Abstract
The immature flavivirus particle contains two envelope proteins, prM and E, that are associated as a heterodimer. Virion morphogenesis of the flaviviruses occurs in association with endoplasmic reticulum (ER) membranes, suggesting that there should be accumulation of the virion components in this compartment. This also implies that ER localization signals must be present in the flavivirus envelope proteins. In this work, we looked for potential subcellular localization signals in the yellow fever virus envelope proteins. Confocal immunofluorescence analysis of the subcellular localization of the E protein in yellow fever virus-infected cells indicated that this protein accumulates in the ER. Similar results were obtained with cells expressing only prM and E. Chimeric proteins containing the ectodomain of CD4 or CD8 fused to the transmembrane domains of prM or E were constructed, and their subcellular localization was studied by confocal immunofluorescence and by analyzing the maturation of their associated glycans. Although a small fraction was detected in the ER-to-Golgi intermediate and Golgi compartments, these chimeric proteins were located mainly in the ER. The C termini of prM and E form two antiparallel transmembrane α-helices. Interestingly, the first transmembrane passage contains enough information for ER localization. Taken altogether, these data indicate that, besides their role as membrane anchors, the transmembrane domains of yellow fever virus envelope proteins are ER retention signals. In addition, our data show that the mechanisms of ER retention of the flavivirus and hepacivirus envelope proteins are different.
At the end of their life cycle, enveloped viruses leave their host cell after having acquired an envelope derived from a cellular membrane. Virus budding can occur at the plasma membrane or at intracellular organelles such as the endoplasmic reticulum (ER), the ER-to-Golgi intermediate compartment (ERGIC), or the Golgi complex. In most cases, when budding occurs at an intracellular compartment, particles released into the lumen of the organelle follow the secretory pathway to leave the host cell. Whatever the site of budding, all the components of the viral particle have to be transported to the site of virion formation.
Flaviviruses belong to the Flavivirus genus within the Flaviviridae family, which also comprises the Hepacivirus and Pestivirus genera (56). They include arthropod-borne human pathogens such as yellow fever virus (YFV), Japanese encephalitis virus, dengue viruses, West Nile virus, and tick-born encephalitis virus (TBE). Flaviviruses are small enveloped plus-strand RNA viruses. The flavivirus particle is made of an envelope, containing 180 copies of E and M proteins, that surrounds a nucleocapsid composed of genomic RNA and multiple copies of the C protein (29). The M protein is synthesized as a precursor called prM that associates with E to form heterodimers (1, 66). These heterodimers are organized in 60 trimeric spikes on the immature viral particle (69). Heterodimeric interactions between prM and E are important for proper folding of E (1, 28, 37). Interestingly, expression of E and prM of several flaviviruses in the absence of other viral proteins results in the secretion of virus-like particles called recombinant subviral particles, which have structural and functional features of the envelope of the virion (reviewed in reference 24).
Virion morphogenesis of the flaviviruses occurs in association with intracellular membranes. Electron microscopic studies of flavivirus-infected cells have consistently demonstrated the presence of virions within the lumen of the ER (reviewed in reference 32). However, budding intermediates at the ER membrane have not been clearly observed, suggesting that the process of assembly is rapid. Assembly of subviral particles following the expression of prM and E in the absence of other viral components suggests that lateral interactions between these envelope proteins are a major driving force leading to particle assembly (20). However, there is growing evidence that in the context of the expression of all the flavivirus components, some nonstructural proteins are also required for virus assembly (30, 34, 35), suggesting that flavivirus assembly is a tightly regulated process. Ultrastructural studies and the use of drugs that inhibit protein and/or membrane traffic throughout the cell have shown that flavivirus particles are transported through the normal secretory pathway (40). Similar observations have been made for subviral particles (38). Shortly before release from the cell, immature virions are converted to the mature form by cleavage of prM by a cellular furin protease (61). Cleavage leads to the dissociation of prME heterodimers and a major reorganization of the virion surface (29, 55, 62, 69). Besides its role in assisted folding of E (1, 28, 37) and potentially in virus budding, the prM interaction with E probably functions to prevent the E protein from prematurely undergoing conformational changes that trigger fusion in endosomal vesicles (24).
Since assembly of the flavivirus particle occurs in the ER, the virion components should accumulate in this compartment. This suggests that ER localization signals must be present in the flavivirus envelope proteins. It has been demonstrated with hepatitis C virus (HCV), another member of the Flaviviridae family, that the transmembrane (TM) domains of the envelope proteins are ER retention signals (reviewed in reference 46). In this work we looked for potential ER localization signals in the TM domains of the flavivirus envelope proteins. By making chimeric proteins containing the ectodomain of CD4 or CD8 fused to the C-terminal hydrophobic sequences of YFV envelope proteins, we showed that the TM domains of prM and E are able to retain these ectodomains in the ER.
MATERIALS AND METHODS
Cell culture.
HeLa, HepG2, SW13, CV-1, and 143B (thymidine kinase-deficient) cell lines were obtained from the American Type Culture Collection, Manassas, Va. Cell monolayers were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and 50 μg of gentamicin per ml.
Plasmid constructions.
Plasmids expressing chimeric proteins were constructed by using standard methods (58). They were constructed in two steps by successively introducing the sequences of domains from two different proteins. A unique restriction site was introduced between the sequences of the protein domains. YFV sequences were amplified from plasmid pAP5, which contains the sequence encoding the structural proteins of the YFV 17D-204-Pasteur strain (16), kindly provided by A. Cahour (Hôpital de la Pitié-Salpétrière, Paris, France). DNA sequences of the TM domains of YFV envelope protein were first introduced into plasmid pTM1 (42) by PCR with an oligonucleotide introducing a BamHI site at the 5′ end and a stop codon followed by a XhoI site at the 3′ end. The sequence of the ectodomain of CD4 or CD8 was then introduced into these intermediate constructs after digestion of pTM1/CD4-E2 (13) or pTM1/CD8-E2 (12) with NcoI and BamHI. Plasmids pTM1/CD4(1-371)-prM(241-285), pTM1/CD4(1-371)-prM(249-285), pTM1/CD8(1-159)-prM(241-285), pTM1/CD8(1-159)-prM(249-285), pTM1/CD4(1-371)-E(731-778), pTM1/CD4(1-371)-E(739-778), pTM1/CD8(1-159)-E(731-778), and pTM1/CD8(1-159)-E(739-778) contain the signal sequence of CD4 or CD8 followed by the sequence of their ectodomain fused with the C-terminal sequences of prM or E. Between these sequences, there is a junction sequence encoding two additional amino acids (Gly and Ser). CD4-TM chimeric proteins containing the Arg-to-Ala mutation (Arg270 in prM or Arg754 in E) or Ala insertion immediately downstream of positions 260 (mutant A261' of prM), 274 (mutant A275' of prM), 748 (mutant A749' of E), or 762 (mutant A763' of E) were obtained by introducing, after PCR amplification, the sequence of mutated TM domains (48) between the BamHI and XhoI restriction sites. CD4-TM chimeric proteins with the second TM domain of prM or E deleted (ΔCD4-ETM1 and ΔCD4-prMTM1) were obtained after introduction of a stop codon immediately downstream of Arg270 in prM and Arg754 in E. The ΔCD4-ETM1 chimeric protein with of the second TM domain of E deleted and containing three additional hydrophobic amino acid (Ala-Leu-Ala) in the middle of the TM domain were obtained by introducing the sequence of these residues immediately downstream of amino acid 747. To construct all of the CD4-TM chimeric proteins with of the second TM domain deleted, synthetic oligonucleotides corresponding to coding and antisense sequences were annealed and cloned between the BamHI and XhoI restriction sites. The nucleotide sequences of all synthetic and PCR-derived DNA fragments were confirmed by sequencing. Plasmids pTM1/prME (48), pTM1/CD8 (12), and pTM1/CD4 (13) and plasmids pTM1/CD4-E1NK and pTM1/CD4-E1 (containing the signal sequence and the ectodomain of CD4 fused with the TM domain of HCV envelope protein E1 in which the charged residues have been mutated or not) (15) have been described previously.
