Abstract
DNA damage induces p53 DNA binding activity, which affects tumorigenesis, tumor responses to therapies, and the toxicities of cancer therapies (B. Vogelstein, D. Lane, and A. J. Levine, Nature 408:307-310, 2000; K. H. Vousden and X. Lu, Nat. Rev. Cancer 2:594-604, 2002). Both transcriptional and transcription-independent activities of p53 contribute to DNA damage-induced cell cycle arrest, apoptosis, and aneuploidy prevention (M. B. Kastan et al., Cell 71:587-597, 1992; K. H. Vousden and X. Lu, Nat. Rev. Cancer 2:594-604, 2002). Small-molecule manipulation of p53 DNA binding activity has been an elusive goal, but here we show that NAD+ binds to p53 tetramers, induces a conformational change, and modulates p53 DNA binding specificity in vitro. Niacinamide (vitamin B3) increases the rate of intracellular NAD+ synthesis, alters radiation-induced p53 DNA binding specificity, and modulates activation of a subset of p53 transcriptional targets. These effects are likely due to a direct effect of NAD+ on p53, as a molecule structurally related to part of NAD+, TDP, also inhibits p53 DNA binding, and the TDP precursor, thiamine (vitamin B1), inhibits intracellular p53 activity. Niacinamide and thiamine affect two p53-regulated cellular responses to ionizing radiation: rereplication and apoptosis. Thus, niacinamide and thiamine form a novel basis for the development of small molecules that affect p53 function in vivo, and these results suggest that changes in cellular energy metabolism may regulate p53.
About one-half of all human tumors contain mutations in p53 (20), and tumor-derived p53 mutations interfere with p53 sequence-specific DNA binding. Consensus p53 binding sites comprise a 20-bp DNA sequence that is functionally organized as two consecutive 10-bp half-sites (13, 16, 25, 26). The tetramerization domain organizes p53 as a dimer of dimers (2, 23, 29), and the two dimers in a p53 tetramer bind cooperatively to adjacent DNA half-sites in a consensus binding site (35).
Genotoxic stresses, including ionizing radiation, induce p53 binding to many consensus DNA sites and enhance transcription of p53 target genes (25, 54, 55). The product of one such target gene, p21, induces cell cycle arrest and may prevent endoreduplication and rereplication (12, 52, 56). However, p21−/− cells fail to undergo DNA damage-induced G1 arrest even though p21−/− mice are not tumor prone, suggesting that p21-dependent processes, including G1 arrest, do not mediate p53 tumor suppressor activity (3, 11).
There are also many potential gene products that are transcriptionally activated by p53 and that are involved in apoptosis (55). p53 can not only activate transcription of various apoptosis-promoting genes but also repress transcription and can promote apoptosis independently from transcriptional effects (4, 8, 10, 18, 27, 37). The multifaceted nature of p53 activity is highlighted by the findings that tumor-derived mutant p53 proteins that are defective in inducing apoptosis not only fail to sequence-specifically bind DNA but also fail to bind ASPP1/2 (the full-length form of 53BP2) and bclxL (37, 47). Moreover, an unusual p53 mutation that inhibits p53-dependent apoptosis but still does not lead to the early onset of spontaneous lymphomas observed in p53 null mice was recently found. This suggests that functions other than apoptosis must be able to contribute to p53 tumor suppressor activity (30).
Despite uncertainty about what constitutes p53 apoptotic and tumor suppressor activity, another p53 target, the negative-feedback regulator mdm2 (or hdm2 in human cells) (24), binds to the p53 protein and both inhibits p53 transactivation functions (38, 42) and facilitates p53 degradation (17, 28). Thus, hdm2 attenuates p53 function by limiting the duration and extent of DNA damage-induced p53 protein accumulation. One approach to enhance p53 activity is to interfere with the p53-hdm2 interaction (6, 51). Another conceptual approach is to inhibit p53 binding to and activating transcription from the hdm2 DNA binding site.
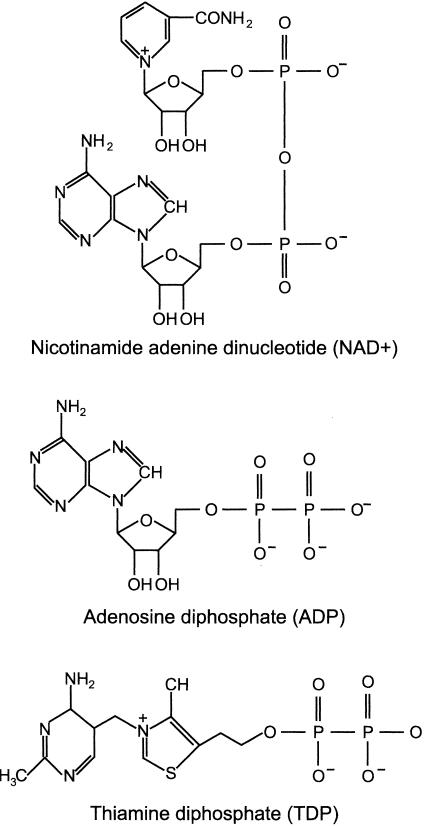
Molecules that modulate p53 binding to the hdm2 DNA binding site could alter the spectrum of p53 targets induced by DNA damage, change the duration of the p53 response, and alter the outcome of cellular responses to DNA damage. The sequence-specific DNA binding activity of p53 in vitro was reported to be affected by ADP (41), and we noted that the metabolic energy cofactor NAD+ contains an ADP moiety (Fig. 1). Since NAD+ is an enzymatic substrate of poly(ADP-ribose) polymerase (PARP), an enzyme that uses NAD+ to poly-ADP ribosylate substrates in response to DNA damage, including p53 (19), and, for Sir2, an NAD+-dependent histone deacetylase that can deacetylate p53 (33, 53), it seemed reasonable to query whether NAD+ also affected p53 DNA binding activity.
FIG. 1.
Structures of NAD+, ADP, and TDP.
MATERIALS AND METHODS
In vitro DNA binding.
The wild-type human p53 protein was prepared as described previously (39). DNA binding proceeded at 23°C in 10 μl of buffer (1 μl of p53, 1 ng of 32P-DNA, 0.1 μg of nonbinding competitor DNA, 0.2 μg of sheared salmon sperm DNA, 3 mM dithiothreitol, 50 mM KCl, 100 mM HEPES [pH 7.5], 12% glycerol, 0.5 μg of antibody, 5 mM nucleotide [determined by measuring A260]). NADH was prepared weekly and stored at 4°C, and NAD+ was stored at −20°C. Complexes were resolved on native 0.5× or 1× Tris-borate-EDTA-polyacrylamide gels and then dried and exposed to film (Kodak) or a PhosphorImager screen (Molecular Dynamics) for quantification.
NAD+ binding and protease sensitivity.
