Abstract
Autism spectrum disorder (ASD) is known to be a heritable neurodevelopmental disorder affecting more than 1% of the population but in the majority of ASD cases, the genetic cause has not been identified. Parent-of-origin effects have been highlighted as an important mechanism in the pathology of neurodevelopmental disorders such as Prader–Willi and Angelman syndrome, with individuals with these syndromes often exhibiting ASD symptoms. Consequently, systematic investigation of these effects in ASD is clearly an important line of investigation in elucidating the underlying genetic mechanisms. Using estimation of maternal, imprinting and interaction effects using multinomial modelling (EMIM), we simultaneously investigated imprinting, maternal genetic effects and associations in the Autism Genome Project and Simons Simplex Consortium genome-wide association data sets. To avoid using the overly stringent genome-wide association study significance level, we used a Bayesian threshold that takes into account the sample size, allele frequency and any available prior knowledge. Between the two data sets, we identified a total of 18 imprinting effects and 68 maternal genetic effects that met this Bayesian threshold criteria, but none met the threshold in both data sets. We identified imprinting and maternal genetic effects for regions that have previously shown evidence for parent-of-origin effects in ASD. Together with these findings, we have identified maternal genetic effects not previously identified in ASD at a locus in SHANK3 on chromosome 22 and a locus in WBSCR17 on chromosome 7 (associated with Williams syndrome). Both genes have previously been associated with ASD.
Introduction
Autism spectrum disorder (ASD) is a complex and heterogeneous neurodevelopmental disorder characterised by patterns of repetitive behaviours and deficits in language and social behaviour. ASD is known to be heritable, with earlier heritability estimates ranging from 0.85 to 0.921 to a more current heritability estimate of 0.524,2 with most of this believed to be due to common rather than rare variation.2 Gaugler et al2 also estimated that 41% of the risk for ASD is due to environmental factors, which include prenatal, perinatal and postnatal environmental factors. Several genome-wide association studies (GWASs) have been performed on common variants in ASD using family studies, but the most significant results from these studies show modest effect size3, 4, 5 and therefore, have low power to replicate.4 Despite growing evidence from investigations of rare and de novo structural and sequence variation in ASD,6 the aetiology of the majority of ASD cases remains unexplained, suggesting that there are complex genetic mechanisms underlying the disorder. There is also evidence to suggest that there is a complex relationship between these underlying genetic factors and environmental factors.7
Broadly speaking, parent-of-origin effects consist of genetic effects on the phenotype of an offspring that are dependent on the parental origin of the associated genetic variant(s). Parent-of-origin effects can occur through numerous mechanisms such as genomic imprinting and certain trans-generational effects (for example, maternal genetic effects).8 Genomic imprinting occurs when the allele from a particular parent is silenced and the gene is expressed only by the remaining allele that has been inherited from the other parent. Evidence for imprinting has been shown in Prader–Willi syndrome and Angleman syndrome,9, 10 both syndromes having autistic features and diagnoses.11 There has been strong suggestive evidence for imprinting in ASD in the 7q and 15q regions which warrant further investigation.12
When statistically investigating GWAS data for imprinted genes, we examine the transmission of alleles from the parent to the offspring. If there is an over-transmission of the variant allele by a particular parent, then this might suggest that there is a difference in the expression of the alleles through epigenetic mechanisms (heritable changes that do not cause changes in the DNA sequence). For example, if the variant allele is over-transmitted from mothers only (ie, maternal over-transmission), this might suggest evidence for nonexpression of the paternally derived allele.
In addition to imprinting, maternal genetic effects can also occur when the maternal genotype exerts an influence on the offspring's phenotype regardless of what genetic material has been passed from the mother to the offspring. One way in which this can occur is when the mother's genotype affects the development of the foetus through the intrauterine environment. There has been evidence of maternal genetic effects in ASD, for example, at the GSTP*A gene13 and the HLA-DR4 gene.14 There has also been some evidence of possible prenatal environmental factors influencing the risk of ASD. For example, ASD is more likely in half siblings with a shared mother (recurrence rate estimates: 5.2 and 7.3%) compared with half siblings with a shared father (estimates: 0 and 3.2%, although sample sizes were small for paternal half siblings).15 However, more recent estimates based on a larger cohort from Sweden have not shown as large a difference16 (recurrence rates: 3.3% for half siblings with shared mother; 2.9% for half siblings with shared father).
