Weinberg, University of Michigan.
Plasmid constructions.
A plasmid, pAS2E1A, containing the full-length MAV-1 E1A cDNA fragment from plasmid Z112.F (4) was generated through several intermediate vectors with an adaptor (5′-GACATGCTCATGAGCATGTCGC-3′). pAS2E1A has the full-length E1A cDNA fragment plus an additional 66 nucleotides (5′-GAATTCATGG CTTACCCATA CGATGTTCCA GATTACGCTA GCTTGGGTGG TCATATGGCC ATGAGC-3′) at the 5′ end of the E1A cDNA sequence. The E1A cDNA and linker were gel purified from EcoRI-digested pAS2E1A and ligated to EcoRI-digested vector pGEX-4T-1 (Amersham Biosciences) to generate plasmid pGST-mE1A. The correct open reading frame of pGST-mE1A was confirmed by DNA sequencing.
Plasmids pGST-wtE1A, pGST-Nter1, pGST-Nter2, pGST-Cter1, pGST-Cter2, and pGST-Cter3 were made by PCR with primers containing a 5′ EcoRI site and a 3′ SalI site to amplify MAV-1 E1A fragments encompassing full-length protein or residues 1 to 45, 1 to 113, 157 to 200, 125 to 200, and 90 to 200, respectively, from plasmid pCME1A (53) as a template (Fig. 1). The same primers as used in pGST-wtE1A cloning were used to construct pGST-CR1Δ (deletion of amino acids 36 to 77 of E1A), pGST-CR2Δ (deletion of amino acids 112 to 128) and pGST-CR3Δ (deletion of amino acids 136 to 153) by PCR with pCMV-CR1Δ, pCMV-CR2Δ, and pCMV-CR3Δ as templates, respectively (53) (Fig. 1). The reaction mix contained the 5′ primer (1 ng/μl), 3′ primer (1 ng/μl), plasmid template (1 ng/μl), 1.25 units of Pfu polymerase (Stratagene), 1× Pfu reaction buffer, and 200 nM each dATP, dCTP, dGTP, and dTTP in 100 μl. After 30 cycles (melting at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 45 s), the PCRs were subjected to an additional 7-min incubation at 72°C (GeneAmplifer 9600, Applied Biosystems). Each PCR yielded a single product that was subsequently purified with a PCR purification kit (Qiagen), digested with EcoRI and SalI, gel purified, and ligated to pGEX-4T-1 that had been digested with EcoRI and SalI. pGST-E3gp11Κ contains the full-length MAV-1 E3 gp11K gene (8).
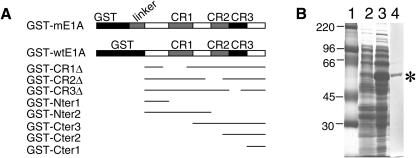
FIG. 1.
Constructs and expression of GST fusion proteins. (A) Schematic diagram of GST fusion protein constructs. GST-mE1A contains the full-length MAV-1 E1A as well as an additional 22-amino-acid linker between GST and the E1A protein. Full-length MAV-1 E1A protein (amino acids 1 to 200) without any linker is shown as GST-wtE1A. The GST fusion proteins had deletions of amino acids 35 to 78 (GST-CR1Δ), amino acids 111 to 129 (GST-CR2Δ), amino acids 135 to 154 (GST-CR3Δ), amino acids 1 to 45 (GST-Nter1), amino acids 1 to 113 (GST-Nter2), amino acids 90 to 200 (GST-Cter3), amino acids 125 to 200 (GST-Cter2), or amino acids 157 to 200 (GST-Cter1). For the deletion constructs, thin lines indicate the portion of the protein contained in the constructs. (B) Expression and purification of GST-mE1A fusion proteins from E. coli. Samples were electrophoresed on an SDS-10% polyacrylamide gel and stained with Coomassie blue. Lane 1, molecular size standards (Rainbow marker from Amersham Biotech); lane 2, crude bacterial extract, without IPTG induction; lane 3, crude bacterial extract, with IPTG; lane 4, purified GST-mE1A fusion protein after elution from glutathione-Sepharose 4B beads.
Purification of GST fusion proteins.
Plasmid pGST-mE1A was transformed into Escherichia coli BL21+ cells (Stratagene). Vector plasmid pGEX-4T-1 was used as a control. A single colony was inoculated into 5 ml of 2-YT-G medium (1.6% tryptone, 1% yeast extract, 0.5% NaCl, and 2% glucose) with 100 μg of ampicillin per ml and cultured overnight at 37°C. Overnight-cultured E. coli cells were diluted 1:100 into fresh medium. GST-mE1A fusion protein expression was induced by 1 mM isopropylthiogalactopyranoside (IPTG) when the optical density at 600 nm reached 0.5, and incubation continued for an additional 4 to 6 h with vigorous agitation at 37°C. Cells were harvested by centrifuging. Pellets were stored at −70°C until use.
Pellets were thawed on ice and resuspended (for 200 ml of original liquid culture) in 10 ml of ice-cold STE buffer (10 mM Tris-HCl, 1 mM EDTA, 150 mM NaCl) with 100 μl of lysozyme solution (30 mg/ml), 1 μl of 100 mM phenylmethylsulfonyl fluoride, and 2 μl of protease inhibitor cocktail (Sigma). Samples were incubated on ice for 15 min before addition of 100 μl of 1 M dithiothreitol and 1.4 ml of 10% Sarkosyl. Samples were sonicated for a total time of 30 s and then centrifuged at 15,000 × g for 30 min to pellet debris. Supernatants were transferred to a fresh 50-ml tube, and 4 ml of 10% Triton X-100 was added. The samples were diluted with STE buffer to a final 20-ml volume and incubated at room temperature for 30 min. The GST fusion proteins were mixed gently overnight at 4°C with a 1-ml bed of prepared glutathione-Sepharose 4B (Pharmacia Biotech) in phosphate-buffered saline. The beads were washed four times with 25 ml of ice-cold phosphate-buffered saline. The GST fusion proteins were eluted with three successive 1-ml volumes of elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM reduced glutathione, 0.1% Sarkosyl). The eluates were pooled, and the reduced glutathione and Sarkosyl were removed by overnight dialysis against phosphate-buffered saline (pH 7.4). The purified GST fusion proteins were stored at −70°C until use.
Large-scale GST pulldown assays.
Equal amounts of purified GST fusion proteins or GST proteins were mixed with 100 μl of glutathione beads and incubated for 2 h at 4°C. Nuclear extraction of 109 3T6 cells or 5 × 108 MBMECs was performed according to the method of Dignam et al. (22); 15 ml of the 3T6 or MBMEC nuclear extracts was preabsorbed against 266 μl of glutathione beads and then with 500 μl of GST-glutathione beads (loaded with 250 μg of GST) at 4°C for 4 h in GST binding buffer (125 mM NaCl, 50 mM Tris-HCl [pH 7.4], 0.1% NP-40). Equal aliquots of the preabsorbed nuclear extracts were then added to glutathione beads bound to 25 μg of GST or GST-mE1A and rocked overnight at 4°C. The beads were washed twice with GST binding buffer and twice with GST wash buffer (250 mM NaCl, 50 mM Tris-HCl [pH 7.4], 0.1% NP-40). The beads were washed once again with GST binding buffer just before they were eluted with 30 μl of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (37) and boiled for 15 min. The bound proteins were separated by SDS-10% PAGE and either Coomassie stained for mass spectrometry analysis or transferred to polyvinylidene difluoride membranes for immunoblotting.
Mass spectrometry analysis.
Bands of interest were cut out of the Coomassie-stained protein gels, digested in-gel with trypsin, and used for tandem mass spectrometry-mass spectrometry analysis with a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) machine (Micromass Company) by the Michigan Proteomic Consortium at the University of Michigan. Two search engines, MS-FIT (http://prospector.ucsf.edu) and Mascot (http://www.matrixscience.com), were used to search databases to identify the proteins.
Antibodies.
Monoclonal antibody against human Sur2 (BD Pharmingen) cross-reacts with mouse Sur2 and was used at a 1:1,000 dilution in Western blot assays. Anti-p107 (sc-318) and anti-p130 (sc-317) polyclonal antibodies were from Santa Cruz Biotechnology. Anti-murine Rb and anti-MAV-1 E1A (AKO7-147) rabbit polyclonal antibodies were described (53). AKO7-147 and normal rabbit serum were purified by DEAE Affi-Gel blue (Bio-Rad) affinity chromatography according to the manufacturer's instructions. Normal rabbit serum was used as a control where indicated. Mouse monoclonal antibodies against MAV-1 E1A (monoclonal antibody10B10) and MAV-1 E3gp11K (monoclonal antibody 11H9) were generated at the University of Georgia Monoclonal Facility with purified GST-mE1A and GST-gp11K fusion proteins as the antigen, respectively. Both monoclonal antibody 10B10 and monoclonal antibody 11H9 were purified with an Affi-Gel protein A MAPS II kit (Bio-Rad) according to the manufacturer's instructions; a 1:500 dilution of monoclonal antibody 10B10 or 1:1,000 dilution of monoclonal antibody 11H9 was used in Western blots. Monoclonal antibody against β-actin was from Sigma (A5441) and used at a 1:5,000 dilution in Western blots.