RNA was translated in vitro (wheat germ extract; Promega). For NAD+ binding, products were diluted in phosphate-buffered saline (PBS), immunoprecipitated (mixture of DO1, 1801, and 1-393; Santa Cruz Biotechnology), and washed three times with PBS-5 μM NAD+. Immunoprecipitates were incubated for 10 min at 25°C in 10 μl of a mixture of PBS and 10 μCi of 32P-NAD+ (Amersham), washed five times with PBS, resuspended in PBS, spotted on 3mm paper (Whatman), and visualized by autoradiography. For protease assays, [35S]methionine-labeled translation reaction mixtures were diluted (0.1 M Tris [pH 7.5], 0.1 M KCl, 0.1% NP-40, 10% glycerol), incubated with NAD+ or NADH (5 mM), and digested with trypsin (sequencing grade; Boehringer-Mannheim) at 37°C for 10 min. Products were denatured in protein sample buffer (5 min, 95°C), resolved on sodium dodecyl sulfate (SDS)-polyacrylamide gels, and visualized by autoradiography.
Nucleotide analysis.
Niacinamide was added to MCF7 cells (600 mg/liter, 90 min). 2-Deoxyglucose (10 mM) was added for 10 min to limit reduction of NAD+ to NADH, and then cells were irradiated (6 Gy of ionizing radiation, ∼1 Gy/min). After 3 h, cells were washed and suspended in PBS. Protein was extracted in radioimmunoprecipitation assay (RIPA) buffer (see “Western blotting” section) from one aliquot, while nucleotides were acid extracted from another aliquot and analyzed by high-performance liquid chromatography (HPLC) essentially as described previously (36). The HPLC column was an LC-18 (3 μm, 7.5 cm by 4.6 mm; Supelco) and was preceded by a LC-18 guard column. Mobile phase A was 0.1 M potassium phosphate-8 mM tetrabutylammonium phosphate (pH 5.5); mobile phase B was 70% mobile phase A-30% methanol, and the gradient was from 10 to 100% mobile phase B. Retention times were 3.8 min for NAD+ and 8.7 min for NADP+. Peak identities were confirmed by matching UV spectra from the diode array with authentic standards.
ChIP.
Chromatin immunoprecipitation (ChIP) was essentially as described previously (49). One percent formaldehyde in PBS was added for 10 min at 25°C to cross-link protein to DNA. Cells were lysed for 20 min in RIPA buffer (as described in the “Western blotting” section), passed five times through a 25-gauge needle, sonicated (Branson) to yield 0.5- to 1-kb DNA fragments, and centrifuged (15 min, 8,000 × g), and then 1 mg of soluble protein (Bio-Rad; DC protein assay) was immunoprecipitated for 3 to 16 h (anti-p53 DO1, PAb1801, and 1-393 or anti-p21 or antibax [Santa Cruz Biotechnology]). Bound DNA from 1 to 10 μg (protein) of cell extract was amplified (Ready-to-go beads [Pharmacia] and GeneAmp 9700 [Applied Biosystems]) in the linear range with 100 ng of primers (49) to yield products containing the genomic p53 binding site. Amplification was linear to 26 to 28 cycles (hdm2), 28 to 30 cycles (p21/GADD45), and 30 cycles (PIG3/bax). The PCR profile was 2 min at 94°C and then 12 s at 94°C, 6 s at 60°C, and 12 s at 72°C. Products were visualized on ethidium bromide-2% agarose gels (Gel Doc 2000; Bio-Rad).
Semiquantitative reverse transcription-PCR.
Total cytoplasmic RNA was isolated (RNeasy kit; QIAGEN), reverse transcribed (Tth polymerase or Superscript II; Roche), amplified to a linear cycle (15 to 24 cycles, determined by quantifying aliquots of PCR products at increasing cycle numbers), and visualized on ethidium bromide-agarose gels (Gel Doc 2000; Bio-Rad).
Western blotting.
MCF7 cells were rinsed and scraped in cold PBS, pelleted, and lysed for 20 min in RIPA buffer (PBS containing 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM NaF, 10 mM β-glycerophosphate, 5 μM trichostatin A, 1 mM phenylmethylsulfonyl fluoride, and 1% mammalian protease inhibitor cocktail σ). The protein concentration of cleared lysates (12 min at 16,000 × g and 4°C) was determined (Bio-Rad; DC). Eighty micrograms of lysate was heated in protein sample buffer at 95°C for 5 min and then electrophoresed (NuPAGE 4 to 12% bis-Tris polyacrylamide gel-MES [morpholineethanesulfonic acid]-SDS running buffer; Invitrogen). Proteins were transferred to nitrocellulose (Schliecher & Schuell) and then blotted (anti-p21, anti-p53 [amino acids 1 to 393], and anti-hdm2 [SMP14] [Santa Cruz Biotechnology]; anti-p53 phosphoserine 15 [Cell Signaling]; anti-p53 acetyl 373/382 [Upstate Biotechnology]; and Ku70 [Ab-4; NeoMarkers]).
Rereplication.
MCF7 cells were treated (0.6 g of niacinamide/liter for 90 min or 4 g of thiamine/liter for 60 min in Dulbecco's modified Eagle medium [DMEM]-10% fetal bovine serum [FBS]) and then irradiated (8 Gy of ionizing radiation, ∼1 Gy/min). After 3 days fresh DMEM-10% FBS was added, and 2 days later cells were treated for 5 h with Colcemid (Gibco) and then harvested and prepared for mitotic spreads.
Apoptosis.
Thymocytes were explanted from 5- to 6-week-old C57BL6 mice into RPMI 1640-10% FBS. p53−/− mice (T. Jacks) were backcrossed onto a C57BL6 background. Samples from individual mice were divided into equal aliquots; aliquots were untreated or treated with niacinamide (0.6 g/liter) or thiamine (4 g/liter) with or without a subsequent ionizing radiation dose. For in vivo studies, 5- to 6-week-old C57BL6 mice were maintained and treated under approved institutional guidelines. Niacinamide was administered by intraperitoneal injection of 0.2 ml of a 0.1-g/ml solution 10 to 15 min before irradiation. A 137Cs source was used to deliver 2 Gy at 0.5 Gy/min. After 18 h, thymocytes were collected by centrifugation, fixed in 70% ethanol, and incubated for 30 min with 10 μg of RNase A/ml and propidium iodide. Dexamethasone was added to 1 μM for 8 h. Cells were analyzed on a FACSCalibur (Becton Dickenson), and apoptotic cells were routinely scored as those with sub-G1 DNA content. Similar data were obtained by DiOC6 (3,3′-dihexyloxacarbocyanine iodide) fluorescence.
RESULTS
NAD+ binds p53 tetramers, induces conformational change, and inhibits DNA binding.
