Abstract
Despite recent advances in linear whole genome amplification of intact DNA/RNA, amplification of degraded nucleic acids in an unbiased fashion remains a serious challenge for genetic diagnosis. We describe a new whole genome amplification procedure, RCA–RCA (Restriction and Circularization-Aided Rolling Circle Amplification), which retains the allelic differences among degraded amplified genomes while achieving almost complete genome coverage. RCA–RCA utilizes restriction digestion and whole genome circularization to generate genomic sequences amenable to rolling circle amplification. When intact genomic DNA is used, RCA–RCA retains gene-amplification differences (twofold or higher) between complex genomes on a genome-wide scale providing highly improved concordance with unamplified material as compared with other amplification methodologies including multiple displacement amplification. Using RCA–RCA, formalin-fixed samples of modest or substantial DNA degradation were successfully amplified and screened via array-CGH or Taqman PCR that displayed retention of the principal gene amplification features of the original material. Microsatellite analysis revealed that RCA–RCA amplified genomic DNA is representative of the original material at the nucleotide level. Amplification of cDNA is successfully performed via RCA–RCA and results to unbiased gene expression analysis (R2 = 0.99). The simplicity and universal applicability of RCA–RCA make it a powerful new tool for genome analysis with unique advantages over previous amplification technologies.
Formalin-fixed, paraffin-embedded (FFPE) specimens in the archives of departments of pathology represent a unique source of histologically classified material derived from normal and diseased tissues for which extensive clinical data are available. Extraction of DNA and RNA from these specimens provides an opportunity for retrospective analysis using microarray-based genomic or gene expression profiling that may accelerate the discovery of associations between gene-expression signatures and the biology and outcome of disease (Perou et al. 1999; Alizadeh and Staudt 2000; Alizadeh et al. 2000, 2001; Perou et al. 2000; Ross and Perou 2001; Sorlie et al. 2001; West et al. 2001; Pomeroy et al. 2002; van't Veer et al. 2002).
However, technical hurdles persist. First, DNA and RNA in FFPE biopsies are often moderately to highly degraded (Lewis et al. 2001) and second, many specimens have very small amounts of tissue, necessitating a whole genome amplification step, often performed via PCR (Nelson et al. 1989; Telenius et al. 1992; Zhang et al. 1992; Klein et al. 1999). However, the introduction of genetic bias during PCR amplification is a serious concern since the quantitative relation among alleles before and after PCR amplification is usually different. In an effort to overcome PCR-introduced bias during whole genome or transcriptome amplification, we recently developed balanced-PCR (Makrigiorgos et al. 2002), a whole genome amplification method that eliminates the effect of saturation and impurities. Balanced PCR has allowed for unbiased gene expression (Makrigiorgos et al. 2002) and genomic analyses (Wang et al. 2004). Because DNA is digested with a restriction enzyme during balanced-PCR, the method enables whole genome amplification when the starting DNA material is modestly degraded (Wang et al. 2004). On the other hand, because of the inefficiency of thermostable polymerases in amplifying DNA fragments >1 kb, balanced-PCR usually amplifies only a minor portion of the entire genome, a genomic representation (Lucito et al. 1998). This incomplete genome coverage may result in the loss of vital genetic information. Lizardi and co-workers introduced rolling circle amplification (Lizardi et al. 1998), an approach that subsequently led to an isothermal whole genome amplification method known as Multiple Displacement Amplification (MDA) (Dean et al. 2002; Lage et al. 2003). MDA operates on very long DNA templates (>10 kb), thereby allowing an almost complete genome coverage (Dean et al. 2002; Lage et al. 2003). MDA produces linearly amplified genomic DNA when starting from intact genomes obtained from cells or fresh tissue and is widely used for genomic profiling and large-scale genotyping (Lovmar et al. 2003; Paez et al. 2004; Rook et al. 2004; Wong et al. 2004). However, the amplification efficiency of MDA rapidly diminishes as the molecular weight of the starting material decreases, thus making it unsuitable for amplification of FFPE DNA or low molecular weight DNA from deteriorated forensic samples (Lage et al. 2003). In addition, MDA may not be applied on cDNA.
Here we describe RCA–RCA (Restriction and Circularization-Aided Rolling Circle Amplification), a new amplification methodology that overcomes problems associated with nucleic acid degradation and retains the allelic differences among amplified genomes while simultaneously achieving almost complete genome coverage. Formalin fixation of tissue results in DNA strand breaks, base damage, and DNA–protein crosslinks, all of which inhibit amplification (Lehmann and Kreipe 2001; Lewis et al. 2001). The principle of RCA–RCA is that fragmentation of the genome with an appropriate restriction enzyme that cuts at least twice between successive DNA damage sites in FFPE samples generates intact DNA fragments that can be circularized (Fig. 1A). Following circularization and elimination of noncircular DNA via exonuclease, the circles are denatured to enable initiation of exponential, hyper-branched rolling circle amplification using random primers and phi29 polymerase, as described by Dean (Dean et al. 2001). Exponential amplification is enabled even if one of the two circularized DNA strands remains intact (Fig. 1). Our data demonstrate that this new adaptation of isothermal rolling circle amplification is simple, robust, and reproducible, allowing unbiased amplification of degraded FFPE samples by several thousandfold without production of primer-associated artifacts. RCA–RCA is superior to other amplification methods when either fresh or degraded FFPE samples are used and provides a general protocol for complex nucleic acid amplification from intact or degraded genomic DNA, as well as from cDNA.
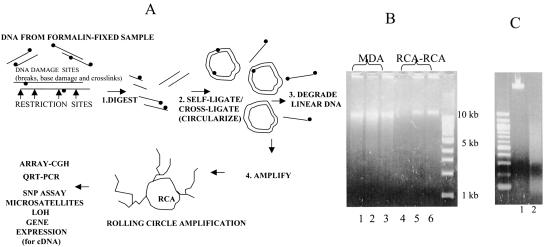
Figure 1.
(A) Outline of whole genome amplification of partially degraded FFPE samples via RCA–RCA (Restriction and Circularization-Aided Rolling Circle Amplification). DNA damage sites (•) are indicated. The same approach may also be used to amplify intact or degraded genomic DNA or cDNA. (B) Comparison of RCA–RCA amplification products with MDA amplification products. MDA amplification products from H2O—no template present— (lane 1), 50 ng intact reference (lane 2), or FFPE #19 (lane 3) genomic DNA; RCA–RCA amplification products from H2O—no template present—(lane 4), 50 ng intact reference (lane 5) or FFPE #19 genomic DNA (lane 6). No Nla-III digestion has been applied at this stage following RCA–RCA. (C) Reference genomic DNA undigested (lane 1) or Nla-III-digested (lane 2) prior to RCA–RCA amplification.