Vaccinia viruses.
Vaccinia virus recombinants were generated by homologous recombination essentially as described previously (48). The genes of YFV proteins expressed in this work are under the control of a T7 promoter, and expression of the proteins of interest was achieved by coinfection with vTF7-3 (a vaccinia virus recombinant expressing the T7 DNA-dependent RNA polymerase) (22). Vaccinia virus recombinants expressing CD4 (vCD4) (13), CD8 (vCD8) (12), and prME (vprME) (48) have been described previously.
YFV.
YFV strain 17D was obtained from Philippe Despres (Institut Pasteur, Paris France). Viral stocks were grown on SW-13 cells and subjected to titer determination on the same cells by the 50% tissue culture infective dose method. For immunofluorescence experiments, HeLa cells were infected at a multiplicity of infection of about 5.
Antibodies.
Mouse monoclonal antibodies (MAbs) 2D12 (anti-E, ATCC CRL-1689), OKT4 (anti-CD4, ATCC CRL-8002), and OKT8 (anti-CD8, ATCC CRL-8014) were produced in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer. Mouse anti-ERGIC-53 MAb and rabbit anti-mannosidase II polyclonal antibody were kindly provided by H.-P. Hauri (University of Basel, Basel, Switzerland) and by K. Moremen (University of Georgia), respectively. Rat anti-CD4 MAb MCA484 was purchased from Serotec (Oxford, United Kingdom). Mouse anti-GM130 MAb was purchased from BD Biosciences. Rabbit antibodies to protein disulfide isomerase (PDI) and calnexin were from Stressgen and Dako, respectively. Alexa-488- and Alexa-546-conjugated goat anti-rabbit and anti-mouse immunoglobulin G (IgG) were purchased from Molecular Probes. FITC-conjugated goat anti-mouse IgG and Cy3-conjugated goat anti-rat IgG were purchased from Jackson Immunoresearch (West Grove, Pa.).
Indirect immunofluorescence microscopy.
Cells grown on 12-mm glass coverslips were fixed with 3% paraformaldehyde and then permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS). Both primary- and secondary-antibody incubations were carried out for 30 min at room temperature with PBS containing 10% goat serum. The coverslips were mounted on slides by using Immunofluor mounting medium (ICN). Confocal microscopy was performed with an SP2 confocal laser-scanning microscope (Leica) using a 100×/1.4 numerical aperture oil immersion lens. Double-label immunofluorescence signals were sequentially collected using single-fluorescence excitation and acquisition settings to avoid crossover. Images were processed using Adobe Photoshop software.
Cell surface labeling.
For cell surface labeling, cells were incubated for 1 h on ice with the primary antibodies. Anti-CD4 MAb OKT4 was used to detect the proteins expressed at the cell surface, together with a rabbit polyclonal anti-calnexin antiserum, as a control for the lack of permeabilization of cell membranes. The cells were then washed three times with cold PBS and fixed with 3% paraformaldehyde. Alexa-488- and Alexa-546-labeled goat anti-mouse and rabbit secondary-antibody incubation were carried out for 30 min at room temperature with PBS containing 10% goat serum.
Intracellular transport assay.
HeLa cells were first infected for 1 h with vTF7-3 in serum-free medium and then transfected with 2.5 μg of pTM1-derived plasmid complexed with 5 μl of DOTAP reagent (Roche). After 1 h of contact, the medium was replaced with fresh medium containing 5 mM hydroxyurea to avoid cytopathic effects, and the cells were allowed to express the fusion proteins for another 1 h. The medium was then complemented with 50 μg of cycloheximide (CHX) per ml to block any further protein synthesis, and the cells were cultured for up to 4 or 6 h. They were fixed in 3% paraformaldehyde in PBS and processed for double-label immunofluorescence.
Metabolic labeling and immunoprecipitation.
Cells expressing the proteins of interest were metabolically labeled with 35S-protein-labeling mix (100 μCi/ml; DuPont NEN) as previously described (18). The cells were then lysed with 0.5% Triton X-100 in TBS (50 mM Tris-HCl [pH 7.5], 150 mM NaCl). Immunoprecipitations were carried out as described previously (19). For in vivo labeling of glycan moieties, HepG2 cells were infected with the appropriate vaccinia virus recombinants and pulse-labeled for 30 min with 100 μCi of [2-3H]mannose (Amersham) per ml in culture medium containing 0.5 mM glucose and 10% dialyzed fetal bovine serum. After 4 h of chase, the cells were lysed in TBS-0.5% Triton X-100 and the lysates were used for immunoprecipitation.
Analysis of oligosaccharide material.
Immunoprecipitated [2-3H]mannose-labeled proteins were digested overnight at room temperature with 0.2 mg of TPCK-treated trypsin in 0.1 M ammonium bicarbonate (pH 7.9). Trypsin-treated proteins were boiled for 10 min to inactivate the trypsin, and the peptides were dried and dissolved in 20 mM sodium phosphate (pH 7.5) containing 50 mM EDTA and 0.2 mg of NaN3 per ml in 50% glycerol. The peptides were incubated overnight at 37°C in the presence of 0.5 U of peptide:N-glycosidase (PNGase) F (New England Biolabs). For sequential enzymatic treatment with β-galactosidase and β-hexosaminidase, oligosaccharides released after PNGase F digestion were dissolved in 20 μl of 100 mM sodium acetate buffer (pH 3.5). Digestion was carried out overnight at 37°C with 0.1 U of β-galactosidase followed by an additional overnight incubation with 0.1 U of β-hexosaminidase. Size analysis of the glycan moieties was achieved by HPLC on an amino-derivatized column ASAHIPAK NH2P-50 (250 by 4.6 mm) (Asahi, Kawasaki-ku, Japan) with a solvent system of acetonitrile-water from 70:30 to 50:50 (vol/vol) at a flow rate of 1 ml/min for 80 min. Oligomannosides were identified as previously described (27) by their retention time. Separation of labeled oligosaccharides was monitored by continuous-flow detection of radioactivity with a Flo-one β detector (Packard).
RESULTS
YFV envelope protein E is localized in the ER.
Immunofluorescence studies of prM and the E proteins of TBE and Kunjin virus have shown that these proteins are localized mainly in the ER (38, 40). In our work, we used YFV to identify the subcellular localization signal(s) in flavivirus envelope proteins, and we first wanted to determine whether YFV envelope proteins have a similar subcellular localization to those of TBE and Kunjin virus.
The subcellular distribution of YFV envelope protein E was examined by confocal immunofluorescence microscopy (Fig. 1). In YFV-infected HeLa cells, E was observed in a network of cytoplasmic membranes (Fig. 1A). This distribution overlapped extensively with the ER marker protein disulfide isomerase (PDI) (Fig. 1A to C). In contrast, there was no apparent overlap with the Golgi marker mannosyl-oligosaccharide-1,3-1,6-α-mannosidase in the juxtanuclear region (Fig. 1D to F). Similar results were obtained at later times after infection (data not shown). Additional dots of E labeling that did not colocalize with the ER and Golgi markers were detected. They probably correspond to incoming or outgoing E-labeled virions, as shown for Kunjin virus (40). Furthermore, when E was expressed, together with prM, by using a vaccinia virus expression system rather than YFV infection, it also colocalized with the ER marker PDI (Fig. 1G to I). Because prM and E assemble rapidly as a heterodimer during their folding (37), the subcellular localization of prME is supposed to be similar to that of E (38).
FIG. 1.