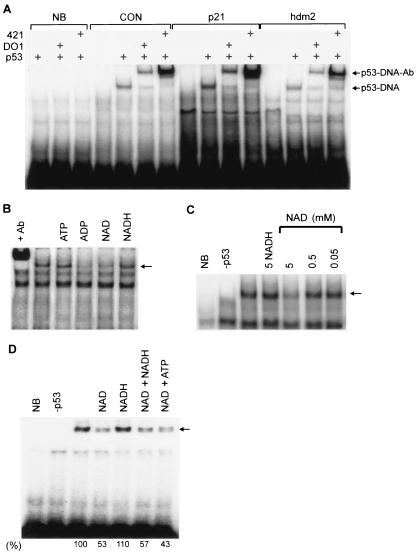
Bacterially produced recombinant p53 binds DNA sequence specifically (Fig. 2A), and this binding was inhibited by ADP and NAD+, but not by ATP or NADH (the reduced form of NAD+) (Fig. 2B). Despite a submicromolar p53 concentration, DNA binding activity was affected only by a millimolar NAD+ concentration (Fig. 2C). Since total free cellular NAD+ is ∼0.4 to 0.5 mM (50) and NAD+ synthesis is nuclear (44), the local nuclear concentration could be in the millimolar range when there is a high rate of NAD+ synthesis. Thus, the concentrations of NAD+ that affect p53 DNA binding in vitro may be physiologically relevant. We note that, although NADP+ affected p53 DNA binding similarly to NAD+, we focused on NAD+ because the cellular concentration of free NAD+ is much higher than that of NADP+ (50).
FIG. 2.
Effect of NAD+ on sequence-specific DNA binding activity of recombinant p53 expressed in bacteria. (A) Wild-type human p53 was incubated with 32P-DNA sequences and electrophoresed under nondenaturing conditions. CON, p53 consensus binding site; NB, p53-nonbinding sequence containing mutations at four critical G/C contact residues; p21 and hdm2, 5′ p21 and hdm2 human genomic binding sites, respectively. Amino- and carboxy-terminal-specific DO1 and PAb421 monoclonal antibodies were added to supershift the p53-DNA complex. (B) p53 was incubated with 32P-hdm2 plus various nucleotides. In lane +Ab antibody PAb421 was added to supershift the p53-DNA complex. (C) NADH (5 mM) or NAD+ at the indicated concentrations were added as indicated. Sequence-specific binding controls include p53 plus the nonbinding control DNA sequence (lane NB) and hdm2 DNA without p53 (lane −p53). (D) Nucleotides were added as indicated to p53 and 32P-hdm2 DNA.
The high concentration of NAD+ required to affect p53 DNA binding activity could reflect either an alteration of the p53 redox state or a low-affinity interaction between p53 and NAD+. In vitro, p53 DNA binding requires reducing conditions (15), but NAD+ modulation of the redox state of p53 was an unlikely cause of the binding inhibition since reducing equivalents of dithiothreitol were present in excess over NAD+ and further increasing the dithiothreitol concentration had no effect (data not shown). Additionally, redox-inactive ADP inhibited DNA binding (Fig. 2B), and the effect of NAD+ was not counteracted by addition of NADH to the reaction (Fig. 2D).
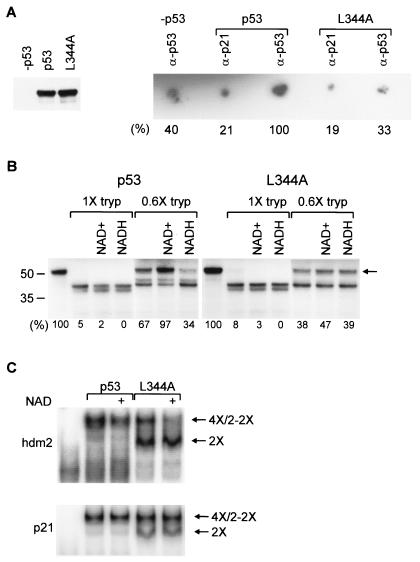
An alternative explanation for the inhibition of p53 DNA binding by NAD+ was suggested by the specific binding of radiolabeled NAD+ to p53 (Fig. 3A, right). NAD+ bound to the inherently tetrameric wild-type p53 protein but did not bind above background to the L344A mutant protein (referred to as L344A) (Fig. 3A, protein expression shown in left panel), which forms only dimers (57). The interaction between p53 and NAD+ is specifically increased in the p53 immunoprecipitation compared to the negative controls. The relatively low degree of binding above nonspecific background further supports the low-affinity binding suggested by the high NAD+ concentration required to affect p53 DNA binding activity in vitro. The interaction between NAD+ and p53 does not occur via the isolated tetramerization domain, as assessed by nuclear magnetic resonance and Biacore analysis (C. Galea and R. Kriwacki, personal communication). NAD+ can also decrease the DNA binding activity of p53 that has PAb421 bound to an epitope at the carboxy-terminal end of the tetramerization domain, suggesting that p53 might bind NAD+ via an extended interface created between two dimers of a tetramer.
FIG. 3.
NAD+ binding, affecting p53 conformation and specifically inhibiting p53 tetramer DNA binding in vitro. (A, left) Equal amounts of p53 and L344A were incubated with 32P-NAD+. (Right) Bound 32P-NAD+. Signal strength is indicated below each spot; signal strength for anti-p53 is arbitrarily 100%. (B) [35S]methionine-labeled proteins were incubated with NAD+ or NADH and then digested with trypsin (tryp). The percentage of undigested p53 (arrow) is indicated below each lane. (C) NAD+ was added with 32P-hdm2 (top) or 32P-p21 (bottom) to p53 tetramers or dimers. DNA-bound proteins are tetramers (4×), single L344A dimers (2×), or pairs of L344A dimers (2-2×).
Moreover, NAD+ caused a conformational change in tetrameric p53. A high concentration of trypsin cleaved [35S]methionine-labeled wild-type p53 and L344A to a pair of bands that migrate at ∼40 and 42 kDa on an SDS gel (Fig. 3B, lanes 1× tryp). NAD+ inhibited cleavage of wild-type p53, but not L344A, at a lower concentration of trypsin (Fig. 3B, lanes 0.6× tryp). The lower concentration of trypsin appears to produce a partially digested fragment that is completely digested at a higher trypsin concentration. That there is a difference between dimers and tetramers is not surprising, as tetramers differ not only in the tetramerization domain per se (34) but also in an extended protein-protein interaction domain that is created between the two dimers of a tetramer and that could confer differential protease sensitivity. NADH increased p53 tetramer, but not dimer, sensitivity to trypsin, suggesting that NADH might also affect p53 tetramer conformation in such a way as to prevent the ADP portion of NADH from affecting p53 DNA binding activity. The lack of effect of NAD+ and NADH on L344A dimer sensitivity to trypsin demonstrates both that dimer conformation and that trypsin activity were unaffected by NAD+.
We therefore compared the effect of NAD+ on the DNA binding activity of p53 tetramers versus dimers. L344A dimers bind DNA because they retain the wild-type conformation (35, 57). L344A bound the hdm2 and p21 DNA sites as two species (Fig. 3C). The lower complex represented a single dimer bound to one of the two half-sites in the full DNA site, and the higher complex represented a pair of dimers bound to the two adjacent DNA half-sites in a full DNA site (35). Strikingly, NAD+ specifically decreased the binding of the pair of dimers, but not the single dimer, to the hdm2 DNA site (Fig. 3C, 2-2× and 2×, respectively). Interestingly, in these DNA binding conditions the effect of NAD+ on the hdm2 DNA site was more pronounced than its effect on the p21 DNA site (Fig. 3C). This suggests that NAD+ does not inactivate p53, but rather alters the conformation of p53 in such a way as to differentially affect the affinity of p53 tetramers for different binding sites.