Results
RCA–RCA amplification product
Figure 1B demonstrates the amplification products obtained following whole genome amplification via RCA–RCA or MDA, using intact reference DNA (lanes 2,5) or DNA obtained from a formalin-fixed, paraffin-embedded sample (paraffin #19, lanes 3,6). High molecular weight DNA is generated as a result of isothermal strand displacement amplification in both cases. However, MDA produces nonspecific amplification artifacts (lane 1) in the absence of input genomic DNA, as was also previously reported (Lage et al. 2003). Therefore the MDA electropherogram is not indicative of successful whole genome amplification. In contrast, RCA–RCA is free of such nonspecific amplification products (lane 4) and the presence of high molecular weight DNA is a clear indication of template-specific amplification (vide infra). Furthermore, in comparison to MDA, the RCA–RCA amplification product is more viscous and produces no evidence of low DNA fragment sizes (Fig. 1B, cf. lanes 5,6 and lanes 2,3). Figure 1C depicts genomic DNA undigested (lane 1) or Nla-III-digested (lane 2) prior to RCA–RCA amplification.
Real-time PCR examination of RCA–RCA amplified, intact genomic DNA
To estimate the extent of amplification obtained following RCA–RCA of BT474 genomic DNA, a copy number comparison was made between unamplified and RCA–RCA-amplified DNA. To enable a direct comparison, 4 μL containing a total of 3 ng genomic DNA were diluted to 100 μL and directly tested via Taqman QRT-PCR using 2 μL per reaction. A further 4 μL containing a total of 3 ng DNA were processed for RCA–RCA amplification, diluted to 100 μL and again tested via Taqman QRT-PCR using 2 μL per reaction. Figure 2A depicts real-time PCR profiles starting from unamplified or amplified genomic DNA for three genes (GAPDH, IL9R and CYC). The resulting PCR threshold is about 10–13 cycles different between amplified and non-amplified material for all three genes, which corresponds to a genomic DNA amplification of ∼1,000–8,000 times. A first principles calculation of the maximum amount of DNA that one could make given the amount of components in the RCA–RCA reaction yields about 10 μg DNA, assuming that the dNTPs are the limiting factor. Given the DNA amplification derived from the data in Figure 2A from an initial material was ∼3 ng, it appears that the RCA–RCA reaction approached the theoretical maximum amplification in these reactions.
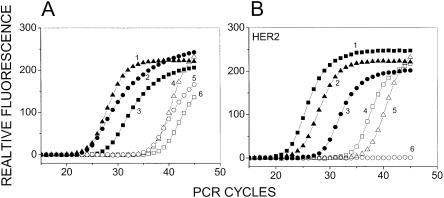
Figure 2.
(A) Quantification of RCA–RCA amplification-fold in genomic DNA. QRT-PCR was performed either directly from unamplified BT474 genomic DNA or from the RCA–RCA-amplified product for three single copy genes. Curves 1–3 GAPDH, IL9R, and CYC respectively, using RCA–RCA-amplified DNA as starting material. Curves 4–6 GAPDH, IL9R, and CYC respectively, using unamplified DNA as starting material. (B) Determining the lower limit of starting material for which RCA–RCA is possible. A comparison of QRT-PCR for HER2 performed directly from unamplified BT474 genomic DNA or from the RCA–RCA-amplified product is depicted, following serial dilution of the DNA. Curves 1–3: 0.3, 0.03, and 0.003 ng respectively, using RCA–RCA-amplified DNA as starting material. Curves 4–6: 0.3, 0.03, and 0.003 ng respectively, using unamplified DNA as starting material. HER2 is amplified in BT474 genomic DNA and 3 pg genomic DNA (curves 3 and 6) are expected to contain ∼15–30 HER2 molecules.
To examine RCA–RCA amplification of limiting starting material, serial dilution of the DNA was performed prior to the amplification. The HER2 gene copy number was examined via QRT-PCR and compared to QRT-PCR from an equal amount of unamplified DNA using the same experimental approach described above. Figure 2B demonstrates the ability of RCA–RCA to amplify from ∼3 pg of DNA, which is of the same order as a single cell. Because of HER2 amplification in BT474 cells, HER2 is expected to have an abundance of 15–30 molecules at the lowest dilution of ∼3 pg (Forozan et al. 2000). The amplified oncogene was clearly detectable following RCA–RCA amplification from ∼3 pg genomic DNA (Fig. 2B). In contrast, QRT-PCR performed directly from unamplified DNA yields PCR thresholds 10–13 units higher than RCA–RCA-amplified DNA, and did not yield a signal for the lowest dilution.
Real-time PCR examination of RCA–RCA amplified, FFPE genomic DNA
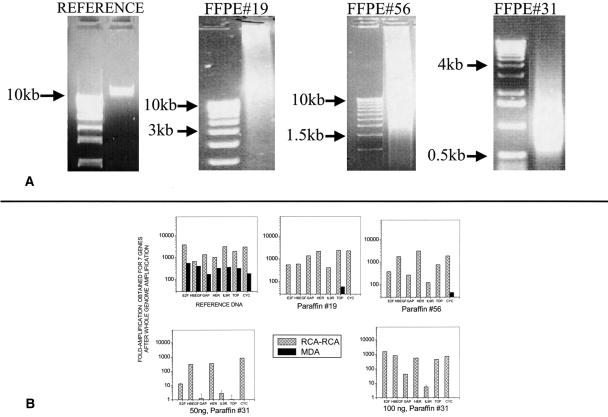
DNA extracted from formalin-fixed, paraffin-embedded tissue was used for amplification via RCA–RCA or alternatively via MDA. The electropherograms in Figure 3A depict an increasing degree of DNA degradation in three FFPE samples, #19, #56, and #31, respectively. Following RCA–RCA (alternatively: MDA) of the reference and FFPE samples, a gene copy number comparison was made between unamplified and amplified DNA in order to estimate the overall amplification obtained. The design of this experiment was similar to the one described in Figure 2 that employed Taqman QRT-PCR. Figure 3B depicts the increase in relative copy number obtained between unamplified and RCA–RCA-amplified genomic DNA for seven genes (E2F, HBEGF, GAPDH, HER2, IL9R, TOP, and CYC). For reference DNA, with a starting material of ∼50 ng, both RCA–RCA and MDA demonstrated significant amplification, ranging from ∼200–800 times (MDA) to ∼600–4,000 times (RCA–RCA). RCA–RCA was also able to amplify most of the genomic regions in the FFPE samples, depending on the degree of degradation. In contrast, the amplification via MDA was unsuccessful for all three paraffin samples (starting material = 20–50 ng for #19, #56 and 50–100 ng for #31). As the degradation of the samples increases, per Figure 3A, the amplification obtained for elected genes becomes smaller. Sample #31, which has the highest degradation among the three samples, yields significant amplification for three out of seven genes for 50 ng starting material and for five out of seven genes for 100 ng starting material. The data indicate that, unlike MDA, which produces no template-specific amplification, RCA–RCA can readily amplify FFPE samples of modest degradation and can also recover information from highly degraded samples.
Figure 3.