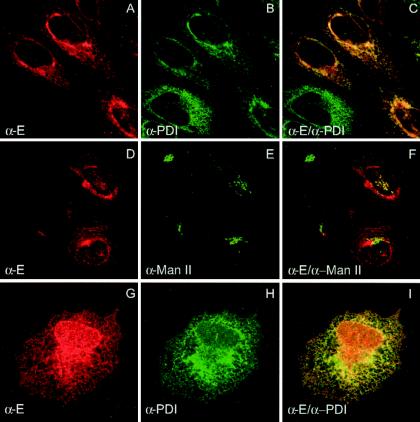
Confocal immunofluorescence analysis of the localization of YFV envelope protein E. HeLa cells were infected with YFV (A to F) or coinfected with VTF7-3 and a recombinant vaccinia virus expressing YFV envelope proteins prM and E (G to I). At 24 h (YFV) or 8 h (vaccinia virus recombinants) after infection, the cells were fixed, permeabilized, and processed for double-label immunofluorescence detection of E (A, D, and G) and PDI isomerase (PDI) (B and H), or mannosidase II (Man II) (E), as ER and Golgi markers, respectively. The merged images are shown in panels C, F, and I.
From these data, we conclude that, similarly to what has been shown for other flaviviruses (38, 40), YFV envelope protein E is localized in the ER. In addition, this localization is not dependent on a productive viral infection but, rather, is an intrinsic property of the envelope proteins, suggesting the presence of ER localization signal(s) in their structure.
Analysis of the cell surface expression of CD4 and CD4-TM fusion proteins.
YFV envelope proteins contain a large N-terminal ectodomain and a C-terminal TM region (Fig. 2). Since the TM domains of HCV envelope proteins contain an ER retention signal (reviewed in reference 46) and because flaviviruses and HCV belong to the same viral family, we hypothesized that ER localization signals might also be present in the TM domains of the flaviviruses.
FIG. 2.
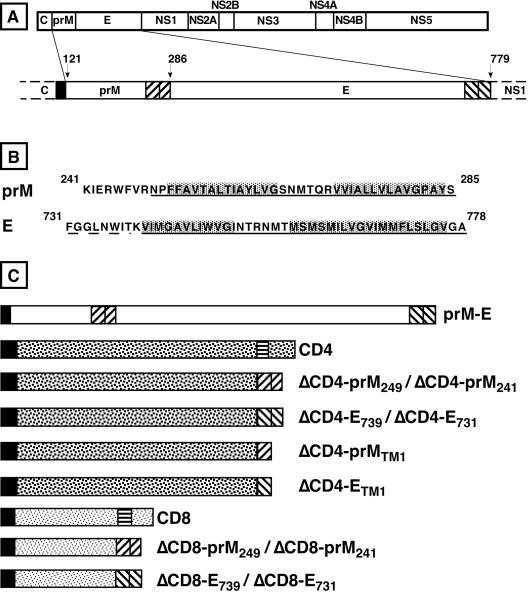
YFV envelope proteins and TM fusion proteins used in this study. (A) Schematic representation of the YFV polyprotein. Signal peptide and double-spanning TM domains are indicated by solid and striped boxes, respectively. The arrows indicate signal peptidase cleavage sites. Numbers indicate the positions of N-terminal residues of prM, E, and NS1. (B) Sequences of the TM domains of prM and E from the YFV 17D strain. The putative double-spanning TM domains (as predicted in reference 15) are underlined. The dotted line indicates the putative N terminus of the first TM sequence of E as predicted previously (2, 62). The two stretches of hydrophobic residues of each TM domain are indicated in grey boxes. (C) Schematic representation of prME, CD4, CD8, and the TM fusion proteins. Signal peptides and TM domains are indicated by solid and striped boxes, respectively. The names of the fusion proteins are indexed according to the first residue of the TM domain fused to the ectodomain of CD4 or CD8.
To assess whether the TM domains of YFV envelope proteins contain ER localization signals, TM domains of E and prM were fused to the ectodomains of human CD4. As shown in Fig. 2, two constructs were made for each TM domain, on the basis of two different predictions concerning the N-terminal boundary of the double-spanning TM domain of E (Fig. 2B) (15, 62). The C termini of the domains fused to the ectodomain of CD4 were defined as the signal peptidase cleavage sites in the precursor polyprotein. The four CD4 fusion proteins were expressed in HepG2 cells with vaccinia viruses, and their expression was compared with that of CD4. At 8 h after infection, cell surface expression was determined by surface labeling using an anti-CD4 MAb. Cell surface expression levels of TM fusion proteins were very close to background and much lower than those of CD4, despite similar levels of expression, as seen by indirect immunofluorescence after permeabilization of cell membranes with Triton X-100 (Fig. 3). For the four TM fusion proteins, the pattern observed after permeabilization was reminiscent of localization of the protein in the ER. For CD4, in addition to the cell surface expression, an ER-like localization was obvious for a fraction of the protein. Since TM sequences are supposed to fold autonomously as α-helix structures in the lipid environment of the ER membrane (54) and the antibody used to detect the fusion proteins is conformation specific, it is unlikely that the lack a cell surface expression of the fusion proteins would be due to misfolding. In addition, similar fusions between the ectodomain of CD4 and TM domains of HCV envelope proteins mutated at the charged residues were exported to the cell surface (15), confirming that fusing the ectodomain of CD4 to a TM domain does not alter the folding CD4. Two constructs were made for each protein, one containing a slightly shorter putative TM region than the other. Nevertheless, no difference was observed between the two sequences, indicating that the intracellular retention signals are contained in the shortest sequences.
FIG. 3.
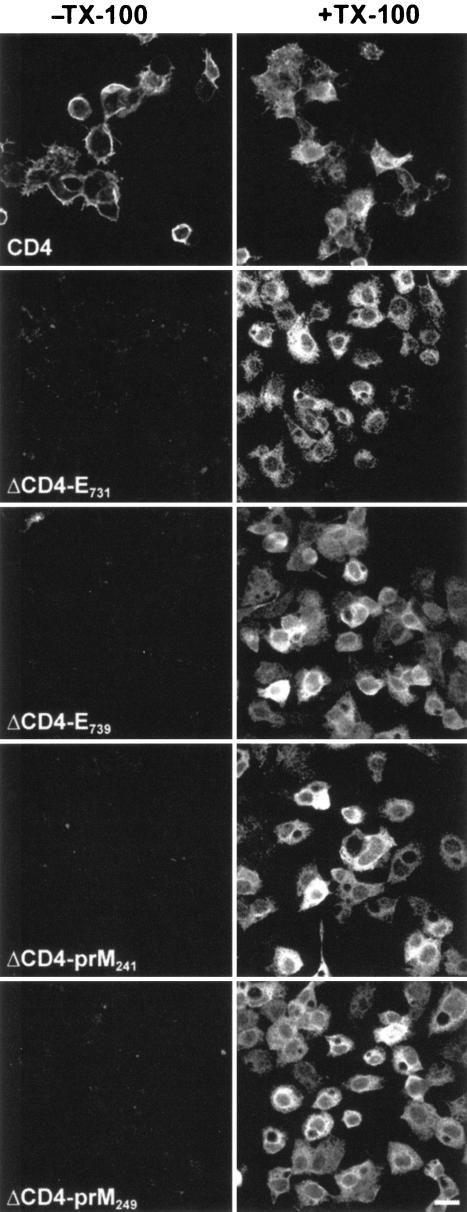
Cell surface expression of CD4 and TM fusion proteins. HepG2 cells were infected with vTF7-3 and the appropriate vaccinia virus recombinant. At 8 h after infection, cell surface labeling (−TX-100, left panels) was carried out on ice with anti-CD4 MAb, before the fixation with 3% paraformaldehyde and the incubation with Alexa-488-labeled secondary antibody. Another set of cells was fixed and processed for immunofluorescence labeling after permeabilization with TX-100 (+TX-100, right panels). All images were acquired and processed with the same settings. Bar, 20 μm.
Taken together, these data indicate that the fusion proteins were expressed normally but not transported to the cell surface as efficiently as CD4, suggesting that the TM domains of prM and E are intracellular retention signals.
The TM domains of YFV envelope proteins are able to target a reporter protein to the ER.
