Abstract
We compared the infection of bone marrow macrophages by the DA and GDVII strains of Theiler's virus and by two viruses constructed by exchanging the DA and GDVII capsids. The replication of the GDVII strain and of both chimeric viruses was restricted in macrophages. Therefore, the infection of macrophages requires both capsid and noncapsid viral determinants.
The DA strain of Theiler's murine encephalomyelitis virus (TMEV) causes a biphasic disease of the central nervous system (11). During the first phase TMEV infects neurons, causing an acute encephalomyelitis, whereas during the second phase it infects mainly macrophages and microglia (5, 14, 20), and also oligodendrocytes and astrocytes (1), in the white matter of the spinal cord. This second phase persists for life and causes primary demyelination, which is studied as a model for multiple sclerosis. In contrast, the GDVII strain kills most mice during the acute phase and does not persist or induce demyelination (10). Attenuated GDVII mutants do not persist, indicating that attenuation and persistence are distinct phenotypes (9, 13).
DA virus persistence has been shown, by using mutant and recombinant viruses, to be linked to capsid and noncapsid determinants (12). One of the noncapsid determinants might be L*, a nonstructural protein expressed only by persistent strains (18, 19, 21, 22). However, the relative importance of capsid and noncapsid determinants for the infection of macrophages, the main reservoir during persistence, has not been studied in detail. In a previous publication, our investigators described infection by the DA strain of macrophages derived from the bone marrow of SJL/J mice (16). Because these cells come from a mouse strain susceptible to persistent infection and are not immortalized, they are an advantageous alternative to macrophage-like cell lines. In the present work, we infected them with the DA and GDVII viruses and with two chimeric viruses obtained by exchanging the capsid coding regions of these two viruses.
The replication of GDVII virus is restricted in macrophage primary cultures.
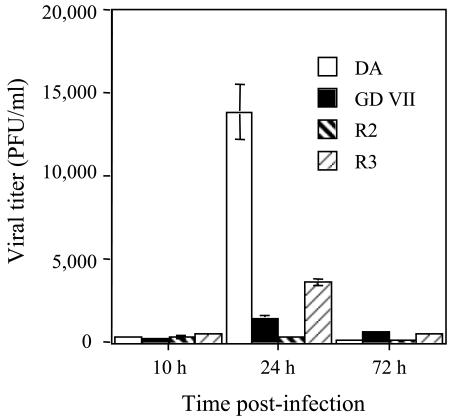
Bone marrow-derived macrophages were prepared from 8- to 9-week-old female SJL/J mice as described elsewhere (16). Briefly, red blood cell-depleted bone marrow cells were grown in RPMI 1640, 10% fetal bovine serum (FBS), and 10% L929 cell-conditioned medium as a source of macrophage colony-stimulating factor. After 6 days, clusters of nonadherent cells were harvested, and 2 × 105 cells were plated on 12-mm-diameter glass coverslips and incubated in RPMI 1640 medium, 10% FBS, and 2.5% L929-conditioned medium. After at least 24 h, each coverslip was washed with medium without serum, incubated for 2 h at 37°C with 106 PFU of virus, washed three times with warm medium, and then incubated with 500 μl of RPMI 1640, 2% FBS, and 2.5% L929-conditioned medium. Virus infectivity was measured in the medium by a standard plaque assay on BHK-21 cells. Figure 1 shows that the titer of DA virus increased about 10-fold between 10 and 24 h postinfection (p.i.) and then declined. The GDVII titers were considerably lower. We suspect that part of this infectivity corresponded to residual input virus, since similar titers were detected as early as 5 h p.i. (data not shown). The restricted expression of GDVII virus is striking, since in BHK-21 cells and in primary cultures of neurons its yield is 5 to 10 times higher than that of the DA or BeAn virus (8; I. Mena, unpublished observations).
FIG. 1.
Viral production in bone marrow-derived macrophages. Primary cultures were infected with viruses DA, GDVII, R2, and R3. Infectivity in the medium was measured by a plaque assay on BHK-21 cells. Each point represents the mean ± standard error of the mean of 10 individual samples from three independent experiments.
