Abstract
Within the cell, several mechanisms exist to maintain homeostasis of the endoplasmic reticulum (ER). One of the primary mechanisms is the unfolded protein response (UPR). In this review, we primarily focus on the latest signal webs and regulation mechanisms of the UPR. The relationships among ER stress, apoptosis, and cancer are also discussed. Under the normal state, binding immunoglobulin protein (BiP) interacts with the three sensors (protein kinase RNA-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1α (IRE1α)). Under ER stress, misfolded proteins interact with BiP, resulting in the release of BiP from the sensors. Subsequently, the three sensors dimerize and autophosphorylate to promote the signal cascades of ER stress. ER stress includes a series of positive and negative feedback signals, such as those regulating the stabilization of the sensors/BiP complex, activating and inactivating the sensors by autophosphorylation and dephosphorylation, activating specific transcription factors to enable selective transcription, and augmenting the ability to refold and export. Apart from the three basic pathways, vascular endothelial growth factor (VEGF)-VEGF receptor (VEGFR)-phospholipase C-γ (PLCγ)-mammalian target of rapamycin complex 1 (mTORC1) pathway, induced only in solid tumors, can also activate ATF6 and PERK signal cascades, and IRE1α also can be activated by activated RAC-alpha serine/threonine-protein kinase (AKT). A moderate UPR functions as a pro-survival signal to return the cell to its state of homeostasis. However, persistent ER stress will induce cells to undergo apoptosis in response to increasing reactive oxygen species (ROS), Ca2+ in the cytoplasmic matrix, and other apoptosis signal cascades, such as c-Jun N-terminal kinase (JNK), signal transducer and activator of transcription 3 (STAT3), and P38, when cellular damage exceeds the capacity of this adaptive response.
Keywords: Unfolded protein response, Endoplasmic reticulum (ER) stress, Mechanism, Signal networks, Homeostasis
1. Introduction
In human and animal cells, many stringent quality control systems are employed to ensure the normal metabolism of cells. The endoplasmic reticulum (ER) is responsible for the folding and maturation of newly synthesized transmembrane and secretory proteins in the Golgi compartment. Alterations in ER homeostasis cause accumulation of misfolded/unfolded proteins in the ER, which activates signaling pathways to orchestrate adaptive cellular responses. However, if homeostasis fails to be restored, the ER will initiate cell death signaling pathways, chronic inflammation, or tumorigenesis. The mechanisms that couple protein translation with protein folding in the ER have been reviewed (Kaufman, 2004) as well as the basic stress signal pathway of the ER (Shen et al.,2004; Rutkowski and Kaufman,2007). Some papers have discussed the function of unfolded protein response (UPR) signaling pathways in disease, and concluded that persistent ER stress, as well as inflammation, triggers the basic mechanisms involved in the development or pathology of metabolic disease, neurodegenerative disease, and inflammatory disease (Wang and Kaufman, 2012; Chaudhari et al.,2014; Zhu et al.,2014). Most studies have focused on the relation between UPR and cancer, and concluded that ER stress and UPR activation in both tumor cells and endothelial cells stimulate tumor angiogenesis, and the activity of different UPRs involved in tumorigenesis has been described (Nagelkerke et al.,2014; Wang and Kaufman,2014; Wang et al.,2014; Yadav et al.,2014). In this review, we primarily focus on our understanding of the network of ER stress signaling pathways and its regulatory mechanisms revealed in recent years. The latest signal webs of ER stress are summarized and their regulatory mechanisms discussed. These webs and mechanisms form the basis for a more thorough understanding and further investigation of the functions of the UPR, as well as its relationship with other cellular signaling cascades that could modulate its output.
2. Endogenous homeostasis and the surveillance mechanism
2.1. ER functions in protein production
The functions of the ER in protein processing are sensitive to environmental conditions and cellular changes. Protein production rates are primarily regulated by the mammalian target of rapamycin (mTOR) pathway and the UPR. As environmental conditions change, mTOR signaling coordinately regulates the rate of global translation initiation and ribosome biogenesis to maintain homeostasis. However, when misfolded or unfolded proteins accumulate in the ER, eukaryotic translation initiation factor 2 α (eIF2α), a downstream component of the UPR, is phosphorylated and inhibits protein expression (Kaufman,2004; DuRose et al.,2009). This is referred to as ER stress.