The study of parent-of-origin effects has the potential to identify genetic and environmental factors that may be contributing to a complex disorder such as ASD.17 Therefore, in order to help elucidate the genetic and epigenetic aetiology of ASD, we investigated two types of parent-of-origin effects, imprinting and maternal genetic effects, in ASD GWAS. Previous studies have investigated parent-of-origin effects in ASD.3, 4, 18, 19 Anney et al3, 4 investigated imprinting in a secondary analysis in the Autism Genome Project (AGP) GWAS data sets using an in-house method reported to be similar to the method of Cordell et al,20 but findings were not considered to be statistically significant after correcting for multiple testing. Chaste et al5 also considered whether the transmission came from the mother or the father in their GWAS analysis of the Simons Simplex Collection (SSC) data although no parental-specific results are reported. Tsang et al18 and Yuan and Dougherty19 used GWAS data to investigate maternal genetic effects in ASD using a case–control type analysis. There were no genome-wide significant findings or replicated findings in either of these studies.18, 19
Our analysis approach differs in that we investigated both imprinting and maternal genetic effects simultaneously as maternal genetic effects are known to mimic imprinting and vice versa.21, 22 We also included offspring genetic effects (associations) in our model to enable us to identify an imprinting effect associated with ASD. These parent-of-origin analyses were investigated using estimation of maternal, imprinting and interaction effects using multinomial modelling (EMIM),23, 24 which in comparison with other tests has been shown to be the most suitable in terms of power and type I errors for this type of data.22 We also adapted a Bayesian method25 to determine an appropriate noteworthy threshold at each locus (taking into account sample size, minor allele frequency (MAF) and prior knowledge) instead of using the genome-wide significance levels, which are known to be stringent.26
Materials and methods
Data
The AGP GWAS family trio data set is described elsewhere,3, 4 and here we are using the Stage 2 data set consisting of 2931 families. This data set contains approximately one million SNPs genotyped on either the Illumina Infinium 1M-single or the Illumina 1M-duo microarray, see Acknowledgements for information on how to obtain the data.
The SSC GWAS consists of data on 2591 simplex families that were genotyped for a million or more SNPs on one of three array versions—Illumina 1Mv1 (333 families), Illumina 1Mv3 Duo (1189 families) or Illumina HumanOmni2.5 M (1069 families) Therefore, since imputation is computationally intensive for trio data sets and we needed the parental genotype data, we combined the three data sets and investigated SNPs common to all three arrays, as was carried out in ref. 5. See refs 5, 27, 28 for further details on the SSC data.
The AGP GWAS data includes families grouped into two nested diagnostic categories, Strict ASD (autism diagnoses met on both ADI and ADOS instruments) and spectrum ASD (autism-spectrum diagnoses met on either the ADI or ADOS instruments), as used in ref. 3. Although not considered in the analyses of the SSC data,5 we applied the same ASD phenotype criteria as was used in the AGP3 to define strict and spectrum ASD phenotypes within the SSC data. We focus our main analyses on the spectrum phenotype which provides the larger sample sizes for the analyses (secondary analyses on the strict phenotype are provided in the Supplementary Information).
Statistical model
Following an extensive review of the parent-of-origin methodology, described elsewhere,22 we used the EMIM23, 24 approach for our analyses. EMIM is a multinomial model that directly maximises the multinomial likelihood to detect parent-of-origin effects and can incorporate missing data (Supplementary Information for further details). We simultaneously investigated offspring genetic effects (associations), maternal genetic effects and imprinting effects using EMIM. We did not include mother/offspring interactions in our model as the inclusion of mother/offspring interaction parameters reduces the power of the model substantially, as is to be expected.22 Instead mother/offspring interaction effects were investigated subsequent to a SNP being identified as having an offspring effect and a maternal genetic effect. A multiplicative model was assumed for offspring and maternal genotype parameters. The benefit of this is twofold; firstly for ease of investigating the effects in an already complex model; and secondly, to reduce the number of parameters in the model to help increase power, see Supplementary Information for more details. EMIM has also been extended to use haplotype estimates to help increase power for detecting imprinting but this method was not used in this paper.29
Quality control procedures
The quality control (QC) follows a standard approach to trio GWAS QC, with individuals and SNPs removed when missingness >0.05, MAF<0.05 and Hardy–Weinberg equilibrium P-value<0.00001. We limited our analyses to complete independent trios (both parents and offspring) to prevent the reduction in power that estimating missing data in our model would cause. Full details of the QC procedures are given in the Supplementary Information. After QC, the AGP with a spectrum phenotype contains 2594 trios and 728 228 SNPs, and the SSC with a spectrum phenotype contains 2433 trios and 483 080 SNPs.