Small molecules related to ADP inhibit p53 DNA binding.
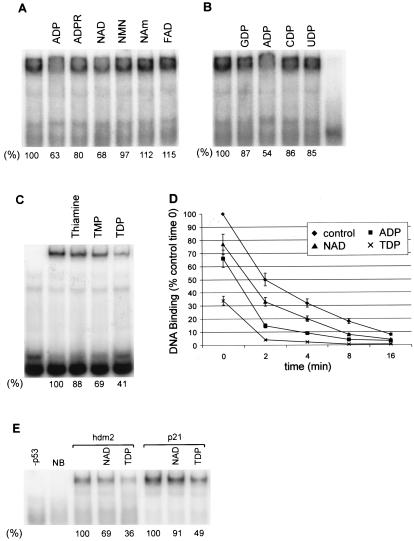
How NAD+ affects p53 tetramers can be further delineated by examining the structural aspects of the NAD+ molecule. Nicotinamide mononucleotide (NMN) is NAD+ without the ADP moiety, and ADP-ribose (ADPR) is NAD+ without the niacinamide moiety. ADP, and to a lesser extent ADPR, but neither NMN nor niacinamide, inhibited p53 DNA binding (Fig. 4A). However, the NMN portion of NAD+ is still important because, when it was reduced in NADH or when it was replaced in oxidized flavin adenine dinucleotide, p53 DNA binding was unaffected (Fig. 2, lanes NADH, and 4A, lane FAD). Thus, the ADP moiety of NAD+ is crucial, while the nicotinamide portion affects whether the ADP portion of NAD+ can regulate p53 DNA binding activity.
FIG. 4.
Effects of related small molecules on p53 DNA binding. (A) Portions of the NAD+ molecule were incubated with p53 and 32P-hdm2. NAm, niacinamide; FAD, flavin adenine dinucleotide. (B) Nucleoside diphosphates were added as indicated to p53-32P-hdm2 DNA. (C) Thiamine, TMP, or TDP was added to p53 DNA binding reactions. (D) Kinetic analysis of the dissociation of p53 from DNA in the presence of nucleotides as indicated. Nucleotides were added to p53-32P-hdm2, and then excess unlabeled hdm2 was added at time zero. The amount of p53 remaining bound to 32P-hdm2 at the indicated times is plotted. (E) The effect of NAD or TDP on p53 binding to p21 versus hdm2 DNA is quantified below each lane. NB and −p53 are as defined for Fig. 2.
Since ADP, but not ATP, affects p53 DNA binding (Fig. 2B), the diphosphate must be important. ADP, but neither GDP, CDP, nor UDP, inhibited p53 DNA binding (Fig. 4B). The feature unique to ADP is the primary amino group on the adenosine ring. We note that TDP (or thiamine pyrophosphate) has weak structural similarity to ADP in that it contains an amino-substituted pyrimidine ring linked to a diphosphate (Fig. 1). Like ADP, TDP inhibited p53 DNA binding in vitro (Fig. 4C), with TDP inhibiting binding to a greater extent than NAD+ (Fig. 4D, time zero). Although NAD+ preferentially inhibited p53 binding to the hdm2 DNA site, TDP inhibited p53 binding to both the hdm2 and p21 DNA sites (Fig. 4E).
Niacinamide increases cellular NAD+ synthesis and regulates p53 DNA binding in vivo.
It was important to determine whether the in vitro observations that NAD+ and TDP affected sequence-specific p53 DNA binding could be replicated in vivo. Although cellular conditions, such as hypoxia, significantly alter the NAD+/NADH ratio, this ratio is preferentially influenced by changes in NADH rather than NAD+, because the free NAD+/NADH ratio is ∼725 (58). We incubated MCF7 cells with a high level of the NAD+ biosynthetic precursor, niacinamide (vitamin B3), which increased the rate of intracellular NAD+ synthesis, resulting in a 30 to 40% increase in total cellular NAD+ (Fig. 5A) (1, 36). Although the majority of cellular NAD+ is mitochondrial or cytoplasmic, NAD+ synthesis is nuclear (44). Thus, especially under nonequilibrium conditions of an increased rate of NAD+ synthesis induced by excess niacinamide, there would be a greater local increase in NAD+ in the nucleus, where p53 DNA binding activity could be affected.
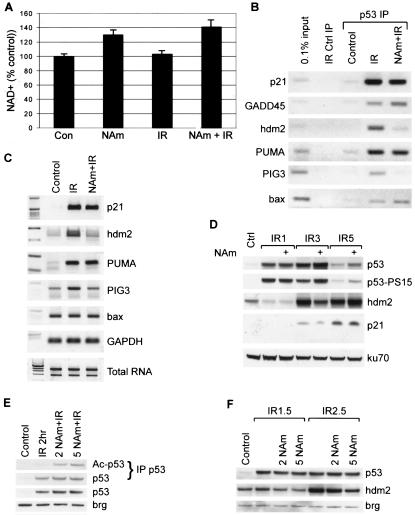
FIG. 5.
Effects of niacinamide on p53 activity. (A) Quantification of intracellular NAD+ levels after treating MCF7 cells with niacinamide (NAm), irradiated (IR) as indicated to induce endogenous, wild-type p53. (B) Cells were treated with niacinamide to increase NAD+ and then irradiated to induce endogenous, wild-type p53. After 3 h (lane IR), genomic DNA sites that were bound to p53 protein were analyzed by ChIP (IP). Ctrl, control. (C) Cells were treated as in panel B, but RNA was extracted and analyzed by semiquantitative reverse transcription-PCR. (D) Cells were untreated or treated with niacinamide and then irradiated and allowed to recover for 1, 3 or 5 h (IR1, IR3, and IR5, respectively). Proteins were then extracted and analyzed by Western blotting. (E) MCF7 cells were incubated with 2 or 5 mM niacinamide and then Western blotted for total p53 or the loading control brg in whole-cell extract (lower two panels) or for immunoprecipitated total or K373/K382 acetylated (Ac) p53 (upper two panels). (F) Western blot of p53 or hdm2 in whole-cell extracts prepared before the onset of p53 degradation due to hdm2 and at 1.5 and 2.5 h after 6-Gy irradiation.
The effects of elevated NAD+ synthesis on cellular p53 DNA binding were analyzed by ChIP of genomic DNA sites bound to endogenous p53 protein. Elevated NAD+ synthesis significantly reduced radiation-induced p53 binding to hdm2 and PIG3 DNA sites, but binding to p21, GADD45, PUMA, and bax sites was unaffected (Fig. 5B, lane NAm+IR compared to IR). This result mirrors the observation that NAD+ preferentially impaired p53 binding to the hdm2, compared to the p21, DNA sites in vitro (Fig. 3C).
