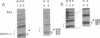Abstract
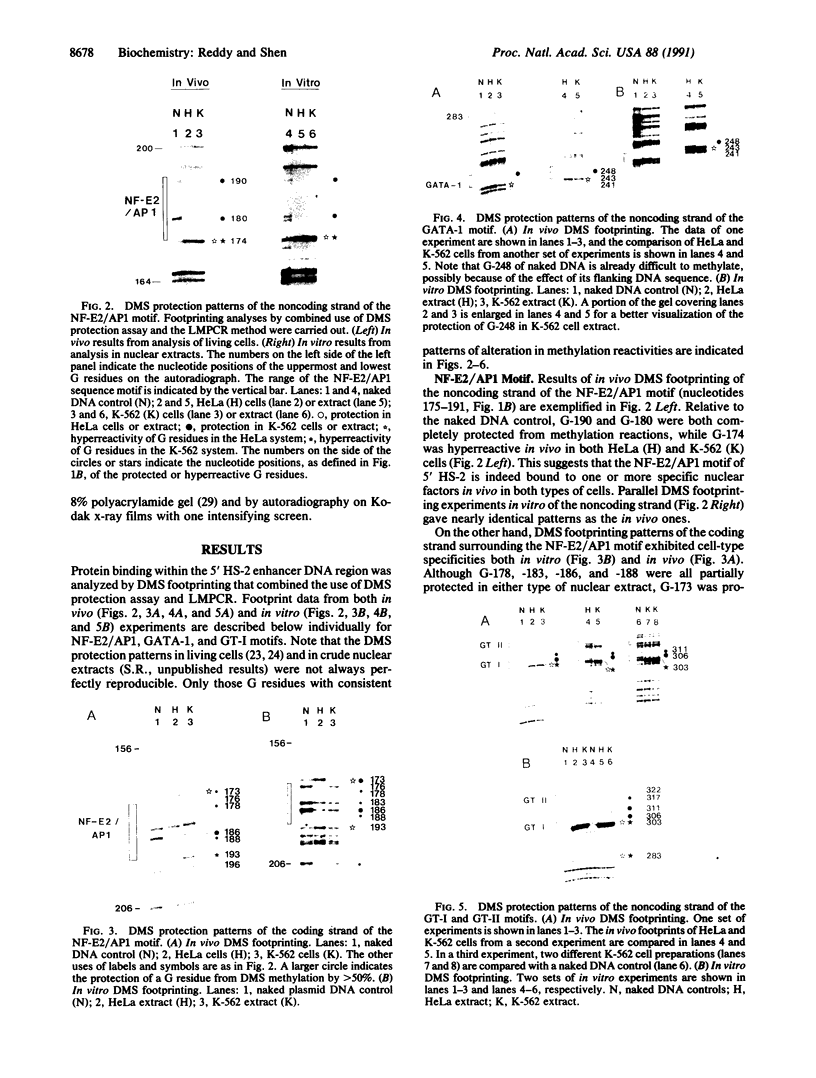
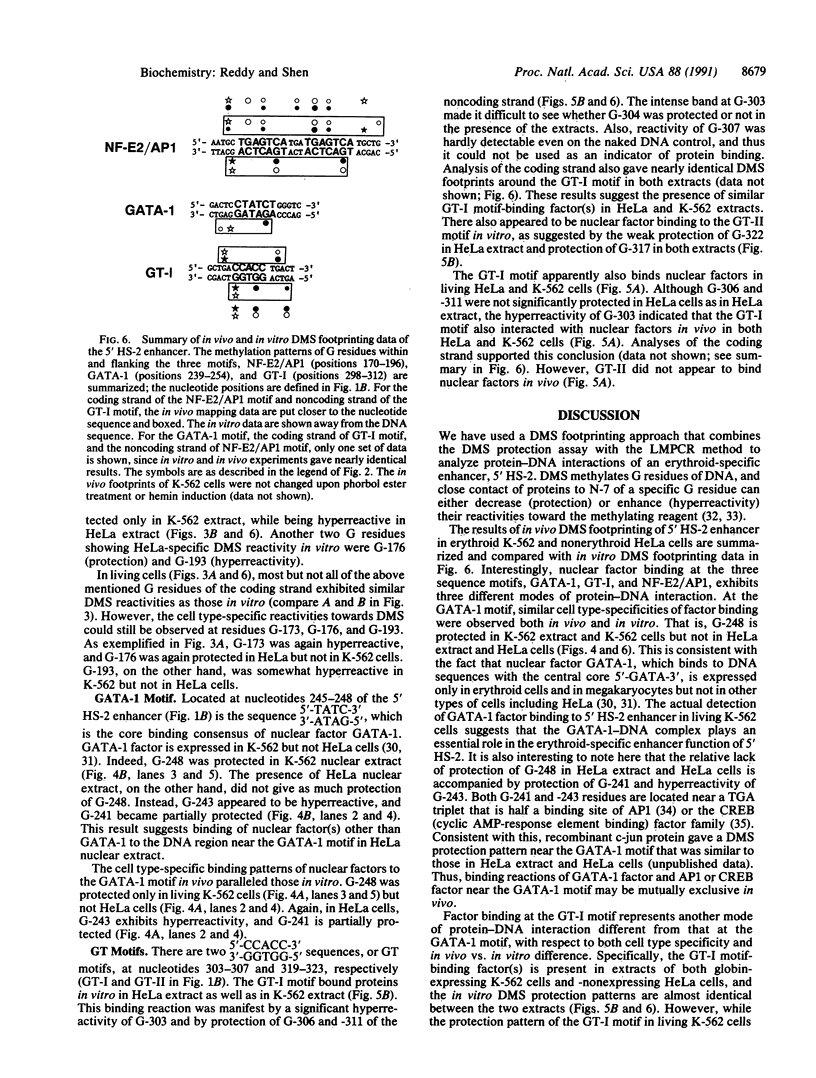
The 5' DNase I-hypersensitive site 2 (5' HS-2) is an erythroid-specific enhancer located 11 kilobases (kb) upstream of the human beta-globin gene cluster. Presence in cis of 5' HS-2 confers a high level of erythroid cell-specific and developmentally regulated promoter activities of human globin genes in transfected cell cultures and in transgenic mice. Combining the use of the methylation protection assay and polymerase chain reaction, we have studied nuclear factor-DNA interactions of the 5' HS-2 enhancer in vitro and in vivo. The data from analyses of three different sequence motifs within 5' HS-2 represent three different modes of protein-DNA interaction with respect to cell-type specificities and in vivo vs. in vitro differences. First, a GATA-1 motif was found to bind nuclear factor(s), presumably the GATA-1 factor, present in K-562 cell extracts and in living K-562 cells. No such binding was seen in nonerythroid HeLa cells or extract. A second motif, NF-E2/AP1 (nuclear factor-erythroid 2/activator protein 1), consists of tandemly arranged dimers of AP1 binding consensus. The presence of either HeLa extract or K-562 extract protects the NF-E2/AP1 motif from methylation, but the footprints are different. This is most likely due to different protein-DNA contacts of the AP1-DNA complex formed in HeLa extract and the NF-E2-DNA complex in K-562 extract. In vivo methylation protection patterns of this motif parallel those observed in vitro, suggesting that it is also bound by NF-E2 in K-562 cells and by AP1 in HeLa cells. Finally, a GT-I motif binds apparently to one or more similar factors in both types of nuclear extracts, but the in vivo methylation protection patterns are not identical between living HeLa and K-562 cells. These data provide direct evidence that specific nuclear factor-DNA complexes form in vivo at functionally important sequence motifs of the 5' HS-2 enhancer in erythroid cells. The detection of conformationally different nuclear factor-DNA complexes at the same sequence motifs in HeLa and Raji cell lines also raises interesting questions regarding the origin and function of these complexes in nonerythroid cells.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker P. B., Ruppert S., Schütz G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell. 1987 Nov 6;51(3):435–443. doi: 10.1016/0092-8674(87)90639-8. [DOI] [PubMed] [Google Scholar]
- Caterina J. J., Ryan T. M., Pawlik K. M., Palmiter R. D., Brinster R. L., Behringer R. R., Townes T. M. Human beta-globin locus control region: analysis of the 5' DNase I hypersensitive site HS 2 in transgenic mice. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1626–1630. doi: 10.1073/pnas.88.5.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi O. R., Engel J. D. A 3' enhancer is required for temporal and tissue-specific transcriptional activation of the chicken adult beta-globin gene. Nature. 1986 Oct 23;323(6090):731–734. doi: 10.1038/323731a0. [DOI] [PubMed] [Google Scholar]
- Church G. M., Ephrussi A., Gilbert W., Tonegawa S. Cell-type-specific contacts to immunoglobulin enhancers in nuclei. 1985 Feb 28-Mar 6Nature. 313(6005):798–801. doi: 10.1038/313798a0. [DOI] [PubMed] [Google Scholar]
