Significance
The wealth of genomic data available for the respiratory pathogen Streptococcus pneumoniae enabled the design of a pangenome-wide proteome microarray. Of over 2,000 pneumococcal proteins, 208 strongly bound antibodies in adult human sera. The vast majority could be classified as either variants of four diverse loci or more conserved proteins involved in adhesion, enzymatic degradation, solute binding, or cell wall synthesis. Detailed analyses of the genomic data revealed some variable antigens rapidly diversified through mechanisms including homologous recombination, mobile genetic element transmission, and phase variation. Other antigens were conserved across the population and may be better candidates for simple vaccine formulations. This raises the question of what evolutionary advantage bacteria derive from altering only a subset of their antigenic loci.
Keywords: genomics, pathogens, evolution, immunology, epidemiology
Abstract
Characterizing the immune response to pneumococcal proteins is critical in understanding this bacterium’s epidemiology and vaccinology. Probing a custom-designed proteome microarray with sera from 35 healthy US adults revealed a continuous distribution of IgG affinities for 2,190 potential antigens from the species-wide pangenome. Reproducibly elevated IgG binding was elicited by 208 “antibody binding targets” (ABTs), which included 109 variants of the diverse pneumococcal surface proteins A and C (PspA and PspC) and zinc metalloprotease A and B (ZmpA and ZmpB) proteins. Functional analysis found ABTs were enriched in motifs for secretion and cell surface association, with extensive representation of cell wall synthesis machinery, adhesins, transporter solute-binding proteins, and degradative enzymes. ABTs were associated with stronger evidence for evolving under positive selection, although this varied between functional categories, as did rates of diversification through recombination. Particularly rapid variation was observed at some immunogenic accessory loci, including a phage protein and a phase-variable glycosyltransferase ubiquitous among the diverse set of genomic islands encoding the serine-rich PsrP glycoprotein. Nevertheless, many antigens were conserved in the core genome, and strains’ antigenic profiles were generally stable. No strong evidence was found for any epistasis between antigens driving population dynamics, or redundancy between functionally similar accessory ABTs, or age stratification of antigen profiles. These results highlight the paradox of why substantial variation is observed in only a subset of epitopes. This result may indicate only some interactions between immunoglobulins and ABTs clear pneumococcal colonization or that acquired immunity to pneumococci is an accumulation of individually weak responses to ABTs evolving under different levels of functional constraint.
The pneumococcus (Streptococcus pneumoniae) is a Gram-positive human commensal and respiratory pathogen commonly carried in the nasopharynx of young infants (1). The prevalence of carriage peaks within the first 3 y of life (2). However, the bacteria are cleared increasingly quickly with age following successive episodes of carriage, a change associated with the development of mucosal immune responses (3). These involve interactions with antibody binding targets (ABTs), recognized by Ig A (IgA) and G (IgG) antibodies, or T-cell receptor targets (TCRTs), recognized by CD4+ TH17 cells (4). The former response leads to targeted opsonophagocytosis of cells bearing ABTs, neutralization of toxins, and inhibition of adhesion to host tissues (5, 6). TCRT-triggered secretion of interleukin-17A by CD4+ TH17 cells results in nonspecific clearance of pneumococci from the nasopharynx through the recruitment of neutrophils and macrophages (7, 8). These interactions with the host immune system are likely to be important in this bacterium’s evolution (9, 10).
The historical focus of pneumococcal immunology has been the polysaccharide capsule, which has over 90 antigenically distinguishable variants (serotypes). Systemic immunization with capsular polysaccharides can stimulate protective levels of serotype-specific antibodies (11, 12). Conjugate vaccines including 7–13 different capsular polysaccharides (13) provide protection against the serotypes included in the vaccine, but a desire for vaccines with broader coverage has caused growing interest in alternative protein- or whole cell-based formulations (14). Previous studies have used ELISA (15), phage display libraries (16), or “antibody fingerprinting” (17) to measure the antibody response to the noncapsular pneumococcal antigens of individual strains. Now, population genomic datasets can be combined with proteome microarrays, an approach with several advantages for studying the host–bacterium interaction. First, this pairing can identify a more complete set of antigens, including proteins that may be absent from any one individual strain, such as the type 1 pilus (18, 19); the prevalence of these proteins can also be ascertained from the genomic data. Second, for highly polymorphic proteins such as pneumococcal surface proteins A (PspA) and C (PspC) (20, 21), this combination allows the immune response to a diverse panel of variants to be assayed, and their distribution across the population to be established. Third, the rate at which different loci diversify through mutation and recombination can be inferred from evolutionary analyses of genomes. This is particularly important in a naturally transformable species such as S. pneumoniae, in which vaccine-escape variants can emerge through recombination, as previously observed for the capsule polysaccharide synthesis (cps) locus (22).
This study used genomic data from 616 nasopharyngeal carriage isolates from young children in Massachusetts (23, 24). The majority of these isolates were classified into 15 monophyletic sequence clusters (SCs), using a core genome alignment; each of these corresponded to a common genotype, the recent diversification of which could be reconstructed based on whole-genome alignments (25). The 1.2 million protein-coding sequences (CDSs) identified in these de novo assemblies were grouped into 5,442 clusters of orthologous genes (COGs) (26); these groupings of similar proteins were used to inform the design of a proteome microarray for measuring IgG binding levels. Here, we characterize the antibody response to thousands of pneumococcal proteins, using healthy adult sera, and describe the distinct evolutionary patterns associated with the most immunogenic sequences.
Results
Extensive Interactions Between the Host and Pneumococcus.
To capture the most common interactions between hosts and pneumococci, representative sequences were included for each COG present in at least 20% of the 616 isolates. Other proteins that proved difficult to assemble from short read data were also included: the pneumococcal serine-rich repeat protein PsrP; the cellular autolytic amidase LytA; a phage amidase; the phage antireceptor PblB; and an oligomer of choline binding domains (CBDs), a common cell-surface attachment motif found in many pneumococcal protein antigens. Rare COGs were included if they were likely to be antigenic: These were type 2 pilus components, zinc metalloprotease ZmpC, and ZmpE, a zinc metalloprotease unique to the atypical unencapsulated pneumococci of SC12 (24, 27). Additionally, multiple variants of some diverse loci were included. Each of the three “clades” of the type I pilus RrgB protein (28) corresponded to a separate COG, as did five variants (one truncated) of the PclA protein. Representatives were added for each of the previously defined variants of the three diverse penicillin-binding proteins (three for Pbp2X and Pbp1A and four for Pbp2B), which were associated with differing levels of β-lactam sensitivity (24). Finally, the manually curated sets of all complete representatives of the “diverse core loci” (encoding PspA, PspC, and the zinc metalloproteases ZmpA and ZmpB) were identified (SI Appendix). Owing to the low similarity between many representatives, these sets were divided into variants, using an alignment-free, kmer-based approach (SI Appendix, Fig. S1). Representatives could be included on the microarray for 36 of the 39 PspA variants, 57 of the 59 PspC variants, and all of the 18 ZmpA variants and 16 ZmpB variants. An independent phylogenetic analysis indicated these sets encompassed the full previously observed diversity of these proteins (20, 21) (SI Appendix, Fig. S2). Overall, a total of 2,190 proteins derived from the Massachusetts pneumococcal population were included on the microarray; also included was the full proteome of S. pneumoniae TIGR4 (29), but these proteins were excluded from the described analyses to maintain unbiased coverage across the systematically sampled bacterial isolates.
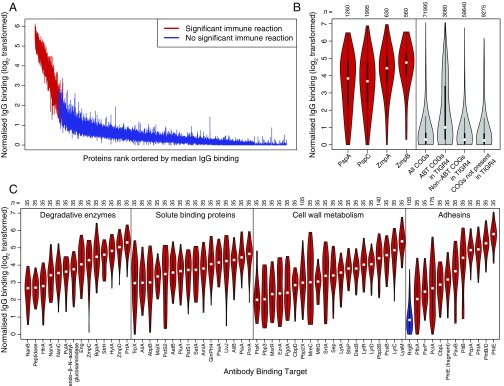
Using the sera of 35 healthy adults, IgG binding responses to all of the Massachusetts-derived proteins on the microarray were normalized and plotted on a logarithmic scale (Fig. 1A). Although the distribution of binding responses was continuous, a subset of the proteins did elicit a consistently elevated antibody response across the serum samples. Dividing these data into two groups classed 208 of the 2,190 proteins as ABTs for subsequent analyses. Approximately half of these ABTs were variants of the four diverse core loci (Fig. 1B and SI Appendix, Figs. S3–S5): These included 29 of the 36 PspA probes [Fisher’s exact test, odds ratio (OR) relative to COGs = 84.9, P < 2.2 × 10−16], 48 of the 57 PspC probes (OR = 109.5, P < 2.2 × 10−16); 17 of the 18 ZmpA probes (OR = 346.0, P < 2.2 × 10−16), and 15 of the 16 ZmpB probes (OR = 304.8, P < 2.2 × 10−16). The IgG binding across the diverse variants of PspA and PspC (SI Appendix, Figs. S3 and S4) is generally substantially higher than that of the CBD (SI Appendix, Fig. S6), the one sequence motif commonly shared between these proteins, indicating that these results reflect a broad immune repertoire spanning the allelic diversity of these proteins.