Consistent with the promoter binding results, transcriptional induction of hdm2 and PIG3, but of neither p21, bax, nor PUMA, was inhibited by elevating intracellular NAD+ (Fig. 5C). Correspondingly, increased NAD+ also reduced radiation induction of hdm2 protein after 3 h, thus slowing degradation of p53 protein and resulting in an increased p53 protein level (Fig. 5D). In the absence of irradiation, niacinamide did not affect hdm2 (data not shown). By 5 h, hdm2 had been activated even in the condition of elevated NAD+, resulting in p53 protein degradation, but with elevated p53 remaining compared to control (Fig. 5D). These effects were not due to defective signaling to p53 or inhibition of translation, as neither p53 serine 15 phosphorylation nor p53 or p21 protein induction was inhibited by niacinamide (Fig. 5D).
Although niacinamide can also inhibit the p53 deacetylase Sir2α (33, 53), we found that lower levels of niacinamide, 2 versus 5 mM, could inhibit p53 deacetylation (Fig. 5E) without significantly altering hdm2 induction (Fig. 5F). Since a higher niacinamide level is required to inhibit hdm2, Sir2α inhibition does not appear to be sufficient to explain the effect of niacinamide on hdm2 induction (Fig. 5F). In addition, although increased p53 acetylation should increase p53 DNA binding activity, niacinamide treatment actually decreased p53 binding to the hdm2 DNA site. This suggests that the negative effect of NAD+ on p53 activities more than compensates for the positive effects of acetylation. Thus, the niacinamide-induced increased rate of NAD+ synthesis correlated with inhibition of p53 binding to and transcriptional activation of hdm2.
Thiamine inhibits p53 DNA binding in vivo.
If the cellular effects of niacinamide were indeed due to a direct effect of increased NAD+ synthesis on p53 DNA binding activity, then our in vitro DNA binding data predict that TDP should also have a direct effect. TDP inhibition of cellular p53 DNA binding was explored by incubating cells with thiamine (vitamin B1), which is rapidly converted to TDP (45). Thiamine treatment inhibited radiation-induced p53 binding to all DNA sites, including hdm2 and p21 sites (Fig. 6A). Similarly, thiamine treatment inhibited radiation-induced hdm2 protein induction (Fig. 6B), allowing enhanced p53 accumulation from 3 to 5 h after irradiation (Fig. 6B). TDP inhibition of both p21 and hdm2 induction (Fig. 6B) again reflected the in vitro DNA binding results, where TDP inhibited p53 binding to both DNA sites (Fig. 4E).
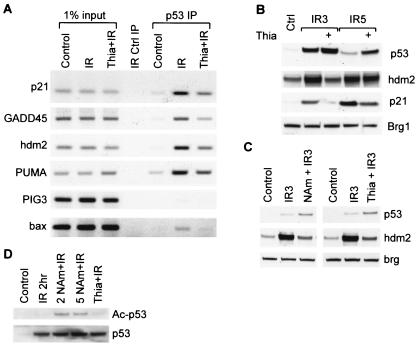
FIG. 6.
Effects of thiamine on p53 activity (A) Cells were treated with thiamine (Thia) to increase TDP and then irradiated (IR) to induce endogenous, wild-type p53. After 3 h (lane IR), genomic DNA sites that were bound to p53 protein were analyzed by ChIP (IP). (B) Cells were untreated or treated with thiamine and then irradiated and allowed to recover for the 3 or 5 h (IR3 and IR5, respectively). Proteins were then extracted and analyzed by Western blotting. (C) In the same experiment, cells were treated with niacinamide (NAm) or thiamine, and then proteins were extracted and analyzed by Western blotting. (D) Immunoprecipitated p53 was Western blotted for total p53 (bottom) or K373/K382 acetylated (Ac) p53 (top) after treatment with thiamine or niacinamide, followed by irradiation.
Furthermore, thiamine had effects virtually identical to those of niacinamide on inhibiting hdm2 and consequently increasing the kinetics of ionizing radiation-induced p53 accumulation (Fig. 6C). In contrast to niacinamide, thiamine treatment did not affect p53 acetylation status (Fig. 6D). These data are most consistent with niacinamide and thiamine elevating the intracellular synthesis of NAD+ and TDP, respectively, which in turn directly affects p53 DNA binding activity.
Niacinamide and thiamine affect p53 physiologic responses.
Treating cells to increase NAD+ or TDP could alter p53-regulated physiologic outcomes following DNA damage. Exposure to ionizing irradiation increased the percentage of highly aneuploid MCF-7 cells to ∼15% of metaphases (Fig. 7A). Treatment to increase NAD+ or TDP augmented this to ∼27% (Fig. 7A). Rereplication occurs when individual DNA replication origins fire more than once without an intervening mitosis, resulting in aneuploid cells, and is a hallmark of solid tumors. Significantly, p53 prevents rereplication, although the relevant molecular targets of p53 have yet to be conclusively elucidated (52). Thus, niacinamide and thiamine inhibited the ability of p53 to repress rereplication or to prevent such cells from progressing into mitosis.
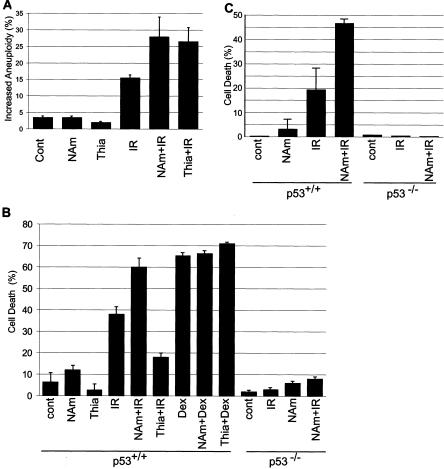
FIG. 7.
Effects of increased NAD+ or TDP on cellular responses to ionizing radiation. (A) MCF7 cells were treated to increase NAD+ or TDP and then irradiated (IR). After 5 days, chromosome aneuploidy in 100 metaphase spreads per sample was analyzed. Bars indicate the numbers of cells displaying aneuploidy greater than that displayed by the average control cell. NAm, niacinamide; Cont, control. (B) Murine thymocytes from wild-type (p53+/+) or p53 knockout (p53−/−) mice were explanted and incubated to increase NAD+ or TDP and then irradiated. Dex, dexamethasone treatment. (C) Mice were treated to increase NAD+ and then irradiated. Thymocytes were collected after 18 h.
Ionizing radiation also induces p53-dependent apoptosis in thymocytes (9, 32). Niacinamide treatment elevated thymocyte NAD+ about twofold ex vivo (data not shown) and enhanced ionizing radiation-induced cell death by ∼50% (Fig. 7B). In contrast to niacinamide, thiamine treatment decreased cell death by ∼50% (Fig. 7B). Specificity for p53-induced apoptosis was demonstrated by the observation that neither treatment altered dexamethasone-induced apoptosis (Fig. 7B), which is p53 independent (9, 32). p53 dependence was further demonstrated by the minimal effects of treatment to increase NAD+ on radiation-induced apoptosis in p53−/− thymocytes (Fig. 7B).
