Abstract
Concerted tandem and traveling wave ion mobility mass spectrometry (CTS analysis) is a unique method that results in a 4-dimensional dataset including nominal precursor ion mass, product ion mobility, accurate mass of product ion, and ion abundance. This non-targeted lipidomics CTS approach was applied in both positive and negative ion mode to phospholipids present in human serum and the data set was used to evaluate the value of product ion mobility in identifying lipids in a complex mixture. It was determined that the combination of diagnostic product ions and unique collisional cross section values of product ions is a powerful tool in the structural identification of lipids in a complex biological sample.
Graphical Abstract
INTRODUCTION
Lipids are a diverse family of biomolecules that play a critical structural role within the cell, are responsible for energy storage, and serve as precursors for a wide variety of biologically active lipid mediators.1,2 Lipidomic studies to investigate disease biology and identify biomarkers are prevalent due to the fact that lipids are known to play a role in many diseases, such as atherosclerosis, obesity, and diabetes.3–5 However, lipidomic studies are challenging due to the diversity of lipid molecular species present in a biological sample. The complexity of lipids in biological samples is attributed to the presence of a variety of lipid classes, such as neutral lipids, phospholipids, and sphingolipids.6,7 Additionally, greater complexity is added due to a variety of possible fatty acyl constituents that differ by both number of acyl carbons and double bonds.
Shotgun lipidomics is one of the most common lipidomic experiments, where a lipid extract is infused using electrospray ionization mass spectrometry and lipids are identified using accurate mass or by using MS/MS to produce diagnostic product ions.8,9 One of the challenges of direct infusion shotgun lipidomics is the accurate determination of ubiquitous isobaric species. Traditionally, liquid chromatography has been used in an attempt to circumvent this issue, but this can be time consuming and reduces the high throughput nature of the experiment. More recently ion mobility of precursor ions of lipid molecular species has been used as a separation parameter to improve the quality of lipidomic data.10 In an ion mobility experiment, ions are separated in the mobility cell according to the size, shape, and charge and the drift time of the lipid can be used to calculate the collisional cross section (CCS). These previous studies using ion mobility of the precursor lipid molecular ions have illustrated an increase in the specificity and selectivity of the lipidomic analysis and an improvement in the confidence in identification of lipid molecular species.11–15
While the ion mobility of lipid molecular species has been utilized as an additional molecular identifier, only limited information is available concerning the ion mobility of the product ions derived from these compounds. In a previous study, Castro-Perez used time-aligned parallel fragmentation in combination with product ion mobility to obtain information about the double bond position and acyl location of fatty acids on phospholipids in plasma.16 The present study was undertaken using a strategy to determine the ion mobility of product ions in order to assist in the structural identification of lipids from a complex biological sample. The general approach was to use concerted tandem mass spectrometry and traveling wave ion mobility (CTS analysis), which is a unique method that results in a 4-dimensional dataset that includes nominal precursor ion mass, product ion mobility, accurate mass of product ion, and ion abundance.17 We have applied this strategy to the study of phospholipids present in a human serum sample and evaluate the value of product ion mobility in identifying lipids in a complex mixture using a non-targeted lipidomics approach.
EXPERIMENTAL
Materials
Lipid ester standards (PC(17:0/20:4), PE(17:0/20:4), PA(17:0/20:4), PS(17:0/20:4), PI(17:0/20:4), and SM(d18:1/18:0)) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Solvents and chemicals used for extraction and mass spectrometry were HPLC grade, purchased from Fisher Scientific Company (Pittsburgh, PA), and were used without further purification. Hanks Balanced Salt Solution (HBSS) was purchased from Mediatech (Manassas, VA). Human serum, SRM 909C, was obtained from the National Institute of Standards and Technology (Gaithersburg MD). Polyalanine was obtained from Sigma (St. Louis MO).
Human serum, extraction protocol
An aliquot (50 uL) of NIST serum was thawed and lipids were extracted by a modified Bligh and Dyer extraction.18 The serum was diluted to 0.4 mL with HBSS followed by the addition of 1 mL of methanol and 0.5 mL of chloroform to make a monophase. After vortexing and sonication, an additional 0.4 mL of HBSS and 0.5 mL of chloroform was added to create a biphasic solution. The mixture was vortexed and centrifuged after which the bottom layer was removed to a separate test tube. The organic layer was dried under nitrogen gas and subsequently resuspended in 5 mL of an infusion solvent (methanol: acetonitrile: 2 mM aqueous ammonium acetate (60:20:20, v/v/v)).