Bayesian noteworthy threshold
Power to detect parent-of-origin effects can be limited22 and the current genome-wide significance threshold guidelines are known to be very stringent.25, 26, 30 These genome-wide significance levels are suggested for all sample sizes (and MAFs) and hence do not take into account the power at individual SNPs.25, 31 To take these factors into account, we adopted a Bayesian method proposed by Wakefield25 to determine an appropriate threshold for identifying noteworthy findings.
This threshold for Z2-score is given by the following:
where Vn is the standard error of the parameter (which is dependent on the sample size, n, and MAF), W is the prior variance for the log of the relative risk Δ (ie, Δ∼N(0,W)), PO is the prior odds (ie π0/(1–π0), where π0 is the prior probability that H0 is true), and R is the ratio of costs of type II to type I errors. See Supplementary Information and refs 25, 31 for more information.
We note that when we are investigating imprinting, it is necessary to have an offspring genotype effect present in addition to an imprinting effect. This is required to ensure that we do not identify non-disease related imprinted regions that would be observed in the general population. For maternal genetic effects, an association is not required and we investigate loci, where the mother's genotype exerts an influence on the offspring's phenotype, regardless of association being present or not. Therefore, we calculated separate Z2-score thresholds for the Wald Z-score for the association parameter (R1) and for the maternal genetic effect parameter (S1). We detect a noteworthy imprinting result when both the association parameter and the imprinting parameter meet the threshold (see Supplementary Figure S2 in Supplementary Information), whereas we detect a noteworthy maternal genetic effect when the maternal genetic effect parameter meets the threshold (see Supplementary Figure S3 in Supplementary Information).
We chose the prior variance for the log of the relative risk to be W=[log(2)/1.645]2=0.422 (interpret this as a 5% chance that the relative risk will be larger than 2) to reflect the low effect sizes in GWAS.32 Because of the evidence that several hundred to thousands of loci are likely to contribute to the complex genetic heterogeneity of ASD,33, 34 we choose π0=1–500/1 000 000=0.9995, which leads to a prior odds of H0 being true of PO=1 999. Since the power is limited to detect parent-of-origin effects in ASD, and this in turn increases type II errors, we chose the ratio of cost of type II errors to type I errors, R, equal to 10, as false negatives cannot be followed up. For more details on the choice of these parameters and the sensitivity of the Bayesian threshold to these choices see Supplementary Information.
Results
Autism Genome Project (AGP)
There were nine noteworthy independent loci showing imprinting effects and there were forty independent loci showing maternal genetic effects, see Supplementary Figure S5 and Supplementary Tables S4 and S5 in the Supplementary Information for all variants that were above the threshold for offspring genetic effect (R1) and imprinting (IM) or were above the threshold for maternal genetic effects (S1). Table 1 gives a summary of the results discussed below. Note: IM>1 indicates a maternal over-transmission of the allele and IM<1 indicates a paternal over-transmission of the allele, whereas IM=1 indicates no imprinting effect.
Table 1. Main results from the AGP and SSC data set.
| AGP results | SSC results | |||||
|---|---|---|---|---|---|---|
| SNP | Effect | P-value | Location | Previous findings | Effect | P-value |
| rs675680 | IM=2.36 | 3.0 × 10−6 | Chr 4p15 | None identified | IM= 0.81 | 2.4 × 10−1 |
| rs10025482 | IM =0.59 | 6.2 × 10−6 | Chr 4, C4orf37 gene | 18 | IM =1.19 | 1.5 × 10−1 |
| rs4516878 | S1=1.40 | 1.2 × 10−5 | Chr 5q34 | None identified | S1=1.07 | 3.7 × 10−1 |
| rs545208 | S1=0.74 | 3.0 × 10−5 | Chr 11, MAML2 gene | 18 | S1=1.02 | 8.1 × 10−1 |
| rs9870610 | S1=1.33 | 7.2 × 10−5 | Chr 3, ROBO2 gene | 18 | S1=1.16 | 5.0 × 10−2 |
| SSC results | AGP results | |||||
| SNP | Effect | P-value | Location | Previous findings | Effect | P-value |