Fig. 1.
Binding of IgG to 2,190 representatives of the pneumococcal pangenome. (A) Distribution of IgG binding responses on a logarithmic scale. A vertical line shows the interquartile range of IgG binding measurements across the 35 serum samples for each protein on the microarray. The proteins are ordered by the rank of the median IgG binding level across sera, with lines for proteins classified as ABTs colored red and those for non-ABTs blue. (B) Validation of IgG binding results. The left four violin plots show the IgG binding elicited across the 35 serum samples by the 36 PspA variants, 57 PspC variants, 18 ZmpA variants, and 16 ZmpB variants. Each violin plot is annotated with the number of datapoints in the distribution. The right four violin plots show, from left to right, the overall IgG binding elicited by the 2,057 proteins selected as representatives of the COGs previously defined using a population of pneumococci from Massachusetts, the IgG binding elicited by the 88 COGs that matched antigens identified in S. pneumoniae TIGR4 using antibody fingerprinting, the IgG binding elicited by the 1,704 COGs matching S. pneumoniae TIGR4 proteins that failed to trigger adaptive immune responses detectable by antibody fingerprinting, and the IgG binding elicited by the 265 COGs that were not orthologous with proteins in S. pneumoniae TIGR4. (C) Red violin plots showing the IgG binding to selected ABTs grouped by function and ordered by median IgG binding level. RrgB, not classed as an ABT in this analysis, is displayed for comparison as a blue violin plot.
These results were compared with a previous “antigenic fingerprinting” study of S. pneumoniae TIGR4 that identified epitopes in 97 annotated CDSs (17). The proteins from the Massachusetts isolates on the array contained 2,039 orthologs of sequences in S. pneumoniae TIGR4 (considering the sets of PspA, PspC, ZmpA, and ZmpB variants each as single orthologs). The 88 COGs from the Massachusetts isolates’ proteome that were orthologous to the antigens identified by fingerprinting had a significantly increased probability of being identified as ABTs in this study relative to the 265 COGs on the microarray not linked to CDSs in TIGR4 (Fisher’s exact test, OR = 8.93, P = 1.81 × 10−11; Fig. 1B). In turn, those TIGR4 proteins not identified through fingerprinting were significantly less likely to be identified as ABTs than those proteins not linked to CDSs in TIGR4 (Fisher’s exact test, OR = 0.45, P = 0.014). Hence, the proteins eliciting the highest antibody binding response comprised a combination of diverse PspA, PspC, ZmpA, and ZmpB variants, and representatives of other proteins that significantly overlap with the antigens identified by a different methodology applied to an individual strain.
Functional Categories Associated with ABTs.
A multivariable logistic regression was used to identify the protein characteristics enriched in ABTs (SI Appendix, Table S1). Both the conventional signal peptide and the secretion-associated YSIRK motif were significantly associated with ABTs, as were the lipid attachment site, which directs lipoproteins to be connected to the cell membrane, and the CBD. Notably, whereas the size of a protein was significantly associated with elevated IgG binding, the number of transmembrane helices was not. Therefore, the ABTs are enriched for large proteins that are secreted or peripherally associated with the cell surface.
Manual annotation of the ABTs revealed they performed diverse functional roles, but most could be grouped into four categories (Fig. 1C and SI Appendix). The first group was degradative enzymes, acting on a wide range of extracellular substrates including proteins, polysaccharides, and their derivatives. The second group was ABC transporter solute binding proteins (SBPs), with representatives specific for amino acids, sugars, nucleotides, and siderophores. The third group was enzymes involved in cell wall metabolism, including proteins required for peptidoglycan synthesis and cell division. The fourth group consisted of adhesins, of which the most immunogenic were the histidine triad proteins (PhtA, PhtB/D, and PhtE), each containing the histidine triad motif identified by the multivariable regression as enriched in ABTs. These divisions are inevitably simplified: There is evidence the histidine triad proteins are important in acquiring divalent cations (30), whereas PsaA is an SBP with a possible role in adhesion (31). Similarly, the transpeptidase domain was enriched in ABTs; this is found in penicillin-binding proteins (PBPs), some of which have been evolving under selection from antibiotic use (24).
A second multivariable regression was conducted within ABTs to identify the characteristics associated with the highest levels of IgG binding (SI Appendix, Table S2). This also found signal peptides, histidine triad motifs, and CBDs to be associated with elevated antibody responses. Additionally, this regression identified the LysM domain, for peptidoglycan binding, and the sortase attachment motif, for covalent attachment to the cell wall. This latter mechanism of surface attachment anchors the two types of pneumococcal pili to the cell surface; whereas the type 2 pilus PitB protein was classed as an ABT, the RrgB backbone of the type 1 pilus was not, despite detectably elevated levels of IgG binding relative to the overall background (SI Appendix, Fig. S6). Hence again, surface-exposed proteins were associated with the strongest antibody response.
Differing Diversity Across ABT Categories.
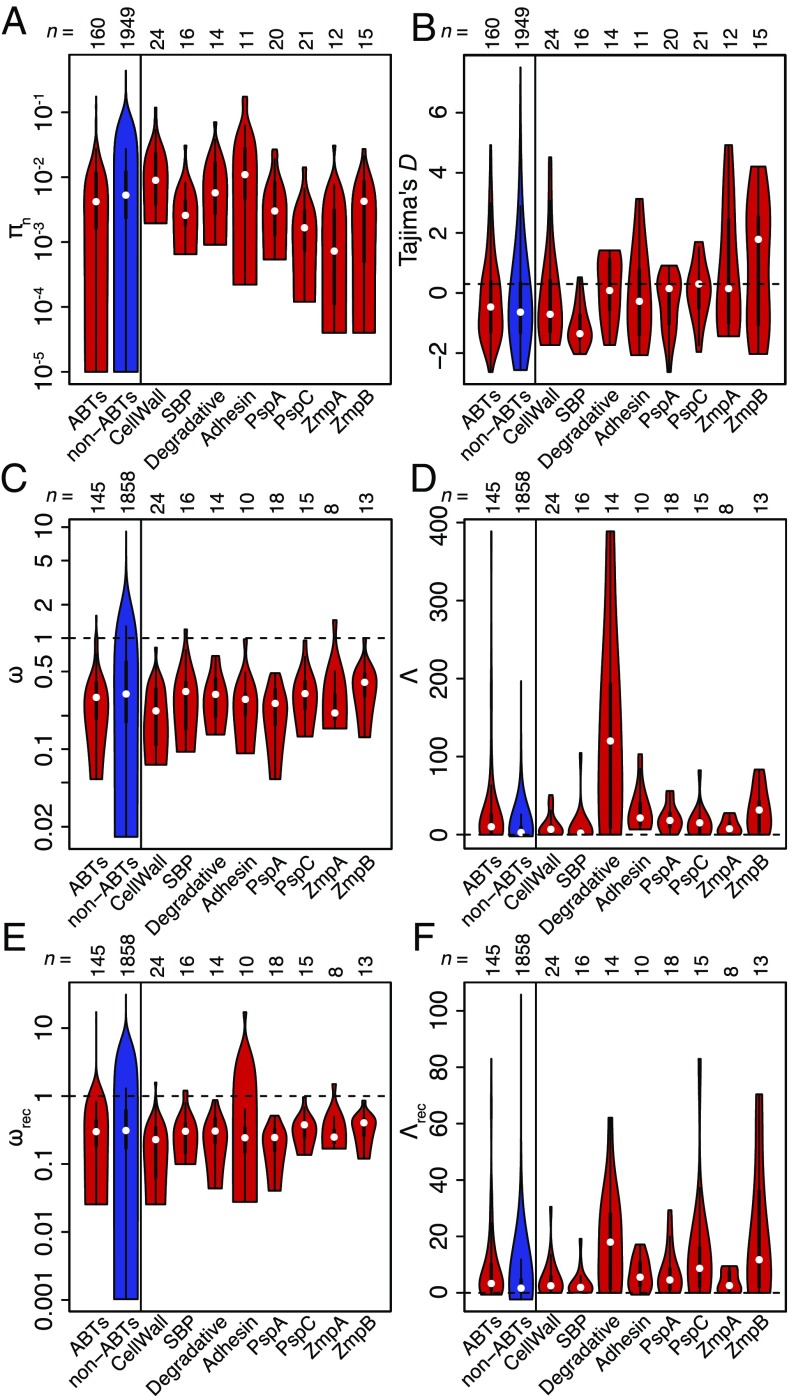
Extensive IgG recognition of pneumococcal proteins suggested immune selection may drive the diversification of targeted loci. To test whether ABTs were more diverse than those proteins that elicited lower IgG binding, codon alignments were generated for individual COGs or, for the diverse loci represented by multiple sequences on the microarray, for individual variants. By the per site πn nucleotide diversity statistic, ABTs were less diverse than non-ABTs (Fig. 2A; Wilcoxon test, W = 191,172, P = 0.00182). However, when the ABTs were categorized as displayed in Fig. 1C, the low πn for ABTs was primarily due to few polymorphisms in the individual variants of the core variable loci (Fig. 2A). As this is not an accurate representation of the overall diversity of these proteins, we used Tajima’s D as a less simplistic test of selection on those codon alignments for which πn was greater than zero (32). As these alignments were drawn from the same population, a detected difference between ABTs and non-ABTs could represent the consequences of immune selection. However, the median Tajima’s D was below zero for both ABTs (−0.762) and non-ABTs (−0.934), a difference that was nonsignificant (Fig. 2B; Wilcoxon test, W = 142,788, P = 0.0762). These negative values suggested purifying selection was stronger than balancing selection across both categories of alignment, although within the ABTs purifying selection appeared weakest within the variants of the core variable loci, with D values around zero, and strongest for SBPs and cell wall synthesis machinery.