Lipid standards and serum analysis
Lipids were diluted in the above infusion solvent system to approximately 1 ng/mL and infused into the electrospray source of the mass spectrometer by a syringe pump at a flow rate of 2 uL/min and data acquisition was begun as previously described.17 Briefly, the sample was infused for 16.7 min while a WREnS (Waters Research Enabled Software, WREnS; Waters Corp) script sequentially ramped the quadrupole precursor mass setting corresponding to each nominal m/z from 200 to 1200 at 1.001 Da/s. The acquisition system was programmed to repeatedly accumulate 200 TOF spectra over the course of the ion mobility drift time (~10 ms). Collisional activation of all ions was carried out in the “trap region” of the triwave sector and ion mobility of all product ions was collected in 200 asynchronous time segments followed by TOF measurements using a combined orthogonal push/pull voltage was around 3300 V (pusher voltage 1900 V and puller voltage 1370 V) at 18.5 KHz.
Mass spectrometry
Mass spectrometry and ion mobility measurements obtained by CTS analysis17 were carried out using a Synapt G2-S instrument (Waters, Manchester, UK) in either negative or positive ion mode. The mass spectrometer parameters used for the negative and positive ion CTS experiments are shown in Table S1. The ion mobility wave velocity was 550 m/s and the ion mobility wave height was 40 V in both polarities. Collision induced dissociation (CID) was carried out with nitrogen as the collision gas and a collision energy of 35 V in negative ion mode and 23 V in positive ion mode. The CTS experiments were carried out in “resolution” mode and a mass defect of 1/1000 Da was added to each mass setting of the ramping quadrupole sector. The total product ion mass spectra obtained from the positive and negative ion CTS experiments were mass measured using the abundant product ion from PC lipids at m/z 184.0734 in positive mode and the abundant product ion from 18:2 fatty acid containing phospholipids at m/z 279.2329 in negative ion mode as lock mass calibrants. This calibration was then applied to each individual product ion spectrum obtained in the CTS experiment.
Data analysis
The data were analyzed using Masslynx and Driftscope software (Waters Corporation). The full set of data (precursor mass, product mass, ion mobility, and intensity) was accessed using Driftscope and transformed to a tabulated spreadsheet with all experimental values. Data was processed using custom code written in Python, version 3 (http://www.python.org) and visualized using Bokeh (http://www.bokeh.pydata.org).
CCS calculations
The ion mobility times of lipid precursor and product ions were determined experimentally at an ion mobility wave height 40 V, ion mobility wave velocity 550 m/sec, and with nitrogen as the ion mobility gas. The centroided ion mobility times were converted into CCS using previously published protocols11,19 with polyalanine (0.1 ng/µL) as a calibrant and are expressed in square Angstrom (Å2). The CCS values obtained from the ion mobility measurements of lipid product ions in the current study were highly reproducible in the traveling wave experiment (Table S2). The T-wave velocity, T-wave height, and IMS gas flow rate in the ion mobility cell were varied when optimizing the parameters for the CTS experiments. The drift times of the product and precursor ions did change with the different T-wave velocity, T-wave height, and IMS gas flow rate values, however the CCS values of the product and precursor ions obtained using polyalanine oligomers as a calibrant did not change. As it is well established that CCS values derived from ion mobility are highly reproducible and uninfluenced by instrument settings.11
RESULTS AND DISCUSSION
CTS analysis of lipid standards and human serum was carried out in both negative and positive ion mode and the resultant 4-dimensional dataset (nominal precursor ion mass, product ion mobility, accurate mass of the product ion, and ion abundance) was obtained for the lipids present in the sample. From the CTS dataset all tandem mass spectral analysis strategies such as precursor and product ion data could be extracted as well as complete MS/MS spectra for each phospholipid under investigation. Additionally, the unique ion mobility drift times of each product ion were also obtained in this experiment.
Phospholipids can be analyzed as either positive or negative molecular ions and CID of these molecular ions yields different information relevant to the nature of the intact precursor ion species and product ions. For example, CID of [M+H]+ of phospholipids yields product ions related to the polar headgroup while the most abundant product ions derived from CID of the [M−H]− of phospholipids correspond to the carboxylate anions and loss of a neutral fatty acid or ketene from one or both of the fatty acyl chains.20 The product ion mobilities in both positive and negative ion mode are very different from that of the precursor phospholipid ions, which is not only due to the lower m/z of the product ions, but also because the product ions tend to have more compact structures. The current study focused on the ion mobility of lipid product ions in negative and positive ion mode and the value of using ion mobility of product ions to identify phospholipids in a complex mixture.