| rs9573533 | IM=0.59 | 8.2 × 10−6 | Chr 13, TBC1D4 gene | None identified | IM=0.98 | 8.3 × 10−1 |
| rs16890706 | IM=1.86 | 1.1 × 10−5 | Chr 6, LRRC16A gene | 35 | IM=1.18 | 2.1 × 10−1 |
| rs6967953 | S1=1.38 | 6.0 × 10−6 | Chr 7, CHRM2 gene | 36 | S1=0.92 | 2.0 × 10−1 |
| rs5770820 | S1=1.25 | 5.5 × 10−5 | Chr 22, SHANK3 gene | 37 | S1=0.98 | 8.2 × 10−1 |
| rs4719103 | S1=1.41 | 5.5 × 10−5 | Chr 7, WBSCR17 gene | 38 | S1=0.96 | 6.3 × 10−1 |
| rs11075447 | S1=0.76 | 9.5 × 10−5 | Chr 22, GNB1L gene | 41 | S1=1.00 | 9.5 × 10−1 |
Autism Genome Project imprinting results
The top imprinting result was a maternal over-transmission on chromosome 4, between LOC391642 and LOC645641 (rs675680, hg18 chr11:g.28082183A>G, allele=G, IM=2.36, Wald P-value=3.02 × 10−6), which has not been previously associated with ASD. We also found a noteworthy paternal over-transmission in SNPs in the STPG2 gene (C4orf37 gene) on chromosome 4 (top SNP, rs10025482, hg18 chr4:g.99272299C>T, allele=T, IM=0.59, Wald P-value=6.21 × 10−6, see Supplementary Figure S7 in the Supplementary Information) and this region was previously implicated in ASD,18 where a mother/offspring interaction effect was identified in the vicinity of the STPG2 gene (rs28539905—not genotyped, R2=0.005). We found no evidence of an interaction effect at rs10025482 (LRT P-value=0.33) or in this region.
Autism Genome Project maternal genetic results
The top result for a maternal genetic effect was on chromosome 5 between the genes LOC391845 and LOC574080 (rs4516878, hg18 chr5:g.164271894T>C, allele=C, S1=1.40, Wald P-value=1.16 × 10−5). This region has not been previously linked with ASD, to our knowledge. In our top results, we also identified two maternal genetic effects that were previously implicated as maternal genetic effects or mother/offspring interactions in the Early Markers for Autism data set.18 The first hit is located on the MAML2 gene on chromosome 11, which was identified as a maternal genetic effect in our analyses and in the same region in Tsang et al18 but in opposite directions (rs545208, hg18 chr11:g.95619756C>T, allele=T, S1=0.74, Wald P-value=3 × 10−5, see Supplementary Figure S8 in the Supplementary Information). We also identified a maternal genetic effect (rs9870610, hg18 chr3:g.95619758C>T, allele=T, S1=1.33, Wald P-value=7.24 × 10−5, see Supplementary Figure S9) on ROBO2 on chromosome 3. An interaction effect was previously identified in the gene ROBO2 in Tsang et al,18 but we found no evidence of an interaction (LRT P-value=0.34) at rs545208.
Simons Simplex Collection (SSC)
There were nine noteworthy imprinting results and there were 28 independent noteworthy loci with a maternal genetic effect in the SSC data, see Supplementary Figure S16 and Supplementary Tables S8 and S9 in the Supplementary Information and Table 1.
Simons Simplex Collection imprinting results
The top imprinting result was a paternal over-transmission on chromosome 13 in the TBC1D4 gene (rs9573533, hg18 chr13:g.74853485G>A, allele=A, IM=0.59, Wald P-value=8.17 × 10−6). To our knowledge, this area has not been previously linked with ASD. We identified a maternal over-transmission in the LRRC16A gene (near the HLA region) (rs16890706, hg18 chr6:g.25628073G>A, allele=A, IM=1.86, Wald P-value=1.09 × 10−5), which was previously implicated in language deficits.35
Simons Simplex Collection maternal genetic results
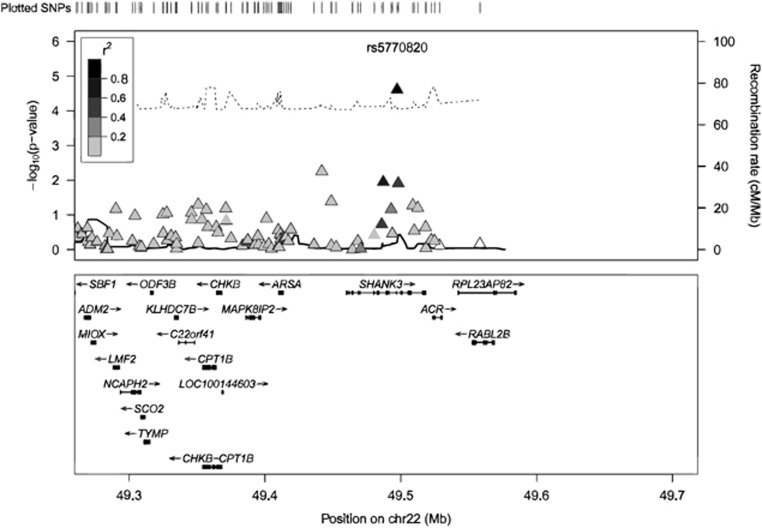
The strongest association for a maternal genetic effect was on chromosome 7 in the CHRM2 gene (rs6967953, hg18 chr7:g.136353916G>A, allele=A, S1=1.38, Wald P-value=6.01 × 10−6). This area has been previously linked with IQ and one of the strongest linkage signals reported for ASD occurred at 7q within 1.6 kb of the CHRM2 gene.36 One of our top hits for maternal genetic effects was identified on chromosome 22 in the SHANK3 gene (rs5770820, hg18 chr22:g.49497339G>A, allele =A, S1=1.25, Wald P-value=5.54 × 10−5, see Figure 1). Disruptions in the SHANK3 gene have been associated with autistic traits and in particular, these disruptions are responsible for the development of Phelan–McDermid syndrome and other non-syndromic ASDs.37 Figure 1 shows that there are no SNPs in high LD with rs5770820 (the SNP in highest LD was rs739365, R2=0.65) due to the limited number of SNPs common to all three arrays in the SSC data set.