- Curtin P. T., Liu D. P., Liu W., Chang J. C., Kan Y. W. Human beta-globin gene expression in transgenic mice is enhanced by a distant DNase I hypersensitive site. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7082–7086. doi: 10.1073/pnas.86.18.7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar V., Nandi A., Schildkraut C. L., Skoultchi A. I. Erythroid-specific nuclease-hypersensitive sites flanking the human beta-globin domain. Mol Cell Biol. 1990 Aug;10(8):4324–4333. doi: 10.1128/mcb.10.8.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A., Church G. M., Tonegawa S., Gilbert W. B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985 Jan 11;227(4683):134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Forrester W. C., Novak U., Gelinas R., Groudine M. Molecular analysis of the human beta-globin locus activation region. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5439–5443. doi: 10.1073/pnas.86.14.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W. C., Takegawa S., Papayannopoulou T., Stamatoyannopoulos G., Groudine M. Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 1987 Dec 23;15(24):10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld F., van Assendelft G. B., Greaves D. R., Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987 Dec 24;51(6):975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Hesse J. E., Nickol J. M., Lieber M. R., Felsenfeld G. Regulated gene expression in transfected primary chicken erythrocytes. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4312–4316. doi: 10.1073/pnas.83.12.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs D. R., Wood W. G., Jarman A. P., Sharpe J., Lida J., Pretorius I. M., Ayyub H. A major positive regulatory region located far upstream of the human alpha-globin gene locus. Genes Dev. 1990 Sep;4(9):1588–1601. doi: 10.1101/gad.4.9.1588. [DOI] [PubMed] [Google Scholar]
- Karlsson S., Nienhuis A. W. Developmental regulation of human globin genes. Annu Rev Biochem. 1985;54:1071–1108. doi: 10.1146/annurev.bi.54.070185.005231. [DOI] [PubMed] [Google Scholar]
- Lee K. A., Hai T. Y., SivaRaman L., Thimmappaya B., Hurst H. C., Jones N. C., Green M. R. A cellular protein, activating transcription factor, activates transcription of multiple E1A-inducible adenovirus early promoters. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8355–8359. doi: 10.1073/pnas.84.23.8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Haslinger A., Karin M., Tjian R. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature. 1987 Jan 22;325(6102):368–372. doi: 10.1038/325368a0. [DOI] [PubMed] [Google Scholar]
- Li Q. L., Zhou B., Powers P., Enver T., Stamatoyannopoulos G. Beta-globin locus activation regions: conservation of organization, structure, and function. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8207–8211. doi: 10.1073/pnas.87.21.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mignotte V., Eleouet J. F., Raich N., Romeo P. H. Cis- and trans-acting elements involved in the regulation of the erythroid promoter of the human porphobilinogen deaminase gene. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6548–6552. doi: 10.1073/pnas.86.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moi P., Kan Y. W. Synergistic enhancement of globin gene expression by activator protein-1-like proteins. Proc Natl Acad Sci U S A. 1990 Nov;87(22):9000–9004. doi: 10.1073/pnas.87.22.9000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon A. M., Ley T. J. Conservation of the primary structure, organization, and function of the human and mouse beta-globin locus-activating regions. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7693–7697. doi: 10.1073/pnas.87.19.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. R., Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989 Nov 10;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- Ney P. A., Sorrentino B. P., McDonagh K. T., Nienhuis A. W. Tandem AP-1-binding sites within the human beta-globin dominant control region function as an inducible enhancer in erythroid cells. Genes Dev. 1990 Jun;4(6):993–1006. doi: 10.1101/gad.4.6.993. [DOI] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. An amino-terminal fragment of lac repressor binds specifically to lac operator. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5851–5854. doi: 10.1073/pnas.75.12.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S. H. Globin gene regulation and switching: circa 1990. Cell. 1990 Nov 16;63(4):665–672. doi: 10.1016/0092-8674(90)90133-y. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Mueller P. R., Wold B., Riggs A. D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989 Nov 10;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Philipsen S., Talbot D., Fraser P., Grosveld F. The beta-globin dominant control region: hypersensitive site 2. EMBO J. 1990 Jul;9(7):2159–2167. doi: 10.1002/j.1460-2075.1990.tb07385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo P. H., Prandini M. H., Joulin V., Mignotte V., Prenant M., Vainchenker W., Marguerie G., Uzan G. Megakaryocytic and erythrocytic lineages share specific transcription factors. Nature. 1990 Mar 29;344(6265):447–449. doi: 10.1038/344447a0. [DOI] [PubMed] [Google Scholar]
- Sorrentino B., Ney P., Bodine D., Nienhius A. W. A 46 base pair enhancer sequence within the locus activating region is required for induced expression of the gamma-globin gene during erythroid differentiation. Nucleic Acids Res. 1990 May 11;18(9):2721–2731. doi: 10.1093/nar/18.9.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos G. Human hemoglobin switching. Science. 1991 Apr 19;252(5004):383–383. doi: 10.1126/science.2017679. [DOI] [PubMed] [Google Scholar]
- Talbot D., Philipsen S., Fraser P., Grosveld F. Detailed analysis of the site 3 region of the human beta-globin dominant control region. EMBO J. 1990 Jul;9(7):2169–2177. doi: 10.1002/j.1460-2075.1990.tb07386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan D. Y., Solomon W. B., London I. M., Lee D. P. An erythroid-specific, developmental-stage-independent enhancer far upstream of the human "beta-like globin" genes. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2554–2558. doi: 10.1073/pnas.86.8.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan D., Solomon W., Li Q., London I. M. The "beta-like-globin" gene domain in human erythroid cells. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. Y., Chen J., Lin L. I., Tam M., Shen C. K. Cell type-specific protein-DNA interactions in the human zeta-globin upstream promoter region: displacement of Sp1 by the erythroid cell-specific factor NF-E1. Mol Cell Biol. 1990 Jan;10(1):282–294. doi: 10.1128/mcb.10.1.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon L. I., Tsai S. F., Burgess S., Matsudaira P., Bruns G. A., Orkin S. H. The major human erythroid DNA-binding protein (GF-1): primary sequence and localization of the gene to the X chromosome. Proc Natl Acad Sci U S A. 1990 Jan;87(2):668–672. doi: 10.1073/pnas.87.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]