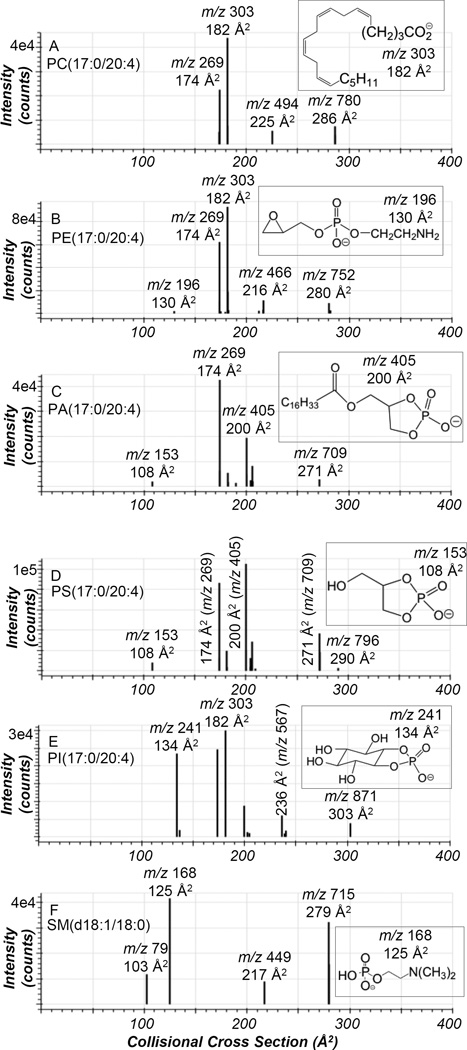
Negative ion CTS analysis of phospholipid standards (PC, PE, PA, PS, and PI) containing heptadecanoic acid (17:0) esterified to the sn-1 position of the glycerol backbone and arachidonic acid (20:4) at the sn-2 position of the glycerol backbone and the sphingomyelin standard, SM(d18:1/18:0), was completed (Figure 1). In the CTS analysis of phospholipid standards, the ions with largest CCS ranging from 271 to 303 Å2 corresponded to the molecular ion [M−H]− for PA, PI, PE, and PS and the [M−15]− ion for PC and SM. Even though each of the phospholipid standards has 17:0 at the sn-1 position and 20:4 esterified to sn-2 position of the glycerol backbone, different ion mobility was observed for each lipid class molecular ion due to the size and shape of the polar headgroup. Additionally, in the CTS analysis of PS(17:0/20:4) a product ion at m/z 709, which corresponded to the neutral loss of serine (87 Da), was observed and the resultant product ion had the same structure as the molecular ion of PA and therefore same CCS of 271 Å2. The dominant product ions in the CID spectra of each of these phospholipid standards obtained during the negative ion CTS analysis were at m/z 269 and 303, which corresponded to the carboxylate anions of 17:0 and 20:4. The CCS of these product ions at m/z 269 and 303 were 174 Å2 and 182 Å2.
Figure 1.
Ion mobilograms of the precursor and product ions obtained from negative ion CTS of (A) PC(17:0/20:4), (B) PE(17:0/20:4), (C) PA(17:0/20:4), (D) PS(17:0/20:4), (E) PI(17:0/20:4), and (F) SM(d18:1/18:0). The insets are structures of various product ions observed in the CTS experiment. The peaks are labeled with their nominal mass values.