Fig. 2.
Evolutionary analysis of codon alignments. In each plot, the left two violin plots compare all ABTs with non-ABTs; the right eight violin plots decompose the ABTs into the COGs corresponding to the four functional classes shown in Fig. 1C and the individual variant alignments of the four variable antigen classes shown in Fig. 1B. Each violin plot is annotated with the number of datapoints in the distribution. (A) Diversity of codon alignments containing sequence variation quantified using the πn metric. (B) Values of Tajima’s D calculated from codon alignments for which πn was above zero. The horizontal dashed line at 0 indicates the neutral expectation. (C) Values of ω calculated by fitting model 0 (single value of ω across all sites) of PAML to individual codon alignments. The horizontal dashed line at 1 represents the neutral expectation. Only analyses for which both dN and dS were greater than 0.25 are shown. (D) Log-likelihood difference (Λ) between fits of PAML evolutionary models 2a (positive selection) and 1a (nearly neutral diversification) to the codon alignments analyzed in C. The dashed line at 0 represents the neutral expectation. (E) Values of ωrec calculated by combining dNrec and dSrec from fitting model 0 of PAML to codon alignments segmented on the basis of recombination breakpoints. Only analyses for which both dNrec and dSrec were greater than 0.25 are shown. (F) Log-likelihood differences (Λrec) between fits of PAML evolutionary models 2a (positive selection) and 1a (nearly neutral diversification) to the segmented codon alignments analyzed in E.
The effects of selection on codon alignments can also be analyzed through phylogenetic reconstruction of sequences’ diversification under different models of evolution, using PAML (33). The ratio of nonsynonymous to synonymous base substitutions (dN/dS or ω) from such a reconstruction estimates whether diversifying (ω > 1) or purifying (ω < 1) selection predominates for an individual protein. The dominance of purifying selection across the ABT codon alignments was observed in all functional categories, suggesting any positive selection was limited only to specific epitopes. Furthermore, the overall distribution of ω was lower for ABTs (median of 0.293) than for non-ABTs (median of 0.314; Fig. 2D; Wilcoxon test, W = 147,248, P = 0.0615); these estimates were robust to different starting values of ω used in independent model fits (SI Appendix, Fig. S7). Despite those alignments containing little diversity (dN or dS < 0.25) being excluded from the PAML analyses, the heightened ω values for non-ABTs were partially driven by alignments containing few polymorphic sites, which had ω estimates strongly influenced by a small number of nonsynonymous mutations (SI Appendix, Fig. S8).
A more specific test for immune-selection–driven diversification is to calculate the log-likelihood difference (Λ) between the fit of PAML model 2a (positive selection) and 1a (nearly neutral evolution) (34) for each codon alignment (Fig. 2D). This revealed consistently more improved fits of model 2a relative to 1a for ABTs (median Λ = 10.1) than for non-ABTs (median Λ = 2.65; Fig. 2D; Wilcoxon test, W = 97,226, P = 2.27 × 10−8). Mirroring the distribution of Tajima’s D, the cell wall metabolism enzymes and SBPs showed the least evidence of immune-driven diversification.
The results of such phylogenetic analyses can be confounded by recombination, which violates the assumptions underlying the phylogenetic reconstruction of sequences (35). To mitigate against such model misspecifications, codon alignments were analyzed with 3SEQ (36) and the maximum-χ2 method (37, 38) to identify recombination breakpoints; each “clonal” alignment segment was then analyzed with PAML independently. In this analysis, the estimates of ω accounting for recombination (ωrec) for ABTs (median of 0.299) and non-ABTs (median of 0.311) were not substantially altered (Fig. 2E). In the comparison of evolutionary model likelihoods, there remained significantly greater evidence for diversifying selection in ABTs (median Λrec = 3.31) than in non-ABTs (median Λrec = 1.65; Fig. 2F; Wilcoxon test, W = 100,785, P = 4.25 × 10−7). Therefore, accounting for recombination diminished the evidence for diversifying selection across the codon alignments, with a particularly pronounced reduction in the evidence for positive selection among surface-associated degradative enzymes, indicating their evolutionary histories may have been substantially affected by exchange of sequence.
Variation in ABT Diversification Through Recombination.
To quantify diversification through recombination, the total number of breakpoints identified in the codon alignments were plotted by protein categorization (Fig. 3A). There was a slightly, albeit significantly, elevated number of recombination breakpoints in ABT alignments (a median of two breakpoints per alignment, rather than one; Wilcoxon test, W = 181,753.5, P = 0.0464). Surface-associated degradative enzymes contained a high number of breakpoints, consistent with the substantial difference between Λ and Λrec for this category (Fig. 2). However, as breakpoints are easier to detect in longer and larger alignments, and as protein size was predictive of being classified as an ABT (SI Appendix, Table S1), the per-base density of recombination breakpoints was plotted (Fig. 3B). The impact of recombination on ABTs was still significantly higher than on non-ABTs (median of 3.50 × 10−6 breakpoints per base pair for ABTs; 2.04 × 10−6 breakpoints per base pair for non-ABTs; Wilcoxon test, W = 184,320, P = 0.0195). This again masked heterogeneity within ABTs: The four core variable antigens and cell wall synthesis machinery were the most mosaic, although the latter category included diversification of penicillin-binding proteins under selection by β-lactam consumption (24), in contrast to the conservation of adhesins and SBPs.
Fig. 3.
Levels of recombination affecting pneumococcal CDSs. In each plot, the left two violin plots compare all ABTs with non-ABTs; the right eight violin plots decompose the ABTs into the COGs corresponding to the four functional classes shown in Fig. 1C and the individual variants of the four variable antigen classes shown in Fig. 1B. (A) Number of recombination breakpoints identified in codon alignments using 3SEQ and the maximum-χ2 approach. (B) Nonzero densities of recombination breakpoints in codon alignments. (C) Number of homologous recombination events, identified by analysis of whole-genome alignments, which overlap the representatives of each COG present in the reference genomes of all 15 monophyletic sequence clusters. As pspA, pspC, zmpA, and zmpB each occupy a single locus in each reference genome, only point estimates are available for each of these ABTs. The variant sets for each of pbp1a, pbp2x, and pbp2b were also considered to be individual loci. The thin dotted horizontal line indicates the number of homologous recombinations overlapping the capsule polysaccharide synthesis (cps) locus based on a previous analysis; the thick dashed horizontal line indicates the number of “serotype-switching” homologous recombinations that cause a functional change at the cps locus. (D) Proportion of homologous recombination events displayed in C occurring on terminal branches of phylogenies. The thin dotted and thick dashed horizontal lines again show the equivalent proportions for all recombinations, and serotype-switching recombinations, affecting the cps locus, respectively.
Analyses of individual sets of orthologs can detect only recombinations that have a breakpoint within a CDS. However, complete replacement of one allele by another, with recombination breakpoints flanking the relevant CDS, can be estimated by identifying recombination events in whole-genome alignments of isolates from the same sequence cluster (24, 25). The number of recombination events affecting the loci encoding proteins ubiquitous throughout the sampled population is shown in Fig. 3C, relative to the number affecting the cps locus, which determines the cell’s serotype. This comparison found ABTs were affected by an elevated number of transformation events (median number of recombinations affecting ABTs of 12; median number of recombinations affecting non-ABTs of 11), although the small difference was not significant (Wilcoxon test, W = 46,711.5, P = 0.182). However, this masked notable heterogeneity between the different categories of ABT once more: Almost all of the genes encoding ABTs were affected by recombination at frequencies typical of non-ABTs, except the pspA and pspC genes, which were affected by much higher numbers of transformation events. This is consistent with previous identification of these genes as “hotspots” of recombination from genome-wide analyses (39–41).