Figure 1.
SSC spectrum chromosome 22, SHANK3 gene, rs5770820 maternal genetic effect. Regional plot of SNPs highlighted in the SSC spectrum analysis for maternal genetic effects (S1, triangles). Markers in linkage disequilibrium with the index SNP are shown and based on 1000 genomes CEU. Recombination rate plotted in black. The black dotted line represents the Bayesian threshold for S1.
We also detected a noteworthy maternal genetic effect on chromosome 7q11.23 in the WBSCR17 gene (rs4719103, hg18 chr7:g.70395849G>A, allele=A, S1=1.41, Wald P-value=5.48 × 10−5, see Supplementary Figure S18 in Supplementary Information), this region is deleted in Williams syndrome38 and it is known that individuals with Williams syndrome exhibit autistic behaviours.39, 40 This region was strongly associated with ASD in a Copy Number Variant (CNV) study carried out on the SSC data set,28 and we acknowledge that there is a large overlap between the samples analysed here and those in the CNV study.
Another noteworthy result was found on chromosome 22q, which is a protective maternal genetic effect in the GNB1L gene (also known as C22orf29 gene) (rs11075447, hg18 chr16:g.60560457A>G, allele=G, S1=0.76, Wald P-value=9.52 × 10−5), this gene has been linked to ASD and schizophrenia.41
Discussion
To our knowledge, this is the first genome-wide study to test for both imprinting and maternal genetic effects simultaneously in ASD. This is also the first study to implement the Bayesian thresholds that take into consideration the sample size and MAF at each SNP, and prior knowledge of effect size and prior odds of finding associations. We analysed the AGP and SSC ASD data sets for parent-of-origin effects, specifically imprinting and maternal genetic effects. Previous studies of parent-of-origin effects in ASD only investigated either imprinting effects or maternal genetic effects3, 4, 18, 19 despite the fact that these effects are known to mimic each other. We identified a total of 18 imprinting effects and 68 maternal genetic effects that met this Bayesian threshold criteria in either the AGP or SSC data sets with a Spectrum phenotype. None of these results were identified in both data sets. The Supplementary Information contains further analyses of parent-of-origin effects in the AGP and SSC data sets for a Strict ASD phenotype, where we identified 10 imprinting effects and 72 maternal genetic effects that met the Bayesian threshold criteria in either data sets. A proportion (10–20%) of the results identified using a Strict ASD phenotype overlap with the results identified using the Spectrum ASD phenotype (see Supplementary Information for further details).
This model is complex as it includes three parameters, offspring genetic effects, imprinting effects and maternal genetic effects, which can reduce power and can lead to the results being harder to interpret. To help identify noteworthy findings, we adopted a Bayesian threshold proposed by Wakefield25 to investigate parent-of-origin effects as it facilitates ease of interpretation of an imprinting effect and a maternal genetic effect. In addition, the Bayesian threshold avoids the use of the overly stringent genome-wide significance threshold.25, 26 The Bayesian threshold takes into account the sample size and MAF as well as other prior knowledge regarding ASD (for example, effect size) to allow for a more appropriate threshold for the model at each locus. The Bayesian threshold does not depend on the number of tests performed but instead depends on the prior odds. If a Bonferroni correction was employed, or the stringent GWAS threshold, then the noteworthy hits we identified would have been missed, but as we have shown, some of these hits show promise by being previously identified in ASD studies (see Table 1). In addition, we have accounted for the rate of true positives and true negatives in the prior odds in the Bayesian threshold, which is a superior method in comparison to using the Bonferroni correction or the GWAS thresholds, which do not account for these.