To further study the putative ER localization function of the TM domains of YFV envelope proteins, the intracellular traffic of CD4 and CD4-based chimeric proteins was compared by using an intracellular transport assay based on indirect immunofluorescence. HeLa cells were infected for 1 h with a vaccinia virus expressing the T7 RNA polymerase, transfected with pTM1-derived expression plasmids for another 1 h, and allowed to express proteins for an additional 1 h. Further protein synthesis was then blocked by CHX for up to 4 h, and the intracellular localization of the expressed protein was analyzed by immunofluorescence with the conformation-specific MAb OKT4.
Before CHX addition, CD4 immunoreactive material was localized in the nuclear envelope and in the ER, as shown by the colocalization with PDI (Fig. 4A). After 1 h of CHX incubation, the protein was still detected in the ER and other peripheral structures, but a larger part of the immunofluorescent material was concentrated in a perinuclear area in most CD4-expressing cells (Fig. 4B). Perinuclear localization was also evident after 2 h of CHX treatment (Fig. 4C). However, in most of the cells, the protein was no longer present in the ER at that time. Some cells were also slightly labeled at the plasma membrane (data not shown). After 4 h of CHX treatment, the labeling was mainly at the plasma membrane, with a minor part of the protein remaining in the perinuclear compartment (Fig. 4D). This perinuclear staining colocalized with GM130, a Golgi marker (data not shown). These data indicate that CD4 was initially synthesized in the ER and transported through the Golgi complex to the plasma membrane, as expected for a cell surface membrane protein.
FIG. 4.
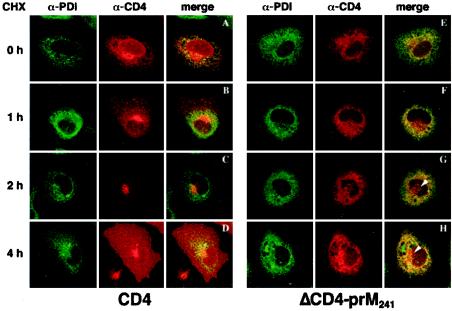
Confocal immunofluorescence analysis of the intracellular traffic of CD4 and ΔCD4-prM241. HeLa cells grown on glass coverslips were infected with vTF7-3 for 1 h and then transfected with 2.5 μg of pTM1/CD4 or pTM1/ΔCD4-prM241. The cells were allowed to express proteins for an additional 1 h and then incubated in the presence of CHX to block any further protein synthesis for 0 to 4 h, as indicated on the left. The cells were fixed and processed for double-label immunofluorescence for CD4 (red) and the ER marker PDI (green). Representative confocal images of individual cells are shown. Arrowheads indicate a perinuclear compartment positive for ΔCD4-prM241 and negative for the ER marker.
Following ΔCD4-prM241 expression, CD4 immunoreactivity was also initially detected in the ER (Fig. 4E). In contrast to CD4, ΔCD4-prM241 was detected in the ER throughout the experiment, up to 4 h of CHX block (Fig. 4F to H). A faint labeling of a perinuclear compartment was also detected after 2 and 4 h, in most of the cells examined (Fig. 4G and H). No staining was observed at the plasma membrane. Similar retention in the ER was observed with ΔCD4-prM249, ΔCD4-E731, and ΔCD4-E739 (data not shown). These data show that the intracellular trafficking of TM fusion proteins was dramatically altered, compared to CD4.
Together, these data suggest that the TM domains of YFV envelope proteins contain ER localization signals and support the notion that the retention of YFV envelope proteins in the ER is mediated by their TM domains.
Intracellular distribution of CD4-TM fusion proteins.
To determine the intracellular distribution of CD4-TM fusion proteins, HeLa cells were infected with a T7 RNA polymerase-expressing vaccinia virus and transfected with plasmid pTM1, which encodes ΔCD4-prM241 (Fig. 5) or ΔCD4-E731 (Fig. 6), as was done for the analysis of the intracellular transport. After 4 h of CHX block, the cells were fixed, permeabilized, and double stained for CD4 and for cellular markers. Anti-GM130 was used as a marker for the Golgi complex, anti-ERGIC-53 was used as a marker for the intermediate compartment, and anti-calnexin was used as an ER marker. Both the mouse MAb OKT4 and the rat MAb MCA484 anti-human CD4 were found to stain the nuclear envelope and a network of cytoplasmic membranes for both ΔCD4-prM241 (Fig. 5B, E, and H) and ΔCD4-E731 (Fig. 6B, E, and H) but not CD4 (data not shown). The distribution of the major part of ΔCD4-prM241 overlapped with calnexin (Fig. 5I). In addition, ΔCD4-prM241 showed some overlap with ERGIC-53 (Fig. 5F) and with GM130 (Fig. 5C). On the other hand, the distribution of ΔCD4-E731 almost completely overlapped with that of calnexin (Fig. 6I) and, to a minor extent, with that of ERGIC-53 (Fig. 6F), but not with that of GM130 (Fig. 6C), in most of the cells. These data indicate that the majority of both fusion proteins were localized in the ER. For ΔCD4-E731, a small part of the protein was also localized in the intermediate compartment, whereas the distribution of ΔCD4-prM241 was more spread out along the secretory pathway, with larger amounts of the protein in the intermediate compartment and the Golgi complex.
FIG. 5.
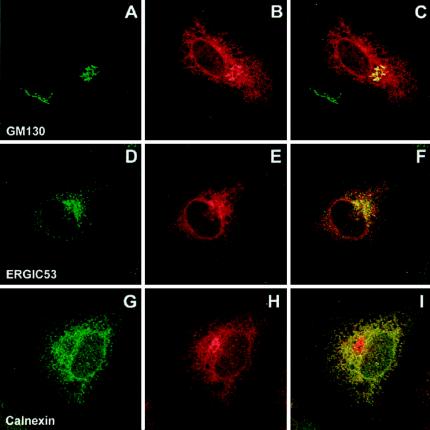
Confocal immunofluorescence analysis of the intracellular distribution of a chimeric protein with the CD4 ectodomain fused to the TM domain of prM. ΔCD4-prM241 was expressed in HeLa cells by the vTF7-3 infection/pTM1 transfection method as in Fig. 4 and incubated for 4 h in medium containing 50 μg of CHX per μl. The cells were fixed and processed for double-label immunofluorescence for CD4 (red) and the following cellular markers (green): GM130, a Golgi matrix protein (top row); ERGIC53, a marker for the ER-to-Golgi intermediate compartment (middle); or calnexin, a chaperone of the ER (bottom). Representative confocal images of individual cells are shown, with the merge images in the right column.
FIG. 6.
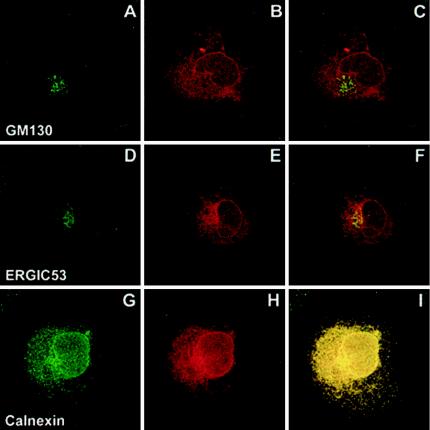
Immunofluorescence analysis of the intracellular distribution of a chimeric protein with the CD4 ectodomain fused to the TM domain of E. ΔCD4-E731 expression and double-label immunofluorescence were carried out as indicated in the legend of Fig. 5.
Analyses of the glycans associated with the fusion proteins.