Another common fragmentation pathway observed in the phospholipid standards during the negative ion CTS experiment corresponded to the loss of one fatty acid from the glycerol backbone. These particular product ions had a smaller CCS than the precursor ions, but a larger CCS than the fatty acids described above. Specifically, the loss of arachidonic acid from the sn-2 position of the glycerol backbone as a neutral ketene [M−H−R2C=O]− was observed leading to the 17:0 lyso product ion with a product ion mobility ranging from 206 to 239 Å2 (Figures 1A–E), depending on the headgroup of the lipid standard (Table S3). Additionally, another product ion that had CCS value in between that of the molecular ion and the fatty acids described above was observed in the negative ion CTS experiment of PI at m/z 567 and in PA and PS standards at m/z 405 (Figures 1C–E). These fragment ions correspond to the loss of arachidonic acid as a free carboxylic acid from the sn-2 position of PI [M−H−R2COO]− with a CCS of 236 Å2 and loss of AA from the sn-2 position of PA [M−H−R2COO]− and PS [M−H−87−R2COO]− with a CCS of 200 Å2. Additionally, the negative ion CTS experiment of the SM(d18:1/18:0) standard revealed a product ion at m/z 449 with a CCS of 217 Å2 (Figure 1F) that arises via cleavage of the amide bond and neutral loss of the 18:0 fatty acid.21
The negative ion CTS analysis of PI, PA, PS, PE, and SM standards also revealed product ions with low CCS values. For PI, the product ion with a low CCS of 134 Å2 corresponded to the characteristic cyclic phosphodiester of inositol (m/z 241), which is unique for PI lipids.22 In the CTS analysis of PA and PS, the product ion at m/z 153, which is a 1,2-cyclic phosphodiester of glycerol, had low CCS value of 108 Å2. This product ion at m/z 153 is observed in the CID of the [M−H]− of all phospholipids at high collision energy (~50 V),23 however the current analyses were performed at a collision energy of 35 V and this product ion was only observed in the PA and PS standards. The CTS analysis of PE in negative ion mode resulted in a product ion at m/z 196 with a CCS value of 130 Å2. This product ion is a diagnostic ion present in the CID of PE lipids in the negative ion mode and corresponds to a dehydrated phosphoethanolamine anion.24 Finally, in the negative ion CTS analysis of the SM standard, the major product ion occurred at m/z 168, which corresponds to the N-dimethylaminoethylphosphate anion with a CCS value of 125 Å2. Another prominent product ion at m/z 79 with a CCS value of 103 Å2 that originated from HPO3− was also observed. Due to the compact nature of these product ions at m/z 153, 168, 196, and 241 and the absence of any fatty acyl chains, low CCS were observed for these product ions.
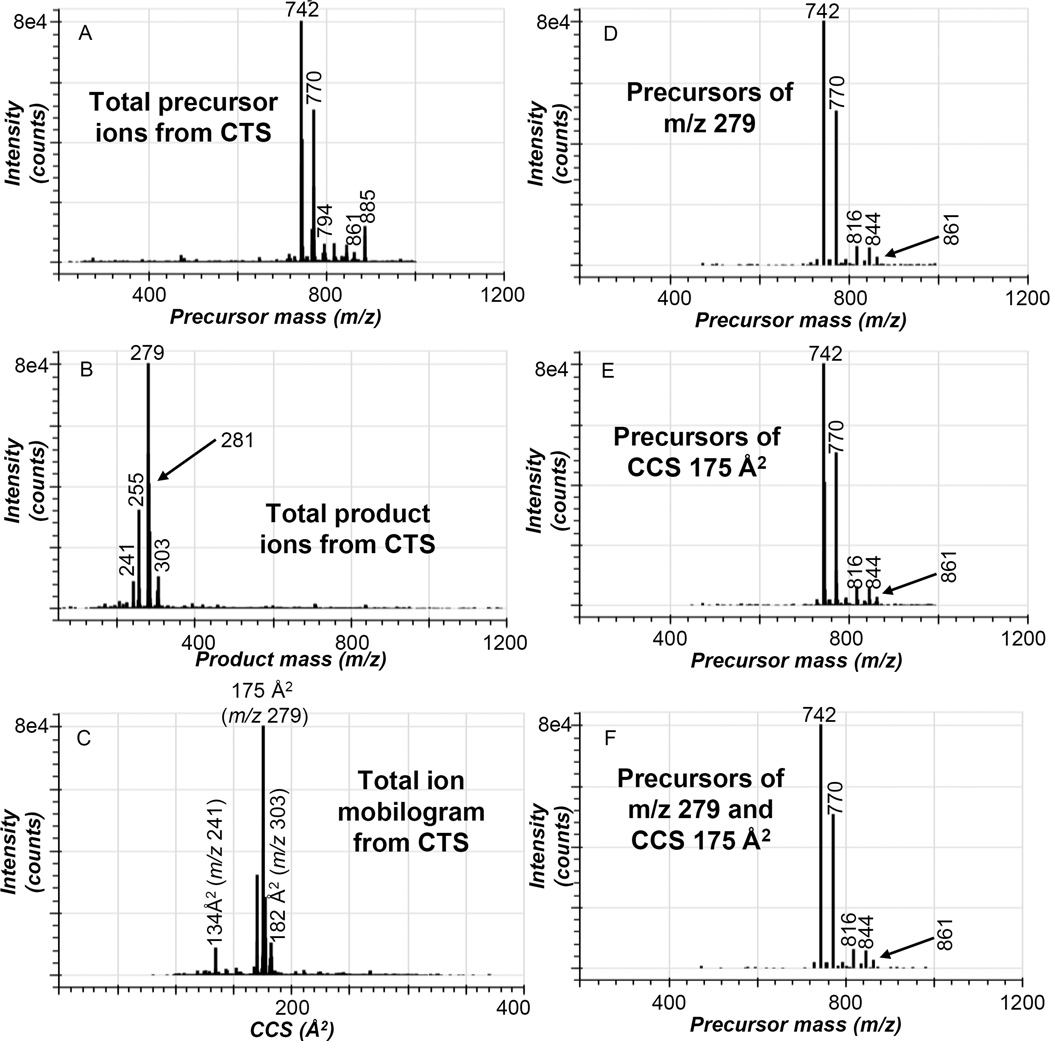