Replicating results identified in a discovery analysis in an independent sample is the gold standard in GWAS analysis as it provides convincing statistical evidence for association, and has the potential to rule out associations due to biases.42 Replication generally involves identifying the significant results in the discovery analysis and examining these in a replication data set that is as close to the ascertainment and design of the original GWAS as possible.43 It is important for a replication data set to be independent of the primary data set, have large enough sample sizes and have the same ascertainment and study design as the discovery GWAS.42, 43
The AGP and SSC data sets did not have the same ascertainment criteria and these differences have led to key differences in the AGP and SSC data sets. The AGP contains both simplex and multiplex families (approximately 38% of families are simplex44) with the aim being to investigate common variation whereas the SSC data contains only simplex families, a design which inherently enriches for rare and de novo mutations.28, 33 In addition, the SSC data set excluded families with parents who met criteria for a spectrum diagnosis based on two instruments, thus further limiting the potential to discover heritable, penetrant genetic risk. This exclusion criterion did not apply in the AGP data set and a small proportion of parents included in the data set who had been screened using these instruments (which is only a small proportion of the sample) meet this criteria.45, 46
From a phenotypic perspective, when compared with multiplex families, simplex family members share less ASD traits.47, 48 Klei et al43 have shown that a lower proportion (<40%) of the heritability of additive effects in ASD is explained in the SSC data set compared with the AGP (55–59%, 65% for AGP multiplex probands) and that family members in the AGP have elevated heritability estimates, which were not seen in the SSC. There has been evidence to suggest that genetic transmission mechanisms differ between multiplex families and simplex families.7, 44, 48 Even though both the AGP and SSC data sets are ASD data sets, and even though we would expect some shared common risk between the two in terms of associations, we do not believe this would be the case for parent-of-origin effects as the transmission mechanisms are the main focus. For these reasons, we felt it was not appropriate to treat either the AGP or SSC as a replication data set of the other. Table 1 and Supplementary Tables S4–S11 in the Supplementary Information show that the effects are often in different directions when comparing the AGP and SSC results to each other, possibly strengthening the theory that this difference in ascertainment leads to a different genetic aetiology for multiplex and simplex ASD families.
Therefore, we have not replicated any of the findings we identified as we did not have an appropriate independent replication data set available. However, we did identify some potential parent-of-origin effects in ASD in regions that have been previously implicated in ASD. For example, one of the imprinting results (in the STPG2 gene) and two of the maternal genetic effects (in the MAML2 gene and the ROBO2 gene) were previously implicated in an ASD study for maternal genetic effects by Tsang et al.18 We also identified a maternal genetic effect at rs5770820 in the SHANK3 gene. SHANK3 (ProSAP2) regulates the structural organisation of dendritic spines and is a binding partner of neuroligins. Mutations in SHANK3 are well known risk factors for ASD.49 Lebold et al50 estimated that 0.69% of cases with ASD had heterozygous truncating mutations in SHANK3. Sanders et al6 has identified SHANK3 as one of the 71 risk loci in ASD, although we note that Sanders et al6 also used the SSC data set. We identified another maternal genetic effect on chromosome 7q11 in the WBSCR17 gene, which is a deleted region in Williams syndrome. Williams syndrome has strong links with ASD as individuals with Williams syndrome often exhibit autistic traits.38 Our findings suggest that mutations in a mother's SHANK3 gene or the WBSCR17 gene could increase the likelihood of the offspring having ASD. Although our findings are very promising, further investigation is necessary.
In conclusion, we set out to detect parent-of-origin effects in ASD using the AGP and the SSC GWAS data sets. We identified many regions with potential parent-of-origin effects. This study has also shown an approach to investigating both imprinting effects and maternal genetic effects in ASD family GWAS data sets using appropriate Bayesian thresholds that take into account the power of the test at each SNP. This approach can be used in future studies of ASD when there are larger and more appropriate replication data sets available in order to produce robust findings. This approach is not limited to ASD but is suitable for the examination of parent-of-origin effects in other phenotypes that have GWAS data sets with parental genotypes available.
