Abstract
Mammalian platelets release platelet microbicidal proteins (PMPs) as components of their antimicrobial armamentarium. The present studies defined the structure of PMP-1 and examined its structure-activity relationships. Amino acid sequencing and mass spectroscopy demonstrated that distinct N-terminal polymorphism variants of PMP-1 isolated from nonstimulated or thrombin-stimulated platelets arise from a single PMP-1 propeptide. Sequence data (NH2-[S]D1DPKE5SEGDL10HCVCV15KTTSL20 . . .) enabled cloning of PMP-1 from bone marrow and characterization of its full-length cDNA. PMP-1 is translated as a 106-amino-acid precursor and is processed to yield 73-residue (8,053 Da) and 72-residue (7,951-Da) variants. Searches with the BLAST program and sequence alignments demonstrated the homology of PMP-1 to members of the mammalian platelet factor 4 (PF-4) family of proteins. On the basis of phylogenetic relatedness, congruent sequence motifs, and predicted three-dimensional structures, PMP-1 shares the greatest homology with human PF-4 (hPF-4). By integration of its structural and antimicrobial properties, these results establish the identity of PMP-1 as a novel rabbit analogue of the microbicidal chemokine (kinocidin) hPF-4. These findings advance the hypothesis that stimuli in the setting of infection prompt platelets to release PF-4-class or related kinocidins, which have structures consistent with their likely multiple roles that bridge molecular and cellular mechanisms of antimicrobial host defense.
Mammalian platelets exhibit structural and functional attributes characteristic of antimicrobial host defense effector cells. Among these, human platelets are now recognized to liberate platelet microbicidal proteins (PMPs) that directly kill microorganisms pathogenic for humans and that mediate chemotaxis of phagocytes. Human PMPs include the CXC chemokine platelet factor 4 (PF-4), platelet basic peptide and its derivatives (e.g., connective tissue activating peptide [CTAP-3]), as well as the CC chemokine RANTES (released upon activation, normal T cell expressed and secreted 47; Y. Q. Tang, M. R. Yeaman, and M. E. Selsted, Blood 86:910a, abstr. 3626, 1995; Y. Q. Tang, M. R. Yeaman, and M. E. Selsted, Blood 86:556a, abstr. 2212, 1995). Such molecules have been termed kinocidins, reflecting their complementary microbicidal and chemokine roles, which likely coordinate molecular and cellular antimicrobial host defenses.
Peptides analogous in source and function to human PMPs exist in platelets from other mammals as well (16, 28, 50; for a review, see reference 58). Most recently, Yeaman et al. (59), Tang et al. (47), and Krijgsveld et al. (32) isolated and characterized a group of PMPs from rabbit and human platelet acid extracts and thrombin-induced releasates. Those studies demonstrated that PMPs from both sources exert rapid and potent microbicidal activities against pathogens that commonly access the bloodstream, including Staphylococcus aureus, viridans group streptococci, Escherichia coli, and Candida albicans. However, the identities, complete cDNA and amino acid sequences, and predicted structures of the predominant rabbit PMPs have not been previously determined. Moreover, the critical structure-activity relationships governing the antimicrobial functions of rabbit or human PMPs have not been previously examined in detail. Thus, the present studies provide new insights into the structural features of novel kinocidins released from platelets and compare these molecules with an analogous human kinocidin. The present studies focused on characterization of native and thrombin-stimulated forms of PMP-1, as these peptides have potent microbicidal activities and are the quantitatively predominant antimicrobial constituents in rabbit platelets (59). In addition, novel structural models were generated to compare these peptides with their human analogues.
MATERIALS AND METHODS
Platelet isolation.
Platelet-rich plasma was isolated from citrated whole blood from New Zealand White rabbits by low-speed centrifugation (250 × g). Platelets were sedimented from platelet-rich plasma by centrifugation (500 × g) for 10 min at 25°C, while the supernatant (platelet-poor plasma) was used in platelet activation assays (see below). Isolated platelets (purity, >95% by microscopy) were washed in Tyrode's salt solution (Sigma Chemical Co., St. Louis, Mo.) prior to use.
Acid extraction of platelets.
Acid extracts of platelets were prepared to examine the PMP-1 present in nonstimulated platelets. Platelets were resuspended in 6 volumes of ice-cold 30% (vol/vol) acetic acid and stirred at 0°C for 18 h. Acid-extracted material was obtained by high-speed centrifugation (18,000 × g) for 30 min at 4°C. The resulting supernatant was lyophilized and stored at −20°C. The lyophylate was dissolved in 15 ml of 30% acetic acid, clarified by centrifugation, and loaded onto a P60 Bio-Gel column (Bio-Rad). Fractions were collected and evaluated for antimicrobial activity by a modification of a well-established bioassay (33), in which Bacillus subtilis ATCC 6633 was used as a sensitive reporter organism for PMP-mediated microbicidal activity. Fractions possessing antimicrobial activity were further purified as described below.
Thrombin stimulation of platelets.
In parallel with acid extraction, thrombin stimulation of platelets was performed essentially as described previously (59). In brief, isolated platelets at a concentration of 108/ml were stimulated with 3 U of bovine thrombin (Sigma Chemical Co.) per ml in minimal essential medium (Irvine Scientific, Santa Ana, Calif.) supplemented with 0.1% (vol/vol) homologous platelet-poor plasma (to facilitate the thrombin activation cascade), obtained as described above, and 2.0 mM CaCl2. Platelets were stimulated with thrombin for 25 min at 37°C, and the supernatant with the releasate was recovered following centrifugation (2,000 × g) for 10 min at 25°C. Thrombin-induced preparations were otherwise prepared in a manner identical to that described below for the acid extracts.
Purification of native and thrombin-induced PMP-1.
The predominant PMP-1 peptides from nonstimulated and thrombin-stimulated platelet preparations were termed native PMP-1 (nPMP-1) and thrombin-induced PMP-1 (tPMP-1), respectively, as observed previously (59). These peptides were purified by reversed-phase high-performance liquid chromatography (RP-HPLC; Beckman, Fullerton, Calif.), as described in detail elsewhere (47, 59).
Verification of antimicrobial activity of purified tPMP-1.
The antimicrobial profiles of purified nPMP-1 and tPMP-1 against a panel of relevant bacterial and fungal pathogens have been well characterized previously (24, 52, 59). Thus, prior to structural characterization the antimicrobial activities of PMP-1 were verified as described above. Consistent with previous results, purified peptides exerted differential cidal activities and kinetics against susceptible S. aureus strain ISP479C compared with those against resistant strain ISP479R (52) and exhibited slightly different conditional optima (59).
SDS-PAGE and electrophoretic transfer.
Purified peptides were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 15% polyacrylamide gel. For transfer, the proteins from SDS-PAGE analyses were electrophoresed horizontally onto polyvinylidene difluoride membranes by established techniques (36). Electrophoresis was conducted in transfer buffer overnight at 30 mA and 14°C, the resulting transfer membrane was stained with Coomassie blue, and the visible bands were excised for sequence analysis.
Sequence analysis and mass spectroscopy.
The amino-terminal amino acid sequences of nPMP-1 and tPMP-1 were identified by automated Edman sequencing (Emory Microchemical Facility), and masses were determined by matrix-assisted laser desorption ionization-time of flight spectroscopy (MALDI-TOF; Protein Microsequence Facility, University of California, Los Angeles). Experimentally determined masses were within the standard confidence intervals of the calculated values.
Cloning of PMP-1 cDNA.