(A) Agarose gel electrophoresis profiles of DNA extracted from reference DNA or from three FFPE samples. The first lane in each gel is a nucleic acid ladder. The reduction in average fragment size indicates that DNA degradation is progressively higher for FFPE samples #19, #56, and #31, respectively. (B) Fold-amplification obtained following whole genome amplification of reference DNA or FFPE tissue-extracted DNA, using RCA–RCA or MDA. QRT-PCR (Taqman assay) was applied to seven genes before and after amplification and the fold-amplification relative to unamplified DNA was determined by threshold comparison. MDA yielded practically no amplification for the three FFPE samples.
Examination of RCA–RCA amplification bias using Taqman QRT-PCR
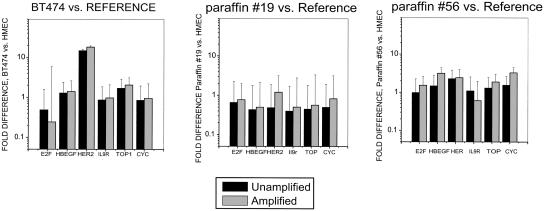
To examine the ability of RCA–RCA to generate unbiased whole genome amplification, we utilized DNA from BT474 cells, which is known to contain well-characterized genetic amplifications and deletions (Forozan et al. 2000; Wang et al. 2004), to compare with DNA from normal cells after amplification in parallel. Figure 4 demonstrates that RCA–RCA retains the previously described (Forozan et al. 2000) HER2 amplification in BT474 cells well relative to reference DNA when amplified and unamplified samples are compared (R2 = 0.98). The relative ratios for the same genes in DNA from FFPE samples #19 and #56 relative to reference DNA are also in concordance (R2 = 0.56–0.65) when amplified and unamplified samples are compared. Concordance in gene dosage from amplified and unamplified samples was low (R2 < 0.1) for the highly degraded paraffin #31 sample (not shown). The error bars in these figures represent real-time PCR performed from three independent RCA–RCA experiments.
Figure 4.
QRT-PCR (Taqman assay)-based comparison of gene copy number ratios before and after RCA–RCA amplification for DNA extracted from BT474, paraffin #19, and paraffin #56 and from reference DNA. The relative (ΔΔCt) threshold method was used to compare relative gene copy number before amplification and after RCA–RCA amplification using the GAPDH housekeeping gene for normalization. The results are presented as copy number ratio relative to co-amplified reference DNA.
Array-CGH screening of RCA–RCA amplified genomic DNA
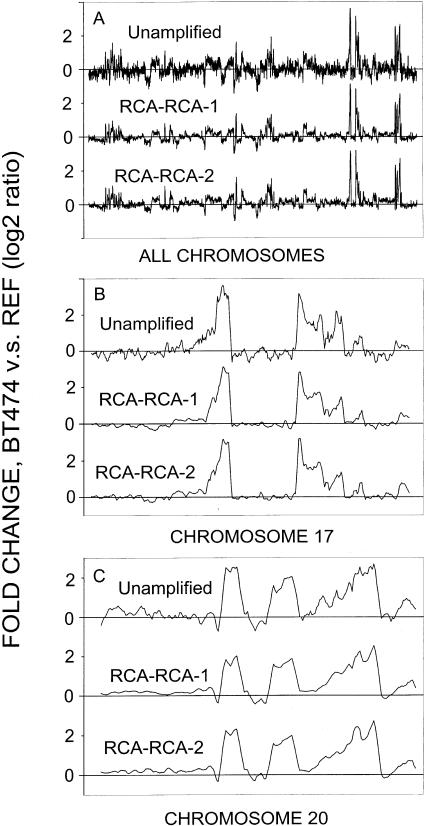
Microarray technology provides an efficient means for identifying gene-dosage alterations across the human genome. To validate the application of RCA–RCA amplified DNA for array-CGH studies, out of an initial 50 ng starting DNA material extracted from BT-474 cells, we used 20 ng for amplification. We then screened the product against reference genomic DNA that had been similarly amplified. The RCA–RCA products were screened in two duplicate independent experiments using the Agilent Human 1 cDNA microarrays. Unamplified BT-474 DNA (4 μg) was also directly screened on microarrays versus unamplified reference DNA. The genomic profiles from the duplicate RCA–RCA amplification products showed a pattern very similar to that from unamplified samples (Fig. 5A). BT-474 cancer cells contain well-known multiple amplification regions in chromosomes 17 and 20, which are depicted in more detail in Figure 5, B and C. For both chromosomes, genetic amplification regions observed in unamplifed BT-474 DNA were also observed in the duplicate amplified samples. The CGH from unamplified samples appears somewhat noisy relative to the RCA–RCA-amplified samples. We consider this a reflection of the relative success of Cy3/Cy5 labeling for the two cases, which was done at different days, that is, for the unamplified samples labeling resulted in lower fluorescent signals, which lead to the observed noise.
Figure 5.
Array-CGH screening for RCA–RCA-amplified BT-474 genomic DNA versus reference DNA. Unamplified represents the array-CGH results from unamplified BT-474 genomic DNA versus reference DNA. RCA–RCA-1 and RCA–RCA-2 are duplicate RCA–RCA amplifications performed in two separate experiments. (A) Comparison for all 23 chromosomes. (B) Comparison for chromosome 17. (C) Comparison for chromosome 20. Five nearest neighbor smoothing was used for these data.
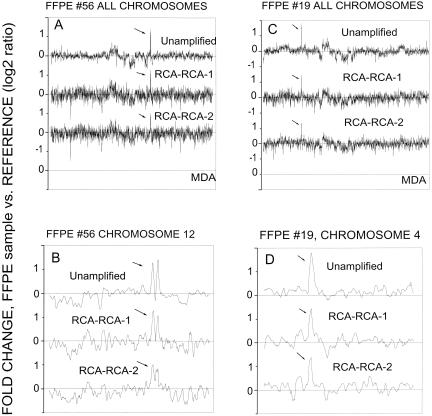
Data analysis demonstrated that the concordance of genedosage profiling between amplified and unamplified DNA increases if nearest-neighbor averaging is employed for data smoothing. We found that increased data smoothing improves R2 but also results in a loss of genome-wide resolution. For example, for chromosome 17, R2 improves from 0.86 to 0.95 when averaging by 2 or 12 nearest neighbors is applied, respectively; concomitantly, the array-CGH resolution decreases from 100 kbp to 1.2 Mbp average spacing along a chromosome. The same observations are valid when MDA is employed instead of RCA–RCA for DNA amplification and array-CGH screening. However, the concordance (R2) between MDA amplified and unamplified DNA is lower relative to RCA–RCA amplified DNA, for example for chromosome 17, R2 ranges from 0.80 to 0.90 when averaging by 2 or 12 nearest neighbors is applied, respectively. Next, array-CGH experiments were performed for the FFPE samples #19 and #56. Array-CGH data obtained for unamplified paraffin #19 or #56 versus reference genomic DNA (4 μg) were compared to that obtained for RCA–RCA-amplified paraffin #19 or #56 versus reference genomic DNA (20 ng DNA used for amplification). Alternatively, MDA-amplified DNA from paraffin #19 or #56 (20 ng DNA used for amplification) was screened. Array-CGH using MDA-amplified DNA produced practically no specific hybridization signals, presumably because of inadequate template-specific amplification for FFPE samples. RCA–RCA amplification of DNA from paraffin #19 and #56 generated array-CGH profiles similar to those obtained when unamplified FFPE samples were directly screened on cDNA microarrays. Figure 6, A–D, demonstrates that RCA–RCA, performed in duplicate independent experiments, reproduced the main amplification (>2 fold) regions in paraffin #19 and #56. In Figure 6, B and D, the main amplified regions on chromosomes 4 and 12 for paraffin #19 and #56 are respectively presented, and a correlation coefficient R2 = 0.59 and 0.49, respectively, was calculated for these data using five nearest neighbor smoothing. The observed concordance of the data for FFPE samples is worse than the one obtained by using intact BT474 DNA (Fig. 5), presumably because of excessive site-specific DNA degradation by the formalin fixation process. Nevertheless, the data indicate that RCA–RCA is capable of successfully identifying the major genetic amplifications occurring in the genome of the FFPE samples by array-CGH. DNA from the highly degraded FFPE sample #31 failed the quality control test for Cy3/Cy5 labeling and was not screened on microarrays.