The high frequency of recombinations affecting pspA or pspC may reflect positive selection for evading adaptive immune responses; alternatively, such recombinations may be transiently beneficial, but later subject to negative selection, if the alteration were to have reduced the functionality of the protein or disrupted epistasis with other loci. The latter scenario should result in recombinations being enriched on the terminal branches of trees, as they occur frequently, but are removed from the population before they leave a large number of descendants (42). This dataset revealed an insignificantly higher proportion of transformation events occurring on terminal branches for non-ABTs (0.444) rather than for ABTs (0.405; Wilcoxon test, W = 37,925, P = 0.143). The pspA and pspC proportions were intermediate between the more slowly diversifying zmpA and zmpB genes. Hence the phylogenetic distribution of the many recombinations affecting pspA and pspC was not detectably skewed by selection.
Distribution of ABTs Across the Pneumococcal Population.
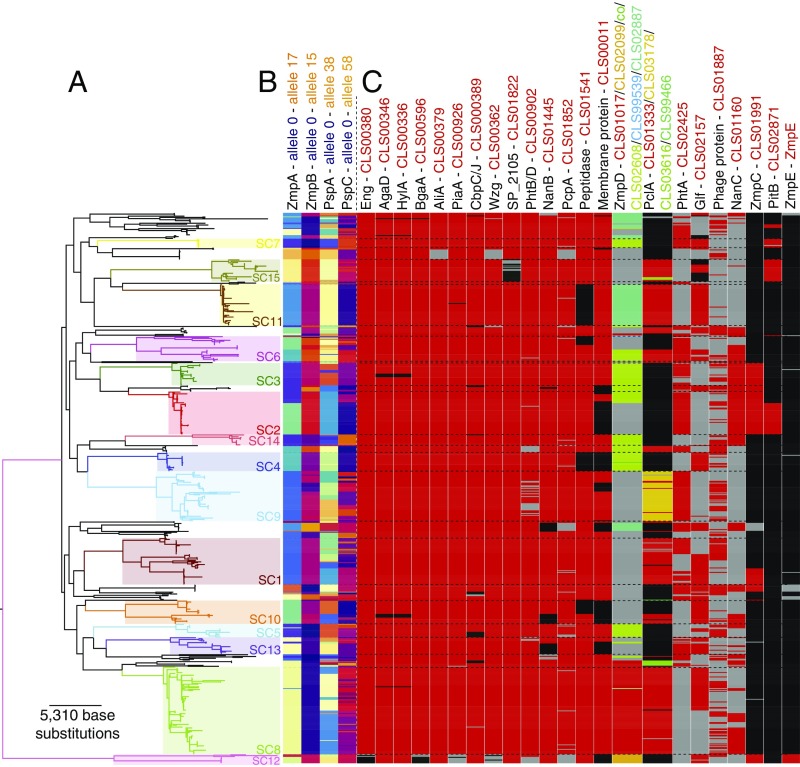
To ascertain the impact of recombination on the distribution of ABTs relative to the overall population structure, the allelic variation of the four core variable antigens was plotted against the core genome phylogeny (Fig. 4 A and B). The linkage between the 15 monophyletic sequence clusters, treated as a discrete genetic trait representing the core genome, and the sequences at polymorphic loci can be quantified by the “index of association” (IA) (43), which increases with higher linkage disequilibrium. In agreement with the estimated recombination rates, IA was highest for zmpA (IA = 0.702) and zmpB (IA = 0.747), strongly rejecting the null hypothesis of linkage equilibrium with the core genome (P < 0.001 in both cases). IA was intermediate for pspA (IA = 0.590; P < 0.001) and lowest for the most highly recombining locus, pspC (IA = 0.404; P < 0.001). Nevertheless, the distribution of variants at all these loci was significant evidence of strong linkage between the ABTs and the core genome.
Fig. 4.
Distribution of ABTs across the pneumococcal population. (A) Maximum-likelihood phylogeny based on polymorphisms within the core genome. Fifteen monophyletic sequence clusters are annotated on the tree; their boundaries are marked by horizontal dashed lines in C. Reproduced from ref. 24. (B) Distribution of the variants of each of the four variable core antigens. The distribution of ZmpA and ZmpB was inferred from the original de novo assemblies; the distribution of PspA and PspC was inferred from a mapping-based approach (SI Appendix) due to the difficulty in identifying and extracting full-length sequences of these genes from de novo assemblies. (C) Distribution of accessory genome ABTs. Each column represents a different antigen and each row a different isolate in the phylogeny. Where an antigen is present, cells are red or, in the case of multiallelic loci, an alternative color indicated by the column heading. Cells are black where an antigen is absent, and using genomic loci flanking a conserved insertion site, it is possible to confirm the relevant gene is missing from the relevant de novo assembly. Cells are gray where the ABT is absent, but assemblies cannot confirm the locus to be missing using flanking genomic loci. With some antigens, it is not possible to confirm the absence of the gene based on flanking genomic loci, due to variation in the surrounding sequence (e.g., zmpD) or the size of the genomic island on which the antigen CDS is found (e.g., CLS01887 and nanC).
Some ABTs were members of the accessory genome, present only in a subset of isolates (Fig. 4C). In the simplest case, a single subpopulation is associated with one variant of an ABT. The unencapsulated SC12 isolates are the most atypical (24, 27) and accordingly lack some antigens conserved throughout the encapsulated pneumococci, such as the glycoside-degrading enzymes Eng and BgaA, and iron SBP PiaA. By contrast, ZmpE was present only in SC12 and elicited high levels of IgG binding (SI Appendix, Fig. S6). Other accessory ABTs were polyphyletically distributed across multiple sequence clusters (24); nevertheless, they were stably associated with clades, indicated by highly significant IA values (P < 0.001). This was true across common antigens, such as neuraminidase NanB (present in 93.0% of isolates) and adhesin PcpA (present in 91.7% of isolates), as well as rarer antigens such as PhtA (present in 46.4% of isolates), neuraminidase NanC (present in 36.0% of isolates), zinc metalloprotease ZmpC (present in 19.0% of isolates), and pilus protein PitB (present in 12.0% of isolates). The ABT least stably associated with sequence clusters was a phage protein (CLS01887; IA = 0.0163; P = 0.011). Phage are the most mobile significant component of the pneumococcal accessory genome (27), and it is likely the phage amidase would show a similar distribution, if it could be efficiently assembled from short read data.
Some accessory ABTs were also present as multiple divergent variants; these could be identified as orthologs based on their shared functional domains and insertion site in the chromosome. A known example is the three clades of the type 1 pilus RrgB protein (18); CLS02942 (clade I; found in 7.8% of isolates), CLS02796 (clade II; found in 5.2% of isolates) and CLS01943 (clade III; found in 8.4% of isolates) have a strong clonal component to their distribution (IA = 0.24; P < 0.001). Clonally distributed allelic variation was also evident in the accessory antigens PclA (44), the four distinct, full-length variants of which were cumulatively found in 51.9% of the population (IA = 0.296; P < 0.001), and zinc metalloprotease ZmpD, the five distinct variants of which were cumulatively found in 52.8% of the population (IA = 0.414; P < 0.001). A single example of a fusion ZmpAD protein, the result of an ∼7.2-kb deletion, was also observed (SI Appendix, Fig. S9). Hence, the allelic variation of both core and accessory antigens was significantly associated with the clonal structure of the bacterial population.
Extensive Variation of the PsrP Locus.
The most variable accessory ABT was PsrP, the pneumococcal representative of the streptococcal serine-rich repeat family of proteins (45) (Fig. 5), the assembly of which from short read data is problematic. Nevertheless, the presence of the protein can be inferred from the SecA2 secretory system (46), hypothesized to allow streptococcal serine-rich proteins to be exported despite their large size and extensive glycosylation (47, 48). The SecA2 protein (CLS01513) is present in 42.0% of the isolates in this collection, exhibiting a strong association with particular sequence clusters (IA = 0.24; P < 0.001). The pore itself is formed by SecY2 proteins, of which three distinct variants were detected (SI Appendix, Fig. S10): group I (CLS01517), group II (CLS02820), and group III (CLS99088). Some of the “accessory secretion proteins,” such as Asp1, Asp2, and Asp3, were multiallelic and supported this split; other components of the machinery, including three glycosylases with orthologs in other streptococci (GtfA, GtfB, and Nss) (49), exhibited similar levels of conservation to SecA2. Far more variable was the complement of type 2 (Pfam domain Glycos_transf_2, accession PF00535) and type 8 (Pfam domain Glyco_transf_8, accession PF01501) glycosyltransferases. This is likely to result in these diverse islands producing antigenically distinct PsrP glycoproteins, potentially analogous to capsule polysaccharide variation.
Fig. 5.
Population-wide distribution of COGs found on the PsrP-encoding island. (A) Annotated maximum-likelihood phylogeny, as displayed in Fig. 4. Reproduced from ref. 24. (B) Each column corresponds to a COG found on the PsrP-encoding island, ordered by the likely functions of each set of orthologs, which are annotated across the top and bottom. Columns connected by inverted U shapes across the top represent genes that were disrupted by frameshift or nonsense mutations in a subset of isolates: The leftmost, black line indicates the full-length, likely functional gene, whereas the brown lines indicate gene fragments generated as a consequence of disruptive mutations. Each cell in the columns corresponds to an isolate in the core-genome phylogeny; red cells indicate the presence of a COG, whereas gray cells indicate it is absent. Glycosyltransferase is abbreviated as “GTase.”