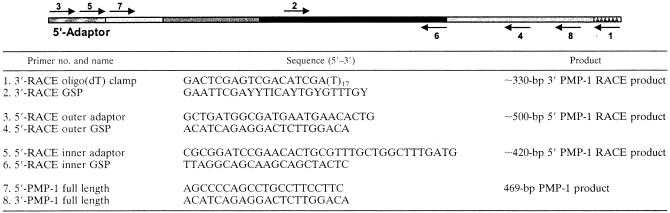
cDNA corresponding to nPMP-1 or tPMP-1 sequence data were isolated by using 3′ and 5′ rapid amplification of cDNA ends (RACE) strategies (Table 1). For the 3′-RACE, total RNA was prepared from rabbit bone marrow (RNA-STAT; ISO-TEX Diagnostics, Friendswood, Tex.), and 1 μg of RNA was used to synthesize oligo(dT)-primed first-strand cDNA (Promega, Madison, Wis.). The 3′-RACE was then performed with a 32-fold degenerate inosine-containing primer [5′-(GAATTC) GAY YTI CAY TGY GTI TGY-3′] whose sequence corresponds to the PMP-1 amino acid sequence DLHCVC paired with a 3′-RACE oligo(dT) primer (Table 1) (17). The PCRs were optimized (FailSafe; Epicenter, Madison, Wis.); and amplifications were carried out by using the following cycling parameters: 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min for 40 cycles. The 3′-RACE product sequence data were used to construct gene-specific primers for the 5′-RACE. The 5′-RACE was conducted according to the directions of the manufacturer (RLM-RACE; Ambion, Austin, Tex.) by using an RNA ligase-mediated protocol in which only full-length RNA can be used as the template. This 5′-RACE method used a primer set specific for the adaptor sequence paired with a corresponding PMP-1 gene-specific primer. Nested PCR was then performed with a second set of adaptor and PMP-1-specific primers.
TABLE 1.
PCR strategy and primers used to clone PMP-1a
The PCR primer pairs were used as indicated by the numbered arrows: 3′-RACE, primers 1 and 2; 5′-RACE, primers 3 to 6; full-length PCR, primers 7 and 8. Note that primer 3 contains deoxyinosine and a degeneracy of 32-fold. 5′-RACE adaptor primers are specific for an adaptor sequence ligated to decapped full-length mRNA sequences. 5′-RACE was conducted with outer and inner (nested) adaptor primers paired with gene-specific primers (GSPs). Shading is as follows: hatched, 5′ adaptor; light gray, untranslated regions; dark gray, signal peptide domain; black, mature peptide domain; AAAAA, poly(A) tail.
cDNA sequence analysis.
PCR-amplified RACE products were subcloned into TA-Cloning vectors (Promega), and the cDNA sequences were determined by automated in-line fluorescent chromatography by standard methods. The cDNA sequence was corroborated by comparison with the corresponding amino acid sequence (see above). Once the 5′- and 3′-end sequences of nPMP-1 and tPMP-1 cDNA were known, a PCR product corresponding to the full-length PMP-1 precursor sequence was generated and characterized by DNA sequence analysis. Sequence data for both strands of clones obtained from three independent PCRs were determined.
Knowledge-based structural characterization of PMP-1 variants.
Robust models of PMP-1 variants were generated through complementary approaches. First, a knowledge-based method, SWISS-MODEL (23, 38, 44), was used to analyze and compare combinatorial extension structural alignments and selected space-filling models of nPMP-1 and tPMP-1 (45). This approach used the BLASTP2 algorithm (1) to search for primary sequence similarities in the ExNRL-3D database. In parallel, the dynamic sequence alignment algorithm SIM (29) was used to select candidate templates with the greatest sequence identity. Subsequently, the ProModII program was used to conduct primary and refined match analyses. In some cases, energy minimization of the models was carried out with the Gromos96 algorithm to optimize template alignment.
In a complementary strategy, the amino acid sequences of nPMP-1 and tPMP-1 were converted to putative solution conformations by the sequence homology method (Composer software) (48) and the threading method (Matchmaker software [21] and Gene-Fold software [19, 20, 22, 30]) with SYBYL software (Tripos Associates, St. Louis, Mo.). The resulting conformers of target PMP-1 peptides were refined by using the AMBER95 force field (9), molecular dynamics, and energy minimization (by the method of Powell [39]). The preferred conformations of the amino-terminal portions of molecules were determined from extended molecular dynamics in an explicit solvent. The torsion angles of the peptide bonds were adjusted to 180 ± 15° with minimal constraints (0.4 kJ). In some cases, molecular dynamics were executed either with no constraints or with the α-helical region constrained by applying a 0.4-kJ penalty to the canonical Ramachandran φ and ψ angles. Final global energy minimizations were performed for each model after removal of all constraints and aggregates. These folds were then prioritized on the basis of three criteria: (i) the most favorable strain energy (molecular mechanics), (ii) empirical positional (pseudo) energy functions, and (iii) preservation of the spatial arrangement of conserved cystine pairs (3, 14, 15, 21, 34).
Comparative molecular modeling of PMP-1 variants and human PF-4 (hPF-4).
Molecular modeling of the predicted PMP-1 structures and the appropriate templates was conducted to enable quantitative structure-activity relationships to be determined with robust correlative accuracy. Homology modeling, as described above, was used to generate three-dimensional models of nPMP-1 and tPMP-1 on the basis of the priority template. The SYBYL suite of programs (version 6.6-6.7) operating on a Silicon Graphics ONYX workstation (SGI, Inc., Mountain View, Calif.) was used for these modeling techniques. Amino-terminal conformations were adopted by using energy-based criteria (9). The side chains of the target nPMP-1 and tPMP-1 models were refined by molecular dynamics and minimization of strain energies, while the backbone trajectory of the template was retained in aggregate (the positions of the peptide backbone atoms were fixed). Subsequently, the torsion angles of all peptide bonds were adjusted to 180 ± 15°, with minimal constraints. Putative differences between the nPMP-1 and tPMP-1 peptides were also quantified by measurement of the partial positive surface area by extended molecular dynamics. Where appropriate, selected model α helices were constrained by applying a 0.4-kJ penalty to the Ramachandran φ and ψ angles. Final global energy minimizations were performed for each model after removal of all constraints and aggregates. Finally, the physicochemical properties of the peptides were visualized with the MOLCAD program (25), as implemented in SYBYL software (version 6.9; Tripos Inc.) and HINT software (31).
RESULTS
Purification of nPMP-1 and tPMP-1.
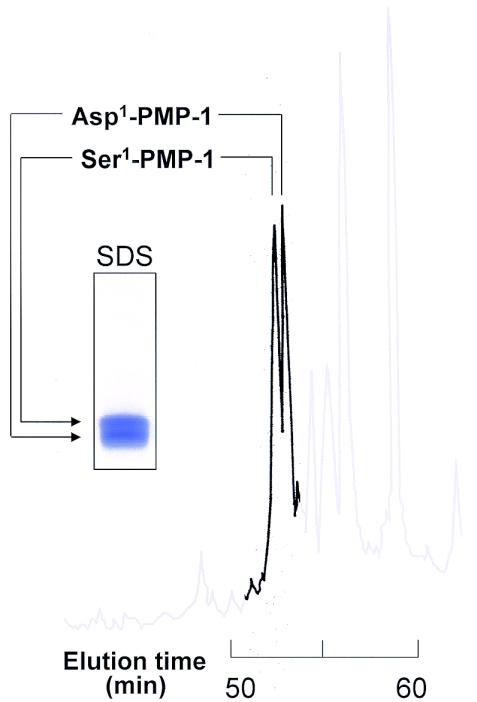
Acid-extracted and thrombin-stimulated platelet preparations contained significant peaks at A220 that were associated with potent antimicrobial activity upon screening. These peaks had very similar, but not simultaneous, elution times of 53.1 and 53.3 min on C18 RP-HPLC (Fig. 1). The respective nPMP-1 and tPMP-1 proteins were purified to homogeneity, as assessed by their elution as single peaks on analytical RP-HPLC.