In order to determine if the identification of phospholipids within a complex biological sample can be confirmed with increased confidence by evaluation of the ion mobility (CCS) of the product ions, negative ion CTS analysis of the Bligh Dyer extract of human serum was performed. The precursor ions generated from this human serum sample revealed abundant ions in the m/z range of 700–900 Da (Figure 2A). The abundant precursor ions present in the negative ion CTS experiment corresponded to the [M−15]− of PC molecular species from m/z 700–800 Da as well as the acetate adducts of PC as [M+OAc]− and the [M−H]− of PI molecular species from m/z 800–900 Da. The carboxylate anion of linoleic acid at m/z 279 was the most abundant product ion in the CTS total product ion spectrum along with other carboxylate anions derived from palmitic (m/z 255), oleic (m/z 281) and arachidonic acid (m/z 303) (Figure 2B). Another fragment ion observed in the CTS total product ion spectrum occurred at m/z 241, which is the cyclic phosphodiester of inositol. The molecular ions of the phospholipids in the human serum sample were not observed in the CTS total product ion spectrum due to the higher collision energy used for these experiments. The total ion mobilogram of the product ions observed in the negative ion CTS experiment had an abundant peak originating from the carboxylate anion of linoleic acid at a CCS value of 175 Å2 (Figure 2C). Additionally the product ions at m/z 241, 255, 281, and 303 were also observed as robust peaks in the total ion mobilogram with CCS values of 134 Å2, 170 Å2, 176 Å2, and 182 Å2.
Figure 2.
Negative ion CTS experiment of human serum. (A) abundance of each precursor ion collected during the CTS experiment as a function of precursor ion m/z, (B) total product ion spectrum collected from the CTS experiment, (C) abundance of all ions collected from the CTS experiment as a function of CCS, (D) abundance of all precursor ions of m/z 279.233±10 ppm collected from the CTS experiment as a function of precursor ion m/z, (E) abundance of precursor ions of product ions with a CCS of 175±1 Å2 collected from the CTS experiment as a function of precursor ion m/z, and (F) abundance of all precursor ions of m/z 279.233±10 ppm and product ions with a CCS of 175±1 Å2 collected from the CTS experiment as a function of precursor ion m/z. Results shown are representative data obtained from four independent experiments.
Extraction of precursors of m/z 279.233±10 ppm from the negative ion CTS data revealed phospholipids containing linoleic acid including the [M−15]− and [M+OAc]− of PC(16:0 /18:2) at m/z 742 and 816, [M−15]− and [M+OAc]− of PC(18:0/18:2) at m/z 770 and 844, and the [M−H]− of PI(18:0/18:2) at m/z 861 (Figure 2D. Previous studies by Quehenberger et al. have reported the lipid composition of an averaged human serum sample made by combining the serum from 100 subjects.25 However, the relative abundances of some of the ions were different in the current study compared to the Quehenberger study because a single human serum sample was used in this study. The phospholipid molecular species present in human serum containing linoleic acid could also be extracted using their unique product ion CCS of 175 Å2. The precursors of product ions with a CCS value of 175±1 Å2 were extracted from the negative ion CTS data (Figure 2E) and that data was nearly identical to the precursors of m/z 279.233±10 ppm (Figure 2D). Additionally when the data is filtered in two dimensions for both precursors of m/z 279.233±10 ppm and precursors of product ions with a CCS value of 175±1 Å2 (Figure 2F), the data looked very similar to the individual extractions of the negative ion CTS data (Figures 2D and 2E).