Variation in PsrP is also observed within sequence clusters, without the involvement of horizontal DNA transfer. Two SC8 isolates are likely to have lost the ability to express the antigen through changes in the PsrP island’s gene content: The asp1 gene was disrupted by an IS element insertion in ATBLM, whereas a large deletion in the 0OH1I isolate eliminated much of the island, including the psrP structural gene (SI Appendix, Fig. S10). Other mutations likely to alter or prevent PsrP’s expression on the cell’s surface were frameshift mutations in core machinery, such as secA2, and several glycosyltransferases (Fig. 5). One particular frameshift of note occurred within a type 2 glycosyltransferase gene labeled pvgP (phase variable glycosyltransferase of PsrP, CLS01518). This reversible mutation, facilitated by an 8- to 9-nt guanine homopolymeric tract, occurred sufficiently frequently that the pseudogene it generated, the distribution of which is shown in the directly adjacent column, did not have a significant index of association (IA = −0.0444; P = 1.00). The pvgP gene was present in all PsrP islands in this collection, but is without close homologs outside of Streptococcus mitis and Streptococcus pseudopneumoniae, suggesting it is a recently emerged mechanism for altering the glycosylation of PsrP over short timescales.
Antigens and Population Structure Determination.
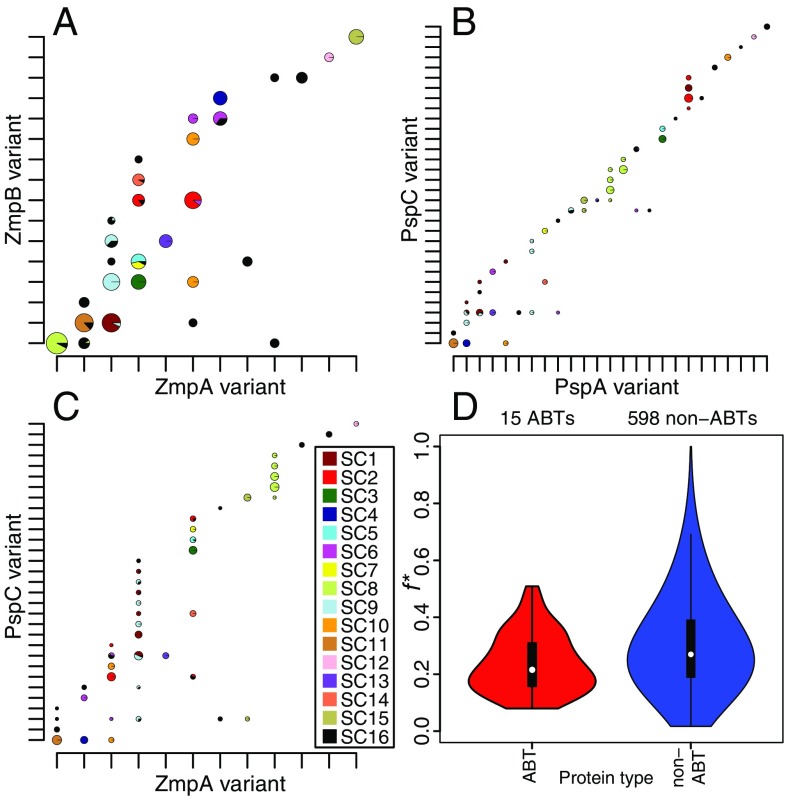
Nevertheless, the highly significant IA values observed between ABTs and sequence clusters indicates that pneumococcal sequence clusters have generally stable antigenic profiles. The neutral hypothesis is that closely related isolates share similar sequences through common descent, consistent with the relatively low estimated recombination rates (Fig. 2) (43). An alternative hypothesis is that the structuring of the population into sequence clusters is a consequence of immune selection on antigenic diversity (50, 51), such that the population is a mixture of nonoverlapping, “discordant” antigenic profiles. This can be quantified by the f* metric, which has a maximum value of one that represents an entirely discordant set of variants (52). The conservation of zmpA and zmpB loci within sequence clusters made them good candidates for having a nonoverlapping distribution across the population. Consistent with both hypotheses, there was clear linkage disequilibrium between the two loci and the core genome (Fig. 6A). However, this comparison found f* to be 0.306, much lower than the value of 0.97 calculated from Neisseria meningitidis Opa sequences, which has been cited as evidence that the meningococcal population structure was shaped by immune selection (53).
Fig. 6.
Cooccurrence of variants of pneumococcal CDSs. (A) Cooccurrence of ZmpA and ZmpB variants. The variants were ordered on the axes to maximize the number of combinations on the “diagonal” of the plot. Observed variant combinations are marked with pie charts, which illustrate the genetic backgrounds in which they were found as segments colored according to the sequence clusters to which the relevant isolates belong (key in C and annotated phylogeny in Fig. 4). The diameter of the pie charts is proportional to the logarithm of the number of isolates with the relevant antigenic profile; only combinations found in >0.5% of the population are shown. Note that the diameters of the pie charts are not directly comparable between panels. (B) Cooccurrence of PspA and PspC variants in the population, displayed as described in A. (C) Cooccurrence of ZmpA and PspC variants in the population, displayed as described in A. (D) Violin plots displaying the f* values for all pairwise comparisons between variable core ABT COGs alongside those for all pairwise comparisons between variable core non-ABT COGs. Only COGs for which at least two variants were defined, with at least two variant combinations both present in the population at greater than 5% frequency, were included in this comparison.
The distribution of antigen profiles is sensitive to the definition of variants; although there was no evidence of more cross-reactivity between genetically similar variants (SI Appendix, Fig. S11), there was the potential for more numerous variant sets to exhibit a more discordant distribution. Hence, this analysis was repeated using the more diverse sets of PspA and PspC proteins (Fig. 6B). However, there was similarly little evidence of a nonoverlapping distribution in this case (f* = 0.285). The most informative comparison was that between PspC, the most rapidly recombining of the four loci (Fig. 2), and ZmpA, which was affected by recombination at a relatively low rate. This revealed a particularly high level of overlap between antigenic profiles (f* = 0.165; Fig. 6C) as a consequence of the vertical “stripes” of isolates that shared the same ZmpA variant, but had diversified through recombination at the PspC locus. To test whether these patterns were broadly representative across the population, ABT and non-ABT COGs were classified into variants based on the proportion of shared 15-mers (SI Appendix, Fig. S12). A higher f* was observed for non-ABTs (0.270) than for ABTs (0.216; Fig. 6D), and therefore there is insufficient evidence to reject the null hypothesis that the linkage disequilibrium observed between antigenic loci represents merely the clonality of pneumococcal populations.
Accessory Antigens Appear Neutrally Distributed.
As the identification of discordant antigen sets is sensitive to the definition of variants, a similar analysis was conducted based on the discrete signal of the presence, or absence, of accessory antigens. In these analyses, high f* metrics can be interpreted in two ways. If the presence of accessory antigens correlates across the population, then this is likely to represent some strains being adapted to infecting immunologically naive hosts, whereas others minimize the number of exposed epitopes to better survive in immunologically mature hosts. However, high f* values resulting from anticorrelation between the presence of accessory antigens may indicate the proteins are functionally redundant. Pairs of structurally similar accessory antigens were used to test these models (SI Appendix, Fig. S13): the zinc metalloproteases ZmpC and ZmpD (f* = 0.177), the neuraminidases NanB and NanC (f* = 0.451), the two pili (f* = 0.521), and the large adhesins PsrP and PclA (f* = 0.228). The low f* values demonstrate these pairs of functionally similar accessory antigens do not conform to either the immunological adaptation or the functional redundancy hypothesis. Additionally, the functional redundancy hypothesis can be definitively rejected, as the modal genotype was the absence of both ABTs in three of these comparisons.
An alternative selective pressure that might shape the distribution of antigens is the age distribution of the host population (54). This hypothesis was tested using the four diverse core loci, but no evidence of heterogeneity in the age distribution of different variants was found (SI Appendix, Fig. S14). Similarly, the only accessory antigen that showed a significant enrichment in younger children was PclA (Wilcoxon test, W = 42,870.5, P = 7.24 × 10−4; SI Appendix, Fig. S15), consistent with one of the PclA variants, CLS01333, showing the strongest age association of all COGs previously assessed across the collection (24). However, there was also no significant evidence for a heterogeneous serotype distribution across ages (SI Appendix, Fig. S16), indicating this sample may have limited power to detect age-based relationships, as other studies have found such a pattern (55).