FIG. 1.
Comparison of Asp1 and Ser1 polymorphic variants of rabbit PMP-1 by analytical RP-HPLC. This representative chromatogram illustrates the similarities in the elution profiles of Asp1-PMP-1 and Ser1-PMP-1 variants from thrombin-stimulated and nonstimulated platelets. RP-HPLC was performed with a water-acetonitrile gradient (5 to 40%) containing 0.1% trifluoroacetic acid developed over 65 min. The inset depicts the results of SDS-PAGE analysis of the corresponding Asp1 and Ser1 variants of PMP-1 stained with Coomassie blue.
N-terminal amino acid sequence and mass spectroscopy analyses of purified PMP-1.
Amino-terminal sequencing identified the N-terminal residues of nPMP-1 and tPMP-1 (Table 2). The two peptide sequences were found to be identical except for their N-terminal amino acid residue sequences. The principal form of PMP-1 isolated from nonstimulated platelets (nPMP-1) contains a serine residue at its N terminus (termed Ser1-PMP-1). In contrast, the relatively predominant form of PMP-1 isolated from thrombin-stimulated platelets (tPMP-1) displayed an aspartic acid residue at its N terminus (termed Asp1-PMP-1). Sequencing data were corroborated by MALDI-TOF analyses, which revealed masses of 8,053 Da (Ser1-PMP-1) and 7,951 Da (Asp1-PMP-1), corresponding to the observed N-terminal polymorphisms of the respective PMP-1 peptides.
TABLE 2.
Comparison of biochemical properties of Ser1-PMP-1, Asp1-PMP-1, and hPF-4
| Peptide | Tissue source | Sequence source | N-terminal 10-amino-acid sequence | Length (no. of residues) | Calculated mass (Da)a | Actual mass (Da)a |
|---|---|---|---|---|---|---|
| Pre-PMP-1 | Bone marrow | cDNA | MSIPEASGAP | 106 | 11,156 | NDb |
| Ser1-PMP-1 | Resting platelet (acid extract) | Amino acid | SDDPKESEGD | 73 | 8,044 | 8,053 |
| Asp1-PMP-1 | Activated platelet (thrombin stimulated) | Amino acid | DDPKESEGDL | 72 | 7,957 | 7,951 |
| Pre-hPF-4 | Erythroleukemic cell line | cDNA | MSSAAGFCAS | 101 | 10,838 | ND |
| hPF-4 | Activated platelet | Amino acid | AEAEEDGDLQC | 70 | 7,765 | 7,756 |
All values are ±0.5%.
ND, not determined.
cDNA cloning and nucleotide sequence characterization of PMP-1.
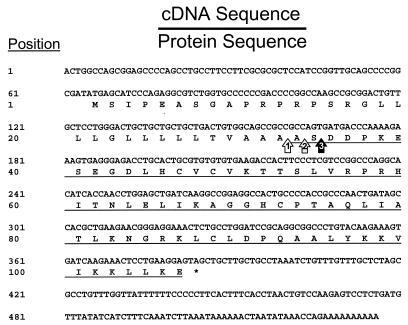
To confirm and extend the molecular characterization of PMP-1, the complete cDNA sequence corresponding to PMP-1 was isolated and analyzed (Fig. 2). The 527-bp cDNA sequence data indicate that PMP-1 is initially translated as a 106-amino-acid propeptide precursor that contains a signal sequence cleavage site at position Ala32 (Fig. 2 and 3), yielding an intermediate protein of 74 residues. Thereafter, PMP-1 is alternately processed to generate the distinct 73-residue (Ser1) or 72-residue (Asp1) derivative peptide.
FIG. 2.
Complete cDNA (top rows) and amino acid (bottom rows) sequences of the PMP-1 precursor (GenBank accession number AY450360). Cleavage of the signal peptide is predicted to occur at the position indicated by arrow 1. The mature PMP-1 sequence is underlined, and the putative Asp1 and Ser1 amino-terminal variant processing sites are indicated with arrows 2 and 3, respectively. The asterisk indicates the UAG termination codon.
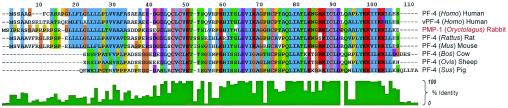
FIG. 3.
Comparative multiple-sequence alignment of PMP-1 and orthologous PF-4 proteins representing various mammalian species. The multiple-sequence alignment was generated with the Clustal W tool (version 1.81) (26, 27), as visualized with Jalview (5). The coloration scheme is formatted to the Clustal W degree of conservation. Individual peptides are designated by the following information series: peptide name (source genus) and common name. Orthologus comparators are peptide name (genus), NCBI accession number, as follows: PF-4 (Homo), NCBI accession number 2144899; vPF-4 (Homo), NCBI accession number 1360695; PMP-1 (Oryctolagus), NCBI accession number AY450360; PF-4 (Rattus), NCBI accession number 92621; PF-4 (Mus), NCBI accession number 9910486; PF-4 (Bos), NCBI accession number 72110; PF-4 (Ovis), NCBI accession number 266800; PF-4 (Sus), NCBI accession number 7441767.
Comparative analysis of the primary structure of PMP-1.
Sequence analyses (with the BLASTX program) (17) revealed that the sequences of the Ser1 and Asp1 forms of PMP-1 are highly homologous to the sequence of PF-4 (Protein Data Bank code 1RHP) from humans and other mammals (Fig. 3). Importantly, sequence alignment and phylogenetic analyses revealed that, among the known PF-4 orthologs, PMP-1 is most similar to hPF-4 (precursor protein identity, 68.2%; mature protein identity, 76%; Fig. 3). The highest degree of homology was present in the carboxy-terminal hemimers of the mature peptide sequences.
Identification of a template for modeling predicted conformations of PMP-1 variants.
All methods of homology and energy-based modeling identified hPF-4 (Protein Data Bank code 1RHP) (62) as the most appropriate modeling template. Of note, threading methods also identified another CXC chemokine, interleukin-8 (IL-8), as a potential fold template. The target peptides were constructed on the template hPF-4 and IL-8 folds and optimized. However, IL-8 was rejected as an optimal template candidate for three reasons: (i) the strain energies of the resulting global minima of the target PMP-1 peptides built on the hPF-4 template were significantly more favorable than those of the corresponding IL-8 fold; (ii) both before and after optimization, the empirical energy functions of the target PMP-1 peptides constructed on the hPF-4 template were more favorable than those of peptides built on the IL-8 template; and (iii) the hPF-4 fold preserved the spatial arrangement of the conserved cystine pairs, whereas the IL-8 fold did not. Therefore, PMP-1 models were constructed on the hPF-4 template.