We then examined apoptosis in vivo. Unfortunately, the maximal tolerated dose of thiamine in mice is much lower than the ex vivo dose used here, and the lower tolerated dose had no effect on thymocyte apoptosis in vivo (data not shown). However, mice and humans tolerate high niacinamide doses, and niacinamide treatment increased thymic NAD+ by ∼40% (data not shown) and increased ionizing radiation-induced thymocyte apoptosis in wild-type, but not p53-null, mice (Fig. 7C). Thus, niacinamide or thiamine can increase cellular NAD+ or TDP and affect p53 DNA binding and p53-dependent functions in cells and in animals, including ionizing radiation-induced rereplication and apoptosis.
DISCUSSION
The experiments presented demonstrate that NAD+ and TDP can directly regulate p53 DNA binding activity in vitro and that treating cells to increase NAD+ or TDP with niacinamide or thiamine affects p53 activity in vivo. The in vitro results have important implications for understanding the mechanism of NAD+ action on p53 DNA binding. The L344A mutation prevents p53 from forming tetramers in solution, and the resultant single-dimer DNA binding activity was unaffected by NAD+ (Fig. 3C, 2×) because dimers cannot bind NAD+ (Fig. 3A). However, the L344A mutation does not directly interfere with NAD+ regulation of p53 binding to the hdm2 DNA site, because the DNA binding activity of pairs of L344A dimers was affected by NAD+ (Fig. 3C, 2-2×). Rather, since pairs of dimers interact cooperatively when bound adjacently on a full DNA site (35), the specificity of NAD+ for pairs of L344A dimers (Fig. 3C, 2-2X) suggests that NAD+ interacts with a new binding surface created between the two dimers of a p53 tetramer (35, 40, 46).
The data do not suggest that NAD+ dissociates tetramers into dimers, otherwise dimers would be seen to bind DNA in the in vitro band shift assay (Fig. 3C). Rather, NAD+ may bind to the dimer-dimer interface in a p53 tetramer, induce a conformational change, and alter the intrinsic DNA binding specificity. As p53 exists in the cell as tetramers, but not dimers (35), NAD+ is able to modulate the relevant cellular form of p53.
It is interesting to consider why TDP and NAD+ differ in their effects on p53 DNA binding activity. One explanation is that TDP simply is a more potent inhibitor of p53 DNA binding activity than NAD+, so that some DNA sites are inhibited by TDP but not NAD+. Alternatively, the ADP-like portion of the molecules may decrease p53 DNA binding, and the NMN portion of NAD+ may further “fine-tune” the effect on p53 DNA binding activity by altering the conformation of p53. When the NMN portion in NADH is reduced, it induces a conformational shift that could prevent the ADP portion of NADH from inhibiting p53 DNA binding activity. When the NMN portion in NAD+ is oxidized, it may induce a conformational shift that differentially affects binding to different DNA target sequences in the presence of the attached ADP portion of the NAD+. TDP, which lacks the NMN moiety of NAD+ and NADH, did not induce a significant conformational change in p53 and therefore did not fine-tune p53 DNA binding specificity.
Treating cells with niacinamide had effects on p53 DNA binding specificity in vivo similar to those of NAD+ in vitro, consistent with niacinamide increasing the rate of NAD+ synthesis and suggesting that NAD+ may directly affect intracellular p53 DNA binding specificity (Fig. 5A and B). Also supporting this interpretation is the observation that thiamine not only inhibits p53 DNA binding but also inhibits ionizing radiation induction of hdm2 to the same extent as niacinamide (Fig. 6C). As p53 is the only known target of both TDP and NAD+, it seems likely that niacinamide and thiamine inhibit intracellular p53 DNA binding to the hdm2 DNA site by direct effects of NAD+ and TDP on p53.
How p53 DNA binding activity is controlled in cells is unknown, including whether p53 DNA binding is even at equilibrium in chromatin in vivo. Even moderate effects on p53 DNA binding in vitro could translate into substantial alteration of p53 activity in cells. As TDP and NAD+ delay, rather than prevent, activation of hdm2, these molecules likely cause a decreased occupancy of p53 on the hdm2 genomic DNA site, resulting in a decreased rate of transcriptional initiation from the hdm2 promoter.
Although niacinamide decreased hdm2 induction to enhance p53 protein following DNA damage, induction of p21 was not enhanced (Fig. 5 and 6). Presumably, p21 is already maximally induced, so increased p53 would not affect p21. This is supported by the ChIP data, where, even though p53 protein level is higher (Fig. 5D), there is no increased binding to the p21 DNA site (Fig. 5B). Neither NAD+ nor thiamine affects cell cycle progression or ionizing radiation-induced cell cycle arrest. As NAD+ does not affect p21 induction, it would not be expected to affect cell cycle arrest. It is important that the p53-induced p21-dependent cell cycle arrest affects only cells that have not yet passed the restriction point in G1 and that it becomes pronounced in an asynchronous cell population only after ∼18 h. Thus, although thiamine delays ionizing radiation induction of p21, this would affect cell cycle arrest in only a very small percentage of cells.
Another physiologic function of p53, which may or may not require p21 induction, is to prevent aneuploidy and rereplication (52). Aneuploidy is augmented by thiamine and NAD+, likely because both molecules inhibit a function of p53 that is required for preventing aneuploidy (Fig. 7A). This function may be performed by a gene or set of genes that are normally activated by p53 in response to ionizing radiation but that are inhibited by NAD+ or TDP (such as the hdm2 or PIG3 gene). However, these molecules may act on p53 independently of effects on p53 transcriptional activity, because it is not known whether the mechanism whereby p53 maintains genomic ploidy requires transcription.
Yet another physiologic function of p53, apoptosis, is affected by niacinamide and thiamine. Since the mechanism of p53-induced apoptosis has not been elucidated, it is impossible to identify the precise mechanism by which niacinamide and thiamine modulate apoptosis. The net effect of changing the level of various apoptosis-related proteins is likely dictated by which p53 apoptotic pathways are able to affect the pro- versus antiapoptotic balance in any given cell type. Whether p53-induced apoptosis even requires transcriptional activation of target genes is debatable (4, 8, 10, 37, 48). It is possible that niacinamide and thiamine could both inhibit one aspect of p53 proapoptotic activity but that niacinamide increases apoptosis by inhibition of Sir2α, because thymocytes lacking Sir2α undergo enhanced ionizing-radiation-induced apoptosis (7). The relationship between Sir2α, which uses NAD+ as a cofactor in deacetylating p53 and histones, NAD+, and p53 will be interesting to pursue. It has been previously proposed that DNA damage induces a cell cycle arrest to allow time for DNA repair but that excessive damage that cannot be repaired may result in apoptosis. However, the cellular choice between DNA damage-induced cell cycle arrest and apoptosis appears to be mainly regulated by other aspects of the cellular state during the DNA damage response, including those that are regulated by growth factors (5, 55). We suggest that one parameter relevant to ionizing radiation's effects on the p53 response is the rate of NAD+ synthesis, which may directly alter p53 DNA binding specificity and function.