As an additional approach to studying the subcellular localization of the CD4-TM fusion proteins, we analyzed the modifications of their associated glycans. The CD4 protein contains two N-linked glycans, and only one of them becomes endo H resistant (60). The glycans associated with the CD4-TM fusion proteins were removed from the fusion proteins by PNGase F treatment and analyzed by affinity chromatography and high-performance liquid chromatography (HPLC). For this approach, the CD4-TM fusion proteins were labeled with [2-3H]mannose and immunoprecipitated with MAb OKT4 before PNGase F treatment and characterization of labeled glycans. Affinity chromatography analyses of these glycans showed that they bound strongly to concanavalin A and eluted in a buffer containing 100 mM α-d-mannoside (data not shown). In addition, HPLC analyses of these glycans demonstrated the presence of three species. Man9GlcNΔc2, Man8GlcNΔc2, and Man7GlcNAc2, respectively (Fig. 7 and data not shown). Since the oligosaccharide precursor which is transferred onto nascent proteins is the Glc3Man9GlcNAc2, this reveals the sequential actions of ER glucosidases I and II and at least the action of ER mannosidase yielding Man8 species. The presence of Man7 is probably due to trimming of mannose residues occurring after prolonged residence in the ER (23). Indeed, the occurence of this latter structure can be explained by the combined action of ER α1,2-mannosidases such as ER mannosidases I and II or Man9 mannosidase (7, 8, 65). As shown in Fig. 7, the retention time of the oligosaccharides associated with full-length CD4 revealed the presence of processed species different from those associated with CD4-TM chimeric proteins. To precisely identify the species associated with full-length CD4, an additional digestion with exoglycosidases (galactosidase and hexosaminidase) had to be performed. The HPLC profile obtained after such a digestion revealed the presence of Man3 (derived from complex-type glycans), Man5 (derived from hybrid-type glycans), and Man7 (derived from high mannose-type glycans) species (Fig. 7). Galactosidase and hexosaminidase digestions did not change the HPLC profiles of CD4-TM chimeric proteins (data not shown). The nature of the glycans observed in this work is consistent with the bulk of CD4-TM fusion proteins being localized in the ER compartment.
FIG. 7.
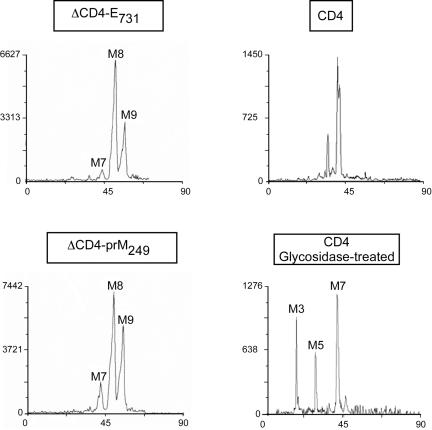
HPLC analysis of the oligosaccharides bound to ΔCD4-prM249 and ΔCD4-E731. Wild-type CD4 was used as a control. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinants. At 4.5 h postinfection, infected cells were pulse-labeled for 30 min with [2-3H]mannose, chased for an additional 4 h, and lysed with Triton X-100. Cell lysates were used for immunoprecipitation with MAb OKT4. Labeled glycans were removed by PNGase F treatment and analyzed by HPLC as described in Materials and Methods. M3, M5, M7, M8, and M9 indicate oligosaccharide species possessing two GlcNAc residues at their reducing end and three, five, seven, eight, or nine mannose residues, respectively.
Since immunofluorescence studies showed some colocalization of ΔCD4-prM241 with the ERGIC and Golgi compartment (Fig. 5C and F) and ΔCD4-E731 showed some penetration into the ERGIC compartment (Fig. 6F), we wanted to further investigate the penetration into these compartments by using a more sensitive approach. For this purpose, the ectodomain of CD8 was fused to the TM domains of YFV envelope proteins. CD8 is a class I TM protein that is transported to the cell surface and has been extensively used as a reporter protein to study localization signals (12, 33, 41, 43). Maturation of its O-linked glycans has been well characterized (50, 51), and it has been established that sugar modifications mark transport in different compartments of the secretory pathway. CD8 is synthesized as a 27-kDa species (u), which is converted to a transient and initially glycosylated 29-kDa (i) form before the full maturation to a completely glycosylated 32-34-kDa doublet (m). The intermediate form is generated in an early Golgi compartment, and the mature form is generated in the trans-Golgi region. When expressed in HepG2 cells with the vaccinia virus-T7 expression system, the intermediate 29-kDa form was barely detectable (Fig. 8). This is probably due to a faster processing of the glycans in HepG2 cells. When the TM and cytosolic domains of CD8 were replaced by the TM domain of prM or E, most of the chimeric proteins remained of the same size in pulse-chase experiments (Fig. 8 and data not shown), suggesting that these proteins are retained in the ER. This is consistent with the observations made on the subcellular localization of the CD4-TM fusion proteins. Interestingly, the mature form was detected earlier and was more intense for ΔCD8-prM249 than ΔCD8-E731 (Fig. 8), indicating some penetration in the ERGIC and Golgi compartment, especially for ΔCD8-prM249. These data are consistent with the immunofluorescence studies (Fig. 5 and 6) and suggest some leakiness in the ER retention signals.
FIG. 8.
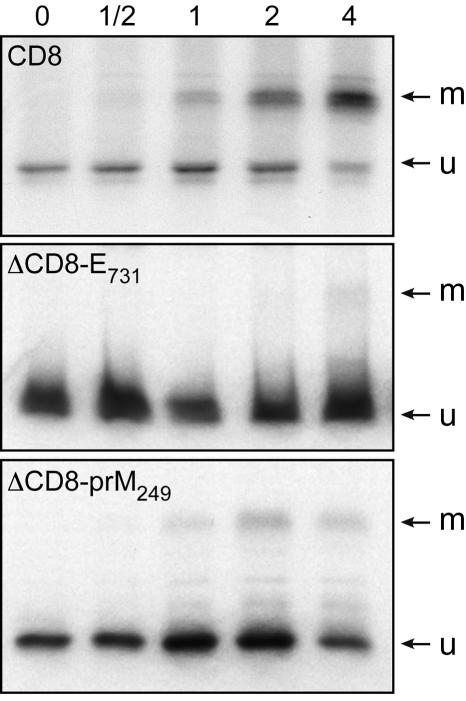
Expression of CD8, ΔCD8-prM249, and ΔCD8-E731 analyzed in pulse-chase experiments. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. At 4.5 h postinfection, infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb OKT8 (anti-CD8). Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% polyacrylamide). u, unglycosylated precursor; m, mature form.
The first TM sequence is sufficient for ER retention.
Replacement of the charged residues between the two hydrophobic stretches by alanine residues alters the ER retention function of the TM domains of HCV envelope proteins (15) (compare Fig. 9A and B). We therefore suspected that the Arg residue similarly located between the two stretches of hydrophobic residues of the C termini of prM and E might also be involved in a similar function in YFV. To test this hypothesis, the Arg residues at positions 270 (for prM) and 754 (for E) were replaced by an Ala in ΔCD4-prM249 and ΔCD4-E731 and the effects of these mutations (ΔCD4-ER ΔCD4-prMR) on the subcellular localization of these chimeric proteins in our intracellular transport assay were analyzed. Surprisingly, these mutations did not modify the subcellular localization of the CD4-TM chimeric proteins. Indeed, after 6 h of CHX block, these mutant proteins showed an ER-like pattern of fluorescence similar to that observed for CD4-TM chimeric proteins and no staining was detected at the plasma membrane (Fig. 9C and E and data not shown). As expected, full-length CD4 and ΔCD4-E1NK were present at the plasma membrane when analyzed under the same conditions (Fig. 9B and D). Taken together, these data indicate that the ER retention mechanism of the flavivirus envelope proteins is different from that of HCV envelope glycoproteins.
FIG. 9.