The hPF-4 template (Protein Data Bank code 1RHP) contains two disulfide bonds that stabilize the overall conformation of the molecule (Fig. 4 and 5 ). Although it is highly probable that the Asp1 and Ser1 PMP-1 homologues have equivalent cross-link structures, this hypothesis was tested by computational methods. The Ser1 (nPMP-1) and Asp1 (tPMP-1) models of PMP-1 exhibited one pair of cysteines (Cys13-Cys39) within the distance and torsional constraints of a disulfide bond (Fig. 5). However, the stereogeometry of the other putative cysteine pairing (Cys15-Cys55) required further analysis. Therefore, two versions of the target PMP-1 peptides were compared to identify optimal models for nPMP-1 and tPMP-1. In the first case, reduced PMP-1 peptides were analyzed by extended molecular dynamics analysis. Studies of equilibrium (assessed by constant potential energy, total energy, radius of gyration, and intersulfur distance) demonstrated that while the secondary structural elements remained intact, the intersulfur distance of cysteine pairs increased beyond that characteristic of a disulfide. Thus, the average conformation at equilibrium was determined and minimized. In the second model, the sulfur-sulfur distances between cysteine pairs were constrained, the peptides were subjected to molecular dynamics analysis, and the conformations were minimized as in the first model. The constrained and relaxed models of hPF-4 were then compared. Although the relaxed peptides revealed more favorable strain energies, their statistical energy functions overwhelmingly indicated that the constrained conformers (reflecting the oxidized forms) are the most probable (+0.10 and −0.20 kT, respectively).
FIG. 4.
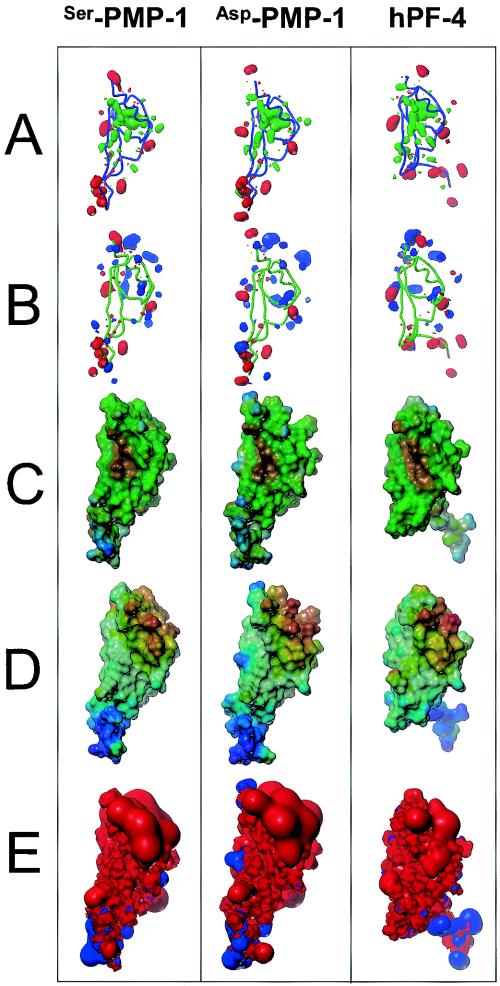
Molecular models comparing PMP-1 variants and hPF-4. PMP-1 structures were predicted from homology- and energy-based space-filling and molecular surface models. The conserved C-terminal helices are shown at the front and top of each of the structures, which are aligned in identical orientations. The flexible N-terminal tails (bottom) are shown in the most common conformations. (A) Comparison of the polar (red; contoured at a level of −30) and hydrophobic (green; contoured at a level of +20) moments (calculated with the HINT program). The peptide trajectory is represented as a cylinder for reference. Note the organization of the polar function to the exterior of each of the peptide surfaces compared with the organization of their hydrophobic cores. (B) Comparison of the acid (red, e.g., glutamyl residues; contour −10) and base (blue, e.g., lysyl residues; contour +30) moments. Note the overall bipolar distribution of acidic and basic residues. (C) Comparison of hydrophobicity projected onto peptide solvent accessible surface area (visualized with the graphic program MOLCAD; most hydrophilic, blue; most hydrophobic, brown, intermediate values, green). Note the hydrophilic propensity of the N-terminal region and hydrophobic centralization in the peptideglobular domains. (D) Comparison of peptide electrostatic field strength (most positive surface, red; most negative surface, violet; intermediate values follow the order of the colors in the spectrum). Note the striking segregation of surface charge in each molecule. (E) Comparison of the electrostatic (Coulombic) fields surrounding the peptides. Each field is contoured at 30 kcal/mol for negative (blue) and positive (red) electrostatic energy. All peptides have a net positive electrical potential and behave much like a macrocation. Note that the removal of the N-terminal Ser1 alters the electric field in the C-terminal globular region of the Asp1 variant of PMP-1. The cationic domain is clearly evident by red protrusions in the C-terminal regions (top) of each peptide. The structural organization and conservation in these peptides are consistent with the presence of biochemically distinct functional domains separated in space (e.g., the anionic N-terminal chemokine domains versus the cationic C-terminal microbicidal domains). These findings support our hypothesis that kinocidins such as PMP-1 and hPF-4 are molecular effectors of innate immunity (e.g., they direct microbicidal function) that also coordinate cell-mediated antimicrobial host defense (e.g., they potentiate the antimicrobial mechanisms of neutrophils).
FIG. 5.
Structural homology of PMP-1 and hPF-4. The predicted three-dimensional structure of PMP-1 was projected by using SWISS-MODEL (23, 38, 44). The predicted PMP-1 conformation and the known hPF-4 conformation (Protein Data Bank accession number 1RHP) were aligned by the combinatorial extension method (45) and visualized by using Protein Explorer software (35). (A) Clustal W secondary structure conservation coloration scheme; (B) DRuMS [Driscoll, Reichsman, (u), Martz, Sayle] polarity two-color scheme, in which hydrophobic residues are represented as gray, while hydrophilic residues are represented as purple; note that, by convention, cysteine residues are indicated as hydrophilic, although in these peptides, they are oxidized (cystine) and colored gray, indicating hydrophobicity; (C) alignment between PMP-1 and hPF-4, with the coloration as described for panel A; (D) alignment between PMP-1 and hPF-4, with the coloration as described for panel B. Amino (N) and carboxy (C) termini for comparative peptides are shown. The relative positions of the disulfide bonds are indicated as dotted yellow lines.
The known crystal structure of hPF-4 (Protein Data Bank code 1RHP) was extended with respect to the initial hexapeptide segment of the N terminus. In the published form (Protein Data Bank code 1RHP), the first 6 amino acids of hPF-4 are not resolved in the crystal structure. Therefore, we created a model of hPF-4 suitable for comparison with the three-dimensional models of PMP-1 peptides described in this study. This first complete model of hPF-4 was generated by appending N-terminal amino acids to the known crystal structure of hPF-4, and a preferential conformation was identified by extended molecular dynamics in explicit solvent (see below). Statistical energy functions indicated that addition of the N-terminal portion is energetically favorable (Table 3), providing a likely conformation for this portion of the molecule. Therefore, this extension of the known crystal structure of the hPF-4 conformation served as the basis for comparison with the structures of PMP-1 target molecules.
TABLE 3.
Quantitative comparison of predicted PMP-1 variant models versus hPF-4
| Peptide | PEFa | PPSAb | MPCAc | ln Pd | PSAe |
|---|---|---|---|---|---|
| Ser1-PMP-1 | −0.23 | 5,142 ± 62 | 0.006 | −15.76 | 2,600 |
| Asp1-PMP-1 | −0.21 | 5,142 ± 151 | 0.007 | −18.28 | 2,900 |
| hPF-4 | −0.22 | 4,034 ± 67 | 0.007 | −15.76 | 2,100 |
PEF, positional energy function (in kiloteslas). Mean residue energies are indicated for peptides in their monomeric forms. See Materials and Methods.