2.2. ER functions in protein modification and misfolded/unfolded protein degradation
In addition to protein processing, the ER functions as a sensitive surveillance system that attempts to monitor and prevent the accumulation of misfolded or unfolded proteins. This system prevents thesemisfolded/unfolded proteins from entering the secretory pathway and directs persistently misfolded proteins towards a degradative pathway (Cao et al.,2013).
Translation of secretory proteins is initiated in the cytoplasm followed by translocation to the ER membrane via signal-recognition particle (SRP) receptor interactions. These nascent polypeptide chains enter the ER lumen with the help of the SEC61 translocon after their signal peptide is cleaved by a signal peptidase on the ER membrane. Once in the ER lumen, (glucose)3(mannose)9-N-(acetylglucosamine)2 (Glc3Man9GlcNAc2) is added to a Asn-X-Ser/Thr consensus sequence, where X is any amino acid except proline (Fig. 1: A1–A3). As part of the protein modification and folding process, the two terminal glucoses are cleaved by glucosidase I and glucosidase II, and then calnexin (CNX) and calreticulin (CRT) recognize and bind with glucose-(1,3)-mannose glycosidic bond, which is present on high mannose-containing asparagine-linked oligosaccharides. This promotes proper folding and disulfide bond formation with the help of protein disulfide isomerase (PDI) and other molecular chaperones (Shen et al., 2004; Chaudhari et al., 2014).
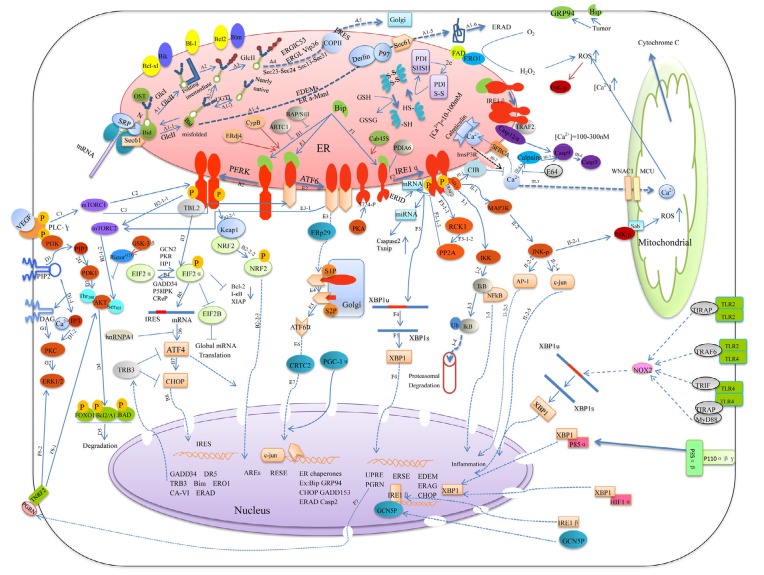
Fig. 1.
Signal network and regulating mechanism of UPR
Proteins that are not properly assembled are subjected to reglycosylation by uridine diphosphate (UDP)-glucose: glycoprotein glucosyltransferase (UGT) and rebind CNX/CRT, until they attain the correct conformation for secretion (Fig. 1: A1-1–A1-3). The retained misfolded proteins are tagged by ER degradation-enhancing α-mannosidase-like protein (EDEM) for ER-associated degradation (ERAD) via the ubiquitin proteasome degradation system or autophagy (Chaudhari et al., 2014) (Fig. 1: A1-4–A1-6). Correctly folded/assembled proteins are transported to the cis-Golgi compartment by coat protein complex II (COPII)-coated vesicles that form at ribosome-free ER exit sites (ERESs) (Shen et al., 2004; Braakman and Bulleid, 2011). COPII assembled at ERES is initiated by the activation of Sar1, a small GTPase, followed by the successive recruitment of the Sec23-Sec24 dimer and Sec13-Sec31 tetrameric complex to the scaffold protein Sec16. These vesicles undergo fusion with the ER membrane and then fission as a 60–80 nm diameter vesicle and bud off (Farhan et al., 2008). These chaperones and oxidoreductases with Lys-Asp-Glu-Leu (KDEL) sequences in the budding of COPII vesicles are continually recycled back to the ER by interacting with the KDEL receptor in the ER membrane (Malhotra and Kaufman, 2007). Under the UPR, ER export ability is augmented by the merger, within minutes, of ERES and the formation of additional ERES after a period of hours to days (Farhan et al., 2008).