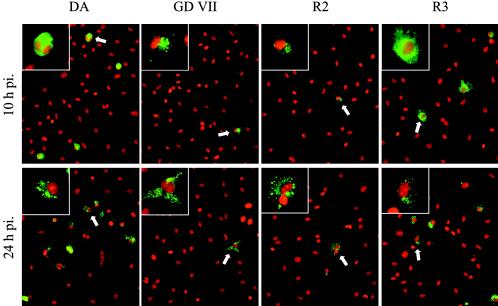
The cells on the coverslips were fixed in 4% paraformaldehyde (PFA), washed in phosphate-buffered saline (PBS), treated for 5 min with 0.1% Triton X-100 in PBS, and then blocked in 10% normal goat serum. Viral antigens were detected with a rabbit polyclonal serum (2) and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin (Jackson Immuno-Research Laboratories). Cell nuclei were counterstained with 0.2 μM ethidium homodimer 1 (Sigma-Aldrich). The slides were examined with a Zeiss Axioplan 2 microscope equipped with a Coolsnap HQ charge-coupled device camera, and the images were acquired by using Simple PCI software (Hamamatsu). A minimum of 5,000 cells from at least 10 individual microscopic fields from four independent experiments were examined. More than 6% of the macrophages contained DA antigens 10 h p.i., whereas this value was 3% for the GDVII virus (Table 1; Fig. 2). The percentage of DA antigen-positive macrophages increased between 10 and 24 h p.i. (P < 0.005), whereas that of GDVII-positive cells did not. The intracellular distribution of DA antigens changed with time (Fig. 2) (16). At 10 h p.i., DA virus fluorescence was intense and uniformly distributed throughout the cytoplasm, whereas at 24 h p.i. and later, viral antigens often formed cytoplasmic inclusions or showed a punctate pattern. Interestingly, this dichotomy has been described in macrophages in vivo during persistent infection (4, 14). GDVII antigens, on the other hand, gave only a weak and punctate fluorescence.
TABLE 1.
Virus-positive macrophages in primary culturesa
| Virus strain | % Virus-positive macrophages
|
|
|---|---|---|
| 10 h p.i. | 24 h p.i. | |
| DAb | 6.22 ± 0.57 | 10.89 ± 1.18 |
| GD VIIc | 3.28 ± 0.65 | 1.17 ± 0.18 |
| R2c | 2.17 ± 0.28 | 1.14 ± 0.25 |
| R3b | 3.72 ± 0.45 | 5.17 ± 1.05 |
Viral antigens were detected by immunofluorescence (see Fig. 2). Nuclei were stained with ethidium homodimer 1. The percentage of virus positive-cells was measured 10 and 24 h p.i. All cells containing green fluorescent dots were counted as virus positive. The table shows the results of four independent experiments. Values are means ± standard errors of the means.
At 10 h p.i., positive cells had intense, uniform cytoplasmic labeling (TMEVhi).
The fluorescent signal at 10 h p.i. was mainly in a weak, punctate pattern (TMEVlo).
FIG. 2.
Detection of viral capsid antigens by immunofluorescence. Cell nuclei were counterstained with ethidium homodimer 1 (red). A representative field, observed at an original magnification of 25×, is shown for each viral strain. One representative infected cell, identified by a white arrow, is shown at higher magnification (×63) in the upper left quadrant to show the difference in intracellular distribution of the viral capsid proteins. Longer exposure times were needed to photograph the R2- and GDVII-infected samples; therefore, fluorescence intensities cannot be compared directly.
The ability to infect macrophages is due to both capsid and noncapsid determinants.