PPSA, partial positive surface area (in angstroms squared) (25). Peptide (monomeric form) motion was simulated by molecular dynamics at 310 K. The mean partial positive surface area ± standard deviation was computed for the resulting ensemble of peptide conformers. The partial positive surface area is a measure of the amount of peptide surface that carries a net positive charge. Since the side chains of the peptides are in motion at realistic temperatures, the use of molecular dynamics provides a realistic estimate of the properties of the peptides.
MPCA, most positive charged atom, defined as the charge on the most positively charged atom divided by the sum of all positive charges. The quantity is a unitless ratio.
ln P, logarithm of the partition coefficient based on Hansch relationships. The HINT program was used to estimate this parameter by the dictionary method (31).
PSA, polar surface area (in angstroms squared).
Predicted conformations of PMP-1 variants compared with that of hPF-4.
The high level of consistency of homology and threading methods suggest that the hPF-4 fold is the most likely conformation for both the Ser1 and the Asp1 forms of PMP-1. Thus, PMP-1 is predicted to be a multidomain peptide characterized by the following features (Fig. 4 and 5): (i) a relatively unstructured N-terminal region (approximate residues, Ser1 or Asp1 to Arg25/24) that has a net anionic charge (e.g., −2 at pH 7.2) and that contains a CXC motif characterstic of α-chemokines, including hPF-4; (ii) an interposing domain (approximate residues, His26/25 to Arg52/51) that conforms to an antiparallel β-sheet motif; (iii) a cationic C-terminal domain (approximate residues Lys53/52 to Glu73/72) that contains an α-helical motif consistent with peptides that exert direct microbicidal activity (11, 56); and (iv) an overall three-dimensional conformation predicted to be stabilized by two disulfide bridges, similar to the conformation in hPF-4. These structural assignments are strongly supported by favorable empirical energy functions, equivalent to those of the hPF-4 template peptide (Table 3).
The unstructured N-terminal section of the hPF-4 template is mirrored in the predicted structures of the PMP-1 variants. These equivalent degrees of structural flexibility are consistent with the large variations in the lengths of the N-terminal domains in this family of peptides (Fig. 3, 4, and 5). However, although the N-terminal loop may be mobile, there is a decided conformational preference. This conformation is supported by data generated by extended molecular dynamics analysis in explicit water. For example, the time-weighted position of the N-terminal section of the equilibrated peptide is in close agreement with that identified by the statistical energy function (Table 3). Thus, the highly similar scores obtained with the hPF-4 template, along with the subtle but detectable changes in the statistical energies of the amino acid, offer considerable support for the accuracy of the predicted conformations of the PMP-1 peptides.
Comparative electrostatic and hydrophobic features of PMP-1 and hPF-4.
The electrostatic and hydrophobic surface areas of the Ser1 and the Asp1 forms of PMP-1 compared with those of hPF-4 offered insights into the structural relatedness of these peptides (Fig. 4). As expected from their nearly identical amino acid sequences, relatively few differences in the physicochemical properties of the N-terminal variants of PMP-1 were seen. For example, the partial positive surface areas, and the relative hydrophobicities of these peptides were highly congruent with the properties of hPF-4 (Table 3). Differences were detected in the polar surface areas of the peptides (Table 3). These variances, although modest, likely arise from differences in the respective hydrophobic side chain rotomers, which have a discernible degree of conformational variability in the two forms of PMP-1 (Fig. 4 and 5). In contrast, electrostatic differences between the PMP-1 variants express contributions from their different N-terminal amino acids (Fig. 4). Moreover, the Ser1 and Asp1 forms of PMP-1 are clearly distinguishable from hPF-4.
Stereogeometry of conserved residues in PMP-1 and hPF-4.
The uniformity of the disulfide bonds in the predicted structures of Asp1-PMP-1 and Ser1-PMP-1 are consistent with the strict conservation of cysteine residues in these molecules. Thus, changes in properties induced by the amino acid substitutions compared with the sequence of hPF-4 do not disrupt the positioning of the cysteine residues for oxidation and folding to the mature peptides.
The strictly conserved double Lys pairs (Lys64-Lys65 in mature Ser1-PMP-1 or Lys63-Lys64 in mature Asp1-PMP-1; Fig. 2 and 3) are predicted to stabilize highly ordered regions of PMP-1 structures. These residues face outward into the solvent, likely facilitating ionic interactions between the peptide and target surfaces. In contrast, the strictly conserved Leu-Ile-Ala (L-I-A) triplet (residues 44 to 46 in mature Ser1-PMP-1 and residues 43 to 45 in mature Asp1-PMP-1; Fig. 2 and 3) is located in the interior of a well-ordered region of these PMP-1 variants. This structural configuration segregates charged and hydrophobic facets of PMP-1, since this three-dimensional stereogeometry creates a hydrophobic core relatively central to each peptide (Fig. 4). Thus, hydrophobic residues have reduced exposure to the aqueous environment, while hydrophilic residues are available to interact with hydrophilic surfaces and aqueous solvents.
DISCUSSION
Activated platelets likely play multiple roles in antimicrobial host defense (54, 58). Human and rabbit platelets liberate an array of microbicidal proteins upon activation with thrombin, a platelet agonist generated at sites of endovascular infection (47, 59). Multiple PMPs have been identified to be released from thrombin-stimulated human platelets (32, 47), including platelet basic protein and its derivative, connective tissue activation peptide 3, thymosin beta-4, fibrinopeptides A and B, RANTES, and PF-4. Likewise, rabbit platelets elaborate an array of PMPs upon activation with thrombin or microorganisms (59) and appear to contribute significantly to antimicrobial host defense in vivo (12, 13, 57). However, the antimicrobial functions of PMPs in vivo have been assessed by determination of outcomes in experimental models of infection due to isogenic microbial strain pairs that are susceptible and resistant to PMPs in vitro. Moreover, analogues of human PMPs have not been identified or characterized, and the structure-activity correlates of their roles in antimicrobial host defense have not been defined.
The present results reveal that rabbit PMP-1 is a microbicidal chemokine that is homologous in structure to hPF-4, a quantitatively predominant human kinocidin (47). Prior reports have offered fragmentary information regarding a rabbit homologue of hPF-4 (18, 43). However, to our knowledge, the present report is the first to describe PMP-1 variants that have been characterized in terms of complete DNA and amino acid sequences, modeled for three-dimensional structure-activity correlates, and thus, identified as rabbit analogues of hPF-4. The present data parallel previous results (59) regarding predominant acid-extracted and thrombin-induced rabbit PMPs but extend the prior data in terms of primary sequence determination and corresponding mass analyses, as well as structure-activity relationships. The potential significance of this new information is relevant to important lines of investigation: (i) these findings offer a structural basis for the activities of kinocidins and the related effector molecules that likely coordinate molecular and cellular immunity, and (ii) the host defense role(s) of PMP-1 and other kinocidin PMPs has been most extensively studied in the rabbit model of infective endocarditis; thus, characterization of a rabbit homologue of a human host defense peptide (hPF-4) emphasizes the relevance of the rabbit model and will enable future experimentation with this model in studies that cannot be performed with humans.