Acknowledgments
The authors are grateful to all of the families for participating in the AGP, as well as the principal investigators, AGRE and the AGP funders: Autism Speaks (USA), the Medical Research Council (UK), the Health Research Board (Ireland), the National Institutes of Health (USA), Genome Canada, the Canadian Institutes of Health Research and the Hilibrand Foundation. The AGP data sets can be obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap. The authors are grateful to all of the families at the participating SSC sites, as well as the principal investigators (A Beaudet, R Bernier, J Constantino, E Cook, E Fombonne, D Geschwind, R Goin-Kochel, E Hanson, D Grice, A Klin, D Ledbetter, C Lord, C Martin, D Martin, R Maxim, J Miles, O Ousley, K Pelphrey, B Peterson, J Piggot, C Saulnier, M State, W Stone, J Sutcliffe, C Walsh, Z Warren and E Wijsman), and appreciate obtaining access to phenotypic data on SFARI Base. Approved researchers can obtain the SSC population data set described in this study by applying at https://base.sfari.org. In particular, We acknowledge S Sanders for his help with the SSC data. We acknowledge the support of the Trinity Centre for High Performance Computing. SC acknowledges the funding received from a Trinity College Dublin Studentship Award.
Summary data results are available at http://www.gwascentral.org/study/HGVST1842.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Hallmayer J, Cleveland S, Torres A et al: Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry 2011; 68: 1095–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaugler T, Klei L, Sanders SJ et al: Most genetic risk for autism resides with common variation. Nat Genet 2014; 46: 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anney R, Klei L, Pinto D et al: A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet 2010; 19: 4072–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anney R, Klei L, Pinto D et al: Individual common variants exert weak effects on the risk for autism spectrum disorderspi. Hum Mol Genet 2012; 21: 4781–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaste P, Klei L, Sanders SJ et al: A genomewide association study of autism using the Simons Simplex Collection: does reducing phenotypic heterogeneity in autism increase genetic homogeneity? Biol Psychiatry 2014; 77: 775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S, He X, Willsey JA et al: Insights into Autism Spectrum Disorder genomic architecture and biology from 71 risk loci. Neuron 2015; 87: 1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaste P, Leboyer M: Autism risk factors: genes, environment, and gene-environment interactions. Dialogues Clin Neurosci 2012; 14: 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilmatre A, Sharp A: Parent of origin effects. Clin Genet 2012; 81: 201–209. [DOI] [PubMed] [Google Scholar]
- Falls J, Pulford D, Wylie A et al: Genomic imprinting: implications for human disease. Am J Pathol 1999; 154: 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurrier F, Accadia M: Genetic imprinting: the paradigm of prader-willi and angelman syndromes. Endocr Dev 2009; 14: 20–28. [DOI] [PubMed] [Google Scholar]
- Veltman MWM, Craig EE, Bolton PF: Autism spectrum disorders in Prader-Willi and Angelman syndromes: a systematic review. Psychiatr Genet 2005; 15: 243–254. [DOI] [PubMed] [Google Scholar]
- Schanen NC: Epigenetics of autism spectrum disorders. Hum Mol Genet 2006; 15: R138–R150. [DOI] [PubMed] [Google Scholar]
- Williams TA, Mars AE, Buyske SG et al: Risk of autistic disorder in affected offspring of mothers with a glutathione S-transferase P1 haplotype. Arch Pediatr Adolesc Med 2007; 161: 356–361. [DOI] [PubMed] [Google Scholar]
- Johnson WG, Buyske S, Mars AE et al: HLA-DR4 as a risk allele for autism acting in mothers of probands possibly during pregnancy. Arch Pediatr Adolesc Med 2009; 163: 542–546. [DOI] [PubMed] [Google Scholar]
- Constantino JN, Todorov A, Hilton C et al: Autism recurrence in half siblings: strong support for genetic mechanisms of transmission in ASD. Mol Psychiatry 2013; 18: 137–138. [DOI] [PubMed] [Google Scholar]
- Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, Reichenberg A: The familial risk of autism. JAMA 2014; 311: 1770–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson Ha, Cheverud JM, Wolf JB: Genomic imprinting and parent-of-origin effects on complex traits. Nat Rev Genet 2013; 14: 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang KM, Croen LA, Torres AR et al: A genome-wide survey of transgenerational genetic effects in autism. PloS One 2013; 8: e76978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Dougherty JD: Investigation of maternal genotype effects in autism by genome- wide association. Autism Res 2014; 7: 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell H, Barratt B, Clayton D: Case/pseudocontrol analysis in genetic association studies: a unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet Epidemiol 2004; 26: 167–185. [DOI] [PubMed] [Google Scholar]