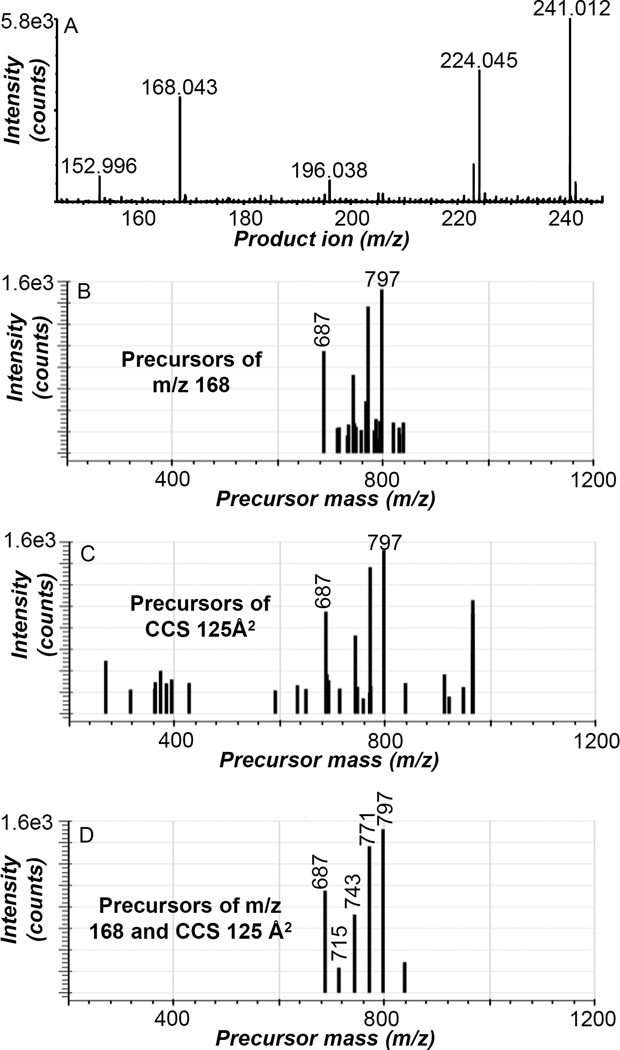
While the product ion CCS and precursor ion extracted data were very similar for the identification of phospholipids containing linoleic acid in human serum (Figures 2D–F), the product ion CCS became a valuable filter when the precursors of other product ions of less abundant lipids are extracted from the negative ion CTS data (Figure 3). The mass range of the negative ion CTS total product ion spectrum (Figure 2B) was expanded from m/z 145–245 Da and revealed various phospholipid-derived product ions observed during the negative ion CTS experiment (Figure 3A). For example, the product ion at m/z 168.043 (N-dimethylaminoethylphosphate anion) is a CID product of all SM lipids as described above. The precursor ions of m/z 168.043±10 ppm present in the negative ion CTS experiment of human serum is shown in Figure 3B with the most abundant ions at m/z 687, 771, and 797 along with various lower abundance ions from m/z 700–850 Da. The fragment ion at m/z 168.043 had a CCS of 125 Å2 and the precursors of product ions with a CCS of 125±1 Å2 were extracted from the negative ion CTS data (Figure 3C), which revealed prominent precursor ions at m/z 687, 771, and 797 and many other low abundance precursor ions from m/z 300–900 Da. The resultant plot was simplified when the data was filtered in two dimensions for both precursors of m/z 168.043±10 ppm and precursors of product ions with a CCS of 125±1 Å2 (Figure 3D) and enabled for the identification of six SM molecular species including SM(d18:1/16:0) at m/z 687, SM(d18:1/18:0) at m/z 715, SM(d18:1/20:0) at m/z 743, SM(d18:1/22:0) at m/z 771, and SM(d18:1/24:1) at m/z 797. It is evident that filtering the data by both product ion m/z and product ion CCS was advantageous for the identification of lipids in human serum. Additionally, the value of filtering the negative ion CTS data in two dimensions for both product ion m/z (241.012±10 ppm) and product ion CCS (134±1 Å2) was further demonstrated with phosphatidylinositol lipids in human serum (Figure S1) and allowed for the identification of PI(16:0/18:2) at m/z 833, PI(16:0/20:4) at m/z 857, PI(18:0/18:2) at m/z 861, and PI(18:0/20:4) at m/z 885 from the negative ion CTS data.
Figure 3.