Discussion
Combining population genomic data with clinical information on the human IgG response to pneumococcal proteins provides a unique insight into the complex immunological interaction between host and pathogen. The continuous distribution of IgG binding values suggests that adaptive immunity responds to many antigens in both the core and the accessory genome. The most immunogenic proteins had physical properties expected to be associated with antibody binding: signal peptides, including those with a YSIRK motif, for secretion out of the bacterial cell; large size; and a lipoprotein processing motif, LPXTG sortase processing motif or CBD for surface attachment. The great majority of the ABTs either were variants of the diverse PspA, PspC, ZmpA, and ZmpB proteins or could be grouped into surface-exposed roles in host molecule adhesion or degradation, cell wall metabolism, or solute binding for transport. Despite the proteins on the microarray being expressed in vitro, detected ABTs included examples with transmembrane helices, such as Wzg; proteins normally subject to posttranslational modifications, such as glycosylation of PsrP or lipid attachment to solute-binding proteins; and individual components of multimeric structures, such as PitB of the type 2 pilus. It remains possible that some immunogenic proteins were not strongly bound by IgG due to their unmodified or misfolded form on the microarray or that infants, the typical hosts of pneumococci, may recognize different ABTs. However, the overall IgG binding response was consistent both with functional analyses of protein sequences and with previous analyses of pneumococcal immunogenicity (17).
In contrast to the consistent physical properties across the functional categories associated with ABTs, there was extensive heterogeneity in their patterns of diversification. There was little difference between ABTs and non-ABTs in their levels of sequence conservation; notable exceptions were pspA and pspC, previously identified hotspots of transformation events that exhibited relatively low levels of clonal association, as measured by IA. Another antigen with an even lower IA was CLS01887, a protein of unknown function that moved rapidly through phage infection (27). However, neither PblB nor any of the other 18 phage COGs on the microarray were classed as ABTs, and the movement of these “selfish” elements is unlikely to represent adaptive evolution in response to immune selection pressures. Similarly, the most rapid variation associated with an ABT was the phase variation of the gene encoding the PsrP glycosyltransferase pvgP. The expression of some pneumococcal capsules is also affected by phase variation (56, 57), although this mechanism is also not uniquely associated with antigens, as phase variation of pneumococcal restriction modification systems is also highly frequent (27). Hence, multiple genetic mechanisms can vary cells’ antigenic profiles, but analysis of genetic diversity could not consistently distinguish ABTs from non-ABTs.
The extensive diversity and rapid variation of several ABTs seems paradoxical in the context of the large number of pneumococcal epitopes recognized by IgG, as it is unclear what benefit cells derive from varying only a small proportion of their overall antigenic profile. Theoretical models have assumed a small number of diverse antigens to be immunodominant, such that strong selection pressures acting on these proteins would shape the bacterial population (50, 51). For this model to be correct, all ABTs that could trigger immune clearance should exhibit significant signs of diversification. The model could be reconciled with our findings if some IgG binding represents antibodies that do not impede pneumococcal colonization, either because they are intrinsically ineffective or because other mechanisms prevent their activity. Other IgG may be more active than their affinities suggest if they effectively block a bacterial protein’s biological function. Notably, some of the most highly variable surface structures inhibit the antipneumococcal activity of antibody and complement: the polysaccharide capsule (58); PspA and PspC, which inhibit complement-mediated opsonophagocytosis (59, 60); and ZmpA, which degrades IgA (61). It may be that these structures are able to nullify the host immune responses, as long as they themselves are not neutralized by antibodies. These few antigens may be more important to the bacterium’s ability to elude innate or acquired immunity than other immunogenic proteins and hence evolve under stronger selective pressure to evade antibody responses. Alternatively, an explanation for the higher level of variability at these loci could be that only larger proteins, which often show extensive evidence of diversification (e.g., PsrP and PclA), are readily accessible to antibodies in encapsulated pneumococci (62). However, the study did not find ABT distributions across the population consistent with a small number of immunodominant proteins.
Additionally, it is difficult to conclude from experimental evidence that all proteins targeted by effective antibody responses are highly variable. The proposed ineffectiveness of the immune response to conserved antigens is not consistent with previous work with some SBPs that found evidence that they can be the targets of functional antipneumococcal antibody responses. For example, IgG targeting the ABC transporter lipoproteins PiuA and PiaA can trigger opsonophagocytic activity in human cell lines (63), and immunization of mice with these, or another conserved ABC transporter protein, is protective against invasive (64, 65) or respiratory (66) challenge with pneumococci. One study found immunization with the conserved PsaA protein actually elicited better protection against carriage than PspA (67). It remains possible these experimental models do not reflect the activity of the human immune system in vivo. Alternatively, the overall B-cell response to pneumococcal carriage may consist of many individually weak interactions. This would be consistent with the continuous range of IgG binding values observed in this dataset. The differing levels of diversification observed across the ABTs would then be explained by variation in the functional constraints on the proteins. PspA and PspC appear to be recently evolved proteins, with many different sequences apparently able to perform their activities (20, 68); low levels of functional constraint would then explain how they are able to switch variants so frequently. By contrast, ABTs with homologs across distantly related bacterial species, such as cell wall synthesis machinery and SBPs, are likely subject to much stronger functional constraint, and hence their greater conservation between pneumococci.
One argument against this latter model is that the polysaccharide capsule exhibits similar levels of diversity to PspA and PspC, but experimental and theoretical analyses show that this variation strongly affects strain fitness (10, 69). However, the immune responses to proteins and polysaccharides are distinct, and it may be that there is stronger selection for switching of serotype, rather than some protein antigens; additionally, serotype switching is not free from selective constraints (22). Resolving the paradox of why extensive variation is observed in only a subset of ABTs will be usefully informed by both experimental and clinical data. The combination of population genomic datasets with pangenome-wide information on antibody binding promises to be a valuable tool for addressing such unresolved questions about the immunological response to infections.
Methods
Analysis of Proteome Microarray Data.
The design and construction of the proteome microarray, and the processing and analysis of the raw data, are described in SI Appendix. Serum samples came from the VAC-002 study approved by the Western Institutional Review Board. For each person, each protein was represented by the probe recording the highest normalized, log2-transformed antibody response score. These IgG binding values across all individuals were used to generate an all-vs.-all distance matrix between proteins, which allowed the proteins to be classified into 208 ABTs and the remaining non-ABT, using the R function “hclust” (70). The annotation of the proteins and the associated functional domain information used in regression analyses are detailed in Dataset S1.
Determining Patterns of Presence and Absence.
The pattern of presence and absence of each COG across isolates was determined by the clustering of sequences defined and made freely available previously (24, 26), combined with the information from scaffolded assemblies, as described elsewhere (27). For each ABT, “flanking” COGs were defined as the nearest COGs, both upstream and downstream, that were found only in single copy in each genome in which they were present and therefore could be used to define a unique locus in the chromosome. The presence of both an ABT’s flanking COGs on the same scaffold, in the absence of the ABT itself, was taken to represent stronger evidence that the ABT was genuinely missing from an isolate (Fig. 4). Multiple COGs were considered to be orthologous representatives of the same antigen if they were present at equivalent positions in different genomes, as judged by sharing the same flanking COGs; if they had similar functional domain structures, as judged by sharing the majority of motifs identified by Pfam in both sequences; and if they never cooccurred in the same strain, to exclude paralogs. These criteria identified the four functional PclA variants (CLS01333, CLS03616, CLS99466, and CLS03178), the three clades of RrgB (CLS01943, CLS02796, and CLS02942), ZmpD (CLS02608, CLS02099, CLS02887, CLS01017, and CLS99539), and PhtD/E (CLS00904 and CLS02083). However, poor assembly precluded further analysis of the repetitive PhtD/E CDSs.
A similar approach was also used to identify the COGs present on PsrP islands. The boundary of these islands was defined as the nearest core COG, CLS01506 (present in a single copy in all isolates), upstream of the SecA2 protein. Therefore, in all cases where CLS01513 (SecA2) was present, the CDSs on the same contig as, and downstream of, the core COG CLS01506 were treated as members of the PsrP island; the other boundary was not defined, as the psrP gene never assembled completely. This set was then manually curated to remove fragments of the psrP CDS and noncore CDSs that were not part of the island.
Detection of Recombination and Selection.
All COGs included on the microarray, and alignments of the individual variants of the core diverse loci, were filtered to remove sequences less than 75%, or more than 125%, of the median sequence length. Protein sequences were aligned with MUSCLE (71), and the corresponding DNA sequences were back translated to give a codon alignment. Population genetic statistics, including πn and Tajima’s D, were calculated using Bioperl (72) and FAST (73). Duplicate sequences were removed, and then a maximum-likelihood phylogeny was generated using RAxML v7.0.3 with a general time-reversible substitution model and a gamma model of rate heterogeneity (74). Evolutionary models were then fitted to the alignments and phylogenies (with fixed branch lengths), using the codeml software of PAML v4.8 (33), applying site models 0 (single ω ratio across the alignment), 1a (nearly neutral diversification), and 2a (evolution under positive selection). Independent runs were conducted with starting ω values of 0.1, 1, and 10 to test for convergence. The ω and Λ results were reported only if dN and dS estimated from an alignment were both greater than 0.25. All amino acids were considered equally dissimilar from one another.