Although we and others have not yet isolated the predicted 106-residue preprotein, processing of this putative precursor has implications regarding the maturation of PMP-1 variants. For example, the predicted signal peptide cleavage site for the PMP-1 precursor occurs between residues Ala32 and Ala33, and cleavage at this position would generate a predicted product of 74 amino acids. However, the mature forms of PMP-1 characterized in these studies are either 73 or 72 residues in length and have N-terminal Ser1 and Asp1 residues, respectively. These data suggest that alternate and/or successive posttranslational modifications of PMP-1 occur after cleavage of the signal peptide. Of note, peptide bonds adjacent to Asp, Ser, and Thr residues are susceptible to acid-catalyzed hydrolysis (10). As these residues have interactions between side chains and the peptide bond, under favorable conditions the side chain can act as a general acid catalyst. Interestingly, PMP-1 has an H2N-Ser1-Asp2-Asp3-Pro4 motif that could be subject to such proteolysis. However, since both the nPMP-1 and the tPMP-1 forms of PMP-1 have intact Asp3-Pro4 bonds, loss of Ser1 is not likely due to autoproteolysis. These differential processing steps at the amino terminus may occur during delivery or packaging of PMP-1 in the platelet α granule or by proteases generated by tissue injury or upon platelet activation. Thus, it is conceivable that the Ser1 and Asp1 versions of PMP-1 may be produced by context-specific proteolysis of the PMP-1 precursor as it is elaborated by activated platelets. Consistent with this hypothesis, other platelet-generated antimicrobial peptides exhibit thrombin-cleavable [Pro]-Arg-Gly motifs at their N termini (47). However, conversion from the 73-amino-acid form (N-Ser) of PMP-1 to the 72-amino-acid form (N-Asp) is probably not catalyzed by thrombin, as PMP-1 lacks the [Pro]-Arg-Gly motif, the consensus cleavage amino acid sequence rules known for thrombin would be violated. Therefore, a protease(s) other than thrombin is likely responsible for generation of the 72-residue form of PMP-1.
PMP-1 variants share a marked degree of amino acid sequence identity with PF-4 proteins from other mammalian species and, on the basis of close structural and functional homologies, are rabbit immunologues of hPF-4. Sequence alignments indicate that the region spanning the CXC domain and the carboxy terminus of PMP-1 variants and PF-4 proteins is highly conserved. The PMP-1 variants share a high level of sequence conservation with hPF-4, with 68% identity over the entire precursor and ∼75% identity in residues beyond the CXC motif. Overall, this rate of amino acid identity is similar to that seen among other α chemokines, including GRO-α, MIP-2, and KC (37). However, the pattern of sequence conservation in the PF-4 family differs somewhat from that in other chemokines. For example, the IL-8, GRO, MIP-2, and KC families of chemokines exhibit the greatest degree of identity in the region encompassing the signal peptide and the ensuing 40 residues (37). It is conceivable that these distinct configurations have derived from evolutionary pressure to optimize the direct (e.g., microbicidal) and indirect (e.g., chemotactic) host defense functions of these peptides (60).
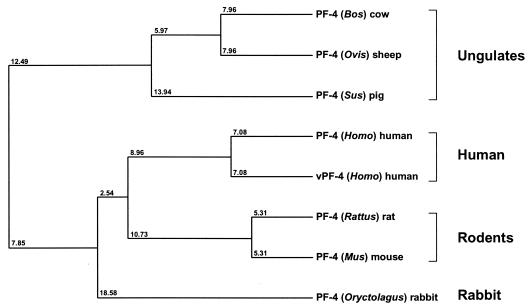
Phylogenetic analyses indicate that PMP-1 is most closely related to PF-4 proteins from humans (Fig. 6). For example, PF-4 members from pigs and rats appear to be posttranslationally glycosylated at threonine residues near the N terminus of the mature peptides (40, 41). However, PF-4 proteins isolated from human, bovine, murine, and ovine platelets do not have this modification. Quantitative agreement was found between the calculated and the spectrometrically measured masses of the Ser1 and the Asp1 versions of PMP-1. Thus, Ser1-PMP-1 and Asp1-PMP-1 appear to lack glycosylation, consistent with PF-4 from human platelets.
FIG. 6.
Phylogenetic relationship among PMP-1 and members of the mammalian PF-4 protein family. Relative evolutionary distances are indicated at branch nodes in this average-distance dendrogram (42). Representative PF-4 species are (descending order) bovine, sheep, pig, human, human variant, rat, mouse, and rabbit (NCB accession numbers are as described in the legend to Fig. 5). Note that PMP-1 (rabbit PF-4; Oryctolagus) maps to the same clade as hPF-4 as well as PF-4 from rodents.
The novel three-dimensional models of Ser1-PMP-1, Asp1-PMP-1, and hPF-4 reveal important insights into conserved antimicrobial features of the PF-4 family of proteins. For example, the PMP-1 variants and hPF-4 exhibit three structural modules (Fig. 4 and 5): (i) an extended N-terminal region containing the CXC motif, (ii) an interposing core domain consisting of triple-stranded antiparallel β sheets, and (iii) an α-helical C-terminal domain characterized by cationic charge. The high density of the positive charge in the α-helical domain imposes a strong electrostatic field upon this domain of PMP-1 and hPF-4, distinct from the anionic potentials of their N-terminal domains (Fig. 4). On the contrary, the greatest hydrophobic density is condensed within a central core in each peptide. Thus, charge and hydrophobicity are segregated and polarized in PMP-1 and hPF-4. This striking degree of three-dimensional partitioning of biophysical properties strongly supports our hypothesis that the N-terminal and C-terminal domains of PMP-1 and hPF-4 mediate distinct, yet complementary, chemotactic and microbicidal functions, respectively. The differential chemokine and antimicrobial activities of the N- and the C-terminal domains of native and chimeric forms of PMP-1 were reported previously (M. R. Yeaman, Innate Immunity Symposium, 43rd Intersci. Conf. Antimicrob. Agents Chemother., 2003). Consistent with the present structural findings, peptides encompassing the N-terminal CXC chemokine motif of PMP-1 were strong chemoattractants for rabbit and human neutrophils in vitro but had little or no direct antimicrobial activity. In contrast, peptides encompassing the N-terminal antimicrobial motif of PMP-1 exerted potent and direct microbicidal activity and enhanced the neutrophil phagocytosis of organisms exposed to these domains but had minimal chemoattractant function.
Conservation in the structural domains of PMP-1 and other PF-4 homologues suggests potential structural correlates of antimicrobial function among this group of proteins. Of note, the C-terminal α-helices in both PMP-1 and hPF-4 are amphipathic. These characteristics are consistent with those of many α-helical antimicrobial peptides (55) and likely contribute to the antibacterial activity of hPF-4 (11, 47). Moreover, synthetic peptide chimeras modeled upon the C-terminal sequences of mammalian PF-4 proteins exert direct antimicrobial activity in human blood, plasma, and serum (56). Thus, structural constraints facilitating the separation of properties in PMP-1 and hPF-4 are consistent with their distinct roles in host defense.
The present results support our hypothesis that PMP-1 and related kinocidins are multifunctional effector molecules integral to antimicrobial host defense. It is now clear that classic chemokines may exhibit direct antimicrobial activity, and known antimicrobial peptides exert chemotactic activity (6-8, 47, 53, 59, 60, 61). The convergence of these complementary functions offers intriguing new possibilities regarding the evolutionary procession of kinocidins and their roles in coordinating molecular and cellular antimicrobial host defenses.