Western blot assays.
Samples were dissolved in SDS-PAGE sample buffer and resolved on SDS-polyacrylamide gels. The proteins were electrotransferred onto polyvinylidene difluoride membranes at 14 V for 16 h at 4°C, and the membranes were blocked by incubation at room temperature for 1 h in TBS-Tween (150 mM NaCl, 10 mM Tris [pH 7.4], 0.1% Tween 20) containing 5% nonfat dry milk. Anti-rabbit (1:12,000 from Amersham Biosciences) or anti-mouse (1:12,000 from Amersham Biosciences) immunoglobulin G-horseradish peroxidase-conjugated antibody was used as the secondary antibody. Immunoblots were developed with SuperSignal West Pico chemiluminescent substrate (Pierce Biotechnology, Inc.).
Immunoprecipitations.
3T6 cells or MBMECs were lysed in E1A lysis buffer (250 mM NaCl, 50 mM Tris [pH 7.4], 0.1% NP-40) containing protease inhibitor cocktail (1:50) (Sigma). The lysates were diluted in E1A binding buffer (125 mM NaCl, 50 mM Tris [pH 7.4], 0.1% NP-40) and then preabsorbed simultaneously against rabbit normal immunoglobulin G (IgG) and protein A-agarose beads (Pharmacia Biotech) by rocking at 4°C for 2 h. The lysates were then incubated with polyclonal antibody against MAV-1 E1A (AKO7-147) (53) or rabbit normal IgG, followed by incubation with 30 μl of protein A-agarose (Oncogene Research Products). The agarose beads were pelleted, washed twice with E1A binding buffer and twice with E1A lysis buffer, and once again with E1A binding buffer. The immunoprecipitated proteins were eluted by boiling in SDS-PAGE sample buffer and subjected to SDS-PAGE and immunoblotting.
Viral growth curves.
Sur2+/+ and Sur2−/− MEFs were infected with wild-type MAV-1 at a multiplicity of infection (MOI) of 0.05, 0.1, 1, or 5 or with mouse gammaherpesvirus 68 at an MOI of 0.01. MAV-1 plaque assays were carried out on 3T6 cells as described previously (18). Briefly, cells were harvested at various times post infection by scraping in their medium. The cell suspensions were subjected to three cycles of freezing and thawing, and the cell debris was spun out of the supernatant. Tenfold serial dilutions of supernatants were plated in triplicate on 3T6 cells, and plaques were counted at day 9 after plating. For the mouse gammaherpesvirus 68 plaque assay, only supernatants were harvested, and 10-fold serial dilutions of supernatants were plated in duplicate on NIH 3T3 cells as described (47).
Southern blots.
Sur2+/+ and Sur2−/− MEFs were infected at an MOI of 0.05, 0.1, 1, or 5 and harvested at various times post infection by scraping the cells off the plates. Viral DNA was isolated by the method of Hirt (28). Equal amounts of DNA samples were digested with HindIII and RNase A and electrophoresed on a 0.7% agarose gel. After staining with ethidium bromide and photography, the gel was soaked in 0.8 M NaCl-0.4 M NaOH for 30 min, rinsed with water, and then soaked in 1.5 M NaCl-0.5 M Tris (pH 7.5) for 30 min. The DNA was transferred to a positively charged nylon membrane (Boehringer) by capillary transfer. The DNA was UV cross-linked to the membrane by incubation for 12 s in a Fisher UV cross-linker.
The membrane was prehybridized in 10 ml of PerfectHyb solution (Sigma) at 65°C for 2 h. Plasmids pMBA, pMBB, and pMBC contain MAV-1 genome sequence from 31.7 to 64.2 map units, 64.2 to 94.5 map units, and 6.5 to 31.7 map units, respectively. They were pooled, digested with HinfI, and labeled with Klenow DNA polymerase with [γ-32P]ATP. The free label was removed by passage over a G25 Sepharose spin column, and 106 to 107 cpm of the probe was added to 10 ml of PerfectHyb solution to hybridize at 65°C for 5 to 18 h. The membranes were washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at room temperature for 15 min, twice with 2× SSC-0.1% SDS at 65°C for 30 min, and once with 0.5x SSC-0.1% SDS at 65°C for 15 min. A phosphorimager was used to quantify the signals.
RNase protection assays.
Sur2+/+ and Sur2−/− MEFs were infected at an MOI of 0.05, 0.1, 1, or 5. Total RNA was extracted with TRI Reagent (Molecular Research Center, Inc.) following the manufacturer's instructions. An equimolar pool of linearized plasmids were used as templates to make an [α-32P]UTP-labeled multiplex RNase protection assay probe set by T7 or T3 polymerase transcription. prK+7, a genomic E1A plasmid containing MAV-1 genome sequence nucleotides 1 to 820 in a pBluescript(+) vector, was digested with BamHI at nucleotide 360 in the MAV-1 sequence (GenBank accession no. NC_000942). The full-length probe was 489 nucleotides (MAV-1 nucleotides 360 to 820 plus 29 nucleotides of vector), and the length protected from RNase digestion after hybridization to an E1A mRNA was 392 nucleotides.
A genomic hexon plasmid, pHEX, was constructed by ligation of MAV-1 genome sequence nucleotides 16432 to 16769 with vector pBS2SK−. pHEX was digested at nucleotide 16432 in the viral sequence at a BamHI site. The full-length probe was 395 nucleotides (MAV-1 nucleotides 16432 to 16769 plus 58 nucleotides of vector) and the protected size was 337 nucleotides. The protected length of L32 was 80 nucleotides and it was used as an internal loading control (29). pZU14 is an E3gp11K cDNA clone used to make an E3 probe (7). The full-length probe is 714 nucleotides, and the protected E3 signal is 645 nucleotides. pH61 (MAV-1 E2A clone) (61) was digested with SmaI, and the full-length probe is 470 nucleotides. The protected E2A signal is about 440 nucleotides. pZ571 is an E4 cDNA clone (36) and was digested with BamHI. The full-length probe is 767 nucleotides, and the protected E4 signal is about 700 nucleotides. The linearized plasmid mouse β-actin was from the Maxiscript in vitro transcription kit (Ambion Inc.). The full-length probe of β-actin is 276 nucleotides with T3 transcriptase, and the protected length is 245 nucleotides.
The RNase protection assays were carried out as described by Hobbs et al. (29). Briefly, 7.5 μg of total RNA was hybridized with the combined MAV-1 probe set and either the L32 or β-actin probe overnight at 56°C. Saccharomyces cerevisiae tRNA was used as a negative control in RNase protection assays. After digestion with RNase A and T1, samples were ethanol precipitated and electrophoresed on 5% polyacrylamide-8 M urea gels. After drying, protected mRNA signals were visualized with a phosphorimager, and quantitation was performed by normalizing the mRNA species of interest to L32 or β-actin signals.
RESULTS
Feasibility of GST pulldown assay for screening cellular proteins that interact with MAV-1 E1A.
In order to screen for potential cellular proteins that interact with MAV-1 E1A, we used an unbiased GST pulldown approach. First, we subcloned full-length MAV-1 E1A into a GST expression vector, resulting in plasmid pGST-mE1A (Fig. 1A). The GST-mE1A fusion protein was overexpressed by inducing the cells with IPTG (Fig. 1B, lane 3) and purified with glutathione beads (Fig. 1B, lane 4).
Differences in posttranslational modifications between proteins expressed in bacterial versus mammalian cells might affect the interactions of MAV-1 E1A with cellular proteins. We previously showed that MAV-1 E1A binds to mouse pRb and p107 proteins in an in vitro mixing experiment (53). We reasoned that if the GST-mE1A fusion protein is capable of binding to these known interacting proteins, GST pulldown assays with the GST-mE1A fusion protein would be a valid experimental approach to screen for cellular proteins that interact with MAV-1 E1A. Since the pRb and p107 used in those experiments were in vitro-translated proteins, we first further verified that MAV-1 E1A interacts with endogenous Rb family proteins in virus-infected cells.
MBMECs were infected with wild-type MAV-1 at an MOI of 5 and harvested at 40 h postinfection. Mock and pmE109 (E1A null mutant) infections were used as negative controls. Coimmunoprecipitation experiments were carried out with antibodies to the Rb family proteins to show the protein-protein interactions. Western blots of the immunoprecipitates were probed with antibody against MAV-1 E1A. As expected, MAV-1 E1A signals were only detected in wild-type MAV-1-infected samples that had been immunoprecipitated with specific antibodies against p130, p107, or pRb (Fig. 2A, lanes 5, 8, and 11) and not with control normal rabbit serum (lane 2), demonstrating that MAV-1 E1A binds to Rb family proteins in infected cells. Coimmunoprecipitation experiments performed by immunoprecipitation with anti-MAV-1 E1A antibodies (rabbit polyclonal and mouse monoclonal antibodies) and detection on Western blots with antibodies against pRb, p107, and p130 confirmed the MAV-1 E1A-pRb, E1A-p107, and E1A-p130 interactions (data not shown). The interaction between MAV-1 E1A and p130 had not been demonstrated previously.