Though thiamine is not reported to modulate DNA damage responses, p53 activates transcription of a murine thiamine transporter (31). However, in addition to regulating p53, as reported here, niacinamide affects other proteins that are involved in cellular decisions of apoptosis and senescence, including PARP (19) and SIR2 (1, 33, 53). Although PARP can deplete cellular NAD+, this is limited to extremely high doses of ionizing radiation (50 to 120 Gy) and is not observed at the doses we used (2 to 6 Gy; Fig. 5A). The relationship between PARP, NAD+, and p53 will be very important to delineate, and it will require substantial investigation to sort out the intertwined effects of PARP using NAD+ as a substrate, NAD+ and/or PARP binding to p53, and PARP ADP-ribosylating p53 (using NAD+!) and other proteins relevant to DNA damage detection and repair. Perhaps this type of role of NAD+ in cellular DNA damage responses provides a selective advantage for tumor cells which utilize anaerobic glycolysis even in the presence of oxygen, known as the Warburg effect (43).
Whether TDP or NAD+, or molecules based on them, can be used for therapeutic benefit remains to be determined. Niacinamide radiosensitizes tumors and prevents UV radiation-induced skin cancer in mice (14, 21, 22). The data presented here suggest that small molecules that specifically affect p53 protein tetramers can be designed or identified, thus forming a novel basis for the development of molecules that affect p53 function in vivo. Such compounds could potentially either enhance apoptosis of tumor cells that retain p53 function or inhibit p53-dependent apoptosis of normal tissues to reduce toxicities of therapy.
Acknowledgments
We thank Mary Relling, Jean Cai, and Yi Su (Pharmaceutical Sciences, St. Jude Children's Research Hospital) for HPLC analysis of NAD+, Vincent Kidd and Susan Ragsdale (Cancer Core Cytogenetics Laboratory, St. Jude Children's Research Hospital) for analysis of mitotic spread chromosome content, and Gerald Zambetti (Biochemistry, St. Jude Children's Research Hospital) for helpful comments after reading the manuscript.
This work was supported by grants from the NIH (ES05777 and CA21765) and by the American Lebanese Syrian Associated Charities of the St. Jude Children's Research Hospital.
REFERENCES
- 1.Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik, and D. A. Sinclair. 2003. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423:181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balagurumoorthy, P., H. Sakamoto, M. S. Lewis, N. Zambrano, G. M. Clore, A. M. Gronenborn, E. Appella, and R. E. Harrington. 1995. Four p53 DNA-binding domain peptides bind natural p53-response elements and bend the DNA. Proc. Natl. Acad. Sci. USA 92:8591-8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brugarolas, J., C. Chandrasekaran, J. I. Gordon, D. Beach, T. Jacks, and G. J. Hannon. 1995. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377:552-557. [DOI] [PubMed] [Google Scholar]
- 4.Caelles, C., A. Helmberg, and M. Karin. 1994. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature 370:220-223. [DOI] [PubMed] [Google Scholar]
- 5.Canman, C. E., T. Gilmer, S. Coutts, and M. B. Kastan. 1995. Growth factor modulation of p53-mediated growth arrest vs. apoptosis. Genes Dev. 9:600-611. [DOI] [PubMed] [Google Scholar]
- 6.Chene, P. 2004. Inhibition of the p53-MDM2 interaction: targeting a protein-protein interface. Mol. Cancer Res. 2:20-28. [PubMed] [Google Scholar]
- 7.Cheng, H. L., R. Mostoslavsky, S. Saito, J. P. Manis, Y. Gu, P. Patel, R. Bronson, E. Appella, F. W. Alt, and K. F. Chua. 2003. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA 100:10794-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chipuk, J. E., U. Maurer, D. R. Green, and M. Schuler. 2003. Pharmacologic activation of p53 elicits Bax-dependent apoptosis in the absence of transcription. Cancer Cell 4:371-381. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, A. R., C. A. Purdie, D. J. Harrison, R. G. Morris, C. C. Bird, M. L. Hooper, and A. H. Wyllie. 1993. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 362:849-852. [DOI] [PubMed] [Google Scholar]
- 10.Crook, T., N. J. Marston, E. A. Sara, and K. H. Vousden. 1994. Transcriptional activation by p53 correlates with suppression of growth but not transformation. Cell 79:817-827. [DOI] [PubMed] [Google Scholar]
- 11.Deng, C., P. Zhang, J. W. Harper, S. J. Elledge, and P. Leder. 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82:675-684. [DOI] [PubMed] [Google Scholar]
- 12.El-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 13.Funk, W. D., D. T. Pak, R. H. Karas, W. E. Wright, and J. W. Shay. 1992. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol. Cell. Biol. 12:2866-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gensler, H. L., T. Williams, A. C. Huang, and E. L. Jacobson. 1999. Oral niacin prevents photocarcinogenesis and photoimmunosuppression in mice. Nutr. Cancer 34:36-41. [DOI] [PubMed] [Google Scholar]
- 15.Hainaut, P., and J. Milner. 1993. Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res. 53:4469-4473. [PubMed] [Google Scholar]
- 16.Halazonetis, T. D., and A. N. Kandil. 1993. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J. 12:5057-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 18.Haupt, Y., S. Rowan, E. Shaulian, K. H. Vousden, and M. Oren. 1995. Induction of apoptosis in HeLa cells by trans -activation-deficient p53. Genes Dev. 9:2170-2183. [DOI] [PubMed] [Google Scholar]
- 19.Herceg, Z., and Z. Q. Wang. 2001. Functions of poly(ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat. Res. 477:97-110. [DOI] [PubMed] [Google Scholar]
- 20.Hollstein, M., B. Shomer, M. Greenblatt, T. Soussi, E. Hovig, R. Montesano, and C. C. Harris. 1996. Somatic point mutations in the p53 gene of human tumors and cell lines: updated compilation. Nucleic Acids Res. 24:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horsman, M. R., D. J. Chaplin, and J. M. Brown. 1987. Radiosensitization by nicotinamide in vivo: a greater enhancement of tumor damage compared to that of normal tissues. Radiat. Res. 109:479-489. [PubMed] [Google Scholar]
- 22.Horsman, M. R., D. W. Siemann, D. J. Chaplin, and J. Overgaard. 1997. Nicotinamide as a radiosensitizer in tumours and normal tissues: the importance of drug dose and timing. Radiother. Oncol. 45:167-174. [DOI] [PubMed] [Google Scholar]
- 23.Jeffrey, P. D., S. Gorina, and N. P. Pavletich. 1995. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science 267:1498-1502. [DOI] [PubMed] [Google Scholar]
- 24.Juven, T., Y. Barak, A. Zauberman, D. L. George, and M. Oren. 1993. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene 8:3411-3416. [PubMed] [Google Scholar]