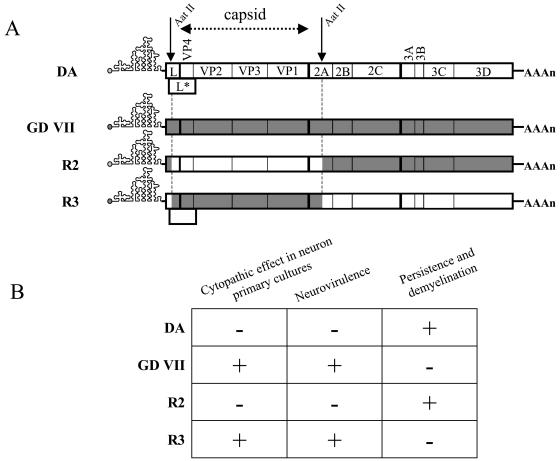
Viruses R2 and R3 were obtained by exchanging an AatII/AatII restriction fragment, containing the entire capsid coding region and small parts of the flanking regions, between the DA and GDVII infectious cDNA (Fig. 3) (17). The 5′ crossover point of these chimera is located 122 nucleotides downstream from the start codon of the L* open reading frame. Therefore, virus R2 has the DA capsid but does not express L*, whereas virus R3 has the GDVII capsid and expresses the L* protein. Virus R3 spreads efficiently in neurons in vitro, is highly neurovirulent in vivo, and does not persist. Virus R2 infects neurons much less efficiently, has low neurovirulence in vivo, and persists (8). Bone marrow-derived macrophages were infected in parallel with viruses DA, GDVII, R2, and R3 (Table 1; Fig. 1 and 2). Virus R2 had essentially the same phenotype as virus GDVII, whereas the phenotype of virus R3 was intermediate between that of its parental viruses. The titers of viruses R3 and DA increased significantly between 10 and 24 h (P < 0.005), whereas there was no increase for viruses R2 and GDVII. At 24 h p.i., the yield of virus R3 was lower than that of virus DA but higher than those of viruses R2 and GDVII (P < 0.005). Immunofluorescence results were consistent with these data (Table 1; Fig. 2). Ten percent and 5% of the cells were positive for DA and R3 viral antigens, respectively, at the peak of virus production, whereas the percentages were lower for viruses GDVII and R2 at all times examined. DA and R3 antigens had a uniform, intense, fluorescence pattern at early times and a punctate distribution later on. In contrast, GDVII and R2 antigens showed only the punctate pattern.
FIG. 3.
Characteristics of the parental and chimeric viruses. (A) Genetic organization of the viral strains used in this study. Viruses R2 and R3 were obtained by exchanging the AatII-AatII restriction fragment between the cDNAs of the parental DA and GDVII strains. (B) Phenotype of the four viruses in neuron primary cultures and in vivo.
Flow cytometric analysis of macrophage permissiveness to viral replication.
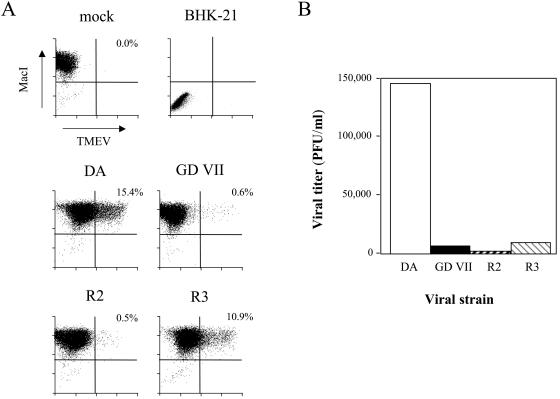
The expression of capsid antigens was quantified by flow cytometry. Single-cell suspensions of infected macrophages were obtained using Dispase II (Roche) (15), fixed in 4% PFA, incubated in PBS, 0.5% saponin, and 0.5% normal goat serum, and then reacted with the anti-TMEV polyclonal rabbit serum and a biotin-conjugated rat monoclonal antibody against the CD11b macrophage marker (Pharmingen). FITC-conjugated goat anti-rabbit immunoglobulin (Jackson Immuno-Research Laboratories) and streptavidin-CyChrome (Pharmingen) were used as secondary reagents. More than 99% of the cells expressed CD11b. BHK-21 cells, a control, were entirely CD11b negative (Fig. 4). DA and R3 viral antigens were expressed by 15.4 and 10.9% of the cells, respectively, while GDVII and R2 antigens were expressed by less than 1% of the cells. The mean fluorescence intensity per positive cell was higher for R3 virus than for DA virus. Therefore, although the percentage of infected cells and the viral yield were lower for virus R3 than for virus DA, on a single-cell basis, the expression of capsid protein was similar for both viruses or even higher for virus R3.
FIG. 4.
Analysis of TMEV-infected macrophage primary cultures by flow cytometry. Macrophages were seeded on six-well plates and infected at a multiplicity of infection of 5 PFU/cell. (A) Cells were harvested 24 h p.i. with Dispase II, fixed, permeabilized, and labeled with an anti-TMEV rabbit serum and a biotinylated rat monoclonal anti-MacI (CD11b) antibody. (B) The medium was harvested 24 h p.i. for viral titration.