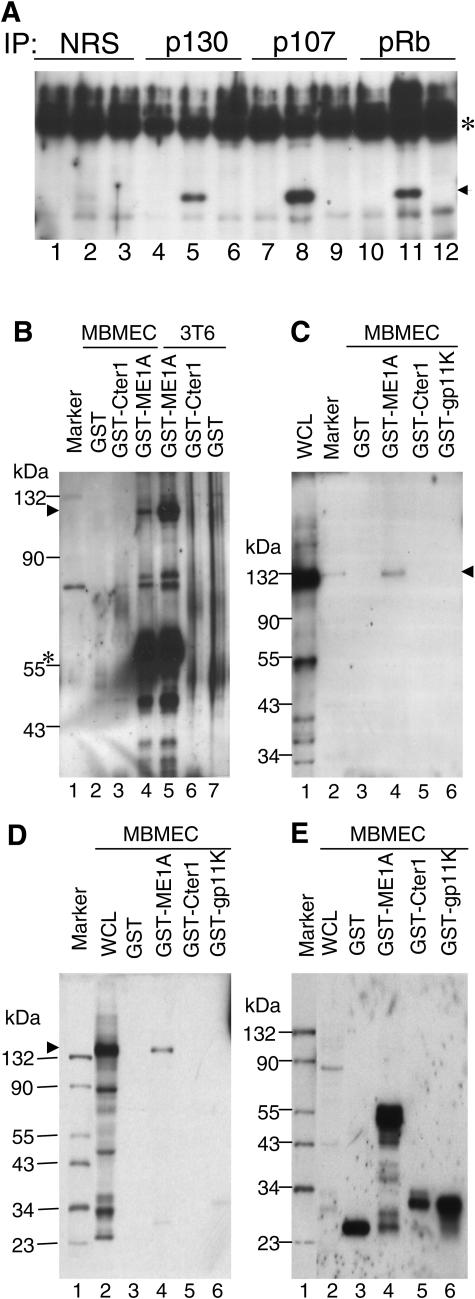
FIG. 2.
Interactions between MAV-1 E1A and Rb family proteins. (A) MBMECs were either mock infected (lanes 1, 4, 7, and 10) or infected with wild-type MAV-1 (lanes 2, 5, 8, and 11) or pmE109 (lanes3, 6, 9, and 12) at an MOI of 5 and harvested at 40 h postinfection. Rabbit polyclonal antibody against p130 (lanes 4 to 6), p107 (lanes 7 to 9), and Rb (lanes 10 to 12) was used to immunoprecipitate the whole-cell lysates. Normal rabbit serum (NRS, purified by DEAE Affi-Gel blue chromatography) was used as a negative control (lanes 1 to 3). Western blots were carried out by probing the membranes with monoclonal antibody against MAV-1 E1A (10B10) (1:1,000). The arrowhead indicates MAV-1 E1A. The asterisk indicates the IgG heavy chain. (B) Equivalent amounts of purified GST, GST-mE1A, GST-Cter1, and GST-gp11K proteins were bound to glutathione beads and then mixed with an equal amount of mammalian whole-cell lysates (WCL) from either MBMECs or 3T6 cells, as indicated. After washing, bound proteins were eluted and electrophoresed on SDS-10% polyacrylamide gels and transferred to polyvinylidene difluoride membranes. The membrane was probed with polyclonal antiserum against mouse Rb protein. Lane 1, protein molecular size standards (sizes indicated on the left side); lanes 2 to 4, proteins bound and eluted from GST beads, GST-Cter1 beads and GST-mE1A beads, respectively, mixed with MBMEC lysates; lanes 5 to 7, proteins bound and eluted from GST-mE1A beads, GST-Cter1 beads, and GST beads, respectively, mixed with 3T6 cell lysates. The arrowhead shows the position of mouse Rb. The asterisk shows the detection of GST-mE1A due to the cross-reaction of Rb antibody with GST protein (it was raised against a GST-Rb fusion protein). (C) GST pulldown assay and Western blot carried out as in B. The membrane was probed with p107 antibody. Lane 1, whole-cell lysates from MBMECs; lane 2, protein molecular size standards; lanes 3 to 6, proteins bound and eluted from GST beads, GST-mE1A beads, GST-Cter1 beads, and GST-gp11K beads, respectively, mixed with MBMEC lysates. The arrowhead shows the p107 position. (D) GST pulldown assay and Western blot carried out as in B. The membrane was probed with p130 antibody. Lane 1, protein molecular size standards (the size is shown at the left); lane 2, whole-cell lysates from MBMECs; lanes 3 to 6, proteins bound and eluted from GST beads, GST-mE1A beads, GST-Cter1 beads, and GST-gp11K beads, respectively, mixed with MBMEC lysates. The arrowhead shows the position of p130. (E) Western blot probed with antibody against GST protein. Same loading order as in panel D.
Since MAV-1 E1A interacts with pRb, p107, and p130 in virus-infected cells, we tested whether the bacterially expressed GST-mE1A fusion proteins would bind to mouse pRb, p107, and p130. In addition to the full-length GST-mE1A protein, we tested a GST fusion construct containing only the C-terminal 157 to 200 amino acids (GST-Cter1) of E1A (Fig. 1A) because we were interested in identifying cellular proteins that specifically bind to the MAV-1 E1A C terminus due to its unique sequence. As controls, we used the GST protein and a GST fusion construct (GST-E3gp11K) containing another MAV-1 early viral protein, E3gp11K (8). As shown in Fig. 2B, 2C, and 2D, mouse pRb, p107, and p130, respectively, bound to GST-mE1A but not to GST or the GST-Cter1 or GST-E3gp11K fusion proteins. Figure 2E was a control to show that approximately equivalent amounts of GST fusion proteins were present in the pulldown assays. Conserved region 2 (CR2) of MAV-1 E1A is critical for binding to pRb and p107 (53). Consistent with those data, the GST-Cter1 fusion protein lacking the CR2 domain did not bind to pRb family proteins. The bacterially expressed GST fusion proteins maintained binding properties similar to those of viral E1A, indicating the suitability of screening for E1A-interacting proteins with GST pulldown assays.
Sur2 interaction with MAV-1 E1A CR3 domain.
Nuclear extracts from two cell lines, MBMECs and 3T6 cells, were used to screen for potential additional interacting proteins in GST pulldown assays. 3T6 cells or MBMECs were used in a single GST pulldown assay and visualized in a Coomassie-stained gel (Fig. 3A; 3T6 cell data not shown). The GST fusion proteins alone were used as a negative control. The purified GST-Cter1 protein (Fig. 1A) was used to screen for proteins that specifically bind to the unique C terminus of MAV-1 E1A. The protein bands that were only present in the GST-mE1A sample (Fig. 3A, lane 6) and not in the GST sample (Fig. 3A, lane 5) were considered potential specific MAV-1 E1A binding proteins. Several specific bands were cut out of the gels, and the identification of proteins was carried out by tandem mass spectrometry-mass spectrometry analysis. One band, indicated by the arrowhead at approximately 150 kDa, was identified as mouse Sur2. We identified mouse Sur2 as a GST-mE1A-interacting protein from both MBMEC (Fig. 3A) and 3T6 (data not shown) cells. Several additional bands were identified by mass spectrometry-mass spectrometry analysis. The characterization of these as potential MAV-1 E1A-interacting proteins will be described elsewhere following verification with specific antibodies (L. Fang and K. R. Spindler, unpublished data).
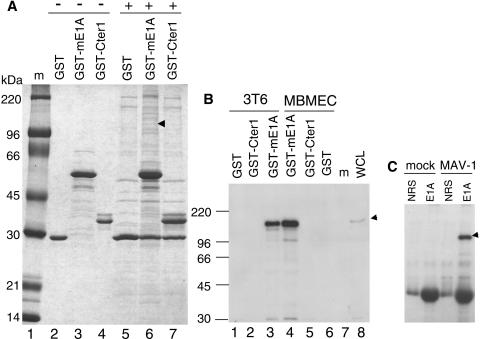
FIG. 3.
Identification of mouse Sur2 as a specific MAV-1 E1A-interacting protein. (A) Large-scale GST-mE1A pulldown assay. Lane 1, protein molecular size standards; lanes 2 to 4, purified GST, GST-mE1A, and GST-Cter1 fusion proteins, respectively, were bound to glutathione-Sepharose beads without mixing with any mammalian cell nuclear extracts (−); lanes 5 to 7, nuclear extracts from 5 × 108 mouse MBMECs were precleared sequentially against glutathione-Sepharose beads and GST beads, and then an equal amount of the precleared mammalian nuclear extracts was added (+) to GST beads, GST-mE1A beads, and GST-Cter1 beads, respectively. Beads were washed, and the bound proteins were eluted off the beads by boiling in protein loading buffer. Proteins binding specifically to GST-mE1A were identified by comparing lane 6 with lanes 3 and 5. The arrowhead shows Sur2 protein. An independent duplicate analysis of both MBMEC and 3T6 cell lines gave similar results, with identification of Sur2 (data not shown). (B) Sur2 protein interacts with GST-mE1A (full-length E1A) in GST pulldowns. GST pulldown assays were performed by incubating the nuclear extracts from 3T6 cells (lanes 1 to 3) or MBMECs (lanes 4 to 6) with GST beads, GST-Cter1 beads, or GST-mE1A beads, as indicated. Monoclonal antibody against Sur2 (BD Pharmingen) (1:1,000) was used for Western blots. Whole-cell lysates (WCL) were used as a positive control (lane 8). Lane 7, protein molecular size standards. The arrowhead shows the Sur2 position. (C) MAV-1 E1A protein interacts with Sur2 in virus-infected cells. MBMECs were mock or MAV-1 infected at an MOI of 5 and harvested at 40 h postinfection. Normal rabbit serum or AKO7-147 (E1A) (both purified by DEAE Affi-Gel blue chromatography) was mixed with whole-cell lysates of MBMECs to carry out the immunoprecipitation. The immunoprecipitates were electrophoresed on 8% polyacrylamide-SDS gels and transferred to polyvinylidene difluoride membranes. Monoclonal antibody against Sur2 was used in Western blots. The arrowhead shows the Sur2 position.
