Abstract
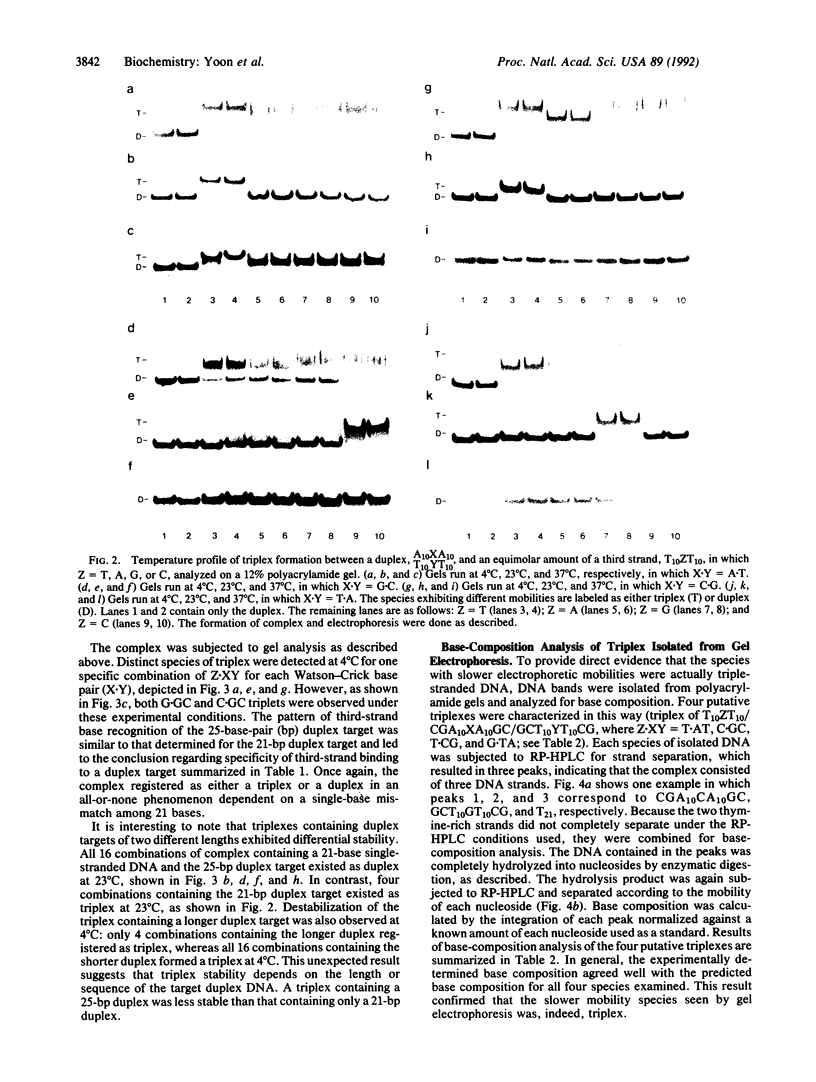
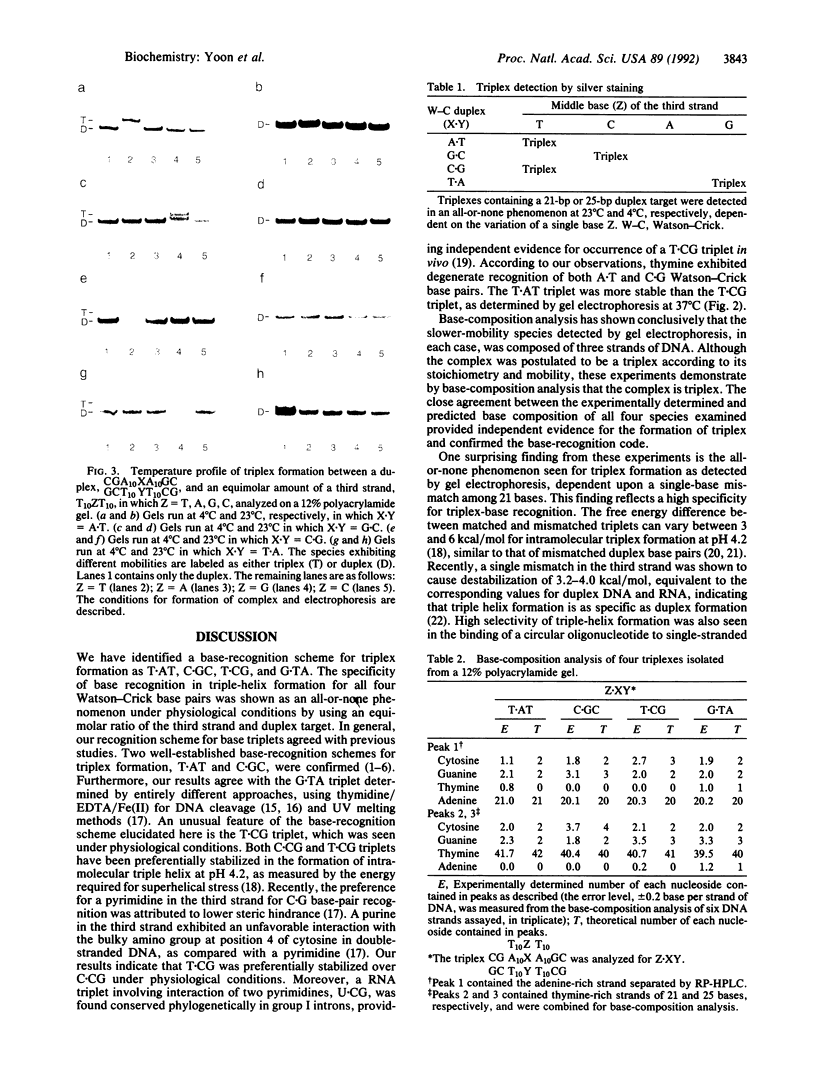
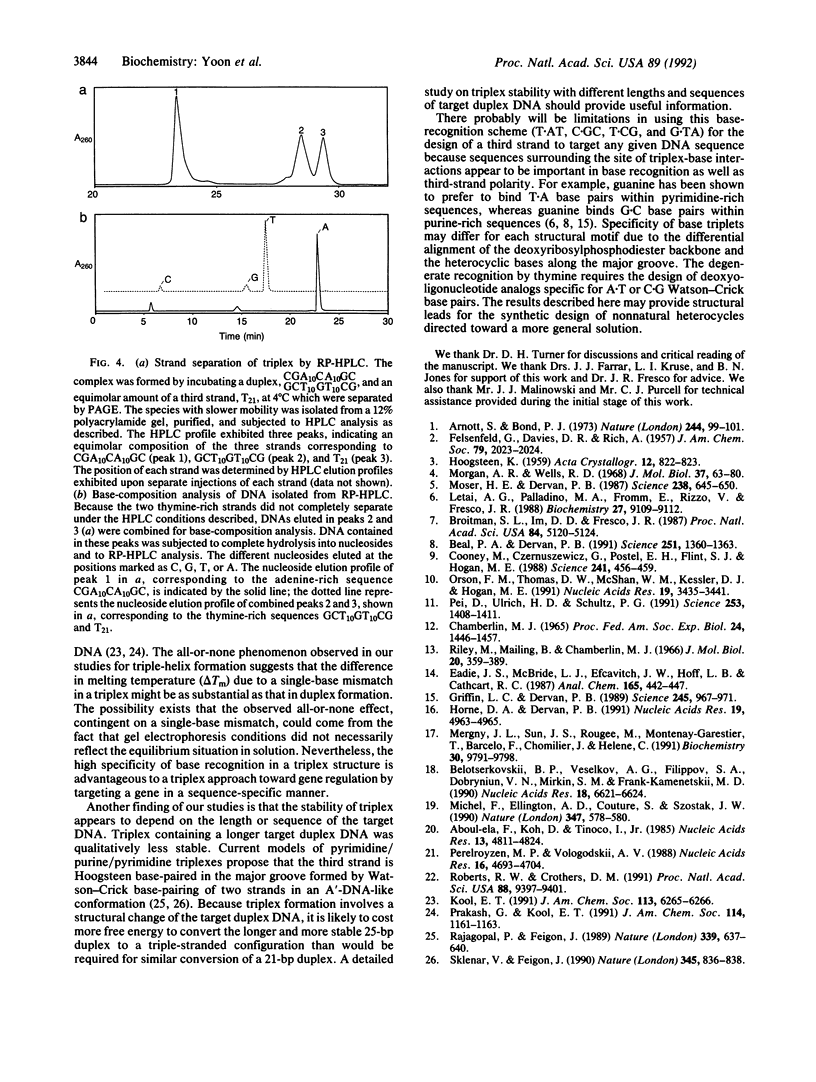
We report a specific pattern of recognition by third-strand bases for each of the four Watson-Crick base pairs within a pyrimidine triple-helix motif as determined by PAGE: T.AT, C.GC, T.CG, and G.TA. Our recognition scheme for base triplets is in agreement with previous studies. In addition, we identified another triplet, T.CG, under physiological conditions, in which formation of triple helix was observed at equimolar ratios of the third strand and duplex target. Although different nearest-neighbor effects are expected, this finding extends the base-recognition code to all 4 base pairs in double-stranded DNA under physiological conditions. Base-composition analysis of putative triplex species provided independent evidence for the formation of triplex and confirmed the base-recognition code determined by PAGE. Moreover, the formation of triplex, as detected by gel electrophoresis, was seen to be an all-or-none phenomenon, dependent upon a single-base mismatch among 21 nucleotides. This result suggests a high specificity for the recognition of double-stranded DNA by a third strand. In addition, we report the surprising finding that triplex stability depends on the length and sequence of the target duplex DNA.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Koh D., Tinoco I., Jr, Martin F. H. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res. 1985 Jul 11;13(13):4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Bond P. J. Structures for Poly(U)-poly(A)-poly(U)triple stranded polynucleotides. Nat New Biol. 1973 Jul 25;244(134):99–101. doi: 10.1038/newbio244099a0. [DOI] [PubMed] [Google Scholar]