To reduce any biases introduced by recombination, the codon alignments were scanned for breakpoints. The initial scan was performed with 3SEQ (36), which is able to detect exchanges between sequences in an alignment, using the “fullrun” mode. Each segment defined by the first set of breakpoints was then iteratively scanned with 3SEQ for further evidence of recombination. This set of segments was then iteratively scanned for imports of divergent tracts of sequence, using the maximum-χ2 approach (37, 38). Following the convergence of these methods, each segment of the alignment was analyzed with RAxML and PAML as described for the whole alignment analysis above.
To calculate the level of recombination affecting ABTs based on the analyses of whole-genome alignments (24–26), the loci within each sequence cluster’s representative reference genome (26) corresponding to each COG in the complete set of isolates had to be identified. As there should be no genuine genetic variation between different assemblies of the same sets of sequence reads, except for potential variance in the start codon selection, the middle segment of each COG was used to select identical matches in the reference sequences of the monophyletic sequence clusters. The corresponding CDSs were then used to calculate the number of recombination events that affected the evolution of these proteins. Only COGs found in all 15 reference sequences were analyzed, to avoid unequal sample sizes. The loci encoding zmpA, zmpB, pspA, and pspC were manually identified (24), and recombinations affecting cps loci were described previously (22).
Analyses of Variant Sequence Distributions.
Indexes of association were calculated using the R package PoppR (75). The f* metric was calculated using the approach described by Buckee et al. (52),
where xij is the frequency of variant combination i (at the first locus) and j (at the second locus) across the population; pi is the frequency of variant i at the first locus; qj is the frequency of variant j at the second locus; and k is the value of j that maximizes xij, whereas h is the value of i that maximizes xij. The comparison of f* between ABTs and non-ABTs used the COG sequences analyzed in Fig. 2. Variants were defined by generating pairwise similarities between sequences within the same alignment based on the number of shared identical 15-mers from the full set of 15 amino acid patterns present in either sequence. Based on the overall distribution of pairwise similarities (SI Appendix, Fig. S12), a threshold similarity of 0.75 was used to define discrete variants within each COG alignment. The f* value was calculated for pairs of ABT or non-ABT COGs if both were divided into two or more variants and more than one combination was present at greater than 5% frequency in the population; only these common combinations were used in the calculation of f*.
Supplementary Material
Acknowledgments
We thank the participants in the phase I trial of the whole-cell vaccine. Research reported in this publication was supported by The Bill & Melinda Gates Foundation, PATH, the National Institute of Allergy and Infectious Diseases of the US National Institutes of Health Grant R01AI066304, and the Wellcome Trust Grant 098051. N.J.C. is funded by a Sir Henry Dale Fellowship, jointly funded by the Wellcome Trust and Royal Society (Grant 104169/Z/14/Z).
Footnotes
Conflict of interest statement: J.J.C., T.Q.L., and X.L. are employees of Antigen Discovery, Inc. In addition, X.L. has an equity interest in Antigen Discovery, Inc. N.J.C., S.D.B., W.P.H., and M.L. were consultants asked to design the proteome array by Antigen Discovery, Inc. In addition, M.L. has received consulting fees from Pfizer and Affinivax, travel reimbursement from GlaxoSmithKline, and grant funding through his institution from Pfizer and PATH Vaccine Solutions.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the European Nucleotide Archive under project code PRJEB2632 and accession codes LT669625–LT669755, and in the Dryad repository under doi.org/10.5061/dryad.t55gq.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613937114/-/DCSupplemental.
References
- 1.Gratten M, et al. Colonisation of Haemophilus influenzae and Streptococcus pneumoniae in the upper respiratory tract of neonates in Papua New Guinea: Primary acquisition, duration of carriage, and relationship to carriage in mothers. Biol Neonate. 1986;50(2):114–120. doi: 10.1159/000242576. [DOI] [PubMed] [Google Scholar]
- 2.Gray BM, Turner ME, Dillon HC., Jr Epidemiologic studies of Streptococcus pneumoniae in infants. The effects of season and age on pneumococcal acquisition and carriage in the first 24 months of life. Am J Epidemiol. 1982;116(4):692–703. doi: 10.1093/oxfordjournals.aje.a113452. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Q, et al. Serum and mucosal antibody responses to pneumococcal protein antigens in children: Relationships with carriage status. Eur J Immunol. 2006;36(1):46–57. doi: 10.1002/eji.200535101. [DOI] [PubMed] [Google Scholar]
- 4.Holmgren J, Czerkinsky C. Mucosal immunity and vaccines. Nat Med. 2005;11(4) Suppl:S45–S53. doi: 10.1038/nm1213. [DOI] [PubMed] [Google Scholar]
- 5.Anttila M, Voutilainen M, Jäntti V, Eskola J, Käyhty H. Contribution of serotype-specific IgG concentration, IgG subclasses and relative antibody avidity to opsonophagocytic activity against Streptococcus pneumoniae. Clin Exp Immunol. 1999;118(3):402–407. doi: 10.1046/j.1365-2249.1999.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janoff EN, et al. Killing of Streptococcus pneumoniae by capsular polysaccharide-specific polymeric IgA, complement, and phagocytes. J Clin Invest. 1999;104(8):1139–1147. doi: 10.1172/JCI6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z, Clarke TB, Weiser JN. Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J Clin Invest. 2009;119(7):1899–1909. doi: 10.1172/JCI36731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malley R, et al. CD4+ T cells mediate antibody-independent acquired immunity to pneumococcal colonization. Proc Natl Acad Sci USA. 2005;102(13):4848–4853. doi: 10.1073/pnas.0501254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, et al. Distinct effects on diversifying selection by two mechanisms of immunity against Streptococcus pneumoniae. PLoS Pathog. 2012;8(11):e1002989. doi: 10.1371/journal.ppat.1002989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobey S, Lipsitch M. Niche and neutral effects of acquired immunity permit coexistence of pneumococcal serotypes. Science. 2012;335(6074):1376–1380. doi: 10.1126/science.1215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rennels MB, et al. Safety and immunogenicity of heptavalent pneumococcal vaccine conjugated to CRM197 in United States infants. Pediatrics. 1998;101(4 Pt 1):604–611. doi: 10.1542/peds.101.4.604. [DOI] [PubMed] [Google Scholar]
- 12.Ghaffar F, et al. Effect of the 7-valent pneumococcal conjugate vaccine on nasopharyngeal colonization by Streptococcus pneumoniae in the first 2 years of life. Clin Infect Dis. 2004;39(7):930–938. doi: 10.1086/423379. [DOI] [PubMed] [Google Scholar]
- 13.Shinefield HR, et al. Safety and immunogenicity of heptavalent pneumococcal CRM197 conjugate vaccine in infants and toddlers. Pediatr Infect Dis J. 1999;18(9):757–763. doi: 10.1097/00006454-199909000-00004. [DOI] [PubMed] [Google Scholar]
- 14.Moffitt KL, Malley R. Next generation pneumococcal vaccines. Curr Opin Immunol. 2011;23(3):407–413. doi: 10.1016/j.coi.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldblatt D, et al. Antibody responses to nasopharyngeal carriage of Streptococcus pneumoniae in adults: A longitudinal household study. J Infect Dis. 2005;192(3):387–393. doi: 10.1086/431524. [DOI] [PubMed] [Google Scholar]
- 16.Beghetto E, et al. Discovery of novel Streptococcus pneumoniae antigens by screening a whole-genome λ-display library. FEMS Microbiol Lett. 2006;262(1):14–21. doi: 10.1111/j.1574-6968.2006.00360.x. [DOI] [PubMed] [Google Scholar]
- 17.Giefing C, et al. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med. 2008;205(1):117–131. doi: 10.1084/jem.20071168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barocchi MA, et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci USA. 2006;103(8):2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagnoli F, et al. A second pilus type in Streptococcus pneumoniae is prevalent in emerging serotypes and mediates adhesion to host cells. J Bacteriol. 2008;190(15):5480–5492. doi: 10.1128/JB.00384-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollingshead SK, Becker R, Briles DE. Diversity of PspA: Mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect Immun. 2000;68(10):5889–5900. doi: 10.1128/iai.68.10.5889-5900.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks-Walter A, Briles DE, Hollingshead SK. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect Immun. 1999;67(12):6533–6542. doi: 10.1128/iai.67.12.6533-6542.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Croucher NJ, et al. Selective and genetic constraints on pneumococcal serotype switching. PLoS Genet. 2015;11(3):e1005095. doi: 10.1371/journal.pgen.1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang SS, et al. Continued impact of pneumococcal conjugate vaccine on carriage in young children. Pediatrics. 2009;124(1):e1–e11. doi: 10.1542/peds.2008-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croucher NJ, et al. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet. 2013;45(6):656–663. doi: 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croucher NJ, et al. Population genomic datasets describing the post-vaccine evolutionary epidemiology of Streptococcus pneumoniae. Sci Data. 2015;2:150058. doi: 10.1038/sdata.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Croucher NJ, et al. Diversification of bacterial genome content through distinct mechanisms over different timescales. Nat Commun. 2014;5:5471. doi: 10.1038/ncomms6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moschioni M, et al. Streptococcus pneumoniae contains 3 rlrA pilus variants that are clonally related. J Infect Dis. 2008;197(6):888–896. doi: 10.1086/528375. [DOI] [PubMed] [Google Scholar]