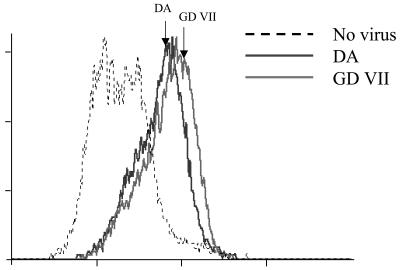
DA and GDVII viruses bind to macrophages with similar efficiency.
These results suggested that both capsid and noncapsid determinants are important for the infection of SJL/J macrophages. We examined the role of the capsid more directly by comparing the binding of the viruses to macrophages. Single-cell suspensions of bone marrow-derived macrophages were incubated for 1 h at 4°C with 100 PFU of virus/cell, washed with ice-cold PBS, and fixed in 4% PFA. Bound virus was detected by flow cytometry as described above, but without permeabilization of the cells. Figure 5 shows that the DA and GDVII viruses bound equally well to macrophages. Similar results were obtained for the R2 and R3 viruses (data not shown). Binding to BHK-21 cells was the same for the four viruses (data not shown). Therefore, restricted expression of the GDVII and R3 viruses in macrophages is not due to poor attachment to the cells. This is congruent with the observations reported above, which suggest that noncapsid determinants contribute more to the infection of macrophages than capsid determinants.
FIG. 5.
Virus cell binding assay. Macrophage single-cell suspensions were obtained from primary cultures, incubated for 1 h at 4°C with 100 PFU of virus/cell, washed extensively with ice-cold PBS, and fixed with 4% PFA. Fixed cells were reacted with an anti-TMEV polyclonal serum followed by a FITC-conjugated goat anti-rabbit immunoglobulin. Labeled cells were analyzed using a FACSCalibur flow cytometer and the CellQuest software. The abscissa is fluorescence intensity, and the ordinate is the number of events.
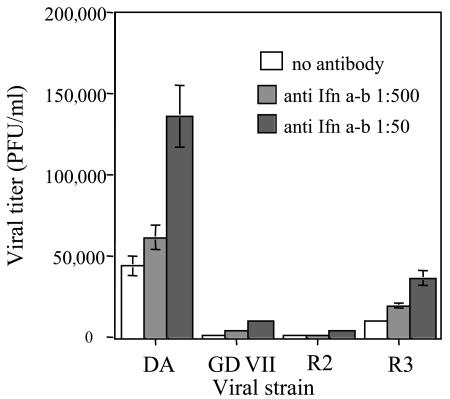
Neutralizing the alpha/beta interferon response does not overcome the restricted replication of the GDVII, R2, and R3 viruses. The L protein of DA virus inhibits the production of α4 and β interferons (23). L is the most polymorphic gene between the DA and GDVII viruses, and we do not know if the L protein of GD VII virus has an anti- interferon activity. Therefore, we examined if restricted replication of viruses GDVII, R2, and R3 (which have hybrid L genes) in macrophages could be due to inefficient inhibition of interferon production by their L proteins. Macrophages were infected with the different viruses and washed three times, and a 1:50 or 1:500 dilution of a neutralizing anti-alpha/beta interferon serum (I. Gresser, Institut Curie, Paris, France) was added to the medium. Viral yields were measured 24 h p.i., at the peak of virus production. For the four viruses, the presence of the antibody increased the titer in a dose-dependent manner (Fig. 6). Neutralizing interferon did not change the ratios between the infectivity titers produced by each strain. Therefore, the production of interferon and/or the susceptibility to interferon was similar for the four viruses. The more-restricted yields of viruses GD VII, R2, and R3 are not explained by an increased sensitivity to the interferon response.
FIG. 6.
Effect of alpha/beta interferon neutralization on viral yield. Macrophages were infected with the four viral strains. After washing the inoculum, the medium was replaced with 400 μl of medium containing the indicated dilutions of neutralizing anti-alpha/beta interferon antibody. The medium was harvested 24 h p.i., and the infectivity was measured by plaque assay. Each point represents the mean and standard error of the mean of five individual samples from two independent experiments.
R2 virus does not infect macrophages efficiently in vivo.
