Abstract
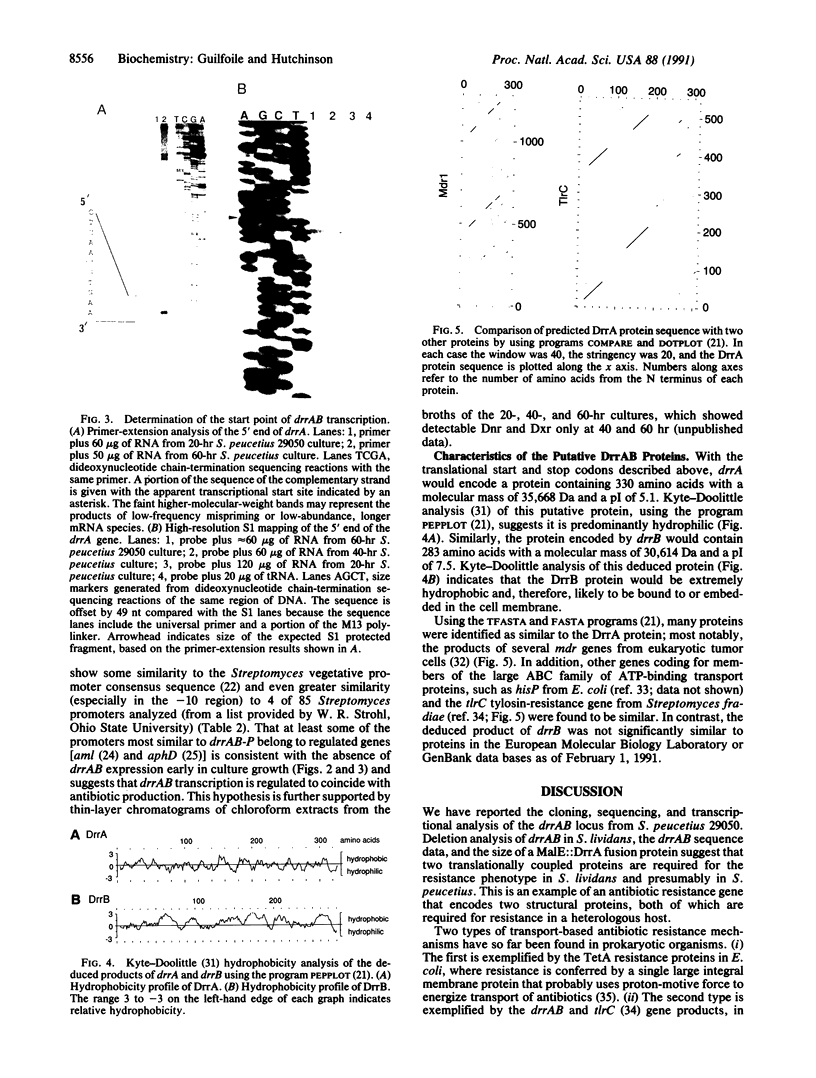
Sequence analysis of the drrAB locus from Streptomyces peucetius (American Type Culture Collection 29050) reveals the presence of two genes, drrA and drrB, both of which are required for daunorubicin and doxorubicin (Adriamycin) resistance in the heterologous host Streptomyces lividans. The DrrA protein is similar to a large family of ATP-binding transport proteins, including the proteins encoded by the mdr genes from mammalian tumor cells, which confer resistance to daunorubicin, doxorubicin, and some other structurally unrelated chemotherapeutic agents. The DrrB protein shows no significant similarity to other known proteins but is probably very hydrophobic, suggesting that it is located in the bacterial membrane. These two proteins may act jointly to confer daunorubicin and doxorubicin resistance by an analog of the antiport mechanism established for mammalian tumor cells that contain amplified or overexpressed mdr genes. Transcriptional analysis of the drrAB region supports the presence of one transcript containing drrA and drrB and indicates that these genes are expressed only during antibiotic production.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Hanamura A., Yamano H. Transcriptional terminator is a positive regulatory element in the expression of the Escherichia coli crp gene. J Biol Chem. 1991 Jan 25;266(3):1721–1727. [PubMed] [Google Scholar]
- Benveniste R., Davies J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2276–2280. doi: 10.1073/pnas.70.8.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormann C., Aberle K., Fiedler H. P., Schrempf H. Genetic complementation of Streptomyces tendae deficient in nikkomycin production. Appl Microbiol Biotechnol. 1990 Jan;32(4):424–430. doi: 10.1007/BF00903777. [DOI] [PubMed] [Google Scholar]
- Chen C. J., Chin J. E., Ueda K., Clark D. P., Pastan I., Gottesman M. M., Roninson I. B. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell. 1986 Nov 7;47(3):381–389. doi: 10.1016/0092-8674(86)90595-7. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler J., Ebert A., Mansouri K., Pissowotzki K., Stockmann M., Piepersberg W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 1987 Oct 12;15(19):8041–8056. doi: 10.1093/nar/15.19.8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt K., Wagner C. Biosynthesis of anthracyclinones. J Basic Microbiol. 1988;28(1-2):137–144. doi: 10.1002/jobm.3620280117. [DOI] [PubMed] [Google Scholar]
- Endicott J. A., Ling V. The biochemistry of P-glycoprotein-mediated multidrug resistance. Annu Rev Biochem. 1989;58:137–171. doi: 10.1146/annurev.bi.58.070189.001033. [DOI] [PubMed] [Google Scholar]
- Ferro-Luzzi Ames G. The basis of multidrug resistance in mammalian cells: homology with bacterial transport. Cell. 1986 Nov 7;47(3):323–324. doi: 10.1016/0092-8674(86)90585-4. [DOI] [PubMed] [Google Scholar]
- Fisher S. H., Wray L. V., Jr Regulation of glutamine synthetase in Streptomyces coelicolor. J Bacteriol. 1989 May;171(5):2378–2383. doi: 10.1128/jb.171.5.2378-2383.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii M., Ishizaki T., Paik S. Y., Manome T., Murooka Y. An operon containing the genes for cholesterol oxidase and a cytochrome P-450-like protein from a Streptomyces sp. J Bacteriol. 1990 Jul;172(7):3644–3653. doi: 10.1128/jb.172.7.3644-3653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E., Thompson C. J., Hopwood D. A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983 Sep;129(9):2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- Kraft R., Leinwand L. A. Sequence of the complete P protein gene and part of the M protein gene from the histidine transport operon of Escherichia coli compared to that of Salmonella typhimurium. Nucleic Acids Res. 1987 Oct 26;15(20):8568–8568. doi: 10.1093/nar/15.20.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Larson J. L., Hershberger C. L. The minimal replicon of a streptomycete plasmid produces an ultrahigh level of plasmid DNA. Plasmid. 1986 May;15(3):199–209. doi: 10.1016/0147-619x(86)90038-7. [DOI] [PubMed] [Google Scholar]