- Wolf J, Wade M: What are maternal effects (and what are they not)? Philos Trans R Soc Lond B Biol Sci 2009; 10: 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly S, Heron EA: Review of statistical methodologies for the detection of parent- of-origin effects in family trio genome-wide association data with binary disease traits. Brief Bioinform 2014; 16: 429–448. [DOI] [PubMed] [Google Scholar]
- Ainsworth H, Unwin J, Jamison D et al: Investigation of maternal effects, maternal-fetal interactions and parent-of-origin effects (imprinting), using mothers and their offspring. Genet Epidemiol 2011; 35: 19–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howey R, Cordell H: PREMIM and EMIM: tools for estimation of maternal, imprinting and interaction effects using multinomial modelling. BMC Bioinformatics 2012; 13: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield J: Commentary: genome-wide significance thresholds via Bayes factors. Int J Epidemiol 2012; 41: 286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JPA, Thomas G, Daly MJ: Validating, augmenting and refining genome-wide association signals. Nat Rev Genet 2009; 10: 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach GD, Lord C: The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron 2010; 68: 192–195. [DOI] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek GA, Hus V et al: Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 2011; 70: 863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howey R, Mamasoula C, Töpf A et al: Increased power for detection of parent-of-origin effects via the use of haplotype estimation. Am J Hum Genet 2015; 97: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen CT, Wang J, McIntyre AD et al: Excess of rare variants in non-genome-wide association study candidate genes in patients with hypertriglyceridemia. Circ Cardiovasc Genet 2012; 5: 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield J, Skrivankova V, Hsu FC, Sale M, Heagerty P: Detecting signals in pharmacogenomic genome-wide association studies. Pharmacogenomics J 2014; 14: 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Newton-Cheh C: Genome-wide association study in humans. Methods Mol Biol 2009; 573: 231–258. [DOI] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR et al: De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 2012; 485: 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D et al: De novo gene disruptions in children on the autistic spectrum. Neuron 2012; 74: 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudel R, Simpson NH, Baird G et al: Genome-wide association analyses of child genotype effects and parent-of-origin effects in specific language impairment. Genes Brain Behav 2014; 13: 418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosso MF, van Belzen M, de Geus EJC et al: Association between the CHRM2 gene and intelligence in a sample of 304 Dutch families. Genes Brain Behav 2006; 5: 577–584. [DOI] [PubMed] [Google Scholar]
- Peça J, Feliciano C, Ting JT et al: Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 2011; 472: 437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke U: Williams-Beuren syndrome:genes and mechanisms. Hum Mol Genet 1999; 8: 1947–1954. [DOI] [PubMed] [Google Scholar]
- Klein-Tasman BP, Phillips KD, Lord C, Mervis CB, Gallo FJ: Overlap with the autism spectrum in young children with Williams syndrome. J Dev Behav Pediatr 2009; 30: 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordjman S, Anderson GM, Botbol M et al: Autistic disorder in patients with Williams-Beuren syndrome: a reconsideration of the Williams-Beuren syndrome phenotype. PloS One 2012; 7: e30778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YZ, Matsushita M, Girirajan S et al: Evidence for involvement of GNB1L in autism. Am J Med Genet B Neuropsychiatr Genet 2012; 159B: 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P, Zeggini E, Ioannidis JP: Replication in genome-wide association studies. Stat Sci 2010; 24: 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush WS, Moore JH: Chapter 11: Genome-wide association studies. PLoS Comput Biol 2012; 8: e1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klei L, Sanders SJ, Murtha MT et al: Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism 2012; 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L et al: Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 2012; 485: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L et al: Functional impact of global rare copy number variation in autism spectrum disorders. Nature 2010; 466: 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts JA, Bernier R, Dawson G, Estes A: The broader autism phenotype in simplex and multiplex families. J Autism Dev Disord 2013; 43: 1597–1605. [DOI] [PubMed] [Google Scholar]
- Bernier R, Gerdts J, Munson J, Dawson G, Estes A: Evidence for broader autism phenotype characteristics in parents from multiple-incidence autism families. Autism Res 2012; 5: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM et al: Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet 2007; 39: 25–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond CS, Nava C, Polge A et al: Meta-analysis of SHANK Mutations in Autism Spectrum Disorders: a gradient of severity in cognitive impairments. PLoS Genet 2014; 10: e1004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.