However, in a solid tumor, chaperones and oxidoreductases, such as binding immunoglobulin protein (BiP), also known as 78 kDa glucose-regulated protein (GRP78), PDI, ER oxidoreductin 1α (ERO1α), and GRP94, are not partially recycled back to the ER, but instead are translocated to the plasma membrane. The localization of these chaperones and oxidoreductases in the plasma membrane can in some cases serve in tumor immunorecognition or promote tumor proliferation (Malhotra and Kaufman, 2007). For example, BiP/GRP78 overexpression has been found to correlate with both the rate of patient survival and the depth of tumor invasion (Yadav et al., 2014).
2.3. ER functions in mRNA quality control and RIDD and rRNA synthesis
Another buffering system is nonsense-mediated mRNA decay (NMD) (Mühlemann and Lykke-Andersen, 2010). Nonsense mRNAs contain an intact open reading frame (ORF) with a premature stop codon that would generate a misfolded polypeptide. NMD is important in preventing the generation of misfolded proteins by capturing the nonsense mRNA and repressing initiation of its translation. It differs from ERAD which primarily functions in the degradation of misfolded proteins. The mechanism of NMD includes the capture of substrate mRNA by a decay-inducing complex, its subsequent release from the ribosome and eukaryotic release factors (eRFs), nonsense mediated mRNA decay associated PI3K related kinase 1 (SMG1)-mediated RNA helicase and ATPase 1 (UPF1) phosphorylation and then SMG6-dependent dephosphorylation of UPF1 by protein phosphatase 2A. Finally, SMG6-mediated cleavage of mRNA occurs at a premature termination codon (PTC)-proximal site in the cytoplasm followed by exosomal degradation (Eberle et al., 2009; Hwang et al., 2010). Though ER stress does not affect UPF1 phosphorylation and NMD complex assembly, it increases exosome endoribonuclease and 3'-5' exoribonuclease 3 (DIS3) and exosome component 3 (EXOSC3), two exosomal subunits, and SMG6 expressions. Also, during ER stress, the regulated inositol-requiring enzyme 1 (IRE1)-dependent decay (RIDD) pathway promotes the degradation of ER membrane-localized mRNAs, which limits the synthesis of secretory proteins on the ER membrane (Sakaki et al., 2012; Maurel et al., 2014). Apart from inducing NMD and ERID for mRNA degradation, the UPR also down-regulates the synthesis of rRNA by inactivation of the RNA polymerase I basal transcription factor RNA polymerase I transcription factor (RRN3/TIF-IA) (DuRose et al., 2009).
3. BiP/GRP78-dependent or -independent UPR activation mechanisms
3.1. BiP/GRP78-dependent UPR activation mechanisms
When ER homeostasis is perturbed by intraluminal calcium, improper glycosylation, nutrient deprivation, pathogen infection, accumulation of misfolded proteins, or changes in redox status, the UPR is activated in an attempt to maintain protein fidelity and minimize unfolded protein accumulation (Malhotra and Kaufman, 2007; Sakaki et al., 2012). The UPR comprises three basic pathways, which are controlled by three respective ER sensors: protein kinase RNA-like ER kinase (PERK), activating transcription factor 6 (ATF6), and IRE1α. The three sensors form stable complexes with immunoglobulin proteins (BiP/GRP78) and prevent the UPR under normal cellular conditions. When misfolded proteins accumulate, the BiP/GRP78s are released from the three sensors. Consequently, the three sensors homodimerize and subsequently autophosphorylate and initiate UPR signaling cascades as a transcription factor (Nagasawa et al., 2007).