We verified the binding specificity of mouse Sur2 to GST-mE1A with Western blots. GST pulldown assays were carried out as in Fig. 3, and samples were transferred to a polyvinylidene difluoride membrane and probed for the presence of mouse Sur2 with a monoclonal antibody against human Sur2. Anti-Sur2 antibody was developed against the human protein but cross-reacts with mouse Sur2. Mouse Sur2 protein is almost identical to its human homologue, and human Sur2 can rescue the phenotypic defects in mouse Sur2−/− embryonic stem cells (56). In both MBMECs and 3T6 cells, we identified mouse Sur2 specifically bound to the GST fusion protein containing the full-length MAV-1 E1A but not to control GST proteins or the C-terminal 157 to 200 amino acids of E1A in GST-Cter1 (Fig. 3B). This demonstrates that mouse Sur2 interacts specifically with GST-mE1A. It is not surprising that mouse Sur2 did not bind to the GST-Cter1 fusion protein because it has been shown that human Sur2 interacts with human adenovirus E1A through the CR3 domain (13, 57). The CR3 domain of MAV-1 E1A is absent in the GST-Cter1 construct.
We investigated whether MAV-1 E1A binds to mouse Sur2 in virus-infected cells (Fig. 3C). Whole-cell lysates from either MAV-1-infected or mock-infected cells were subjected to immunoprecipitation with rabbit polyclonal anti-E1A antibody. The anti-E1A immunoprecipitates were analyzed for the presence of mouse Sur2 with anti-Sur2 monoclonal antibody in Western blots. Mouse Sur2 was only detected from the MAV-1-infected sample that was immunoprecipitated with anti-E1A antibody (Fig. 3C). The results demonstrate that the interaction between MAV-1 E1A and mouse Sur2 occurred in MAV-1-infected cells.
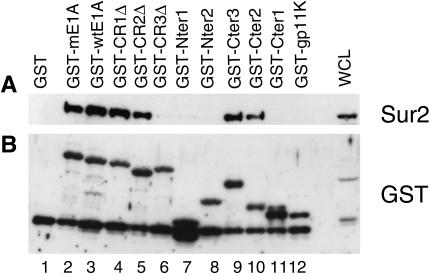
CR3 of human adenovirus E1A is necessary and sufficient for binding to Sur2 (13, 56, 57). Despite the presence of the CR3 domain in MAV-1 E1A, there is no significant similarity to human adenovirus E1A outside of the conserved regions (6), making it intriguing to investigate which regions of MAV-1 E1A are important for binding to mouse Sur2. We used two different experimental approaches to map the region required for MAV-1 E1A interaction with mouse Sur2. First, GST pulldown assays were used. In addition to GST-mE1A, containing a 22-amino-acid linker (Fig. 1A), we constructed another full-length MAV-1 E1A directly downstream of GST, GST-wild-type E1A (Fig. 1A). A series of GST fusion proteins were also prepared that contained either truncated or deleted forms of MAV-1 E1A, as shown in Fig. 1A. GST pulldown assays were carried out with these constructs, and the bound proteins were subjected to Western blotting with anti-Sur2 antibody (Fig. 4A). The same membrane was stripped and reprobed with anti-GST antibody to verify equivalent loading of GST fusion proteins (Fig. 4B). The mouse Sur2 protein was detected in samples with GST-E1A constructs containing the CR3 of E1A (Fig. 4A, lanes 2 to 5 and 9 and 10), and absent in samples with GST-E1A constructs lacking CR3 (Fig. 4A, lanes 6 to 8 and 11). Mouse Sur2 did not bind the negative controls GST (Fig. 4A, lane 1) or GST-gp11Κ (Fig. 4A, lane 12). The results indicate that the CR3 domain is required for the protein-protein interaction.
FIG. 4.
MAV-1 E1A CR3 is required for binding to Sur2. The GST fusion proteins were purified from induced E. coli cells. Whole-cell lysates of MBMECs were incubated with different regions of MAV-1 E1A as GST fusion proteins bound to glutathione beads. (A) Bound proteins were analyzed by Western blotting with monoclonal anti-Sur2 antibody. (B) Membrane was stripped and reprobed with anti-GST antibody to show the equivalent loading of various GST fusion proteins.
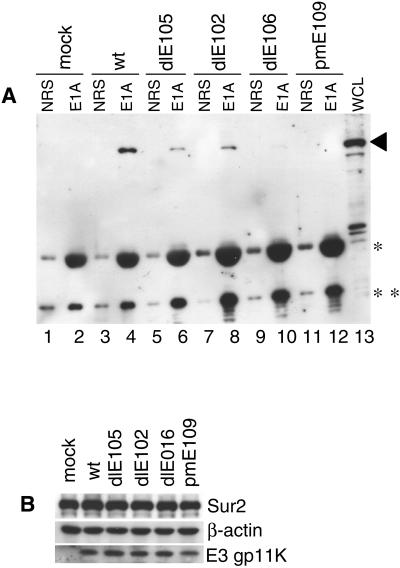
We tested whether CR3 was required for binding to Sur2 in virus-infected cells with MAV-1 E1A mutant viruses. The dlE105, dlE102, and dlE106 viruses are CR1, CR2, and CR3 deletion mutants of E1A, respectively (53). Wild-type MAV-1 and pmE109 (E1A null mutant virus) were used as a positive control and a negative control, respectively. Immunoprecipitation and Western blots were carried out as in Fig. 3C. The anti-E1A rabbit polyclonal antibody AKO7-147 is able to recognize all mutant-form E1A proteins by immunoprecipitation (53). Mouse Sur2 proteins from the cells infected with dlE106 (CR3 deletion mutant) failed to coimmunoprecipitate with E1A (Fig. 5A, lane 10). This not only confirms the GST pulldown data but also demonstrates that CR3 of MAV-1 E1A is required for binding to mouse Sur2 in virus-infected cells. There were slight reductions in the amount of mouse Sur2 coimmunoprecipitated with the CR1 and CR2 deletion mutants relative to wild-type virus (Fig. 5A, compare lanes 6 and 8 to lane 4). The control blots showed that the mouse Sur2 protein levels present in all virus-infected cells were very similar to that in mock-infected cells (Fig. 5B), indicating that there was no mouse Sur2 protein degradation upon viral infection.
FIG. 5.
CR3 of MAV-1 E1A is essential for the interaction between Sur2 and MAV-1 E1A. (A) MBMECs were infected with wild-type MAV-1, dlE105 (CR1 deletion mutant), dlE102 (CR2 deletion mutant), or dlE106 (CR3 deletion mutant) at an MOI of 5, harvested at 40 h postinfection, and aliquoted. Immunoprecipitation and Western blot analysis was carried out as in Fig. 3C on one aliquot of each infection. The IgG used in immunoprecipitation was also detected due to the cross-reaction with the secondary antibody used in Western blotting (*, IgG heavy chain; **, IgG light chain). The arrowhead shows the Sur2 position. (B) Western blots (without immunoprecipitation) of another aliquot of each infection were probed with the indicated primary antibodies.
There is no growth defect in the E1A mutant viruses at an MOI of 5, and E1A proteins are expressed at wild-type levels by the CR1, CR2, and CR3 deletion mutant viruses (61). The viral early gene E3gp11K was monitored to show a comparable efficiency of viral infection by the E1A viral mutants (Fig. 5B). E3gp11K protein levels in E1A mutant virus infections were nearly equivalent to those in wild-type virus infections, as expected, because at this multiplicity E3gp11K mRNA levels are not reduced in E1A mutant virus infections relative to wild-type virus infection (57).
Sur2 is important for MAV-1 replication.
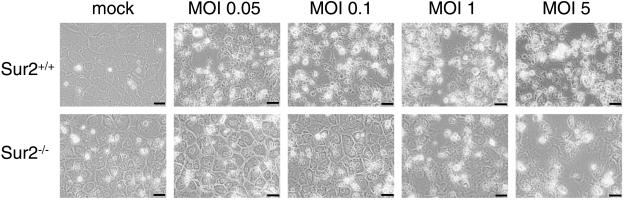
We investigated the functional role of mouse Sur2 in MAV-1 infection with Sur2+/+ and Sur2−/− MEFs. Cytopathic effects after MAV-1 infection were quite different between Sur2−/− and Sur2+/+ MEFs. Wild-type MEFs started to show dramatic cytopathic effect at an MOI of 1 or 5 at 3 days postinfection, with only a slight cytopathic effect at an MOI of 0.05 (data not shown). At 7 days postinfection, the infected wild-type MEFs showed cytopathic effect at all multiplicities, whereas Sur2−/− MEFs showed little or no cytopathic effect except at the higher MOIs of 1 and 5 (Fig. 6). The results showed that there was a delay in the appearance of cytopathic effect in Sur2−/− MEFs upon MAV-1 infection compared to wild-type cells at equivalent MOIs.