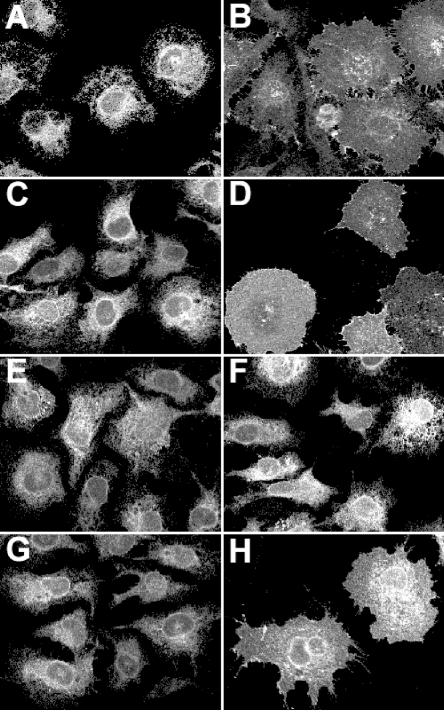
Immunofluorescence analysis of the intracellular distribution of ΔCD4-E731 mutants. ΔCD4-E1 (A), ΔCD4-E1NK (B), ΔCD4-E731 (C), CD4 (D), ΔCD4-ER (E), ΔCD4-EA749 (F), ΔCD4-ETM1 (G), and ΔCD4-ETM1-ALA (H) were expressed in HeLa cells by the vTF7-3 infection-pTM1 transfection method as in Fig. 4 and incubated for 6 h in medium containing 50 μg of CHX per ml. The cells were fixed and processed for immunofluorescence by using an anti-CD4 antibody. ΔCD4-E1 and ΔCD4-E1NK are chimeric proteins made of the ectodomain of CD4 in fusion with the TM domain of HCV envelope protein E1 containing (ΔCD4-E1NK) or not containing (ΔCD4-E1) a mutation of the Asn and Lys residues in the middle of these TM domains.
Recently, alanine scanning insertion mutagenesis has been used to examine the role of the TM domains of prM and E in YFV subviral particle formation. Insertions introduced in the hydrophobic stretches of the TM regions of prM or E had a dramatic effect on the release of YFV subviral particles without affecting prM-E heterodimerization (48). However, the subcellular localization of these mutant proteins was not analyzed in our previous work, and we cannot exclude the possibility that alteration of secretion was due to modification of their subcellular localization. If this is the case, such mutants would be helpful in our understanding of the mechanism of ER retention of prM and E TM domains. To test this hypothesis, some of the alanine insertion mutants were introduced in the context of CD4-TM chimeric proteins and their subcellular localization was analyzed by immunofluorescence. However, the mutants tested (ΔCD4-prMA261, ΔCD4-prMA275, ΔCD4-EA749, and ΔCD4-EA763) had the same fluorescenca pattern as the wild-type CD4-TM chimeric proteins (Fig. 9F and data not shown), indicating that alanine insertion does not alter subviral particle secretion by modifying the subcellular localization of YFV envelope proteins.
The C termini of prM and E form two antiparallel transmembrane α-helices (68, 69), and we wondered whether the full-length TM regions of prM and E were necessary for subcellular retention in an early compartment of the secretory pathway. We therefore designed chimeric CD4-TM proteins with the C-terminal α-helix deleted (ΔCD4-ETM1 and ΔCD4-prMTM1). Such a deletion did not alter the subcellular localization of the chimeric proteins (Fig. 9G and data not shown), indicating that the first TM passage of YFV envelope proteins contains enough information for ER retention.
It has been shown that increasing the length of TM domains can disrupt ER retention of some proteins (52, 63, 67). To test this hypothesis for YFV envelope proteins, we inserted three hydrophobic residues (Ala-Leu-Ala) in the middle of the TM passage of a chimeric CD4-E protein in which the C-terminal α-helix was deleted (ΔCD4-ETM1-ALA). Such an insertion corresponds to an additional turn in the TM α-helix. Interestingly, this insertion led to partial export of the chimeric protein at the plasma membrane (Fig. 9H). Indeed, the immunofluorescent material was clearly detected at the plasma membrane and some staining was also detected in the perinuclear area. The perinuclear fluorescence colocalized with calnexin but did not colocalize with the Golgi marker GM130 (data not shown), indicating that this corresponds to a fraction of proteins still retained in the ER. These data indicate that the length of the first TM passage of E plays a role in the ER retention of YFV E protein.
DISCUSSION
Enveloped viruses acquire their envelope by budding through one of several host cellular membranes. Therefore, viral envelope proteins need to accumulate in the appropriate compartment before budding can take place (53). A strategy developed by most of these viruses is to endow the spike proteins with signals for compartment-specific localization (3, 4, 25, 36, 39, 64). Assembly of the flavivirus particle in the ER has therefore suggested that ER localization signals must be present in the flavivirus envelope proteins (38). Secretion of the TBE virus E protein with its C-terminal anchor domain deleted (1) suggests that there is no ER retention signal in the ectodomain of this protein. Although we cannot exclude the presence of a potential ER retention signal in the ectodomain of prM, as suggested for E1 of HCV (43), we have focused our work on the role of the TM domains of YFV envelope proteins in ER localization. In this study, we showed that proteins containing the ectodomain of CD4 or CD8 fused with the TM domain of prM or E of YFV were retained in the ER, indicating that the TM domains of flavivirus envelope proteins are ER retention signal.
Export of many secretory proteins from the ER relies on signal-mediated sorting into ER-derived vesicles (5). Proteins are transported from the ER to the Golgi complex by carrier vesicles that are formed from the membrane of the ER and that selectively fuse with the ERGIC or the cis-Golgi membranes. These vesicles are coated with a set of proteins known as coatomer protein II (COPII) (6). At least two types of sorting motifs have been identified on proteins leaving the ER compartment. They consist of a diacidic (45, 59) or a dihydrophobic (17, 21, 26) motif found in the cytosolic domain of some cargo proteins. Both the diacidic and dihydrophobic motifs bind to subunits of the COPII coat, providing a direct mechanism for cargo selection. This interaction therefore acts as a crucial element for export from the ER compartment. The C termini of prM and E contain two hydrophobic stretches separated by a short hydrophilic segment (15). Cryoelectron microscopy at high resolution and image reconstruction techniques have allowed workers to determine the disposition of the TM domains of flavivirus envelope proteins in their membrane environment (68, 69). Interestingly, the two hydrophobic stretches of prM and E were shown to form two antiparallel transmembrane α-helices, potentially making coiled-coil structures, leaving the C terminus of each protein in the lumen of the ER. In addition, the linking residues between the TM α-helices was shown to remain associated with the inner phospholipid polar head groups in the mature particle (68), strongly suggesting that the envelope proteins of the flaviviruses have no cytosolic domain. It is therefore unlikely that these proteins can be selectively incorporated into COPII vesicles. This could explain some ER retention of YFV envelope proteins or the fusion proteins containing their TM domains. However, in the absence of this type of signal, we would expect to detect some slow release of YFV envelope proteins out of the ER as observed for mutants of vesicular stomatitis virus G with their cytoplasmic tail deleted (45). In the absence of positive transport signals, localization of a protein in the ER can potentially result from ER retrieval signals. However YFV envelope proteins also lack any of the classical ER retrieval signals, KDEL, KKXX, or RRXX (57).
In the absence of any known signal, localization of flavivirus envelope proteins probably results from the properties of their TM domains and their interaction with the membranes. It has been observed that the presence of one or several hydrophilic residues in the middle of hydrophobic TM domains can play a role in ER retention (9, 10, 14, 15, 31, 67). In the case of HCV envelope proteins, replacement of the charged residues between the two hydrophobic stretches by alanine residues alters the ER retention function of their TM domains (15). However, a similar replacement of the Arg residue located between the two hydrophobic stretches in the C termini of YFV envelope proteins did not lead to an alteration in subcellular localization. As discussed above, the C termini of prM and E do not form classical TM domains. They form intramembrane antiparallel α-helices with the Arg residue probably located in the phospholipid polar head groups of the membrane leaflet in contact with the cytosol (68). This charged residue is therefore not in the middle of the TM passages, which is different from the TM domains of HCV envelope proteins (14). Interestingly, we have also shown that the first hydrophobic stretch in the C termini of prM and E is sufficient for ER retention, indicating that there is enough information in the first TM passage for ER retention. Taken together, our results are not in favor of ER retention mediated by hydrophilic residues located in the middle of a transmembrane passage of flavivirus envelope proteins.