The diversity of the primary and secondary structures of chemokines may also offer insights into the potential roles of PMP-1 in antimicrobial host defense. In contrast to highly conserved C-terminal domains, the least conserved domains within PMP-1 and the PF-4 family occur in their N-terminal hemimers. The present models of PMP-1 and hPF-4 indicate that these domains are relatively extended and contain the CXC motif, similar to that seen in other α-CXC and β-CC chemokines (2). Its high degree of accessibility may explain the fact that this domain is commonly subject to numerous proteolytic events, resulting in the high degree of amino-terminal polymorphisms observed in mature CXC and CC chemokines (4, 46, 49, 51). For example, the mature 79-residue peptide is rarely found in the CXC chemokine IL-8; however, amino-terminal truncation variants with 72, 70, and 68 residues are commonly isolated (2). Structure-activity studies of known chemokines correlate the CXC and CC motifs and flanking sequences with their chemotactic properties and specificities (4, 46). In support of this hypothesis, alternate processing of amino termini has been associated with various affinities and specificities of CXC-receptor binding and ensuing chemotactic function (2). These facts suggest a posttranslational mechanism for modulation of the antimicrobial and/or chemotactic activities of PMP-1, hPF-4, and other platelet kinocidins, depending on the local context of proteases.
The present findings underscore the roles of PMP-1, hPF-4, and other kinocidins in host defense against infection. Thrombin, a serine-protease generated at sites of damaged or infected vascular endothelium, increases platelet adherence to vascular endothelial cells, leading to platelet accumulation at these sites. In this context, adherent platelets degranulate, elaborating PMP-1 and other kinocidins that are believed to exert direct antimicrobial effects. As they diffuse away from these settings, we hypothesize that kinocidins such as PMP-1 function to recruit neutrophils and potentiate their antimicrobial mechanisms (58). While the roles of PMP-1 and hPF-4 in antimicrobial host defense have yet to be fully elucidated, the present evidence reinforces the concept that platelets are integral to preventing and/or limiting infection, likely through multiple and coordinated antimicrobial functions mediated to a large extent by kinocidins.
Acknowledgments
We thank Audree Fowler (Protein Microsequencing Facility, University of California, Los Angeles) and Jan Pohl (Emory University Microchemical Facility), Anastasia Dodson, and Robert Wiese (University of Nevada, Reno) for assistance. The insights of Alan Waring are greatly appreciated.
This work was supported in part by grant AI-48031 from the National Institutes of Health (to M.R.Y. and W.H.W.).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Baggiolini, M., B. Dewald, and B. Moser. 1994. Interleukin-8 and related chemotactic cytokines—CXC and CC chemokines. Adv. Immunol. 55:97-179. [PubMed] [Google Scholar]
- 3.Bowie, J. U., R. Luthy, and D. Eisenberg. 1991. A method to identify protein sequences that fold into a known three-dimensional structure. Science 253:164-170. [DOI] [PubMed] [Google Scholar]
- 4.Brown, K. D., S. M. Zurawski, T. R. Mosmann, and G. Zurawski. 1989. A family of small inducible proteins secreted by leukocytes are members of a new superfamily that includes leukocyte and fibroblast-derived inflammatory agents, growth factors, and indicators of various activation processes. J. Immunol. 142:679-687. [PubMed] [Google Scholar]
- 5.Clamp, M. 1998. Jalview—Java multiple alignment editor, version 1.7b. www.ebi.ac.uk/∼michele/jalview/.
- 6.Cocchi, F., A. L. DeVico, A. Garzino-Demo, S. K. Arya, R. C. Gallo, and P. Lusso. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811-1815. [DOI] [PubMed] [Google Scholar]
- 7.Cocchi, F., A. L. DeVico, R. Yarchoan, R. Redfield, F. Cleghorn, W. A. Blattner, A. Garzino-Demo, S. Colombini-Hatch, D. Margolis, and R. C. Gallo. 2000. Higher macrophage inflammatory protein (MIP)-1alpha and MIP-1beta levels from CD8+ T cells are associated with asymptomatic HIV-1 infection. Proc. Natl. Acad. Sci. USA 97:13812-13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, A. M., T. Ganz, A. M. Liese, M. D. Burdick, L. Liu, and R. M. Strieter. 2001. Cutting edge: IFN-inducible ELR-CXC chemokines display defensin-like antimicrobial activity. J. Immunol. 167:623-627. [DOI] [PubMed] [Google Scholar]
- 9.Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117:5179-5197. [Google Scholar]
- 10.Creighton, T. E. 1993. Proteins: structures and molecular properties. W. H. Freeman & Co., New York, N.Y.
- 11.Darveau, R. P., J. Blake, C. L. Seachord, W. L. Cosand, M. D. Cunningham, L. Cassiano-Clough, and G. Maloney. 1992. Peptides related to the carboxyl terminus of human platelet factor IV with antibacterial activity. J. Clin. Investig. 90:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhawan, V. K., A. S. Bayer, and M. R. Yeaman. 1998. In vitro resistance to thrombin-induced platelet microbicidal protein is associated with enhanced progression and hematogenous dissemination in experimental Staphylococcus aureus infective endocarditis. Infect. Immun. 66:3476-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhawan, V. K., M. R. Yeaman, A. L. Cheung, E. Kim, P. M. Sullam, and A. S. Bayer. 1997. Phenotypic resistance to thrombin-induced platelet microbicidal protein in vitro is correlated with enhanced virulence in experimental endocarditis due to Staphylococcus aureus. Infect. Immun. 65:3293-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenberg, D., R. Luthy, and J. U. Bowie. 1997. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, D., D. Rice, J. U. Bowie, and D. Eisenberg. 1996. Assigning amino acid sequences to 3-dimensional protein folds. FASEB J. 10:126-136. [DOI] [PubMed] [Google Scholar]
- 16.Fodor, J. 1887. Die Fahigkeit des Blutes. Bakterien zu Vernichten. Dtsch. Med. Wochenschr. 13:745-747. [Google Scholar]
- 17.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginsberg, M. H., R. Hoskins, P. Sigrist, and R. G. Painter. 1979. Purification of a heparin-neutralizing protein from rabbit platelets and its homology with human platelet factor 4. J. Biol. Chem. 254:12365-12371. [PubMed] [Google Scholar]
- 19.Godzik, A., A. Kolinski, and J. Skolnick. 1993. De novo and inverse folding predictions of protein structure and dynamics. J. Comput. Aided Mol. Design 7:397-438. [DOI] [PubMed] [Google Scholar]
- 20.Godzik, A., and J. Skolnick. 1992. Sequence-structure matching in globular proteins: application to supersecondary and tertiary structure determination. Proc. Natl. Acad. Sci. USA 89:12098-12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godzik, A., J. Skolnick, and A. Kolinski. 1992. Topology fingerprint approach to the inverse protein folding problem. J. Mol. Biol. 227:227-238. [DOI] [PubMed] [Google Scholar]
- 22.Godzik, A. 1995. In search of the ideal protein sequence. Protein Eng. 8:409-416. [DOI] [PubMed] [Google Scholar]
- 23.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 24.Harwig, S. S., T. Ganz, and R. I. Lehrer. 1994. Neutrophil defensins: purification, characterization, and antimicrobial testing. Methods Enzymol. 236:160-172. [DOI] [PubMed] [Google Scholar]