- Beal P. A., Dervan P. B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991 Mar 15;251(4999):1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- Belotserkovskii B. P., Veselkov A. G., Filippov S. A., Dobrynin V. N., Mirkin S. M., Frank-Kamenetskii M. D. Formation of intramolecular triplex in homopurine-homopyrimidine mirror repeats with point substitutions. Nucleic Acids Res. 1990 Nov 25;18(22):6621–6624. doi: 10.1093/nar/18.22.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broitman S. L., Im D. D., Fresco J. R. Formation of the triple-stranded polynucleotide helix, poly(A.A.U). Proc Natl Acad Sci U S A. 1987 Aug;84(15):5120–5124. doi: 10.1073/pnas.84.15.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M. J. Comparative properties of DNA, RNA, and hybrid homopolymer pairs. Fed Proc. 1965 Nov-Dec;24(6):1446–1457. [PubMed] [Google Scholar]
- Cooney M., Czernuszewicz G., Postel E. H., Flint S. J., Hogan M. E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988 Jul 22;241(4864):456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- Eadie J. S., McBride L. J., Efcavitch J. W., Hoff L. B., Cathcart R. High-performance liquid chromatographic analysis of oligodeoxyribonucleotide base composition. Anal Biochem. 1987 Sep;165(2):442–447. doi: 10.1016/0003-2697(87)90294-6. [DOI] [PubMed] [Google Scholar]
- Griffin L. C., Dervan P. B. Recognition of thymine adenine.base pairs by guanine in a pyrimidine triple helix motif. Science. 1989 Sep 1;245(4921):967–971. doi: 10.1126/science.2549639. [DOI] [PubMed] [Google Scholar]
- Horne D. A., Dervan P. B. Effects of an abasic site on triple helix formation characterized by affinity cleaving. Nucleic Acids Res. 1991 Sep 25;19(18):4963–4965. doi: 10.1093/nar/19.18.4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letai A. G., Palladino M. A., Fromm E., Rizzo V., Fresco J. R. Specificity in formation of triple-stranded nucleic acid helical complexes: studies with agarose-linked polyribonucleotide affinity columns. Biochemistry. 1988 Dec 27;27(26):9108–9112. doi: 10.1021/bi00426a007. [DOI] [PubMed] [Google Scholar]
- Mergny J. L., Sun J. S., Rougée M., Montenay-Garestier T., Barcelo F., Chomilier J., Hélène C. Sequence specificity in triple-helix formation: experimental and theoretical studies of the effect of mismatches on triplex stability. Biochemistry. 1991 Oct 8;30(40):9791–9798. doi: 10.1021/bi00104a031. [DOI] [PubMed] [Google Scholar]