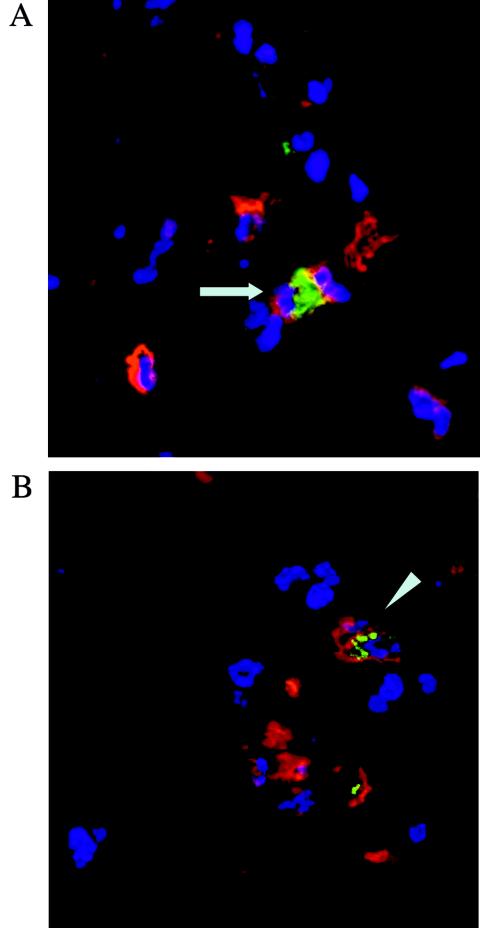
Virus DA persists in vivo in macrophages. Virus R2, which bears the DA capsid but lacks L*, also persists in vivo although it infects macrophages inefficiently in vitro. Therefore, we asked whether the R2 virus persists in macrophages or in another central nervous system cell type. Groups of five SJL/J mice were inoculated intracerebrally with 5 × 104 or 5 × 105 PFU of either DA or R2 virus and sacrificed 45 days later. The mice were perfused with PBS followed by 2% PFA. The spinal cords were dissected, postfixed in 4% PFA, and incubated in 15% sucrose overnight. Tissue blocks were snap-frozen and cut on a cryostat to perform immunofluorescence. CD11b was detected with a rat monoclonal antibody and a Cy3-coupled secondary antibody. Viral capsid antigens were detected with a rabbit polyclonal serum and a goat anti-rabbit antibody coupled to Alexa 488. Figure 7 illustrates the two patterns of intracellular viral antigen (TMEVhi and TMEVlo) observed. TMEVhi corresponds to cells with intense diffuse fluorescence, and TMEVlo corresponds to cells with faint punctate fluorescence. Both patterns have already been observed in vivo (4, 14) as well as in primary bone marrow macrophages in vitro (reference 16 and this study). The sections were scanned systematically, and virus-positive cells were recorded for their CD11b+ or CD11b− phenotype and their TMEVhi or TMEVlo phenotype. The number of infected cells was much larger in DA-infected mice than in R2-infected mice. Table 2 shows that virus R2 persisted mainly in macrophages, almost all of them (97%) with a TMEVlo phenotype, whereas DA virus persisted in macrophages with either a TMEVlo (44%) or a TMEVhi (56%) phenotype. Therefore, both viruses persisted in macrophages, but the number of infected macrophages and the level of expression of viral antigen per macrophage were much lower for virus R2 than for virus DA.
FIG. 7.
Detection of viral capsid antigens by immunofluorescence in frozen sections of spinal cord. Mice were sacrificed 45 days p.i. Sections were reacted with an anti-CD11b antibody (red) and an anti-TMEV antibody (green), and nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (blue). (A) DA virus-infected SJL/J mouse; (B) R2-infected SJL/J mouse. Arrow, TMEVhi cell; arrowhead, TMEVlo cell.
TABLE 2.