- 25.Heiden, W., T. Goetze, and J. Brickmann. 1993. Fast generation of molecular surfaces from 3D data fields with enhanced ′marching cube' algorithm. J. Comput. Chem. 14:246-250. [Google Scholar]
- 26.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 27.Higgins, D. G., and P. M. Sharp. 1989. Fast and sensitive multiple sequence alignments on a microcomputer. Comput. Appl. Biosci. 5:151-153. [DOI] [PubMed] [Google Scholar]
- 28.Hirsch, J. G. 1960. Comparative bactericidal activities of blood serum and plasma serum. J. Exp. Med. 112:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang, X., and M. Miller. 1991. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 12:337-367. [Google Scholar]
- 30.Jaroszewski, L., L. Rychlewski, B. Zhang, and A. Godzik. 1998. Fold prediction by a hierarchy of sequence, threading, and modeling methods. Protein Sci. 7:1431-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kellogg, G. E., S. F. Semus, and D. J. Abraham. 1991. HINT: a new method of empirical hydrophobic field calculation for CoMFA. J. Comput. Aided Mol. Design 5:545-552. [DOI] [PubMed] [Google Scholar]
- 32.Krijgsveld, J., S. A. Zaat, J. Meeldijk, P. A. van Veelen, G. Fang, B. Poolman, E. Brandt, J. E. Ehlert, A. J. Kuijpers, G. H. Engbers, J. Feijen, and J. Dankert. 2000. Thrombocidins, microbicidal proteins from human blood platelets, are C-terminal deletion products of CXC chemokines. J. Biol. Chem. 275:20374-20381. [DOI] [PubMed] [Google Scholar]
- 33.Lehrer, R. I., M. Rosenman, S. S. Harwig, R. Jackson, and P. Eisenhauer. 1991. Ultrasensitive assays for endogenous antimicrobial polypeptides. J. Immunol. Methods 137:167-173. [DOI] [PubMed] [Google Scholar]
- 34.Luthy, R., J. U. Bowie, and D. Eisenberg. 1992. Assessment of protein models with three-dimensional profiles. Nature 356:83-85. [DOI] [PubMed] [Google Scholar]
- 35.Martz, E. 2002. Protein Explorer: easy yet powerful macromolecular visualization. Trends Biochem. Sci. 27:107-109. [DOI] [PubMed] [Google Scholar]
- 36.Matsudaira, P. 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262:10035-10038. [PubMed] [Google Scholar]
- 37.Modi, W. S., and T. Yoshimura. 1999. Isolation of novel GRO genes and a phylogenetic analysis of the CXC chemokine subfamily in mammals. Mol. Biol. Evol. 16:180-193. [DOI] [PubMed] [Google Scholar]
- 38.Peitsch, M. C. 1995. Protein modeling by e-mail. Bio/Technology 13:658-660. [Google Scholar]
- 39.Powell, M. J. D. 1977. Restart procedures for the conjugate gradient method. Math. Program 12:241-254. [Google Scholar]
- 40.Proudfoot, A. E., E. Magnenat, T. M. Haley, T. E. Maione, and T. N. Wells. 1995. The complete primary structure of glycosylated porcine platelet factor 4. Eur. J. Biochem. 228:658-664. [DOI] [PubMed] [Google Scholar]
- 41.Ravanat, C., C. Gachet, J. M. Herbert, S. Schuhler, J. C. Guillemot, F. Uzabiaga, C. Picard, P. Ferrara, M. Freund, and J. P. Cazenave. 1994. Rat platelets contain glycosylated and non-glycosylated forms of platelet factor 4. Identification and characterization by mass spectrometry. Eur. J. Biochem. 223:203-210. [DOI] [PubMed] [Google Scholar]
- 42.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 43.Santoro, M. L., K. C. Barbaro, T. R. F. Rocha, R. J. S. Torquato, I. Y. Hirata, and I. S. Sano-Martins. 2003. Partial sequence rabbit platelet factor 4 (PF-4). Accession number 24638134. http://www.ncbi.nlm.nih.gov/entrez/.
- 44.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shindyalov, I. N., and P. E. Bourne. 1998. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11:739-747. [DOI] [PubMed] [Google Scholar]
- 46.Stoeckle, M. Y., and K. A. Barker. 1990. Two burgeoning families of platelet factor 4-related proteins: mediators of the inflammatory response. New Biol. 4:313-323. [PubMed] [Google Scholar]
- 47.Tang, Y. Q., M. R. Yeaman, and M. E. Selsted. 2002. Antimicrobial peptides from human platelets. Infect. Immun. 70:6524-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Topham, C. M., P. Thomas, J. P. Overington, M. S. Johnson, F. Eisenmenger, and T. L. Blundell. 1990. An assessment of COMPOSER: a rule-based approach to modelling protein structure. Biochem. Soc. Symp. 57:1-9. [PubMed] [Google Scholar]
- 49.Van Damme, J., J. Van Beeumen, R. Conings, B. Decock, and A. Billiau. 1989. Purification of granulocyte chemotactic peptide/interleukin-8 reveals N-terminal sequence heterogeneity similar to that of beta-thromboglobulin. Eur. J. Biochem. 181:337-344. [DOI] [PubMed] [Google Scholar]
- 50.Weksler, B. B., and R. L. Nachman. 1971. Rabbit platelet bactericidal protein. J. Exp. Med. 134:1114-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wuyts, A., C. Govaerts, S. Struyf, J. P. Lenaerts, W. Put, R. Conings, P. Proost, and J. Van Damme. 1999. Isolation of the CXC chemokines ENA-78, GRO alpha and GRO gamma from tumor cells and leukocytes reveals NH2-terminal heterogeneity. Functional comparison of different natural isoforms. Eur. J. Biochem. 260:421-429. [DOI] [PubMed] [Google Scholar]
- 52.Xiong, Y. Q., A. S. Bayer, and M. R. Yeaman. 2002. Inhibition of intracellular macromolecular synthesis in Staphylococcus aureus by thrombin-induced platelet microbicidal proteins. J. Infect. Dis. 185:348-356. [DOI] [PubMed] [Google Scholar]
- 53.Yang, D., Q. Chen, D. M. Hoover, P. Staley, K. D. Tucker, J. Lubkowski, and J. J. Oppenheim. 2003. Many chemokines including CCL20/MIP-3 alpha display antimicrobial activity. J. Leukoc. Biol. 74:448-455. [DOI] [PubMed] [Google Scholar]
- 54.Yeaman, M. R., and A. S. Bayer. 1999. Antimicrobial peptides from platelets. Drug Resist. Updates 2:116-126. [DOI] [PubMed] [Google Scholar]
- 55.Yeaman, M. R., and N. Y. Yount. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55:27-55. [DOI] [PubMed] [Google Scholar]
- 56.Yeaman, M. R., K. D. Gank, A. S. Bayer, and E. P. Brass. 2002. Synthetic peptides that exert antimicrobial activities in whole blood and blood-derived matrices. Antimicrob. Agents Chemother. 46:3883-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeaman, M. R., S. S. Soldan, M. A. Ghannoum, J. E. Edwards, Jr., S. G. Filler, and A. S. Bayer. 1996. Resistance to platelet microbicidal protein results in increased severity of experimental Candida albicans endocarditis. Infect. Immun. 64:1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yeaman, M. R. 1997. The role of platelets in antimicrobial host defense. Clin. Infect. Dis. 25:951-968. [DOI] [PubMed] [Google Scholar]
- 59.Yeaman, M. R., Y. Q. Tang, A. J. Shen, A. S. Bayer, and M. E. Selsted. 1997. Purification and in vitro activities of rabbit platelet microbicidal proteins. Infect. Immun. 65:1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yount, N. Y., and M. R. Yeaman. 2004. Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. USA 101:7363-7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, L., W. Yu, T. He, J. Yu, R. E. Caffrey, E. A. Dalmasso, S. Fu, T. Pham, J. Mei, J. J. Ho, W. Zhang, P. Lopez, and D. D. Ho. 2002. Contribution of human alpha-defensin 1, 2, and 3 to the anti-HIV-1 activity of CD8 antiviral factor. Science 298:995-1000. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, X., L. Chen, D. P. Bancroft, C. K. Lai, and T. E. Maione. 1994. Crystal structure of recombinant human platelet factor 4. Biochemistry 33:8361-8366. [DOI] [PubMed] [Google Scholar]