Figure 6.
Array-CGH screening for RCA–RCA-amplified genomic DNA from FFPE samples #19 or #56 versus reference DNA. Five-nearest-neighbor smoothing has been applied to these data. Unamplified represents the array-CGH results from sample paraffin #19 or #56 versus reference DNA. RCA–RCA-1 and RCA–RCA-2 are duplicate RCA–RCA amplifications performed in two separate experiments. (A,C) Comparison for all 23 chromosomes. (B,D) Comparison for chromosome 4 (#19) or chromosome 12 (#56). Five nearest neighbor smoothing was used for these data.
Another evaluation of the concordance of the CGH data in Figures 5 and 6 is depicted in Table 1, where the fraction of significant genomic amplifications (log2 > 1.5 or, alternatively, log2 > 2) in the unamplified samples, which results in an amplification of at least log2 > 1 in the RCA–RCA-amplified samples, is derived. Log2 > 1 is considered the lowest amplification that can be reliably detected given the average noise (log2 ≅ 0.35) in the CGH profiles. The majority (>76%) of amplifications in the unamplified sample was also detected in the RCA–RCA-amplified samples. Similarly, the paraffin samples demonstrated amplifications in chromosomes 4 and 12 that were efficiently detected in the amplified samples.
Table 1.
Analysis of gene amplifications that are detectable (factor of 2 amplification or more) following RCA-RCA and array-CGH: Comparison to unamplified DNA
| CGH | log2 gene amplifications: Unamplified DNA | log2 gene amplifications: RCA-RCA DNA | Percent gene amplifications detected via CGH after RCA-RCA |
|---|---|---|---|
| BT-474 DNA vs. Reference DNA | >1.5 (189) | >1 (144) | 76 |
| >2 (117) | >1 (98) | 84 | |
| BT-474 DNA vs. Reference DNA: | >1.5 (85) | >1 (74) | 87 |
| Chromosome 17 | >2 (56) | >1 (52) | 93 |
| BT-474 DNA vs. Reference DNA: | >1.5 (60) | >1 (54) | 90 |
| Chromosome 20 | >2 (45) | >1 (41) | 91 |
| Paraffin #19 DNA vs. Reference DNA: | >1.5 (4) | >1 (2) | 50 |
| Chromosome 4 | >2 (2) | >1 (2) | 100 |
| Paraffin #56 DNA vs. Reference DNA: | >1.5 (3) | >1 (3) | 100 |
| Chromosome 12 | >2 (3) | >1 (3) | 100 |
Amplications are quoted as the log2 value. For example, for BT-474 DNA versus Reference, >1.5 (189) means that there were 189 data points with an amplification of more than 2.6-fold relative to reference. In the corresponding CGH from RCA-RCA-amplified DNA there were 144 data points with an amplification of at least two-fold.
Microsatellite instability analysis for RCA–RCA-amplified genomic DNA
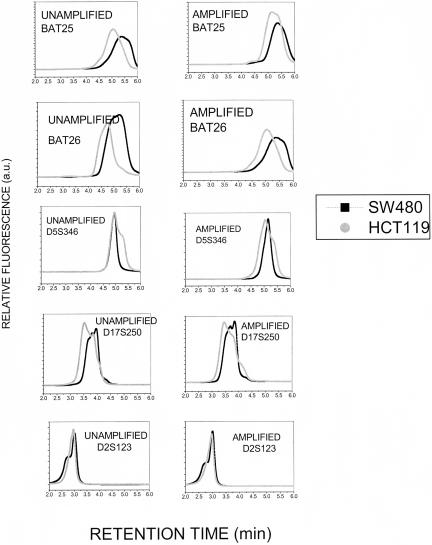
To examine the retention of the ability to detect microsatellite instability (MSI) following RCA–RCA amplification, DNA from two colon cancer cell lines, HCT116 and SW480, with known MSI status (Bhattacharyya et al. 1995) was analyzed before and after RCA–RCA amplification. The dHPLC-based methodology (Kim et al. 2003; Pan et al. 2003) described in detail in our recent work (Liu et al. 2004) was employed to identify MSI as chromatogram shifts between MSI-Stable (SW480) and MSI-High (HCT116) DNA, using the five microsatellite markers recommended by the Bethesda guidelines (Boland et al. 1998). Figure 7 demonstrates that the MSI chromatogram changes detected for HCT116 cells prior to amplification are also detectable after RCA–RCA amplification, demonstrating the fidelity of whole genome amplification at the single nucleotide level. Therefore, RCA–RCA enables MSI examination from limited starting material using a wide array of microsatellite markers.
Figure 7.
dHPLC-based detection of microsatellite instability before and after RCA–RCA amplification. Genomic DNA from two colon cancer cell lines, HCT116 (MSI-H) and SW480 (MSI-S) is examined for five microsatellite markers, BAT25, BAT26, D5S346, D17S250, and D2S123. The chromatogram shifts are retained after RCA–RCA amplification.