It has also been postulated that a characteristic of proteins localized to the membrane of the Golgi apparatus or plasma membrane is the length of their hydrophobic TM domains, with shorter ones being characteristic of the Golgi and longer ones being characteristic of the plasma membrane (11). As a result of a higher content of cholesterol, the plasma membrane is usually thicker than the membranes of the Golgi, and so the longer TM domains would be selected for the transport out of the Golgi to the plasma membrane whereas the shorter TM domains would be retained in the Golgi (44). Interestingly, increasing the length of the TM domains disrupts the ER retention of some proteins (52, 63, 67), suggesting that a similar TM-based sorting might exist in the ER. The intramembrane antiparallel α-helices formed by the TM domains of prM and E are supposed to be short (68, 69). Our data indicate that the first TM passage of E contains enough information for ER localization. In addition, increasing the length of this TM passage, in the context of a chimeric CD4-E protein with the C-terminal α-helix deleted, led to partial export of this protein at the plasma membrane. This observation therefore supports the hypothesis that the length of the TM passages plays a role in ER retention of YFV envelope proteins. Together, these data suggest a lipid-based retention for YFV envelope proteins as proposed for some Golgi proteins (44).
The TM domains of HCV envelope glycoproteins have a similar ER retention function to the TM domains of prM and E (reviewed in reference 46), indicating some conservation in the functions of these TM domains in the Flaviviridae family. Sequence analyses have shown that the C termini of the envelope proteins of all the members of the Flaviviridae family have a similar organization (15): they are all composed of two hydrophobic stretches separated by a small connecting segment containing one or more charged residues. However, the TM domains of the envelope proteins of the hepaciviruses and the flaviviruses do not have the same structure (47). In the case of HCV, the TM domains form a hairpin structure before cleavage of HCV polyprotein by a host signal peptidase, and a reorientation of the second hydrophobic stretch occurs after cleavage to produce a single membrane-spanning domain (14). As discussed above, the TM domains of flavivirus envelope proteins form antiparallel α-helices (68, 69). These structural differences are associated with differences in other functions. The TM domains of HCV envelope glycoproteins have indeed been shown to play a major role in heterodimerization (49), whereas prME association does not involve the TM domains (48).
Recently, we have investigated the role of the TM domains of prM and E in the envelope formation of the YFV by alanine-scanning insertion mutagenesis (48). Most of the alanine insertions tested had a dramatic effect on the release of YFV subviral and viral particles, indicating that the TM domains play a role in flavivirus assembly. The alanine insertions probably led to an alteration of the potential coiled-coil structure of these domains. Here, we show that the TM domains of YFV envelope proteins are also involved in their subcellular localization, indicating that these domains play a major role in the formation of the flavivirus envelope.
Acknowledgments
We thank L. Cocquerel for critical reading of the manuscript and A. Pillez and S. Ung for excellent technical assistance. We are grateful to A. Cahour, P. Despres, H.-P. Hauri, and K. Moremen for providing reagents.
This work was supported by the CNRS and the Pasteur Institute of Lille.
REFERENCES
- 1.Allison, S. L., K. Stadler, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol. 69:5816-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, S. L., K. Stiasny, K. Stadler, C. W. Mandl, and F. X. Heinz. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73:5605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, A. M., L. Melin, A. Bean, and R. F. Pettersson. 1997. A retention signal necessary and sufficient for Golgi localization maps to the cytoplasmic tail of a Bunyaviridae (Uukuniemi virus) membrane glycoprotein. J. Virol. 71:4717-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong, J., and S. Patel. 1991. The Golgi sorting domain of coronavirus E1 protein. J. Cell Sci. 98:567-575. [DOI] [PubMed] [Google Scholar]
- 5.Barlowe, C. 2003. Signals for COPII-dependent export from the ER: what's the ticket out? Trends Cell Biol. 13:295-300. [DOI] [PubMed] [Google Scholar]
- 6.Barlowe, C., L. Orci, T. Yeung, M. Hosobuchi, S. Hamamoto, N. Salama, M. F. Rexach, M. Ravazzola, M. Amherdt, and R. Schekman. 1994. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77:895-907. [DOI] [PubMed] [Google Scholar]
- 7.Bause, E., W. Breuer, J. Schweden, R. Roeser, and R. Geyer. 1992. Effect of substrate structure on the activity of Man9-mannosidase from pig liver involved in N-linked oligosaccharide processing. Eur. J. Biochem. 208:451-457. [DOI] [PubMed] [Google Scholar]
- 8.Bischoff, J., and R. Kornfeld. 1986. The use of 1-deoxymannojirimycin to evaluate the role of various α-mannosidases in oligosaccharide processing in intact cells. J. Biol. Chem. 261:4758-4765. [PubMed] [Google Scholar]
- 9.Bonifacino, J. S., P. Cosson, N. Shah, and R. D. Klausner. 1991. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 10:2783-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonifacino, J. S., C. K. Suzuki, and R. D. Klausner. 1990. A peptide sequence confers retention and rapid degradation in the endoplasmic reticulum. Science 247:79-82. [DOI] [PubMed] [Google Scholar]
- 11.Bretcher, M. S., and S. Munro. 1993. Cholesterol and Golgi apparatus. Science 261:1280-1281. [DOI] [PubMed] [Google Scholar]
- 12.Cocquerel, L., S. Duvet, J.-C. Meunier, A. Pillez, R. Cacan, C. Wychowski, and J. Dubuisson. 1999. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocquerel, L., J.-C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocquerel, L., A. Op de Beeck, M. Lambot, J. Roussel, D. Delgrange, A. Pillez, C. Wychowski, F. Penin, and J. Dubuisson. 2002. Topologic changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J. 21:2893-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cocquerel, L., C. Wychowski, F. Minner, F. Penin, and J. Dubuisson. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a key role in the processing, subcellular localization and assembly of these envelope proteins. J. Virol. 74:3623-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Despres, P., A. Cahour, A. Dupuy, V. Deubel, M. Bouloy, J. P. Digoutte, and M. Girard. 1987. High genetic stability of the region coding for the structural proteins of yellow fever virus strain 17D. J. Gen. Virol. 68:2245-2247. [DOI] [PubMed] [Google Scholar]
- 17.Dominguez, M., K. Dejgaard, J. Fullekrug, S. Dahan, A. Fazel, J. P. Paccaud, D. Y. Thomas, J. J. Bergeron, and T. Nilsson. 1998. gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COP I and II coatomer. J. Cell Biol. 140:751-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubuisson, J., and C. M. Rice. 1996. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J. Virol. 70:778-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7:593-602. [DOI] [PubMed] [Google Scholar]
- 21.Fiedler, K., M. Veit, M. A. Stamnes, and J. E. Rothman. 1996. Bimodal interaction of coatomer with the p24 family of putative cargo receptors. Science 273:1396-1399. [DOI] [PubMed] [Google Scholar]
- 22.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebert, D. N., B. Foellmer, and A. Helenius. 1995. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell 81:425-433. [DOI] [PubMed] [Google Scholar]
- 24.Heinz, F. X., and S. L. Allison. 2000. Structures and mechanisms in flavivirus fusion. Adv. Virus Res. 55:231-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobman, T. C., L. Woodward, and M. G. Farquahr. 1995. Targeting of a heterodimeric membrane complex to the Golgi complex: rubella virus E2 glycoprotein contains a transmembrane Golgi retention signal. Mol. Biol. Cell 6:7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kappeler, F., D. R. Klopfenstein, M. Foguet, J. P. Paccaud, and H. P. Hauri. 1997. The recycling of ERGIC-53 in the early secretory pathway. ERGIC-53 carries a cytosolic endoplasmic reticulum-exit determinant interacting with COPII. J. Biol. Chem. 272:31801-31808. [DOI] [PubMed] [Google Scholar]