There are three kinds of basic elements involved in the transcription of ER chaperone genes: one is the cis-acting ER stress response element (ERSE). ERSE contains a CCAAT-N9-CCACG sequence, which can be bound by ATF6α and X-box binding protein 1 (XBP1) via its CCACG conserved sequence. This is followed by the binding of the general transcription factor, nuclear factor Y (NF-Y), to the CCAAT part of the ERSE, leading to the activation of ER chaperone gene transcription (Yamamoto et al., 2004). The second element is the UPR element (UPRE), which contains a TGACGTGG/A sequence and is preferentially bound by XBP1 compared with ATF6. Activation of transcription from this element does not require the assistance of NF-Y (Yamamoto et al., 2004). The third element, ERSE-II, is very similar to ERSE except that it is separated by a single nucleotide spacer and the CCAAT and CCACG sites are in the opposite orientation, resulting in the sequence ATTGG-N-CCACG. ERSE-II binds ATF6 in an NF-Y-dependent fashion, while XBP1 is bound in an NF-Y-independent fashion (Yamamoto et al., 2004). ERSE regulates the expressions of ER-localized molecular chaperones such as BiP in order to refold unfolded proteins in the ER. UPRE may primarily regulate the expressions of components of the ERAD system in order to degrade unfolded proteins in the ER. However, except for Herp, the target genes mediated by ERSE-II remain unclear (Yamamoto et al., 2004; Renna et al., 2007).
3.2. BiP/GRP78-dependent UPR regulation mechanisms
The interaction between BiP/GRP78 and the three UPR sensors is also regulated by several proteins. The first is ER-localized cyclophilin B (CypB), which interacts with BiP and stabilizes the BiP/UPR sensors complex via its peptidyl-prolyl cis-trans isomerase (PPIase) activity (Kim et al., 2008). The second is ERdj4, which stabilizes the BiP/UPR sensor complex by inhibiting the connection between the nucleotide-binding domain (NBD) and the unfolded/misfolded protein substrate (Shen et al., 2002; Awad et al., 2008; Chen et al., 2014a). Cab45S, the third regulative protein, stabilizes BiP/IRE1 interactions, inhibiting ER stress-induced IRE1-c-Jun N-terminal kinase (JNK) signaling by specifically interacting with the NBD of BiP/GRP78 (Chen et al., 2014a). Finally, BAP/Sil1 dissociates the BiP/three sensors complex by directly interacting with GRP78/BiP (Chung et al., 2002; Chen et al., 2014a). At the same time, the activity of GRP78/BiP is also regulated by mono-ADP-ribosylation by arginine-specific ecto-enzymes (ARTCs) on two arginine residues (R470 and R492) in UPR, which interferes with BiP to bind with the three sensors and augment the UPR signals (Fabrizio et al., 2014). The expressions of all of these interacting proteins are induced through UPR signaling pathways to regulate cooperatively and balance protein processing with the demands of protein synthesis.
3.3. BiP/GRP78-independent UPR activation mechanisms
In addition to ER stress, ATF6 and PERK are also activated by vascular endothelial growth factor (VEGF)-VEGF receptor (VEGFR)-phospholipase C-γ (PLCγ)-mTOR complex 1 (mTORC1) signaling in tumors (Fig. 1: C1–C3). Solid tumors exist in hypoxic environments and rely on adaptive signaling pathways such as hypoxia-inducible factor 1α (HIF-1α), UPR, and macroautophagy to maintain proteostasis and energy balance. VEGF, induced by HIF-1α, XBP1, ATF6, and ATF4, is the most important proangiogenic driver secreted by an autocrine (tumor cell) and paracrine (endothelial cell) effect. VEGFR interacting with VEGF induces PLCγ activation and subsequent mTORC1 phosphorylation, which activates ATF6 and PERK, but not IRE1, as a result, further activating UPR and mTORC2 (Fig. 1: B2-1-1, E3-1). The dissociation of BiP from UPR sensors is not necessary for the activation of VEGF-VEGFR-PLCγ-mTORC1 pathway in tumors (Urra and Hetz, 2014). VEGF interacting with its receptor also induces phosphoinositide-dependent kinase 1 (PDK1)-dependent RAC-alpha serine/threonine-protein kinase (AKT) phosphorylation at Thr308 and mTORC2-dependent phosphorylation at Ser473, which functionally impacts endothelial cell survival and angiogenesis (Urra and Hetz, 2014). However, ER stress leads to Rictor phosphorylation at S1235 via glycogen synthase kinase 3β (GSK-3β), which interferes with AKT/mTORC2 binding and subsequent AKT Ser473 phosphorylation (Karali et al., 2014) (Fig. 1: D1–D3).