Extraction of less abundant product ions from the negative ion CTS experiment of human serum. (A) expanded mass range (m/z 145–245) of the total product ion spectrum collected from the CTS experiment, (B) precursors of m/z 168.043±10 ppm, (C) precursors of product ions with a CCS value of 125±1 Å2, (D) precursors of m/z 168.043±10 ppm and of product ions with a CCS value of 125±1 Å2. The ions at m/z 687, 715, 743, 771, and 797 were identified as SM(d18:1/16:0), SM(d18:1/18:0), SM(d18:1/20:0), SM(d18:1/22:0), and SM(d18:1/24:1), respectively. Results shown are representative data obtained from four independent experiments.
Additionally, positive ion CTS analysis of the Bligh Dyer extract of serum was performed and resulted in an abundance of precursor/product information (Figure S2), including the CCS values of product ions derived from experimental ion mobility times (Table S2). The overwhelming majority of glycerophospholipid in human serum is PC and the abundant precursor ions present in the positive ion CTS experiment correspond to the major PC and lyso PC species found in serum.25 When the positive ion CTS data was filtered in two dimensions specifically for PC lipids (precursors of m/z 184.073±10 ppm and precursors of product ions with a CCS value of 132±1 Å2) (Figure S2F), the data looks very similar to the individual extractions (Figures S2D and S2E). While the product ion CCS and precursor ion extracted data were very similar for the identification of abundant PC molecular ions in serum, the product ion CCS does become a useful filter for phospholipids present in lower abundance in the serum sample. Some of the lower abundance product ions observed in the positive ion CTS of serum are derived from PE plasmalogen lipids, which result in an abundant product ion that provides information about the acyl chain esterified sn-2 position of the glycerol backbone in positive ion mode.26 When the positive ion CTS data was filtered in two dimensions specifically for PE plasmalogens containing AA (precursors of m/z 361.274±10 ppm and precursors of product ions with a CCS value of 197±1 Å2) (Figures S3C–D), the resultant plot is simplified compared to only filtering in one dimension (Figures S3A–B) and allowed for the identification of an AA containing lyso PE, PE(OH/20:4) at m/z 502, and three AA containing plasmalogen PE lipids, PE(16:0p/20:4) at m/z 724, PE(18:0p/20:4) at m/z 752, and PE(20:0p/20:4) at m/z 780 from the positive ion CTS data. Additionally, similar results were obtained for PE plasmalogens containing DHA (Figures S3E–H).
CONCLUSIONS
CTS analysis of phospholipid standards and human serum was carried out in both negative and positive ion mode, which provided a 4-dimensional dataset that included precursor ion, product ion mobility, accurate mass of product ion and ion abundance. This data can be analyzed using typical tandem mass spectral analysis strategies by extraction of precursor ion and product ion data. Additionally, the unique CCS values derived from the ion mobility of each product ion could also be utilized in this experiment. The CCS values obtained from ion mobility measurements of phospholipid product ions were found to be highly reproducible in the traveling wave experiment. The combination of diagnostic product ions and unique CCS values of product ions was demonstrated to be a powerful tool in identifying lipids in a complex biological sample. This additional identifying feature could be especially beneficial in lipidomic infusion type experiments of complex samples that are not subjected to chromatographic separation. One could envision that a database of CCS values of phospholipid fragment ions could be constructed and implemented as an additional classification factor to enhance confidence in the identification of lipids.
Supplementary Material
Acknowledgments
FUNDING SOURCES
This work was supported by a grant from the National Institute of Environmental Health Sciences of the National Institutes of Health (ES022172) (KZB, RMB, and RCM) and does not necessarily represent the official views of NIH. JJB was supported by the National Renewable Energy Laboratory via the U.S. Department of Energy under Contract No. DE-AC36-08GO28308.
ABBREVIATIONS
- CCS
collisional cross section
- CTS
concerted tandem mass spectrometry and traveling wave ion mobility
- CID
collision induced dissociation
- TOF
time of flight
- PC
phosphatidylcholine
- PA
phosphatidic acid
- PI
phosphatidylinositol
- PE
phosphatidylethanolamine
- PS
phosphatidylserine
- SM
sphingomyelin
Footnotes
SUPPORTING INFORMATION
Synapt G2-S experimental parameters, Table S1; Reproducibility of lipid product ion mobility times and CCS values in positive and negative ion mode over 4 months, Table S2; CCS values of [M−H−R2C=O]− product ions derived from PC, PE, PS, PA and PI standards, Table S3; Extraction of PI species from negative ion CTS data from human serum, Figure S1; Positive ion CTS of human serum, Figure S2; Extraction of PE plasmalogen species from positive ion CTS data from human serum, Figure S3. This material is available free of charge via the Internet at http://pubs.acs.org.