- Lee S. Y., Rasheed S. A simple procedure for maximum yield of high-quality plasmid DNA. Biotechniques. 1990 Dec;9(6):676–679. [PubMed] [Google Scholar]
- Maina C. V., Riggs P. D., Grandea A. G., 3rd, Slatko B. E., Moran L. S., Tagliamonte J. A., McReynolds L. A., Guan C. D. An Escherichia coli vector to express and purify foreign proteins by fusion to and separation from maltose-binding protein. Gene. 1988 Dec 30;74(2):365–373. doi: 10.1016/0378-1119(88)90170-9. [DOI] [PubMed] [Google Scholar]
- McMurry L., Petrucci R. E., Jr, Levy S. B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal R. J., Chater K. F. Nucleotide sequence analysis reveals similarities between proteins determining methylenomycin A resistance in Streptomyces and tetracycline resistance in eubacteria. Gene. 1987;58(2-3):229–241. doi: 10.1016/0378-1119(87)90378-7. [DOI] [PubMed] [Google Scholar]
- Otten S. L., Stutzman-Engwall K. J., Hutchinson C. R. Cloning and expression of daunorubicin biosynthesis genes from Streptomyces peucetius and S. peucetius subsp. caesius. J Bacteriol. 1990 Jun;172(6):3427–3434. doi: 10.1128/jb.172.6.3427-3434.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido D., Jiménez A. Optimization of gene expression in Streptomyces lividans by a transcription terminator. Nucleic Acids Res. 1987 May 26;15(10):4227–4240. doi: 10.1093/nar/15.10.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M., Gros P. Mammalian multidrug-resistance gene: correlation of exon organization with structural domains and duplication of an ancestral gene. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6488–6492. doi: 10.1073/pnas.86.17.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynes J. P., Calmels T., Drocourt D., Tiraby G. Cloning, expression in Escherichia coli and nucleotide sequence of a tetracycline-resistance gene from Streptomyces rimosus. J Gen Microbiol. 1988 Mar;134(3):585–598. doi: 10.1099/00221287-134-3-585. [DOI] [PubMed] [Google Scholar]
- Richardson M. A., Kuhstoss S., Solenberg P., Schaus N. A., Rao R. N. A new shuttle cosmid vector, pKC505, for streptomycetes: its use in the cloning of three different spiramycin-resistance genes from a Streptomyces ambofaciens library. Gene. 1987;61(3):231–241. doi: 10.1016/0378-1119(87)90187-9. [DOI] [PubMed] [Google Scholar]
- Rosteck P. R., Jr, Reynolds P. A., Hershberger C. L. Homology between proteins controlling Streptomyces fradiae tylosin resistance and ATP-binding transport. Gene. 1991 Jun 15;102(1):27–32. doi: 10.1016/0378-1119(91)90533-h. [DOI] [PubMed] [Google Scholar]
- Sharrocks A. D., Hornby D. P. S1 nuclease transcript mapping using sequenase-derived single-stranded probes. Biotechniques. 1991 Apr;10(4):426–428. [PubMed] [Google Scholar]
- Stein D. S., Kendall K. J., Cohen S. N. Identification and analysis of transcriptional regulatory signals for the kil and kor loci of Streptomyces plasmid pIJ101. J Bacteriol. 1989 Nov;171(11):5768–5775. doi: 10.1128/jb.171.11.5768-5775.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vara J., Lewandowska-Skarbek M., Wang Y. G., Donadio S., Hutchinson C. R. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J Bacteriol. 1989 Nov;171(11):5872–5881. doi: 10.1128/jb.171.11.5872-5881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virolle M. J., Long C. M., Chang S., Bibb M. J. Cloning, characterisation and regulation of an alpha-amylase gene from Streptomyces venezuelae. Gene. 1988 Dec 30;74(2):321–334. doi: 10.1016/0378-1119(88)90166-7. [DOI] [PubMed] [Google Scholar]
- Vögtli M., Hütter R. Characterisation of the hydroxystreptomycin phosphotransferase gene (sph) of Streptomyces glaucescens: nucleotide sequence and promoter analysis. Mol Gen Genet. 1987 Jun;208(1-2):195–203. doi: 10.1007/BF00330442. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]