Virus-positive macrophages in vivoa
| Virus strain | % Virus-positive macrophages
|
||
|---|---|---|---|
| TMEVhi MacI+ cells | TMEVlo MacI+ cells | TMEV+ MacI− cells | |
| DA | 26 ± 3 | 21 ± 4 | 53 ± 1 |
| R2 | 2 ± 1 | 82 ± 8 | 16 ± 6 |
The mice were sacrificed 45 days after intracerebral inoculation. Viral and CD11b antigens were detected by immunofluorescence in frozen sections of longitudinal spinal cord. Nuclei were stained with 4′,6′-diamidino-2-phenylindole (see Fig. 6). The sections were scanned systematically, and virus-positive cells were recorded for their CD11b+ or CD11b− phenotype and their TMEV hi or lo phenotype. TMEVhi corresponds to cells with intense diffuse fluorescence; TMEVlo corresponds to cells with faint punctate fluorescence. Values are means ± standard errors of the means.
Figure 5 shows that the DA and GDVII strains bind equally well to macrophages. A similar result was reported for the binding of the BeAn and GDVII strains to the macrophage-derived P388D1 line (6). DA and R2 viruses, which have the same capsid, gave the highest and the lowest yields, respectively, of infectivity in macrophages (Fig. 1). Therefore, the capsid does not seem to play an essential role in the tropism of DA virus for macrophages. This contrasts with its major role in tropism for neurons and in in vivo neurovirulence (3, 7, 17). On the other hand, our results corroborate the notion that the L* protein is important for the infection of macrophages, since the DA and R3 viruses, which express L*, replicated in macrophages better than the GDVII and R2 viruses, which lack the L* AUG (Fig. 4). Interestingly, virus R3, whose yield is lower that that of DA virus, has a chimeric L*. The titers produced by viruses R2 and R3 did not add up to the yield of DA virus, suggesting capsid and noncapsid determinants interact to give DA virus its full phenotype. We found that, although it lacks L*, virus R2 persists in vivo in macrophages. However, the number of infected macrophages and the level of capsid expression in these cells were much lower than for virus DA (Fig. 7; Table 2). Therefore, our results confirm that infecting macrophages is important for TMEV's persistence and that L* plays a role in this phenotype, both in vitro and in vivo.
In summary, we showed that the replication of strain GDVII is highly impaired in bone marrow-derived macrophages compared to that of strain DA. The results obtained with two chimeric viruses suggest that genetic determinants in the capsid and the noncapsid regions must coexist in the viral genome to allow efficient infection of SJL/J macrophages and that protein L* has an important role in this phenotype.
Acknowledgments
We thank Emmanuelle Perret from the Centre d'Imagerie Dynamique (Institut Pasteur, Paris, France) for her help with the immunofluorescence imaging, Ion Gresser (Institut Curie) for the neutralizing anti-interferon serum, Galina Karachtchouk for unpublished experimental data, Jean-François Bureau and Thomas Michiels for critically reading the manuscript, and Mireille Gau for secretarial assistance.
I. Mena held fellowships from EMBO, the Fondation pour la Recherche Médicale, and the Association pour la Recherche sur le Cancer. Work on Theiler's virus in the Brahic laboratory is supported by the Institut Pasteur, the Centre National de la Recherche Scientifique, the National Multiple Sclerosis Society, and the Association pour la Recherche sur la Sclérose en Plaques.
REFERENCES
- 1.Aubert, C., M. Chamorro, and M. Brahic. 1987. Identification of Theiler's virus infected cells in the central nervous system of the mouse during demyelinating disease. Microb. Pathog. 3:319-326. [DOI] [PubMed] [Google Scholar]
- 2.Brahic, M., A. T. Haase, and E. Cash. 1984. Simultaneous in situ detection of viral RNA and antigens. Proc. Natl. Acad. Sci. USA 81:5445-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calenoff, M. A., K. S. Faaberg, and H. L. Lipton. 1990. Genomic regions of neurovirulence and attenuation in Theiler's murine encephalomyelitis virus. Proc. Natl. Acad. Sci. USA 87:978-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cash, E., M. Chamorro, and M. Brahic. 1985. Theiler's virus RNA and protein synthesis in the central nervous system of demyelinating mice. Virology 144:290-294. [DOI] [PubMed] [Google Scholar]
- 5.Clatch, R. J., S. D. Miller, R. Metzner, M. C. Dal Canto, and H. L. Lipton. 1990. Monocytes/macrophages isolated from the mouse central nervous system contain infectious Theiler's murine encephalomyelitis virus (TMEV). Virology 176:244-254. [DOI] [PubMed] [Google Scholar]