Furthermore, in the absence of an ER stress response, glucagon markedly increases hepatic IRE1α phosphorylation (Ser724) via protein kinase A (PKA) activation leading to the full activation of AKT (Ser473) and other downstream effectors, such as forkhead box O1 (FOXO1), but not tuberous sclerosis 2 (TSC2) (Karali et al., 2014).
4. PERK, ATF6, and IRE1α signaling and regulatory mechanisms
4.1. PERK signaling and regulatory mechanisms
PERK dimerization and trans-autophosphorylation lead to the activation of eIF2α by phosphorylation at Ser51 (first 3 h). As a result, the phosphorylated eIF2α (eIF2α(P)) prevents the recycling of the eIF2-bound guanosine diphosphate (GDP) to guanosine triphosphate (GTP) and the subsequent formation of the eIF2-GTP-tRNA-Met ternary complex (Sonenberg and Hinnebusch, 2009). eIF2α phosphorylation consequently turns off the synthesis of a large proportion of proteins, but selectively increases the expressions of other proteins that are involved in transcription regulation, oxidative stress, amino acid synthesis, protein folding, and differentiation apoptosis. These mRNAs have an internal ribosome entry site (IRES) sequence in the 5'-untranslated region (Muaddi et al., 2010; Malhotra and Kaufman, 2011; Chaudhari et al., 2014) (Fig. 1: B1–B5).
In addition to PERK, there are three other eIF2α kinases, including double-stranded RNA-activated protein kinase (PKR), general control non-derepressible kinase 2 (GCN2), and heme-regulated inhibitor kinase (HRI), that also phosphorylate eIF2α at Ser51. PKR is activated by ER stress thereby initiating inflammatory response signaling. Along with PERK, it cytoprotectively functions through the activating transcription factor 4 (ATF4)-C/EBP homologous protein (CHOP) pathway in response to a glucose deficiency (Fig. 1: B6–B8). HRI is activated by an iron or heme deficiency as well as oxidative stress. GCN2 is activated by uncharged tRNA caused by an amino acid deficiency and functions proapoptotically through the induction of expression of the X-linked inhibitor of apoptosis protein (XIAP) (Muaddi et al., 2010). However, GADD34, P58IPK, and CReP can promote eIF2α dephosphorylation and function as a negative feedback mechanism of eIF2α phosphorylation (Harding et al., 2009).