- 29.Tettelin H, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293(5529):498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 30.Loisel E, et al. Biochemical characterization of the histidine triad protein PhtD as a cell surface zinc-binding protein of pneumococcus. Biochemistry. 2011;50(17):3551–3558. doi: 10.1021/bi200012f. [DOI] [PubMed] [Google Scholar]
- 31.Anderton JM, et al. E-cadherin is a receptor for the common protein pneumococcal surface adhesin A (PsaA) of Streptococcus pneumoniae. Microb Pathog. 2007;42(5-6):225–236. doi: 10.1016/j.micpath.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 34.Anisimova M, Bielawski JP, Yang Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 2001;18(8):1585–1592. doi: 10.1093/oxfordjournals.molbev.a003945. [DOI] [PubMed] [Google Scholar]
- 35.Anisimova M, Nielsen R, Yang Z. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics. 2003;164(3):1229–1236. doi: 10.1093/genetics/164.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boni MF, Posada D, Feldman MW. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics. 2007;176(2):1035–1047. doi: 10.1534/genetics.106.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34(2):126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 38.Piganeau G, Gardner M, Eyre-Walker A. A broad survey of recombination in animal mitochondria. Mol Biol Evol. 2004;21(12):2319–2325. doi: 10.1093/molbev/msh244. [DOI] [PubMed] [Google Scholar]
- 39.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331(6016):430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Croucher NJ, et al. Evidence for soft selective sweeps in the evolution of pneumococcal multidrug resistance and vaccine escape. Genome Biol Evol. 2014;6(7):1589–1602. doi: 10.1093/gbe/evu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Croucher NJ, et al. Variable recombination dynamics during the emergence, transmission and ‘disarming’ of a multidrug-resistant pneumococcal clone. BMC Biol. 2014;12(1):49. doi: 10.1186/1741-7007-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303(5655):223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 43.Smith JM, Smith NH, O’Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90(10):4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paterson GK, Nieminen L, Jefferies JM, Mitchell TJ. PclA, a pneumococcal collagen-like protein with selected strain distribution, contributes to adherence and invasion of host cells. FEMS Microbiol Lett. 2008;285(2):170–176. doi: 10.1111/j.1574-6968.2008.01217.x. [DOI] [PubMed] [Google Scholar]
- 45.Rose L, et al. Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge. J Infect Dis. 2008;198(3):375–383. doi: 10.1086/589775. [DOI] [PubMed] [Google Scholar]
- 46.Bensing BA, Sullam PM. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol Microbiol. 2002;44(4):1081–1094. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 47.Bensing BA, Gibson BW, Sullam PM. The Streptococcus gordonii platelet binding protein GspB undergoes glycosylation independently of export. J Bacteriol. 2004;186(3):638–645. doi: 10.1128/JB.186.3.638-645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feltcher ME, Braunstein M. Emerging themes in SecA2-mediated protein export. Nat Rev Microbiol. 2012;10(11):779–789. doi: 10.1038/nrmicro2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yen YT, et al. Differential localization of the streptococcal accessory sec components and implications for substrate export. J Bacteriol. 2013;195(4):682–695. doi: 10.1128/JB.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta S, et al. The maintenance of strain structure in populations of recombining infectious agents. Nat Med. 1996;2(4):437–442. doi: 10.1038/nm0496-437. [DOI] [PubMed] [Google Scholar]
- 51.Gupta S, Ferguson N, Anderson R. Chaos, persistence, and evolution of strain structure in antigenically diverse infectious agents. Science. 1998;280(5365):912–915. doi: 10.1126/science.280.5365.912. [DOI] [PubMed] [Google Scholar]
- 52.Buckee CO, Gupta S, Kriz P, Maiden MCJ, Jolley KA. Long-term evolution of antigen repertoires among carried meningococci. Proc Biol Sci. 2010;277(1688):1635–1641. doi: 10.1098/rspb.2009.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Callaghan MJ, et al. The effect of immune selection on the structure of the meningococcal Opa protein repertoire. PLoS Pathog. 2008;4(3):e1000020. doi: 10.1371/journal.ppat.1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Regev-Yochay G, et al. Re-emergence of the type 1 pilus among Streptococcus pneumoniae isolates in Massachusetts, USA. Vaccine. 2010;28(30):4842–4846. doi: 10.1016/j.vaccine.2010.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hausdorff WP, Feikin DR, Klugman KP. Epidemiological differences among pneumococcal serotypes. Lancet Infect Dis. 2005;5(2):83–93. doi: 10.1016/S1473-3099(05)01280-6. [DOI] [PubMed] [Google Scholar]
- 56.Waite RD, Struthers JK, Dowson CG. Spontaneous sequence duplication within an open reading frame of the pneumococcal type 3 capsule locus causes high-frequency phase variation. Mol Microbiol. 2001;42(5):1223–1232. doi: 10.1046/j.1365-2958.2001.02674.x. [DOI] [PubMed] [Google Scholar]
- 57.Waite RD, Penfold DW, Struthers JK, Dowson CG. Spontaneous sequence duplications within capsule genes cap8E and tts control phase variation in Streptococcus pneumoniae serotypes 8 and 37. Microbiology. 2003;149(Pt 2):497–504. doi: 10.1099/mic.0.26011-0. [DOI] [PubMed] [Google Scholar]
- 58.Hyams C, Camberlein E, Cohen JM, Bax K, Brown JS. The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect Immun. 2010;78(2):704–715. doi: 10.1128/IAI.00881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tu AH, Fulgham RL, McCrory MA, Briles DE, Szalai AJ. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect Immun. 1999;67(9):4720–4724. doi: 10.1128/iai.67.9.4720-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Janulczyk R, Iannelli F, Sjoholm AG, Pozzi G, Bjorck L. Hic, a novel surface protein of Streptococcus pneumoniae that interferes with complement function. J Biol Chem. 2000;275(47):37257–37263. doi: 10.1074/jbc.M004572200. [DOI] [PubMed] [Google Scholar]
- 61.Wani JH, Gilbert JV, Plaut AG, Weiser JN. Identification, cloning, and sequencing of the immunoglobulin A1 protease gene of Streptococcus pneumoniae. Infect Immun. 1996;64(10):3967–3974. doi: 10.1128/iai.64.10.3967-3974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gor DO, Ding X, Briles DE, Jacobs MR, Greenspan NS. Relationship between surface accessibility for PpmA, PsaA, and PspA and antibody-mediated immunity to systemic infection by Streptococcus pneumoniae. Infect Immun. 2005;73(3):1304–1312. doi: 10.1128/IAI.73.3.1304-1312.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jomaa M, et al. Antibodies to the iron uptake ABC transporter lipoproteins PiaA and PiuA promote opsonophagocytosis of Streptococcus pneumoniae. Infect Immun. 2005;73(10):6852–6859. doi: 10.1128/IAI.73.10.6852-6859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown JS, Ogunniyi AD, Woodrow MC, Holden DW, Paton JC. Immunization with components of two iron uptake ABC transporters protects mice against systemic Streptococcus pneumoniae infection. Infect Immun. 2001;69(11):6702–6706. doi: 10.1128/IAI.69.11.6702-6706.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saxena S, Khan N, Dehinwal R, Kumar A, Sehgal D. Conserved surface accessible nucleoside ABC transporter component SP0845 is essential for pneumococcal virulence and confers protection in vivo. PLoS One. 2015;10(2):e0118154. doi: 10.1371/journal.pone.0118154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jomaa M, et al. Immunization with the iron uptake ABC transporter proteins PiaA and PiuA prevents respiratory infection with Streptococcus pneumoniae. Vaccine. 2006;24(24):5133–5139. doi: 10.1016/j.vaccine.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 67.Briles DE, et al. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect Immun. 2000;68(2):796–800. doi: 10.1128/iai.68.2.796-800.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iannelli F, Oggioni MR, Pozzi G. Allelic variation in the highly polymorphic locus pspC of Streptococcus pneumoniae. Gene. 2002;284(1-2):63–71. doi: 10.1016/s0378-1119(01)00896-4. [DOI] [PubMed] [Google Scholar]
- 69.Trzciński K, et al. Effect of serotype on pneumococcal competition in a mouse colonization model. MBio. 2015;6(5):e00902–e00915. doi: 10.1128/mBio.00902-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R Core Development Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2011. [Google Scholar]
- 71.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stajich JE, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12(10):1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lawrence TJ, et al. FAST: FAST Analysis of Sequences Toolbox. Front Genet. 2015;6:172. doi: 10.3389/fgene.2015.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stamatakis A, Ludwig T, Meier H. RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21(4):456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 75.Kamvar ZN, Brooks JC, Grünwald NJ. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front Genet. 2015;6(JUN):208. doi: 10.3389/fgene.2015.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.