- 25.Kastan, M. B., Q. Zhan, W. S. El-Deiry, F. Carrier, T. Jacks, W. V. Walsh, B. S. Plunkett, B. Vogelstein, and A. J. Fornace, Jr. 1992. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71:587-597. [DOI] [PubMed] [Google Scholar]
- 26.Kern, S. E., K. W. Kinzler, A. Bruskin, D. Jarosz, P. Friedman, C. Prives, and B. Vogelstein. 1991. Identification of p53 as a sequence-specific DNA-binding protein. Science 252:1708-1711. [DOI] [PubMed] [Google Scholar]
- 27.Koumenis, C., R. Alarcon, H. E. Siliciano, P. Sutphin, W. Hoffman, M. Murphy, J. Derr, Y. Taya, M. Kastan, and A. Giaccia. 2001. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol. Cell. Biol. 21:1297-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 29.Lee, W., T. S. Harvey, Y. Yin, P. Yau, D. Litchfield, and C. H. Arrowsmith. 1994. Solution structure of the tetrameric minimum transforming domain of p53. Nat. Struct. Biol. 1:877-890. [DOI] [PubMed] [Google Scholar]
- 30.Liu, G., J. M. Parant, G. Lang, P. Chau, A. Chavez-Retes, A. K. El-Naggar, A. Multani, S. Chang, and G. Lozano. 2004. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat. Genet. 36:63-68. [DOI] [PubMed] [Google Scholar]
- 31.Lo, P.-K., J.-Y. Chen, P.-P. Tang, J. Lin, C.-H. Lin, L.-T. Su, C.-H. Wu, T.-L. Chen, Y. Yang, and F.-F. Wang. 2001. Identification of a mouse thiamine transporter gene as a direct transcriptional target for p53. J. Biol. Chem. 276:37186-37193. [DOI] [PubMed] [Google Scholar]
- 32.Lowe, S. W., E. M. Schmitt, S. W. Smith, B. A. Osborne, and T. Jacks. 1993. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362:847-849. [DOI] [PubMed] [Google Scholar]
- 33.Luo, J., A. Y. Nikolaev, S. Imai, D. Chen, F. Su, A. Shiloh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107:137-148. [DOI] [PubMed] [Google Scholar]
- 34.McCoy, M., E. S. Stavridi, J. L. Waterman, A. M. Wieczorek, S. J. Opella, and T. D. Halazonetis. 1997. Hydrophobic side-chain size is a determinant of the three-dimensional structure of the p53 oligomerization domain. EMBO J. 16:6230-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLure, K. G., and P. W. K. Lee. 1998. How p53 binds DNA as a tetramer. EMBO J. 17:3342-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Micheli, V., and S. Sestini. 1997. Determining NAD synthesis in erythrocytes. Methods Enzymol. 280:211-221. [DOI] [PubMed] [Google Scholar]
- 37.Mihara, M., S. Erster, A. Zaika, O. Petrenko, T. Chittenden, P. Pancoska, and U. M. Moll. 2003. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 11:577-590. [DOI] [PubMed] [Google Scholar]
- 38.Momand, J., G. P. Zambetti, D. C. Olson, D. L. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237-1245. [DOI] [PubMed] [Google Scholar]
- 39.Muller-Tiemann, B. F., T. D. Halazonetis, and J. J. Elting. 1998. Identification of an additional negative regulatory region for p53 sequence-specific DNA binding. Proc. Natl. Acad. Sci. USA 95:6079-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagaich, A. K., V. B. Zhurkin, S. R. Durell, R. L. Jernigan, E. Appella, and R. E. Harrington. 1999. p53-induced DNA bending and twisting: p53 tetramer binds on the outer side of a DNA loop and increases DNA twisting. Proc. Natl. Acad. Sci. USA 96:1875-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okorokov, A. L., and J. Milner. 1999. An ATP/ADP-dependent molecular switch regulates the stability of p53-DNA complexes. Mol. Cell. Biol. 19:7501-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliner, J. D., K. W. Kinzler, P. Meltzer, D. L. George, and B. Vogelstein. 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358:80-83. [DOI] [PubMed] [Google Scholar]
- 43.Racker, E. 1972. Bioenergetics and the problem of tumor growth. Am. Sci. 60:56-63. [PubMed] [Google Scholar]
- 44.Rechsteiner, M., and V. Catanzarite. 1974. The biosynthesis and turnover of nicotinamide adenine dinucleotide in enucleated culture cells. J. Cell. Physiol. 84:409-422. [DOI] [PubMed] [Google Scholar]
- 45.Rindi, G., and U. Laforenza. 1997. In vitro systems for studying thiamin transport in mammals. Methods Enzymol. 279:118-131. [DOI] [PubMed] [Google Scholar]
- 46.Rippin, T. M., S. M. V. Freund, D. B. Veprintsev, and A. R. Fersht. 2002. Recognition of DNA by p53 core domain and location of intermolecular contacts of cooperative binding. J. Mol. Biol. 319:351-358. [DOI] [PubMed] [Google Scholar]
- 47.Samuels-Lev, Y., D. J. O'Connor, D. Bergamaschi, G. Trigiante, J.-K. Hsieh, S. Zhong, I. Campargue, L. Naumovski, T. Crook, and X. Lu. 2001. ASPP proteins specifically stimulate the apoptotic function of p53. Mol. Cell 8:781-794. [DOI] [PubMed] [Google Scholar]
- 48.Symonds, H., L. Krall, L. Remington, M. Saenz-Robles, S. Lowe, T. Jacks, and T. Van Dyke. 1994. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell 78:703-711. [DOI] [PubMed] [Google Scholar]
- 49.Szak, S. T., D. Mays, and J. A. Pietenpol. 2001. Kinetics of p53 binding to promoter sites in vivo. Mol. Cell. Biol. 21:3375-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tischler, M. E., D. Friedrichs, K. Coll, and J. R. Williamson. 1977. Pyridine nucleotide distributions and enzyme mass action ratios in hepatocytes from fed and starved rats. Arch. Biochem. Biophys. 184:222-236. [DOI] [PubMed] [Google Scholar]
- 51.Vassilev, L. T., B. T. Vu, B. Graves, D. Carvajal, F. Podlaski, Z. Filipovic, N. Kong, U. Kammlott, C. Lukacs, C. Klein, N. Fotouhi, and E. A. Liu. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844-888. [DOI] [PubMed] [Google Scholar]
- 52.Vaziri, C., S. Saxena, Y. Jeon, C. Lee, K. Murata, Y. Machida, N. Wagle, D. S. Hwang, and A. Dutta. 2003. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11:997-1008. [DOI] [PubMed] [Google Scholar]
- 53.Vaziri, H., S. K. Dessain, E. N. Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]
- 54.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 55.Vousden, K. H., and X. Lu. 2002. Live or let die: the cell's response to p53. Nat. Rev. Cancer 2:594-604. [DOI] [PubMed] [Google Scholar]
- 56.Waldman, T., C. Lengauer, K. W. Kinzler, and B. Vogelstein. 1996. Uncoupling of S phase and mitosis induced by anticancer agents in cells lacking p21. Nature 381:713-716. [DOI] [PubMed] [Google Scholar]
- 57.Waterman, J. L., J. L. Shenk, and T. D. Halazonetis. 1995. The dihedral symmetry of the p53 tetramerization domain mandates a conformational switch upon DNA binding. EMBO J. 14:512-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williamson, D. H., P. Lund, and H. A. Krebs. 1967. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem. J. 103:514-526. [DOI] [PMC free article] [PubMed] [Google Scholar]