FIG. 6.
Cytopathic effects in Sur2+/+ and Sur2−/− MEFs upon MAV-1 infection. MEFs were either mock or MAV-1 infected at the indicated MOI. Phase contrast pictures were taken at 7 days postinfection with an Olympus microscope (400×). The scale bar is 50 μm.
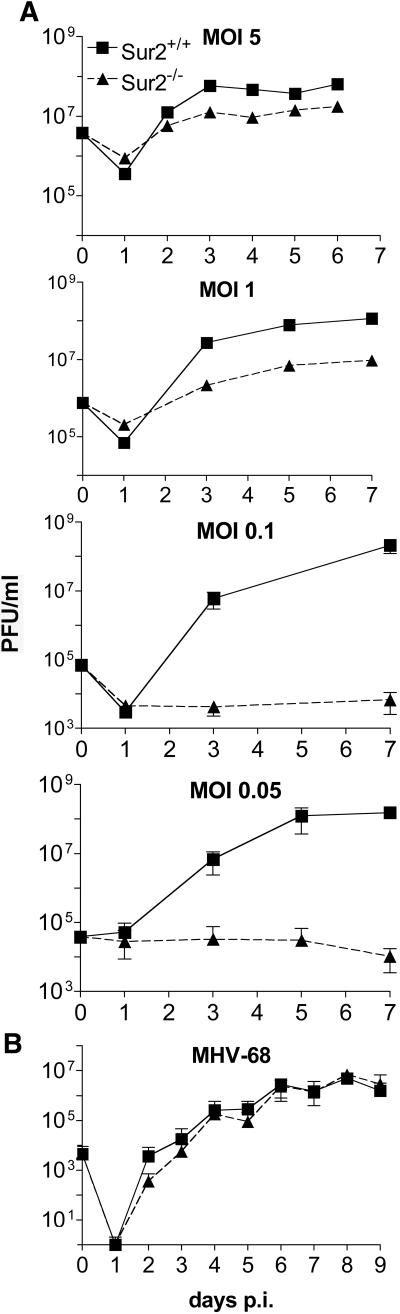
We examined MAV-1 replication in Sur2+/+ and Sur2−/− MEFs in growth curves (Fig. 7A). We observed a multiplicity-dependent virus growth defect upon MAV-1 infection of Sur2−/− MEFs. At an MOI of 5, the viral yields were reduced by a factor of 5 in Sur2−/− MEFs compared with wild-type cells, whereas at an MOI of 1, the yields were reduced by a factor of 10 to 50. Therefore, at high MOIs (1 and 5), MAV-1 was able to replicate and generate mature infectious virions in Sur2−/− MEFs, though less efficiently than in Sur2+/+ MEFs (Fig. 7A). However, at low MOIs (0.05 and 0.1), the viral yields in Sur2−/− MEFs were equivalent to the amount of input virus, indicating that viral replication was severely reduced. These results indicated that the mouse Sur2 protein was required for MAV-1 replication at a low input MOI and important for efficient viral replication at a high input MOI.
FIG. 7.
Multiplicity dependence of MAV-1 viral yields on MEFs. (A) Growth curves of MAV-1 on Sur2+/+ and Sur2−/− MEFs. Sur2+/+ and Sur2−/− MEFs were infected with MAV-1 at the indicated MOIs and harvested at the indicated times. The viral yields were determined by plaque assays on 3T6 cells. Three independently infected cultures were assayed for each MOI. These experiments were repeated three times, with similar results (data not shown). (B) Growth curves of mouse gammaherpesvirus 68 (MHV-68) on Sur2+/+ and Sur2−/− MEFs. Sur2+/+ and Sur2−/− MEFs were infected with mouse gammaherpesvirus 68 at an MOI of 0.01 and harvested at the indicated times. The viral yields were determined by plaque assays on NIH 3T3 cells. The legend in the top panel is the same for all panels.
We tested whether this viral replication defect in Sur2−/− MEFs is a general or virus-specific effect. Mouse gammaherpesvirus 68 showed no significant differences in growth in Sur2+/+ MEFs compared with Sur2−/− MEFs (Fig. 7B). The data indicate that mouse Sur2 is not required for mouse gammaherpesvirus 68 replication and suggest that the requirement of mouse Sur2 for viral replication is a relatively specific effect.
The plaque assays measured the final outcome of viral infection. Every step in the infectious process, such as viral early gene expression, viral genome DNA replication, late gene expression, DNA packaging, viral assembly, and secretion of mature virions, could affect the final viral yield. We examined some of these steps to determine the potential roles of Sur2 in MAV-1 infection.
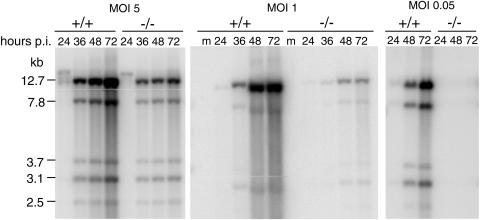
We tested whether viral DNA replication differed in Sur2−/− and Sur2+/+ MEFs by analyzing MAV-1 genome DNA in Southern blots (Fig. 8). At MOIs of 1 and 5, MAV-1 DNA was detected at 24 h postinfection in both cell types. The kinetics of viral DNA accumulation during the course of viral infection were shown by the increasing intensity of viral DNA bands in virus-infected Sur2+/+ MEFs. In contrast, less DNA accumulation was observed in Sur2−/− MEFs. This indicates that there was no defect in the time of onset of viral DNA synthesis at MOIs of 1 and 5, but the accumulation of viral DNA was substantially decreased in Sur2−/− cells. We did not detect any MAV-1 DNA from Sur2−/− MEFs at an MOI of 0.05, whereas in Sur2+/+ MEFs it increased to a level comparable to that seen in Sur2+/+ MEFs at MOIs of 1 and 5 (Fig. 8). The defect in MAV-1 DNA replication in Sur2−/− MEFs, taken together with overall lower virus yield in the same conditions (Fig. 7A), suggested that mouse Sur2 is required for some step at or prior to DNA replication.
FIG. 8.
Multiplicity-dependent defects in viral DNA replication in Sur2−/− MEFs. Viral DNA was isolated from MAV-1-infected Sur2+/+ (+/+) and Sur2−/− (−/−) MEFs at the indicated MOIs and times by the Hirt method. Equal amounts of DNA were digested with HindIII, loaded on 0.7% agarose gels, and transferred to membranes. The membranes were probed with MAV-1-specific DNA probes labeled with [α-32P]dATP. The sizes of the viral DNA fragments are indicated in kilobases. Lanes m, mock infected. These experiments were repeated three times with similar results (data not shown).
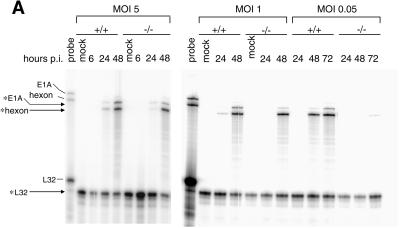
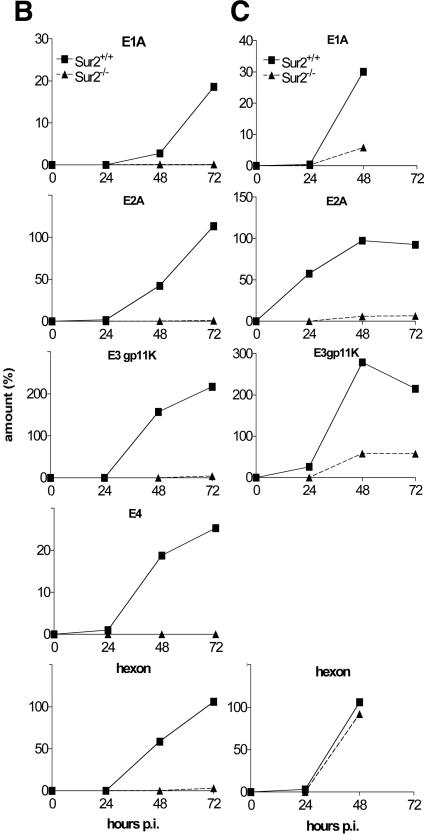
We quantitated the steady-state mRNA levels of MAV-1 early genes E1A, E2, E3, and E4 and one viral late gene, hexon, in Sur2−/− and Sur2+/+ MEFs with RNase protection assays. Representative RNase protection assay gels (Fig. 9A) show the mRNA levels of E1A and hexon. The mRNA levels of each viral gene at MOIs of 0.05 and 1 were normalized to that of the housekeeping gene L32 or to β-actin, and the relative levels are shown in Fig. 9B and 9C. There was a delay in the onset of detectable transcription of all viral early genes tested, and their steady-state mRNA levels were reduced in Sur2−/− MEFs compared with Sur2+/+ MEFs at high MOIs (1 and 5). In particular, E2A and E3 transcription defects were apparent at an MOI of 1 (Fig. 9C), even though at this multiplicity viral replication is only 1 log unit different between Sur2+/+ and Sur2−/− MEFs (Fig. 7). At an MOI of 0.05, the expression of viral early genes was severely diminished or not detectable in Sur2−/− MEFs.