Application of RCA–RCA to cDNA amplification
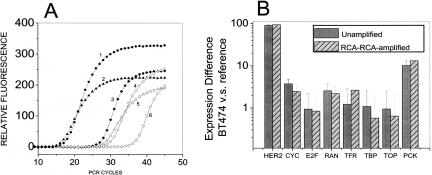
To examine whether RCA–RCA can be used for the unbiased amplification of cDNA, we generated double stranded cDNA from 25 ng total RNA from reference mammary epithelial cells and from the breast cancer cell line BT474. The cDNA was used either directly in a Taqman QRT-PCR reaction to examine gene dosage for seven genes or was first amplified via RCA–RCA and then examined via QRT-PCR. Figure 8A compares copy numbers prior to and after RCA–RCA amplification for three genes. Similar to the results obtained with genomic DNA in Figure 2A, the resulting PCR threshold is about 10–12 cycles different between amplified and non-amplified material. Figure 8B (average of three independent RCA–RCA experiments) demonstrates the upregulation in the HER2 and PCK1 genes as well as a very good retention (R2 = 0.99) of the relative gene expression between reference and BT474 cells following RCA–RCA amplification of total cDNA. It should be noted that with the present approach end-fragments of cDNA sequences will be lost, as only sequences between two restriction sites will be amplified. Nevertheless, the data indicate that a single RCA–RCA protocol can be used for the unbiased amplification of intact or fragmented genomic DNA as well as cDNA.
Figure 8.
(A) Quantification of RCA–RCA amplification-fold in cDNA. QRT-PCR was performed either directly from unamplified BT474 cDNA or from the RCA–RCA-amplified product for three single copy genes. Curves 1–3 CYC, GAPDH, HER2 respectively, using RCA–RCA-amplified DNA as starting material. Curves 4–6 CYC, GAPDH, and HER2 respectively, using unamplified DNA as starting material. (B) Detection of relative gene expression of BT-474 versus reference human mammary epithelial cells before and after RCA–RCA-amplification of cDNA, using Taqman realtime PCR. cDNA used for RCA–RCA amplification was obtained from the reverse transcription of ∼25 ng total RNA starting material.
Discussion
The need for developing RCA–RCA was precipitated by several observations: (1) Ligation-mediated PCR approaches for whole genome amplification, such as SCOMP (Nelson et al. 1989; Klein et al. 1999) or balanced-PCR (Makrigiorgos et al. 2002; Wang et al. 2004), are efficient in amplification of modestly degraded DNA and balanced PCR can also retain the gene-dosage relation among two genomes. However, because of the fragment size limit dictated by the PCR process, these methods result in incomplete genome coverage (Lucito et al. 1998) and potential loss of informative genomic regions for subsequent genetic analysis. (2) Isothermal multiple displacement amplification using random primers produces linearly amplified DNA and almost complete genome coverage (Dean et al. 2002; Hosono et al. 2003). However, as also demonstrated in the present investigation, MDA fails to amplify efficiently fragmented DNA (Lage et al. 2003). (3) Random-primer-based displacement amplification from circular DNA is highly efficient for a much wider range of fragment sizes than conventional PCR (Dean et al. 2001). Accordingly, the current RCA–RCA method was conceived to combine all the advantages of these methods.
RCA–RCA employs restriction enzymes to dissect the genome into smaller fragments and a ligase to circularize these fragments into DNA circles via self-ligation thus generating a complex population of DNA templates of small and big circles that is amenable to exponential rolling circle amplification. Self-circularization of DNA fragments <250 bp has been known to be inefficient (Dallman and Porter 1996). However, in previous studies (Liu et al. 2004) we found that, during whole genome circularization, small DNA fragments (<200 bp) could cross-ligate to form larger fragments that subsequently circularize. At the same time, during the subsequent amplification step, a significantly higher amplification could be expected for the small fragments relative to large fragments as the polymerase completes replication of a smaller circle faster given a constant incorporation of nucleotides per second. Overall, however, this variability is smaller than one would expect, that is, small fragments like the HER2 (92 bp) result in amplification comparable to that of fragments >200 bp long (Fig. 3B). Accordingly, a possible explanation is that the ligation step evens out to some extent the distribution of circular DNA fragments and balances out the resulting RCA–RCA amplification. Furthermore, the data indicate that when a control genome is co-amplified using the same RCA–RCA methodology, the resulting gene amplifications tends to cancel out residual biases related to ligation and uneven rolling-circle replication (Fig. 4). The ability of RCA–RCA to generate an array-CGH profile representative of the original starting material was found to be superior to that of MDA when starting from intact genomic DNA. In a previous report, we showed that balanced-PCR amplification followed by array-CGH provides concordance (R2) similar to that obtained with MDA (Wang et al. 2004). Therefore at the present time the concordance of RCA–RCA products to unamplified samples clearly exceeds that of all other available methods.
There are numerous occasions where the starting DNA material requiring whole genome amplification is degraded to various extents, such as DNA in fluids from cancer patients, DNA in FFPE samples, DNA in forensic applications, and in other instances. RCA–RCA is uniquely suited for high fidelity amplification of such DNA. For example, FFPE samples of low (#19), medium (#56) or high (#31) degradation can be efficiently amplified using RCA–RCA and produce material amenable to PCR-based analysis (Taqman PCR, microsatellite/mutation detection analysis). However, for the sample of highest degradation #31, gene copy ratios relative to reference DNA were not retained, indicating that DNA damages caused by harsh formalin fixation conditions may have impeded the maintenance of gene copy number relations. The success of RCA–RCA relies on generating a double cut by the restriction enzyme between the average spacing of formalin-caused damages (Fig. 1A). Accordingly, it can be expected that, for highly degraded FFPE samples, a more frequent cutter must be chosen in order to achieve maximum sample recovery and amplification. The design of the RCA–RCA protocol allows any single enzyme or combination of enzymes to be substituted in place of Nla-III with no further modification. Therefore, by substituting the restriction enzyme in the first RCA–RCA step it may be possible to recover samples of very high degradation. Array-CGH using RCA–RCA amplified FFPE samples of low and medium degradation (#19 and #56) reliably revealed significant gene copy amplifications. On the other hand, array-CGH data of sample #56 erroneously indicated deletions within 2 chromosomal regions (Fig. 6A). A working hypothesis is that such false chromosomal losses are the consequences of sitespecific DNA damage caused by formalin fixation. Therefore when FFPE samples are used, the confidence level in detecting chromosomal losses is low compared to scoring chromosomal amplifications that generally are true positives.
Another significant feature of RCA–RCA is that, by using Phi29 polymerase, which has a very low error rate of ∼3 – 5 × 10-6 (Nelson et al. 2002) it is possible to utilize the amplified material for performing error-sensitive assays such as microsatellite instability analysis or mutation detection. As Figure 7 demonstrates, the size of five microsatellite markers commonly used for colon cancer diagnosis was unchanged at the single nucleotide level before and after RCA–RCA amplification.