- 6.Fotiadis, C., D. R. Kilpatrick, and H. L. Lipton. 1991. Comparison of the binding characteristics to BHK-21 cells of viruses representing the two Theiler's virus neurovirulence groups. Virology 182:365-370. [DOI] [PubMed] [Google Scholar]
- 7.Fu, J., S. Stein, L. Rosenstein, T. Bodwell, M. Routbort, B. L. Semler, and R. P. Roos. 1990. Neurovirulence determinants of genetically engineered Theiler's virus. Proc. Natl. Acad. Sci. USA 87:4125-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarousse, N., S. Syan, C. Martinat, and M. Brahic. 1998. The neurovirulence of the DA and GDVII strains of Theiler's virus correlates with their ability to infect cultured neurons. J. Virol. 72:7213-7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarousse, N., E. G. Viktorova, E. V. Pilipenko, V. I. Agol, and M. Brahic. 1999. An attenuated variant of the GDVII strain of Theiler's virus does not persist and does not infect the white matter of the central nervous system. J. Virol. 73:801-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipton, H. L. 1980. Persistent Theiler's murine encephalomyelitis virus infection in mice depends on plaque size. J. Gen. Virol. 46:169-177. [DOI] [PubMed] [Google Scholar]
- 11.Lipton, H. L. 1975. Theiler's virus infection in mice: an unusual biphasic disease process leading to demyelination. Infect. Immun. 11:1147-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipton, H. L., and M. L. Jelachich. 1997. Molecular pathogenesis of Theiler's murine encephalomyelitis virus-induced demyelinating disease in mice. Intervirology 40:143-152. [DOI] [PubMed] [Google Scholar]
- 13.Lipton, H. L., A. E. Pritchard, and M. A. Calenoff. 1998. Attenuation of neurovirulence of Theiler's murine encephalomyelitis virus strain GDVII is not sufficient to establish persistence in the central nervous system. J. Gen. Virol. 79:1001-1004. [DOI] [PubMed] [Google Scholar]
- 14.Lipton, H. L., G. Twaddle, and M. L. Jelachich. 1995. The predominant virus antigen burden is present in macrophages in Theiler's murine encephalomyelitis virus-induced demyelinating disease. J. Virol. 69:2525-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luder, C. G., T. Lang, B. Beuerle, and U. Gross. 1998. Down-regulation of MHC class II molecules and inability to up-regulate class I molecules in murine macrophages after infection with Toxoplasma gondii. Clin. Exp. Immunol. 112:308-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinat, C., N. Mena, and M. Brahic. 2002. Theiler's virus infection of primary cultures of bone marrow-derived monocytes/macrophages. J. Virol. 76:12823-12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McAllister, A., F. Tangy, C. Aubert, and M. Brahic. 1990. Genetic mapping of the ability of Theiler's virus to persist and demyelinate. J. Virol. 64:4252-4257. (Author's correction, 67:2427, 1993.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michiels, T., N. Jarousse, and M. Brahic. 1995. Analysis of the leader and capsid coding regions of persistent and neurovirulent strains of Theiler's virus. Virology 214:550-558. [DOI] [PubMed] [Google Scholar]
- 19.Obuchi, M., J. Yamamoto, T. Odagiri, M. N. Uddin, H. Iizuka, and Y. Ohara. 2000. L* protein of Theiler's murine encephalomyelitis virus is required for virus growth in a murine macrophage-like cell line. J. Virol. 74:4898-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pena Rossi, C., M. Delcroix, I. Huitinga, A. McAllister, N. van Rooijen, E. Claassen, and M. Brahic. 1997. Role of macrophages during Theiler's virus infection. J. Virol. 71:3336-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takata, H., M. Obuchi, J. Yamamoto, T. Odagiri, R. P. Roos, H. Iizuka, and Y. Ohara. 1998. L* protein of the DA strain of Theiler's murine encephalomyelitis virus is important for virus growth in a murine macrophage-like cell line. J. Virol. 72:4950-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Eyll, O., and T. Michiels. 2000. Influence of the Theiler's virus L* protein on macrophage infection, viral persistence, and neurovirulence. J. Virol. 74:9070-9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Pesch, V., O. van Eyll, and T. Michiels. 2001. The leader protein of Theiler's virus inhibits immediate-early alpha/beta interferon production. J. Virol. 75:7811-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]