- Michel F., Ellington A. D., Couture S., Szostak J. W. Phylogenetic and genetic evidence for base-triples in the catalytic domain of group I introns. Nature. 1990 Oct 11;347(6293):578–580. doi: 10.1038/347578a0. [DOI] [PubMed] [Google Scholar]
- Morgan A. R., Wells R. D. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J Mol Biol. 1968 Oct 14;37(1):63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Orson F. M., Thomas D. W., McShan W. M., Kessler D. J., Hogan M. E. Oligonucleotide inhibition of IL2R alpha mRNA transcription by promoter region collinear triplex formation in lymphocytes. Nucleic Acids Res. 1991 Jun 25;19(12):3435–3441. doi: 10.1093/nar/19.12.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei D. H., Ulrich H. D., Schultz P. G. A combinatorial approach toward DNA recognition. Science. 1991 Sep 20;253(5026):1408–1411. doi: 10.1126/science.1716784. [DOI] [PubMed] [Google Scholar]
- Perelroyzen M. P., Vologodskii A. V. Theoretical analysis of 'addressed' chemical modification of DNA. Nucleic Acids Res. 1988 May 25;16(10):4693–4704. doi: 10.1093/nar/16.10.4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal P., Feigon J. Triple-strand formation in the homopurine:homopyrimidine DNA oligonucleotides d(G-A)4 and d(T-C)4. Nature. 1989 Jun 22;339(6226):637–640. doi: 10.1038/339637a0. [DOI] [PubMed] [Google Scholar]
- Riley M., Maling B. Physical and chemical characterization of two- and three-stranded adenine-thymine and adenine-uracil homopolymer complexes. J Mol Biol. 1966 Sep;20(2):359–389. doi: 10.1016/0022-2836(66)90069-6. [DOI] [PubMed] [Google Scholar]
- Roberts R. W., Crothers D. M. Specificity and stringency in DNA triplex formation. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9397–9401. doi: 10.1073/pnas.88.21.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenár V., Feigon J. Formation of a stable triplex from a single DNA strand. Nature. 1990 Jun 28;345(6278):836–838. doi: 10.1038/345836a0. [DOI] [PubMed] [Google Scholar]