There are additional practical advantages to RCA–RCA amplification. The success or failure of amplification can be easily determined through measurement of DNA concentration via PicoGreen or alternatively by simple inspection of the viscosity of the reaction mixture because without input DNA virtually no product is detectable by gel electrophoresis. In contrast, conventional PCR often produces primer–dimer artifacts and MDA generates well-documented, primer-mediated, nonspecific amplification products. The presence of nonspecific amplification in MDA-amplified DNA becomes particularly troublesome when starting amounts of less than 1–5 ng DNA are to be used for array-CGH as the percentage of primer-induced artifacts increases as the starting material decreases. Methods that produce primer-mediated nonspecific amplification artifacts (e.g., MDA; random primer labeling) necessitate further downstream characterization of the product, for example via real-time PCR, prior to expensive applications like microarray screening. It is not clear why RCA–RCA does not produce the same primer-artifacts as MDA since both processes utilize random primers and strand displacement for amplification. A potential explanation lies with the difference in buffers employed as in order to achieve optimal amplification we included DMSO and T4 gene 32 in the RCA–RCA reaction. Finally, RCA–RCA is simple, inexpensive to perform, and does not require a thermal cycler.
In summary, we have developed a powerful whole genome amplification method, that has superior features over current technology and is tolerant to degradation of the starting material. A single RCA–RCA protocol can be used for gene copy analysis (genomic DNA), gene expression analysis (cDNA), or genotyping. The simplicity, robustness, and versatility of the procedure will help unveil large amounts of valuable genetic information, especially those locked in archived FFPE samples.
Methods
Cell lines, genomic DNA and cDNA
The breast cancer cell line BT-474 and the colon cancer cell lines HCT116 and SW480 were obtained from the American Type Culture Collection. Total genomic DNA was isolated from cultured cells using the QIAamp DNA Mini Kit (QIAGEN). Human male genomic DNA was obtained from Promega and human mammary epithelial cells were purchased from Cambrex and used as controls (reference DNA). Genomic DNA was prepared from formalin-fixed paraffin-embedded high grade glioma specimens (FFPE#19, #56, and #31) obtained from the Department of Pathology, Brigham and Women's Hospital by modification of standard Qiagen methods. For fixation, a standard procedure was followed. Briefly, tissue samples were harvested, placed in 10% buffered formalin for not longer than 7 h, processed by standard methods and embedded in paraffin. DNA was extracted from paraffin blocks over a range of months after fixation, specifically 17 mo for sample #19, 20 mo for sample #56, and 29 mo for sample #31. Double stranded cDNA was obtained from RNA extracted from reference breast epithelial cells (Stratagene) or from BT-474 cells using the Strategene kit.
RCA–RCA whole genome amplification
Genomic DNA or cDNA were digested with 0.5 μL Nla-III (10 units/μL, New England Biolabs) at 37°C for 2 h in 10 μL of 1 × T4 DNA ligase buffer (New England Biolabs). Each sample was heated at 65°C for 20 min to inactivate Nla-III. The fragmented DNAs were circularized with 0.5 μL T4 DNA ligase (2000 units/μL, New England Biolabs) in a volume of 15 μL at room temperature for 2 h. After inactivation of ligase at 65°C for 10 min, linear DNAs were eliminated with 1.2 μL Lamda Exonuclease (5 units/μL, New England Biolabs) and 0.3 μL Exonuclease I (20 units/μL, New England Biolabs) in a volume of 25 μL at 37°C for 1 h. The circularized DNAs were then purified using a QIAquick PCR Purification Kit (QIAGEN) and eluted in 35 μL of H2O. Four microliters circular DNA was mixed with 0.5 μL hexamers (400 ng/μL, Sigma) and 0.5 μL binding buffer (400 mM Tris-HCl at pH 8.0, 160 mM KCl) and denatured at 95°C for 4 min. Alternatively, the denaturation step was omitted. The denatured DNA was amplified using 0.3 μL Phi29 DNA polymerase (10 units/μL, New England Biolabs) complemented with 2 μL of 10 × Phi29 DNA polymerase buffer, 0.2μL of 100 × BSA, 3.2μL of 2.5mM dNTP (Applied Biosystems), 1 μL of 20% DMSO (Sigma), and 1 μL of 20 ng/μL T4 gene32 (Amersham Biosciences) in a volume of 20 μL at 30°C for 16 h. The Phi29 DNA polymerase was inactivated at 65°C for 10 min and the amplification product was digested with 2.5 μL of Nla-III at 37°C for 3 h in a volume of 100 μL.
Multiple displacement amplification (MDA)
MDA was performed for target (BT474) and reference genomic DNAs using the Repli-g whole genome amplification kit (Molecular Staging) according to kit instructions. Briefly, 5–20 ng of either BT474 or reference genomic DNA was brought to a final volume of 2.5 μL with sterile, distilled water. A reaction master mix was prepared by adding 12.5 ml of 4 × mix, 0.5 ml of DNA polymerase mix, and 34.5 μL of sterile, distilled water. The reaction master mix was added to the DNA, and samples were incubated at 30°C for 16 h, following which the enzyme was heat-denatured at 65°C for 3 min. The concentration of amplified samples was determined using a PicoGreen DNA quantification assay (Molecular Probes). Alternatively, the target DNA used for MDA amplification was DNA (10–50 ng) extracted from paraffin-embedded tissue.
Genomic and gene expression analysis using quantitative PCR
Real-time PCR TaqMan (Holland et al. 1991) assays were performed to determine the relative copy number of specific genes in target DNA (BT474 or DNA from paraffin-embedded tissue) relative to reference DNA for unamplified genomic DNA, RCA–RCA amplified DNA, or MDA-amplified DNA. TaqMan assays were performed as previously described (Wang et al. 2004). Briefly, amplification was performed using AmpliTaq Gold (Applied Biosystems) in a Smart-Cycler (Cepheid). Primers and probes for exonic regions of these genes were obtained from Bioresearch Technologies. Three independent triplicates of quantitative PCR experiments were performed for each gene to generate an average relative copy number and standard deviation. For each triplicate, 3 ng of DNA was added to a final volume of 70 μL with a final concentration of 1 × ABI Taq-Man master mix, 4 μM each primer, and 2 μM probe. This reaction mix was split into three different 20 μL PCR reactions and thermocycled. The cycling program was 50°C 2 min 1 cycle, 95°C for 10 min 1 cycle, and 40 cycles at 95°C for 15 sec and 60°C for 1 min. The relative genomic copy number was calculated using the comparative threshold (Ct) method (Heid 1996). Briefly, the threshold cycle (CT) for each gene was determined using the thermocycler software and the average of three independent Cts/DNA was calculated. The copy number of the target gene normalized to an endogenous reference and relative to calibrator is given by the formula 2-ΔΔCT. GAPDH was used as an endogenous reference, and ΔCT was calculated by subtracting the average GAPDH CT from the average CT of the gene of interest. Reference DNA was used as a calibrator DNA to calculate ΔΔCT (ΔCT DNA of interest – ΔCT calibrator DNA).
Similarly, for gene expression analysis from double stranded cDNA obtained from BT474 and reference cells we utilized probes and primers obtained from Applied Biosystems and analysis approaches similar to those used for gene copy number determination.