The PERK signal cascade includes several primary protein components, such as eIF2α, transducin (β)-like 2 (TBL2), nuclear factor erythroid-derived 2-like 2 (Nrf2), ATF4, CHOP, and kelch-like ECH-associated protein 1 (KEAP1) (Fig. 1: B2-2-1–B2-2-3). TBL2 is an ER-localized type-I transmembrane protein that preferentially binds to p-PERK, but not GCN2 or IRE1. TBL2 serves as a potential regulator of the PERK pathway by interacting with p-PERK via its N-terminus proximal region and with eIF2α via its WD40 domain, which is necessary for phosphorylating eIF2α and ER stress-induced ATF4 expression (Tsukumo et al., 2014). KEAP1 functions as a cytoskeletal anchor to retain Nrf2 in the cytosol of unstressed cells. When PERK is activated, Nrf2/KEAP1 complexes dissociate following Nrf2 phosphorylation by PERK, thereby allowing Nrf2 nuclear translocation and the expressions of antioxidant and detoxification enzymes (Cullinan et al., 2003; Chaudhari et al., 2014). Moreover, ATF4 and Nrf2 have been reported to dimerize and regulate oxidative stress responses (Nagelkerke et al., 2014). Heterogeneous nuclear ribonucleoprotein (hnRNP) A1, induced by ER stress, attenuates IRES-mediated translation of anti-apoptotic mRNAs, including Bcl-2-like protein 1 (Bcl-xL) (Bevilacqua et al., 2010). However, over-expressed Bcl-2-associated athanogene 5 (Bag5) in prostate cancer inhibits ER stress-induced apoptosis by decreasing CHOP and BCL2-associated X (BAX) while increasing Bcl-2 (Li et al., 2014b).
CHOP consists of two functional domains, an N-terminal transcriptional activation domain and a C-terminal basic-leucine zipper (bZIP) domain composed of a basic amino acid-rich DNA-binding region followed by a leucine zipper dimerization motif (Li et al., 2014b). CHOP induces the transcription of many genes, some of which are involved in negative feedback. For example, GADD34 functions in eIF2α dephosphorylation, tribbles homologue 3 (TRB3) represses ATF and CHOP transcriptional activity, histone deacetylase 4 (HDAC4) directly inhibits ATF4 transcriptional activity, and Atg5 is vital for autophagy (Kouroku et al., 2007; Majumder et al., 2012; Li et al., 2014b; Zhang et al., 2014). Some genes, such as stanniocalcin 2 (STC2), which mediates invasion, are up-regulated by hypoxia-inducible factor 1 (HIF1) which is a downstream gene of ATF4/CHOP in tumors (Nagelkerke et al., 2014). Some of these genes are also involved in apoptosis and inflammation when ER stress and UPR signal activation are prolonged. For example, Bax-Bak mediates mitochondrial membrane permeabilization, the ERO1α-IP3R-calcium-calcium/calmodulin-dependent protein kinase II (CaMKII) pathway, caspase 3 pathway, and c-Jun pathway (Li et al., 2014b; Nagelkerke et al., 2014). Bim, essential for apoptotic induction, is usually phosphorylated by ERK/Rsk1/2, followed by ubiquitylation through the E3-ligase bTRCP. CHOP increases Bim transcription. At the same time, Bim is dephosphorylated by activated protein phosphatase 2A (PP2A), which inhibits Bim-ubiquitylation and proteosomal degradation during ER stress (Häcker, 2014). ER stress can also induce the expressions of interleukin 6 (IL-6) and several chemokines by the activation of the Janus kinase 1 (JAK1)/signal transducer and activator of transcription 3 (STAT3) axis (Meares et al., 2014).
Apoptosis is further promoted by BCL-2 transcriptional suppression and the induction of expression of death receptor 5 (DR5) by eIF2α phosphorylation (Karali et al., 2014). Furthermore, PERK down-regulates X-Linked inhibitor of apoptosis (XIAP) synthesis through p-eIF2α and promotes XIAP degradation through ATF4 (Hiramatsu et al., 2014). eIF2α phosphorylation is both necessary and sufficient to activate NF-κB DNA binding and an NF-κB reporter gene. eIF2α phosphorylation-dependent NF-κB activation correlates with decreased levels of inhibitor of κBα (IκBα), an inhibitory protein, by repression and not by increased phosphorylation or decreased stability. This differs from canonical signaling pathways that promote IκBα phosphorylation and degradation (Deng et al., 2004).
4.2. ATF6 signal cascade and regulatory mechanisms
ATF6 is a type II ER transmembrane protein that contains a bZIP domain in the cytosol and a stress-sensing domain in the ER lumen. During ER stress, ATF6 is released from BiP and translocates to the Golgi compartment where it is sequentially cleaved by site-1 and site-2 proteases (Fig. 1: E1–E4). This causes the release of an N-terminal fragment that translocates to the nucleus as a transcription factor to increase XBP1-, BiP/GRP78-, CHOP-, PDI-, and ERAD-associated protein transcriptions (Zhang et al., 2006; Nagelkerke et al., 2014). The activation of ATF6α can also drive lipid biosynthesis and ER expansion (Bommiasamy et al., 2009).