AUTHOR CONTRIBUTIONS
KZB performed and designed experiments, data analysis, and wrote the manuscript. RCM, RMB, and JAH were involved in the experimental design, data analysis, and writing of the manuscript. JJB wrote the Python code to visualize data. EH and JMB wrote the WREnS script.
The authors declare no competing financial interest.
REFERENCES
- 1.van Meer G, Voelker DR, Feigenson GW. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wymann MP, Schneiter R. Nat. Rev. Mol. Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro MD, Fazio S. Circ. Res. 2016;118:732–749. doi: 10.1161/CIRCRESAHA.115.306471. [DOI] [PubMed] [Google Scholar]
- 4.Miyazawa T, Nakagawa K, Shimasaki S, Nagai R. Amino Acids. 2012;42:1163–1170. doi: 10.1007/s00726-010-0772-3. [DOI] [PubMed] [Google Scholar]
- 5.Choi S, Snider AJ. Mediators Inflamm. 2015;2015:12. doi: 10.1155/2015/520618. Article ID 520618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Meer G. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Murphy RC, Raetz CRH, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA. J. Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Simons B, Kauhanen D, Sylvänne T, Tarasov K, Duchoslav E, Ekroos K. Metabolites. 2012;2:195–213. doi: 10.3390/metabo2010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han X, Yang K, Gross RW. Mass Spectrom Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paglia G, Kliman M, Claude E, Geromanos S, Astarita G. Anal Bioanal Chem. 2015;407:4995–5007. doi: 10.1007/s00216-015-8664-8. [DOI] [PubMed] [Google Scholar]
- 11.Paglia G, Angel P, Williams JP, Richardson K, Olivos HJ, Thompson JW, Menikarachchi L, Lai S, Walsh C, Moseley A, Plumb RS, Grant DF, Palsson BO, Langridge J, Geromanos S, Astarita G. Anal. Chem. 2015;87:1137–1144. doi: 10.1021/ac503715v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lintonen TPI, Baker PRS, Suoniemi M, Ubhi BK, Koistinen KM, Duchoslav E, Campbell JL, Ekroos K. Anal. Chem. 2014;86:9662–9669. doi: 10.1021/ac5021744. [DOI] [PubMed] [Google Scholar]
- 13.Basit A, Pontis S, Piomelli D, Armirotti A. Metabolomics. 2016;12:50. doi: 10.1007/s11306-016-0971-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker PRS, Armando AM, Campbell JL, Quehenberger O, Dennis EA. J. Lipid Res. 2014;55:2432–2442. doi: 10.1194/jlr.D051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shvartsburg AA, Isaac G, Leveque N, Smith RD, Metz TO. J. Am. Soc. Mass Spectrom. 2011;22:1146–1155. doi: 10.1007/s13361-011-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castro-Perez J, Roddy TP, Nibbering NMM, Shah V, McLaren DG, Previs S, Attygalle AB, Herath K, Chen Z, Wang S-P, Mitnaul L, Hubbard BK, Vreeken RJ, Johns DG, Hankemeier T. J. Am. Soc. Mass Spectrom. 2011;22:1552–1567. doi: 10.1007/s13361-011-0172-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hankin JA, Barkley RM, Zemski-Berry K, Deng Y, Murphy RC. Anal. Chem. 2016;88:6274–6282. doi: 10.1021/acs.analchem.6b00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bligh EG, Dyer WJ. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 19.Michaelevski I, Kirshenbaum N, Sharon M. J Vis Exp. 2010;41:e1985, 1–10. doi: 10.3791/1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pulfer M, Murphy RC. Mass Spectrom Rev. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- 21.Sullards MC, Liu Y, Chen Y, Merrill AH. Biochim. Biophys. Acta. 2011;1811:838–853. doi: 10.1016/j.bbalip.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu FF, Turk J. J. Am. Soc. Mass Spectrom. 2000;11:986–999. doi: 10.1016/S1044-0305(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 23.Brügger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu FF, Turk J. J. Am. Soc. Mass Spectrom. 2000;11:892–899. doi: 10.1016/S1044-0305(00)00159-8. [DOI] [PubMed] [Google Scholar]
- 25.Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CRH, Guan Z, Laird GM, Six DA, Russell DW, McDonald JG, Subramaniam S, Fahy E, Dennis EA. J. Lipid Res. 2010;51:3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zemski Berry KA, Murphy RC. J. Am. Soc. Mass Spectrom. 2004;15:1499–1508. doi: 10.1016/j.jasms.2004.07.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.