FIG. 9.
Multiplicity-dependent defects in viral mRNA levels in Sur2−/− MEFs. (A) Total RNA was isolated from MAV-1-infected Sur2+/+ (+/+) and Sur2−/− (−/−) MEFs at the indicated MOIs and times. The multiplexed probe (“probe”) shows the full-length probes (no RNase) for E1A, hexon, and L32, as indicated by the tick marks. The size of probe protected from RNase treatment is shown by the arrows and asterisks for each gene. L32 was used as an internal loading control. These experiments were repeated three times with similar results (data not shown). (B) Quantitation of RNase protection assays. Sur2+/+ and Sur2−/− MEFs were infected at an MOI of 0.05 and harvested at the indicated times. Viral genes E1A, E2A, E3gp11K, E4, and hexon were analyzed by RNase protection assays. The amount of mRNA for each gene was determined by quantitation of band intensities in the autoradiograph with a phosphorimager and Imagequant software and then normalized to L32 or β-actin controls, whose levels were set to 100%. (C) Sur2+/+ and Sur2−/− MEFs were infected at an MOI of 1, and viral genes were analyzed as in panel B. E4 mRNAs were not analyzed at an MOI of 1.
The viral late gene hexon had essentially the same expression pattern as viral early genes in Sur2−/− MEFs, i.e., delayed and reduced relative to that of Sur2+/+ MEFs at an MOI of 0.05. However, we also noticed that at high MOIs (1 and 5), the defects in hexon mRNA levels were not as severe as the defects in viral DNA levels. Nevertheless, the results indicate that mouse Sur2 is involved in the transcription regulation of the viral early genes, supporting the proposed human adenovirus infection model in which E1A recruits the Mediator complex through stable binding to the Sur2 subunit to transactivate the transcription of the other viral early genes (57).
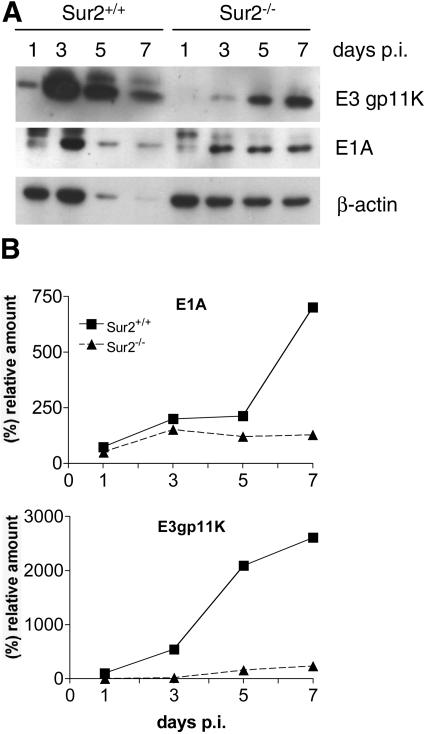
To confirm that the RNase protection assay results for mRNA levels were reflected in protein levels, we analyzed the expression of two MAV-1 early proteins, E1A and E3gp11K, and compared them to that of β-actin (Fig. 10). As expected, the levels of MAV-1 E1A and E3gp11K protein accumulation were higher in Sur2+/+ MEFs than in Sur2−/− MEFs when the cells were infected at an MOI of 1 (Fig. 10). The decreased protein recovery (and loading; see the figure legend) of E3 gp11K, E1A, and β-actin seen at 5 and 7 days postinfection in Sur2+/+ MEFs (Fig. 10A, lane 3 and 4) was likely due to the cytopathic effect at this time in infection (Fig. 6). Quantitation (Fig. 10B) shows that E1A and E3gp11K protein levels were relatively high even when the cytopathic effect was apparent in the Sur2+/+ MEFs. At an MOI of 0.1, E1A and E3gp11K proteins were not detected by Western blots in Sur2−/− MEFs even at 7 days postinfection (data not shown), supporting the hypothesis that mouse Sur2 is required for MAV-1 replication at a low input multiplicity. In contrast, both early viral proteins were readily detected in Sur2+/+ MEFs at 3 days postinfection (data not shown). The correlation of the Western blot data with the RNase protection assay results strongly argues that mouse Sur2 plays a critical role in an early phase of MAV-1 replication.
FIG. 10.
Differences in viral protein levels between infected Sur2+/+ and Sur2−/− MEFs. (A) MEFs were infected with MAV-1 at an MOI of 1 and harvested at the indicated times. Cell pellets were lysed in E3 lysis buffer (420 mM NaCl, 50 mM Tris-HCl [pH 7.4], 1% NP-40). Protein concentrations were measured with a Bio-Rad protein assay kit. Equivalent amounts of protein except for the Sur2+/+ MEF 5 and 7 day postinfection samples were loaded. For those two samples, only 39 and 32%, respectively, were recovered and loaded. Samples were electrophoresed on an 8 to 15% gradient polyacrylamide-SDS gel and analyzed by Western blotting with antibodies as indicated at the right. β-Actin was assayed as a loading control. (B) Quantitation of Western blots. The band intensities were quantitated by densitometry and normalized to β-actin levels for each sample.
DISCUSSION
MAV-1 E1A is involved in viral pathogenesis, undoubtedly through association with cellular proteins. We showed experimentally for the first time that endogenous pRb and p107 proteins interacted with MAV-1 E1A in virus-infected cells, which is consistent with in vitro mixing experiment data (53). In addition, for the first time, p130 was also shown to interact with MAV-1 E1A in virus-infected cells by immunoprecipitation and Western blots. We used a GST pulldown assay and showed that mouse Sur2 protein interacted with GST-mE1A and with viral E1A in virus-infected cells (Fig. 3). We mapped the region of MAV-1 E1A required for binding to mouse Sur2 to CR3 (Fig. 4 and 5). The mutant viruses used in the mapping have no significant growth defects compared to wild-type MAV-1 infection in 3T6 cells at an MOI of 5 (61), demonstrated by similar E3gp11K protein levels (Fig. 5B) and E3gp11K mRNA levels (54).
The polyclonal antibody against MAV-1 E1A (AKO7-147) can recognize the E1A deletion mutant proteins, including the CR3-deleted E1A, in immunoprecipitations (61). Therefore, the absence of the mouse Sur2 signal from the CR3 deletion mutant-infected sample immunoprecipitated with anti-E1A antibody was not due to a reduced efficiency of viral infection or failure of antibody recognition. However, we also noticed that less mouse Sur2 coimmunoprecipitated with MAV-1 E1A from dlE105 and dlE102 virus-infected samples than wild-type MAV-1-infected samples (Fig. 5, lanes 6 and 8). It is possible that the deleted forms of MAV-1 E1A protein might have conformational changes resulting in reduced binding affinity to mouse Sur2. A formal possibility that we believe is less likely is that CR1 and CR2 have direct but small effects on the Sur2-E1A interaction in viral infection.
With a variety of single-amino-acid human adenovirus E1A mutations in GST pulldown assays, an intact zinc finger structure (four cysteine residues that bind a single Zn2+ ion) in human adenovirus E1A CR3 was shown to be important for the Sur2-E1A CR3 interaction (13). MAV-1 E1A CR3 has the same four cysteine residues, and it is likely that this conserved zinc finger structure in MAV-1 E1A is important for binding to mouse Sur2. One question that has not been addressed for human adenovirus or MAV-1 is which regions of Sur2 protein are required for the Sur2-E1A interaction. Human Sur2 protein has been reported to interact with three transcription factors to date, Elk-1 (13, 56), ESX (2), and C/EBPβ (40). Elk-1 is activated by extracellularly regulated kinase (ERK) in the mitogen-activated protein kinase signal transduction pathway and binds serum response elements of targeted genes. The region of Sur2 that binds to Elk-1 is unknown. ESX, an Ets factor, is an epithelial cell-specific transcription factor, and residues 352 to 625 of Sur2 are important for binding to ESX (2). It has been suggested that E1A competes with C/EBPβ to bind to Sur2 (40). Mapping the Sur2 region required for binding to E1A will allow us to postulate whether E1A interferes with the binding of Sur2 to these transcription factors. In turn, it will help us understand the physiological functions of Sur2 and address why adenovirus E1A targets Sur2.
The conservation of adenovirus E1A binding to Sur2 in human adenovirus and MAV-1 indicates its importance for adenovirus replication. However, due to adenovirus species specificity and the availability of only mouse Sur2 knockout cells, Sur2 function in adenoviral replication had not been directly tested prior to this study. No Sur2 protein was detected in Sur2−/− MEFs by Western blots (data not shown). Expressing human Sur2 in Sur2−/− mouse stem cells rescues their defective transcription activation function, showing that Sur2 is the only missing factor in the knockout cells (56). The data in Fig. 6 to 10 suggest that the MAV-1 replication defect is due at least in part to a defect in viral early gene transcription.