Array-CGH using cDNA microarrays
Array-based comparative genomic hybridization (Array-CGH) was performed on Agilent Human 1 cDNA microarrays as previously described (Wang et al. 2004). Briefly, for each labeling reaction, 2 μg of digested DNA (amplified or unamplified) was used. Each sample pair was dye-swap labeled for hybridization. DNA samples (2 μg) were denatured in the presence of Random Primer and Reaction Buffer (Invitrogen BioPrime Labeling Kit) at 98°C for 5 min, and then cooled to 2°C for 5 min. The denatured sample was incubated with Klenow Fragment, dNTP mix (2.0 mM dATP dGTP dTTP, 1.0 mM dCTP in 10 mM Tris at pH 8.0, 1 mM EDTA), and Cy3 or Cy5 dCTP nucleotides (1 mM, Perkin Elmer) for 2 h at 37°C. Reactions were terminated using EDTA (0.5 M, pH 8.0). Cy3 and Cy5 reaction pairs (labeled pair = Cy5-sample:Cy3-reference) were pooled, precipitated, and resuspended in 18.5 μL of 0.514% SDS; the reversed labeled pair (Cy3-sample:Cy5-reference) was also treated in the same manner. Samples were mixed with blocking solution concentrated from 50 μL human Cot-1 DNA (Gibco, 1 mg/ml), 20 μL yeast tRNA (Gibco, 5 mg/ml), and 4 μL (dA)-poly(dT) (Sigma 5 mg/ml). SSC was added to a final concentration of 3.4 × and 2.5 μL of Deposition Control Target (Operon) was added to a final volume of 30 μL. For hybridization, samples are denatured at 98°C for 2 min then cooled at 37°C for 30 min under light-protection with foil. Labeled reactions in a volume of 27.5 μL were pipetted onto Agilent Human 1 cDNA arrays. Hybridization was carried out for 18–20 h in a 65°C water bath. After hybridization was complete, arrays were washed in 2 × SSC–SDS (100 ml 20 χ SSC, 0.03% SDS [10%; v/v]) at 65°C for 5 min, followed by additional 5-min wash steps in 1 χ SSC, then 0.2 χ SSC, each at room temperature. After drying, hybridized arrays are scanned on Axon scanner and spot finding and flagging are accomplished using GenePix software. The value of the two dye-swap hybridization experiments was then combined to give the reported log2 ratios. Custom tools developed at the Belfer Center for Cancer Genomics (C. Brennan and L. Chin, in prep.) including cDNA-to-chromosome mapping, exclusion of non-reporters, ratio calculation, normalization, and visualization were used to compile the CGH profiles from these array data points.
Microsatellite instability analysis
To examine retention of the ability to detect microsatellite instability (MSI) following RCA–RCA amplification, DNA from two colon cancer cell lines, HCT116 and SW480, known to be MSI-High and MSI-Stable, respectively (Bhattacharyya et al. 1995; Liu et al. 2004) was analyzed before and after RCA–RCA amplification. The dHPLC-based methodology (Kim et al. 2003; Pan et al. 2003) described in detail in our recent work (Liu et al. 2004) was employed to identify MSI as chromatogram shifts between MSI-Stable (SW480) and MSI-High (HCT116) DNA, for the five microsatellite markers recommended by the Bethesda guidelines (Boland et al. 1998). As also observed in the detection of MSI using denaturing gel electrophoresis, the dHPLC chromatogram changes can be in either direction relative to the stable cell line, corresponding to sequence repeat expansion or contraction.
Acknowledgments
Funding for this work was provided in part by the JCRT Foundation, by DOD grant BC020504, by NIH P50 CA 93683, and by the Arthur and Rochelle Belfer Cancer Genomics Center.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2813404.
References
- Alizadeh, A.A. and Staudt, L.M. 2000. Genomic-scale gene expression profiling of normal and malignant immune cells. Curr. Opin. Immunol. 12: 219-225. [DOI] [PubMed] [Google Scholar]
- Alizadeh, A.A., Eisen, M.B., Davis, R.E., Ma, C., Lossos, I.S., Rosenwald, A., Boldrick, J.C., Sabet, H., Tran T., Yu, X., et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503-511. [DOI] [PubMed] [Google Scholar]
- Alizadeh, A.A., Ross, D.T., Perou, C.M., and van de Rijn, M. 2001. Towards a novel classification of human malignancies based on gene expression patterns. J. Pathol. 195: 41-52. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, N.P., Ganesh, A., Phear, G., Richards, B., Skandalis, A., and Meuth, M. 1995. Molecular analysis of mutations in mutator colorectal carcinoma cell lines. Hum. Mol. Genet. 4: 2057-2064. [DOI] [PubMed] [Google Scholar]
- Boland, C.R., Thibodeau, S.N., Hamilton, S.R., Sidransky, D., Eshleman, J.R., Burt, R.W., Meltzer, S.J., Rodriguez-Bigas, M.A., Fodde, R., Ranzani, G.N., et al. 1998. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 58: 5248-5257. [PubMed] [Google Scholar]
- Dallman, M.J. and Porter, A.C.G. 1996. PCR1: A Practical Approach. Oxford University Press, Oxford, UK.