- 27.Kmiécik, D., V. Herman, C. J. M. Stroop, J. C. Michalski, A. M. Mir, O. Labiau, A. Verbert, and R. Cacan. 1995. Catabolism of glycan moieties of lipid intermediates leads to a single Man5GlcNAc oligosaccharide isomer: a study with permeabilized CHO cells. Glycobiology 5:483-494. [DOI] [PubMed] [Google Scholar]
- 28.Konishi, E., and P. W. Mason. 1993. Proper maturation of the Japanese encephalitis virus envelope glycoprotein requires cosynthesis with the premembrane protein. J. Virol. 67:1672-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kummerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letourneur, F., and P. Cosson. 1998. Targeting to the endoplasmic reticulum in yeast cells by determinants present in transmembrane domains. J. Biol. Chem. 273:33273-33278. [DOI] [PubMed] [Google Scholar]
- 32.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1042. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 33.Littman, D. R., Y. Thomas, P. J. Maddon, L. Chess, and R. Axel. 1985. The isolation and sequence of the gene encoding T8: a molecule defining functional classes of T lymphocytes. Cell 40:237-246. [DOI] [PubMed] [Google Scholar]
- 34.Liu, W. J., H. B. Chen, and A. A. Khromykh. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 77:7804-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, W. J., P. L. Sedlak, N. Kondratieva, and A. A. Khromykh. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J. Virol. 76:10766-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Locker, J. K., J. Klumperman, V. Oorschot, M. C. Horzinek, H. J. Geuze, and P. J. M. Rottier. 1994. The cytoplasmic tail of mouse hepatitis virus M protein is essential but not sufficient for its retention in the Golgi complex. J. Biol. Chem. 269:28263-28269. [PubMed] [Google Scholar]
- 37.Lorenz, I. C., S. L. Allison, F. X. Heinz, and A. Helenius. 2002. Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J. Virol. 76:5480-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorenz, I. C., J. Kartenbeck, A. Mezzacasa, S. L. Allison, F. X. Heinz, and A. Helenius. 2003. Intracellular assembly and secretion of recombinant subviral particles from tick-borne encephalitis virus. J. Virol. 77:4370-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Machamer, C. E., and J. K. Rose. 1987. A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J. Cell Biol. 105:1205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martire, G., G. Mottola, M. C. Pascale, N. Malagolini, I. Turrini, F. Serafini-Cessi, M. R. Jackson, and S. Bonatti. 1996. Different fate of a single reporter protein containing KDEL or KKXX targeting signals stably expressed in mammalian cells. J. Biol. Chem. 271:3541-3547. [DOI] [PubMed] [Google Scholar]
- 42.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 43.Mottola, G., N. Jourdan, G. Castaldo, N. Malagolini, A. Lahm, F. Serafini-Cessi, G. Migliaccio, and S. Bonatti. 2000. A new determinant of endoplasmic reticulum localization is contained in the juxtamembrane region of the ectodomain of hepatitis C virus glycoprotein E1. J. Biol. Chem. 275:24070-24079. [DOI] [PubMed] [Google Scholar]
- 44.Munro, S. 1995. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 17:4695-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishimura, N., and W. E. Balch. 1997. A di-acidic signal required for selective export from the endoplasmic reticulum. Science 277:556-558. [DOI] [PubMed] [Google Scholar]
- 46.Op De Beeck, A., L. Cocquerel, and J. Dubuission. 2001. Biogenesis of hepatitis C virus envelope glycoproteins. J. Gen. Virol. 82:2589-2595. [DOI] [PubMed] [Google Scholar]
- 47.Op De Beeck, A., and J. Dubuisson. 2003. Topology of hepatitis C virus envelope glycoproteins. Rev. Med. Virol. 13:233-241. [DOI] [PubMed] [Google Scholar]
- 48.Op De Beeck, A., R. Molenkamp, M. Caron, A. Ben Younes, P. Bredenbeek, and J. Dubuisson. 2003. Role of the transmembrane domains of prM and E proteins in the formation of yellow fever virus envelope. J. Virol. 77:813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Op De Beeck, A., R. Montserret, S. Duvet, L. Cocquerel, R. Cacan, B. Barberot, M. Le Maire, F. Penin, and J. Dubuisson. 2000. Role of the transmembrane domains of hepatitis C virus envelope proteins E1 and E2 in the assembly of the noncovalent E1E2 heterodimer. J. Biol. Chem. 275:31428-31437. [DOI] [PubMed] [Google Scholar]
- 50.Pascale, M. C., M. C. Erra, N. Malagolini, F. Serafini-Cessi, A. Leone, and S. Bonatti. 1992. Post-translational processing of an O-glycosylated protein, the human CD8 glycoprotein, during the intracellular transport to the plasma membrane. J. Biol. Chem. 267:25196-25201. [PubMed] [Google Scholar]
- 51.Pascale, M. C., N. Malagolini, F. Serafini-Cessi, G. Migliaccio, A. Leone, and S. Bonatti. 1992. Biosynthesis and oligosaccharide structure of human CD8 glycoprotein expressed in a rat epithelial cell line. J. Biol. Chem. 267:9940-9947. [PubMed] [Google Scholar]
- 52.Pedrazzini, E., A. Villa, and N. Borgese. 1996. A mutant cytochrome b5 with a lengthened membrane anchor escapes from the endoplasmic reticulum and reaches the plasma membrane. Proc. Natl. Acad. Sci. USA 93:4207-4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersson, R. F. 1991. Protein localization and virus assembly at intracellular membranes. Curr. Top. Microbiol. Immunol. 170:67-104. [DOI] [PubMed] [Google Scholar]
- 54.Popot, J.-L. 1993. Integral membrane protein structure: transmembrane α-helices as autonomous folding domains. Curr. Opin. Struct. Biol. 3:532-540. [Google Scholar]
- 55.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 56.Robertson, B., G. Myers, C. Howard, T. Brettin, J. Bukh, B. Gaschen, T. Gojobori, G. Maertens, M. Mizokami, O. Nainan, S. Netesov, K. Nishioka, T. Shini, P. Simmonds, D. Smith, L. Stuyver, and A. Weiner, for the International Committee on Virus Taxonomy. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch. Virol. 143:2493-2503. [DOI] [PubMed] [Google Scholar]
- 57.Rothman, J. E., and F. T. Wieland. 1996. Protein sorting by transport vesicles. Science 272:227-234. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 59.Sevier, C. S., O. A. Weisz, M. Davis, and C. E. Machamer. 2000. Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di-acidic motifs. Mol. Biol. Cell 11:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shin, J., R. L. Dunbrack, S. Lee, and J. L. Strominger. 1991. Signals for retention of transmembrane proteins in the endoplasmic reticulum studied with CD4 truncation mutants. Proc. Natl. Acad. Sci. USA 88:1918-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stiasny, K., S. L. Allison, A. Marchler-Bauer, C. Kunz, and F. X. Heinz. 1996. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J. Virol. 70:8142-8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szczesna-Skorupa, E., and B. Kemper. 2000. Endoplasmic reticulum retention determinants in the transmembrane and linker domains of cytochrome P450 2C1. J. Biol. Chem. 275:19409-19415. [DOI] [PubMed] [Google Scholar]
- 64.Weisz, O. A., A. M. Swift, and C. E. Machamer. 1993. Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J. Cell Biol. 122:1185-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weng, S., and R. G. Spiro. 1993. Demonstration that a kifunensine-resistant alpha-mannosidase with a unique processing action on N-linked oligosaccharides occurs in rat liver endoplasmic reticulum and various cultured cells. J. Biol. Chem. 268:25656-25663. [PubMed] [Google Scholar]
- 66.Wengler, G. 1989. Cell-associated West Nile flavivirus is covered with E+pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J. Virol. 63:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, M., J. Ellenberg, J. S. Bonifacino, and A. M. Weissman. 1997. The transmembrane domain of a carboxy-terminal anchored protein determines localization to the endoplasmic reticulum. J. Biol. Chem. 272:1970-1975. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]