The importance of mouse Sur2 protein for viral replication is specific for MAV-1, because there was no growth defect for mouse gammaherpesvirus 68 in Sur2−/− MEFs (Fig. 7) (13, 40, 56). The transcription activities of many transcription factors are not affected by human Sur2 (2, 13, 56), indicating the narrow target range of Sur2. This raises the interesting question of why the adenovirus E1A protein conserves the ability to bind to such a selective Mediator subunit.
The molecular mechanisms of Sur2 function in MAV-1 infection have not been fully investigated. It has been proposed that the primary mechanism by which E1A recruits the Mediator complex to transactivate the transcription of viral early genes is that E1A stimulates and also stabilizes the assembly of a transcription preinitiation complex on promoter DNA through stable Sur2-E1A CR3 interaction (17, 57). However, the ability of MAV-1 to replicate in Sur2−/− MEFs suggests that there is a Sur2-independent virus replication pathway. This raises many possibilities. First, it is possible that the Mediator complex or transcription preinitiation complex is recruited to viral gene promoters with decreased efficiency in the absence of Sur2. It is possible that E1A binds other components of the Mediator complex, and it will be interesting to see whether E1A can coimmunoprecipitate other Mediator components in Sur2−/− MEFs.
Second, other regions of human adenovirus E1A protein, including CR1 and CR2, also have very strong transcriptional activity, primarily through binding of p300/CBP (10, 58) and Rb family proteins (23), respectively. Binding of Rb family members and binding of p300/CBP are two independent mechanisms utilized by human adenovirus E1A protein to manipulate the cell cycle from G1 to S phase (51). The MAV-1 E1A protein contains all three conserved regions, and interactions between MAV-1 E1A and Rb family members are shown in this work. Another possibility is that E4 gene products also play a role in MAV-1 replication. The human adenovirus E4 ORF6/7 protein has been shown to induce E2F DNA binding to the viral E2A promoter and thus functionally compensate for the loss of E1A in human adenovirus infection (44, 49). MAV-1 E4 ORFd (36) has 17% identity and 43% similarity to human adenovirus E4 ORF6/7 protein (L. Fang and K. R. Spindler, unpublished data). It is reasonable to speculate that MAV-1 uses one or more of these mechanisms to replicate if Sur2 is absent in the infected host cells.
The CR3 deletion mutant virus (dlE106) replicates to a level comparable to wild-type virus in 3T6 cells at an MOI of 5 (61), but its CR3-deleted E1A protein does not bind to mouse Sur2 (Fig. 5), indicating that the MAV-1 E1A-mouse Sur2 interaction itself is not absolutely essential for MAV-1 replication. Moreover, the fact that the human adenovirus E1A null mutant virus (dl312) can replicate at high MOIs clearly demonstrates that E1A is not an essential gene for human adenovirus replication (26, 32, 43, 50). MAV-1 E1A is dispensable for MAV-1 replication in cell culture, demonstrated by growth of the MAV-1 E1A null mutant (pmE109) at MOIs as low as 1 (61). This indicates the existence of an E1A-independent viral replication pathway and may also explain why MAV-1 is able to replicate in the absence of the mouse Sur2-E1A CR3 interaction in Sur2−/− MEFs. We are currently testing the viral replication of E1A null and CR3 deletion mutant viruses in Sur2−/− MEFs. The data in the work presented here clearly showed that mouse Sur2 is a critical factor for efficient MAV-1 replication.
We note that there are differences between MAV-1 infection in cell culture and in mice. The E1A null mutant pmE109 is less virulent than wild-type MAV-1 in mice (52). The 50% lethal dose of pmE109 is 2 and 4 logs higher than that of wild-type MAV-1 in inbred SJL/J and outbred NIHS mice, respectively (55). Both pmE109 and CR3 deletion (dlE106) mutant viruses replicated to significantly lower levels in several inbred stains of mice than did wild-type MAV-1 (L. Fang and K. R. Spindler, manuscript in preparation). This suggests that mouse Sur2-E1A binding may be more important for MAV-1 replication in mice than cells in culture.
A multiplicity-dependent growth phenotype has been observed for many viruses, including human adenoviruses (26, 32, 43, 50), herpes simplex virus type 1 (16, 20, 24), cytomegalovirus (14, 45), and African swine fever virus (41). dl312, an E1A null mutant of human adenovirus, shows the multiplicity-dependent phenotype (26, 32, 43, 50). At low MOIs, there is a delay and reduced expression of viral early genes, but at higher MOIs, the expression of viral early genes can be enhanced (43). The UL82-deficient mutant of human cytomegalovirus displays a multiplicity-dependent phenotype in cell culture (14). The defect of severely restricted viral replication of this UL82-deficient mutant virus at low input MOIs can be rescued by higher input MOIs. Very similarly, the severe replication defect of ICP0 null mutant herpes simplex virus type 1 in Vero or BHK cells at low MOIs can be overcome at higher multiplicities (16, 20, 24). A thymidine kinase gene deletion mutant of African swine fever virus shows a growth defect on swine macrophages at low MOIs, but the defect is not apparent at high MOIs (41).
It is interesting that adenovirus E1A, herpes simplex virus type 1 ICP0, and human cytomegalovirus UL82 all function in the early phase of viral infection and in gene activation (9, 14, 15, 19, 30, 33, 39), and mutations in these genes all show multiplicity-dependent effects. E1A is the first transcribed gene in adenovirus infection (43, 51), and it transactivates the transcription of the other viral early genes, including E4 (9, 33). However, E4 gene expression is seen in the absence of E1A at high MOIs (46), suggesting that there is an E1A-independent pathway to activate the transcription of E4 genes. Therefore, it is possible that E4 gene products play a role in this multiplicity-dependent effect in MAV-1-infected Sur2−/− MEFs. There might be a threshold in Sur2−/− MEFs for MAV-1 replication. At a relatively low MOI, there might not be enough accumulation of E1A or E4 gene products. However, at a higher MOI, due to the increase in copy number of viral promoters in cells, the viral E1A and/or E4 gene products might accumulate to high enough levels to support MAV-1 replication. In any case, MAV-1 does not replicate as well in Sur2−/− MEFs as in Sur2+/+ MEFs, indicating that Sur2 is important for efficient MAV-1 replication.
Acknowledgments
We thank Carol Eng, Gwen Hirsch, Carla Pretto, and Amanda Welton for excellent technical assistance. We thank the monoclonal antibody facility at the University of Georgia for assistance with antibody generation. We thank Mary Lutzke, Jason Weinberg, and Martin Moore for assistance with RNase protection assays. We thank Michael Imperiale and members of the Spindler laboratory for critical reviews of the manuscript.
This work was supported by NIH grants R01 AI023762 to K.R.S. and CA25235 to A.J.B.
REFERENCES
- 1.Ackrill, A. M., G. R. Foster, C. D. Laxton, D. M. Flavell, G. R. Stark, and I. M. Kerr. 1991. Inhibition of the cellular response to interferons by products of the adenovirus type-5 E1A oncogene. Nucleic Acids Res. 19:4387-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asada, S., Y. Choi, M. Yamada, S. C. Wang, M. C. Hung, J. Qin, and M. Uesugi. 2002. External control of Her2 expression and cancer cell growth by targeting a Ras-linked coactivator. Proc. Natl. Acad. Sci. USA 99:12747-12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avvakumov, N., R. Wheeler, J. C. D'Halluin, and J. S. Mymryk. 2002. Comparative sequence analysis of the largest E1A proteins of human and simian adenoviruses. J. Virol. 76:7968-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, A. O., C. W. Beard, S. D. Redick, and K. R. Spindler. 1989. Genome organization of mouse adenovirus type 1 early region 1: A novel transcription map. Virology 170:523-536. [DOI] [PubMed] [Google Scholar]
- 5.Ball, A. O., C. W. Beard, P. Villegas, and K. R. Spindler. 1991. Early region 4 sequence and biological comparison of two isolates of mouse adenovirus type 1. Virology 180:257-265. [DOI] [PubMed] [Google Scholar]
- 6.Ball, A. O., M. E. Williams, and K. R. Spindler. 1988. Identification of mouse adenovirus type 1 early region 1: DNA sequence and a conserved transactivating function. J. Virol. 62:3947-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beard, C. W., A. O. Ball, E. H. Wooley, and K. R. Spindler. 1990. Transcription mapping of mouse adenovirus type 1 early region 3. Virology 175:81-90. [DOI] [PubMed] [Google Scholar]
- 8.Beard, C. W., and K. R. Spindler. 1995. Characterization of an 11K protein produced by early region 3 of mouse adenovirus type 1. Virology 208:457-466. [DOI] [PubMed] [Google Scholar]
- 9.Berk, A. J., F. Lee, T. Harrison, J. Williams, and P. A. Sharp. 1979. Pre-early adenovirus 5 gene product regulates synthesis of early viral messenger RNAs. Cell 17:935-944. [DOI] [PubMed] [Google Scholar]
- 10.Bondesson, M., M. Mannervik, G. Akusjarvi, and C. Svensson. 1994. An adenovirus E1A transcriptional repressor domain functions as an activator when tethered to a promoter. Nucleic Acids Res. 22:3053-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boube, M., L. Joulia, D. L. Cribbs, and H.-M. Bourbon. 2002. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110:143-151. [DOI] [PubMed] [Google Scholar]
- 12.Boyd, J. M., T. Subramanian, U. Schaeper, M. La Regina, S. Bayley, and G. Chinnadurai. 1993. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis, and metastasis. EMBO J. 12:469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyer, T. G., M. E. D. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 14.Bresnahan, W. A., and T. E. Shenk. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 97:14506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai, W., and P. A. Schaffer. 1991. A cellular function can enhance gene expression and plating efficiency of a mutant defective in the gene for ICP0, a transactivating protein of herpes simplex virus type 1. J. Virol. 65:4078-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai, W., and P. A. Schaffer. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66:2904-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantin, G. T., J. L. Stevens, and A. J. Berk. 2003. Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc. Natl. Acad. Sci. USA 100:12003-12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cauthen, A. N., and K. R. Spindler. 1999. Construction of mouse adenovirus type 1 mutants, p. 85-103. In W. S. M. Wold (ed.), Adenovirus methods and protocols. Humana Press, Totowa, N.J.