- Dean, F.B., Nelson, J.R., Giesler, T.L., and Lasken, R.S. 2001. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 11: 1095-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, F.B., Hosono, S., Fang, L., Wu, X., Faruqi, A.F., Bray-Ward, P., Sun, Z., Zong, Q., Du, Y., Du, J., et al. 2002. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. 99: 5261-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forozan, F., Mahlamaki, E.H., Monni, O., Chen, Y., Veldman, R., Jiang, Y., Gooden, G.C., Ethier, S.P., Kallioniemi, A., and Kallioniemi, O.P. 2000. Comparative genomic hybridization analysis of 38 breast cancer cell lines: A basis for interpreting complementary DNA microarray data. Cancer Res. 60: 4519-4525. [PubMed] [Google Scholar]
- Heid, C., Stevens, J., Livak, K., and Williams, P. 1996. Real time quantitative PCR. Genome Methods: Genome Research. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed]
- Holland, P.M., Abramson, R.D., Watson, R., and Gelfand, D.H. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′–3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. 88: 7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono, S., Faruqi, A.F., Dean, F.B., Du, Y., Sun, Z., Wu, X., Du, J., Kingsmore, S.F., Egholm, M., and Lasken, R.S. 2003. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 13: 954-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, I.J., Shin, Y., Kang, H.C., Park, J.H., Ku, J.L., Park, H.W., Park, H.R., Lim, S.B., Jeong, S.Y., Kim, W.H., et al. 2003. Robust microsatellite instability (MSI) analysis by denaturing high-performance liquid chromatography (DHPLC). J. Hum. Genet. 48: 525-530. [DOI] [PubMed] [Google Scholar]
- Klein, C.A., Schmidt-Kittler, O., Schardt, J.A., Pantel, K., Speicher, M.R., and Riethmuller, G. 1999. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc. Natl. Acad. Sci. 96: 4494-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage, J.M., Leamon, J.H., Pejovic, T., Hamann, S., Lacey, M., Dillon, D., Segraves, R., Vossbrinck, B., Gonzalez, A., Pinkel, D., et al. 2003. Whole genome analysis of genetic alterations in small DNA samples using hyperbranched strand displacement amplification and array-CGH. Genome Res. 13: 294-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, U. and Kreipe, H. 2001. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods 25: 409-418. [DOI] [PubMed] [Google Scholar]
- Lewis, F., Maughan, N.J., Smith, V., Hillan, K., and Quirke, P. 2001. Unlocking the archive—Gene expression in paraffin-embedded tissue. J. Pathol. 195: 66-71. [DOI] [PubMed] [Google Scholar]
- Liu, W.H., Kaur, M., Wang, G., Zhu, P., Zhang, Y., and Makrigiorgos, G.M. 2004. Inverse PCR-based RFLP scanning identifies low-level mutation signatures in colon cells and tumors. Cancer Res. 64: 2544-2551. [DOI] [PubMed] [Google Scholar]
- Lizardi, P.M., Huang, X., Zhu, Z., Bray-Ward, P., Thomas, D.C., and Ward, D.C. 1998. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 19: 225-232. [DOI] [PubMed] [Google Scholar]
- Lovmar, L., Fredriksson, M., Liljedahl, U., Sigurdsson, S., and Syvanen, A.-C. 2003. Quantitative evaluation by minisequencing and microarrays reveals accurate multiplexed SNP genotyping of whole genome amplified DNA. Nucl. Acids. Res. 31: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucito, R., Nakimura, M., West, J.A., Han, Y., Chin, K., Jensen, K., McCombie, R., Gray, J.W., and Wigler, M. 1998. Genetic analysis using genomic representations. Proc. Natl. Acad. Sci. 95: 4487-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrigiorgos, G.M., Chakrabarti, S., Zhang, Y., Kaur, M., and Price, B.D. 2002. A PCR-based amplification method retaining the quantitative difference between two complex genomes. Nat. Biotechnol. 20: 936-939. [DOI] [PubMed] [Google Scholar]
- Nelson, D.L., Ledbetter, S.A., Corbo, L., Victoria, M.F., Ramirez-Solis, R., Webster, T.D., Ledbetter, D.H., and Caskey, C.T. 1989. Alu polymerase chain reaction: A method for rapid isolation of human-specific sequences from complex DNA sources. Proc. Natl. Acad. Sci. 86: 6686-6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J.R., Cai, Y.C., Giesler, T.L., Farchaus, J.W., Sundaram, S.T., Ortiz-Rivera, M., Hosta, L.P., Hewitt, P.L., Mamone, J.A., Palaniappan, C., et al. 2002. TempliPhi, phi29 DNA polymerase based rolling circle amplification of templates for DNA sequencing. Biotechniques Suppl: 44-47. [PubMed]
- Paez, J.G., Lin, M., Beroukhim, R., Lee, J.C., Zhao, X., Richter, D.J., Gabriel, S., Herman, P., Sasaki, H., Altshuler, D., et al. 2004. Genome coverage and sequence fidelity of phi29 polymerase-based multiple strand displacement whole genome amplification. Nucleic Acids Res. 32: e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, K.F., Liu, W., Lu, Y.Y., Zhang, L., Li, Z.P., Lu, W.L., Thibodeau, S.N., and You, W.C. 2003. High throughput detection of microsatellite instability by denaturing high-performance liquid chromatography. Hum. Mutat. 22: 388-394. [DOI] [PubMed] [Google Scholar]
- Perou, C.M., Jeffrey, S.S., van de Rijn, M., Rees, C.A., Eisen, M.B., Ross, D.T., Pergamenschikov, A., Williams, C.F., Zhu, S.X., Lee, J.C., et al. 1999. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc. Natl. Acad. Sci. 96: 9212-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou, C.M., Sorlie, T., Eisen, M.B., van de Rijn, M., Jeffrey, S.S., Rees, C.A., Pollack, J.R., Ross, D.T., Johnsen, H., Akslen, L.A., et al. 2000. Molecular portraits of human breast tumours. Nature 406: 747-752. [DOI] [PubMed] [Google Scholar]
- Pomeroy, S.L., Tamayo, P., Gaasenbeek, M., Sturla, L.M., Angelo, M., McLaughlin, M.E., Kim, J.Y., Goumnerova, L.C., Black, P.M., Lau, C., et al. 2002. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 415: 436-442. [DOI] [PubMed] [Google Scholar]
- Rook, M.S., Delach, S.M., Deyneko, G., Worlock, A., and Wolfe, J.L. 2004. Whole genome amplification of DNA from laser capture-microdissected tissue for high-throughput single nucleotide polymorphism and short tandem repeat genotyping. Am. J. Pathol. 164: 23-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, D.T. and Perou, C.M. 2001. A comparison of gene expression signatures from breast tumors and breast tissue derived cell lines. Dis. Markers 17: 99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie, T., Perou, C.M., Tibshirani, R., Aas, T., Geisler, S., Johnsen, H., Hastie, T., Eisen, M.B., van de Rijn, M., Jeffrey, S.S., et al. 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. 98: 10869-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenius, H., Carter, N.P., Bebb, C.E., Nordenskjold, M., Ponder, B.A., and Tunnacliffe, A. 1992. Degenerate oligonucleotide-primed PCR: General amplification of target DNA by a single degenerate primer. Genomics 13: 718-725. [DOI] [PubMed] [Google Scholar]
- van't Veer, L.J., Dai, H., van de Vijver, M.J., He, Y.D., Hart, A.A., Mao, M., Peterse, H.L., van der Kooy, K., Marton, M.J., Witteveen, A.T., et al. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530-536. [DOI] [PubMed] [Google Scholar]
- Wang, G., Brennan, C., Rook, M., Wolfe, J.L., Leo, C., Chin, L., Pan, H., Liu, W.H., Price, B., and Makrigiorgos, G.M. 2004. Balanced-PCR amplification allows unbiased identification of genomic copy changes in minute cell and tissue samples. Nucleic Acids Res. 32: e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, M., Blanchette, C., Dressman, H., Huang, E., Ishida, S., Spang, R., Zuzan, H., Olson Jr., J.A., Marks, J.R., and Nevins, J.R. 2001. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc. Natl. Acad. Sci. 98: 11462-11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, K.K., Tsang, Y.T., Shen, J., Cheng, R.S., Chang, Y.M., Man, T.K., and Lau, C.C. 2004. Allelic imbalance analysis by high-density single-nucleotide polymorphic allele (SNP) array with whole genome amplified DNA. Nucleic Acids Res. 32: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Cui, X., Schmitt, K., Hubert, R., Navidi, W., and Arnheim, N. 1992. Whole genome amplification from a single cell: Implications for genetic analysis. Proc. Natl. Acad. Sci. 89: 5847-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]