- 19.Chau, N. H., C. D. Vanson, and J. A. Kerry. 1999. Transcriptional regulation of the human cytomegalovirus US11 early gene. J. Virol. 73:863-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, J., and S. Silverstein. 1992. Herpes simplex viruses with mutations in the gene encoding ICP0 are defective in gene expression. J. Virol. 66:2916-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deleu, L., S. Shellard, K. Alevizopoulos, B. Amati, and H. Land. 2001. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene 20:8270-8275. [DOI] [PubMed] [Google Scholar]
- 22.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egan, C., T. N. Jelsma, J. A. Howe, S. T. Bayley, B. Ferguson, and P. E. Branton. 1988. Mapping of cellular protein-binding sites on the products of early-region 1A of human adenovirus type 5. Mol. Cell. Biol. 8:3955-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Everett, R. D., C. Boutell, and A. Orr. 2004. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J. Virol. 78:1763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 26.Gaynor, R. B., and A. J. Berk. 1983. Cis-acting induction of adenovirus transcription. Cell 33:683-693. [DOI] [PubMed] [Google Scholar]
- 27.Harlow, E., P. Whyte, B. R. J. Franza, and C. Schley. 1986. Association of adenovirus early-region 1A proteins with cellular polypeptides. Mol. Cell. Biol. 6:1579-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 29.Hobbs, M. V., W. O. Weigle, D. J. Noonan, B. E. Torbett, R. J. McEvilly, R. J. Koch, G. J. Cardenas, and D. N. Ernst. 1993. Patterns of cytokine gene expression by CD4+ T cells from young and old mice. J. Immunol. 150:3602-3614. [PubMed] [Google Scholar]
- 30.Homer, E. G., A. Rinaldi, M. J. Nicholl, and C. M. Preston. 1999. Activation of herpesvirus gene expression by the human cytomegalovirus protein pp71. J. Virol. 73:8512-8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horwitz, M. S. 2001. Adenoviruses, p. 2301-2326. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 32.Imperiale, M. J., H.-T. Kao, L. T. Feldman, J. R. Nevins, and S. Strickland. 1984. Common control of the heat shock gene and early adenovirus genes: evidence for a cellular E1A-like activity. Mol. Cell. Biol. 4:867-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, N. C., and T. Shenk. 1979. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc. Natl. Acad. Sci. USA 76:3665-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kajon, A. E., C. C. Brown, and K. R. Spindler. 1998. Distribution of mouse adenovirus type 1 in intraperitoneally and intranasally infected adult outbred mice. J. Virol. 72:1219-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kajon, A. E., and K. R. Spindler. 2000. Mouse adenovirus type 1 replication in vitro is resistant to interferon. Virology 274:213-219. [DOI] [PubMed] [Google Scholar]
- 36.Kring, S. C., A. O. Ball, and K. R. Spindler. 1992. Transcription mapping of mouse adenovirus type 1 early region 4. Virology 190:248-255. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 38.Lang, S. E., and P. Hearing. 2003. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene 22:2836-2841. [DOI] [PubMed] [Google Scholar]
- 39.Lium, E. K., C. A. Panagiotidis, X. Wen, and S. J. Silverstein. 1998. The NH2 terminus of the herpes simplex virus type 1 regulatory protein ICP0 contains a promoter-specific transcription activation domain. J. Virol. 72:7785-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mo, X., E. Kowenz-Leutz, H. Xu, and A. Leutz. 2004. Ras induces mediator complex exchange on C/EBP beta. Mol. Cell 13:241-250. [DOI] [PubMed] [Google Scholar]
- 41.Moore, D. M., L. Zsak, J. G. Neilan, Z. Lu, and D. L. Rock. 1998. The African swine fever virus thymidine kinase gene is required for efficient replication in swine macrophages and for virulence in swine. J. Virol. 72:10310-10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moran, E., and M. B. Mathews. 1987. Multiple functional domains in the adenovirus E1A gene. Cell 48:177-178. [DOI] [PubMed] [Google Scholar]
- 43.Nevins, J. R. 1981. Mechanism of activation of early viral transcription by the adenovirus E1A gene product. Cell 26:213-220. [DOI] [PubMed] [Google Scholar]
- 44.O'Connor, R. J., and P. Hearing. 2000. The E4-6/7 protein functionally compensates for the loss of E1A expression in adenovirus infection. J. Virol. 74:5819-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oliveira, S. A., and T. E. Shenk. 2001. Murine cytomegalovirus M78 protein, a G protein-coupled receptor homologue, is a constituent of the virion and facilitates accumulation of immediate-early viral mRNA. Proc. Natl. Acad. Sci. USA 98:3237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reichel, R., S. D. Neill, I. Kovesdi, M. C. Simon, P. R. Raychaudhuri, and J. R. Nevins. 1989. The adenovirus E4 gene, in addition to the E1A gene, is important for trans-activation of E2 transcription and for E2F activation. J. Virol. 63:3643-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarawar, S. R., R. D. Cardin, J. W. Brooks, M. Mehrpooya, R. A. Tripp, and P. C. Doherty. 1996. Cytokine production in the immune response to murine gammaherpesvirus 68. J. Virol. 70:3264-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaeper, U., J. M. Boyd, S. Verma, E. Uhlmann, T. Subramanian, and G. Chinnadurai. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. USA 92:10467-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaley, J., R. J. O'Connor, L. J. Taylor, D. Bar-Sagi, and P. Hearing. 2000. Induction of the cellular E2F-1 promoter by the adenovirus E4-6/7 protein. J. Virol. 74:2084-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shenk, T., N. Jones, W. Colby, and D. Fowlkes. 1979. Functional analysis of adenovirus-5 host-range deletion mutants defective for transformation of rat embryo cells. Cold Spring Harb. Symp. Quant. Biol. 44:367-375. [DOI] [PubMed] [Google Scholar]
- 51.Shenk, T. E. 2001. Adenoviridae: The viruses and their replication, p. 2265-2300. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 52.Smith, K., C. C. Brown, and K. R. Spindler. 1998. The role of mouse adenovirus type 1 early region 1A in acute and persistent infections in mice. J. Virol. 72:5699-5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, K., B. Ying, A. O. Ball, C. W. Beard, and K. R. Spindler. 1996. Interaction of mouse adenovirus type 1 early region 1A protein with cellular proteins pRb and p107. Virology 224:184-197. [DOI] [PubMed] [Google Scholar]
- 54.Spindler, K. R., C. Y. Eng, and A. J. Berk. 1985. An adenovirus early region 1A protein is required for maximal viral DNA replication in growth-arrested human cells. J. Virol. 53:742-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spindler, K. R., L. Fang, M. L. Moore, C. C. Brown, G. N. Hirsch, and A. K. Kajon. 2001. SJL/J. mice are highly susceptible to infection by mouse adenovirus type 1. J. Virol. 75:12039-12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevens, J. L., G. T. Cantin, G. Wang, A. Shevchenko, A. Shevchenko, and A. J. Berk. 2002. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296:755-758. [DOI] [PubMed] [Google Scholar]
- 57.Wang, G., and A. J. Berk. 2002. In vivo association of adenovirus large E1A protein with the human mediator complex in adenovirus-infected and -transformed cells. J. Virol. 76:9186-9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, H. G., Y. Rikitake, M. C. Carter, P. Yaciuk, S. E. Abraham, B. Zerler, and E. Moran. 1993. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J. Virol. 67:476-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, L., D. S. E. Rosser, M. C. Smith, and A. Berk. 1987. A TATA box implicated in E1A transcriptional activation of a simple adenovirus 2 promoter. Nature 326:512-515. [DOI] [PubMed] [Google Scholar]
- 60.Yee, S. P., and P. E. Branton. 1985. Detection of cellular proteins associated with human adenovirus type 5 early region 1A polypeptides. Virology 147:142-153. [DOI] [PubMed] [Google Scholar]
- 61.Ying, B., K. Smith, and K. R. Spindler. 1998. Mouse adenovirus type 1 early region 1A is dispensable for growth in cultured fibroblasts. J. Virol. 72:6325-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]