Endoplasmic reticulum protein 29 (ERp29), belonging to a redox-inactive PDI-Db-subfamily of PDI proteins, acts as an escort factor for ATF6 (Hirsch et al., 2014). Various auxiliary proteins also play an important role in ATF6α nuclear translocation and its tissue specificity function. For example, ATF6α interacting with CRTC2 antagonizes the ability of cyclic adenosine 3,5-monophosphate response element-binding protein (CREB) to activate gluconeogenesis in the liver (Malhotra and Kaufman, 2011), and ATF6α associated with the transcriptional coactivator PGC-1α enables ATF6α to regulate several exercise-associated aspects of skeletal muscle function. PGC-1α is also necessary for efficient recovery from acute exercise (Wu et al., 2011) (Fig. 1: E6, E7).
4.3. IRE1α signal cascade and regulatory mechanisms
4.3.1 Different activation mechanisms of IRE1α
The amino terminus of IRE1α resides in the ER lumen to sense the protein folding status, while the carboxyl terminus resides in the cytosol to initiate a unique signaling through its AKT and endoribonuclease (RNase) activities to induce an unconventional splicing reaction. IRE1α activation is mediated by oligomerization and trans-autophosphorylation leading to a conformational change that fully elicits its RNase activity (Poothong et al., 2010). In addition to IRE1α being activated by disassociation from BiP during ER stress, hepatic IRE1α phosphorylation (Ser724) is markedly increased via PKA activation in the presence of glucagon (Mao and Shao, 2011). Furthermore, Toll-like receptor (TLR) 2/4 mediates IRE1/XBP1 activation through the adaptor proteins TIRAP and MyD88 (TLR2), MyD88 or TRIF (TLR4), and TRAF6 (TLR2/4). Intriguingly, this activation appears to depend on the activity of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase NOX2, thus providing further insight into the functional significance of XBP1 in innate immunity (Kaufman and Cao, 2010).
4.3.2 Synthesis and translocation mechanisms of XBP1
Activated IRE1α initiates the splicing of several kinds of mRNA. The unspliced mRNA (XBP1u) of the transcription factor XBP1 is a primary target, from which 26 nucleotides are excised to form the spliced XBP1 mRNA (XBP1s). The protein encoded by the XBP1u is rapidly degraded, while the protein XBP1 from the spliced variant XBP1s induces ER stress-related gene expression (Majumder et al., 2012). XBP1, whose expression is induced by both XBP1 itself and activated ATF6, is an unstable protein with a half-life of 22 min (Majumder et al., 2012) (Fig. 1: F1–F6). Also, phosphorylation of eIF2α and translational repression during early UPR augment the stabilization of the XBP1s (Majumder et al., 2012). All of these increase the expression of XBP1 during UPR.
There have been few recent insights into the mechanisms of XBP1 nuclear translocation under traditional UPR. However, following growth factor stimulation, XBP1 nuclear translocation is facilitated by p85α, but not p85α-p85β heterodimers, with the increased translocation occurring independently of phosphoinositide-3 kinase (PI3K) catalytic activity (Kaufman and Cao, 2010). In addition to p85α, HIF1α increases XBP1 nuclear translocation by interacting with its N-terminal bZIP domain, leading to augmented VEGF-A, PDK1, glucose transporter type 1 (GLUT1), and DNA damage inducible transcript 4 (DDIT4) expressions under normoxic and hypoxic conditions (Chen et al., 2014b).
4.3.3 Regulatory webs of the IRE1α/XBP1 signal cascade
As in the PERK pathways, several proteins function in limiting IRE1α/XBP1 signaling and maintaining it within a physiologically appropriate range. In the luminal domain of IRE1α, IRE1α activity is limited by the direct binding of PDIA6, a resident ER protein disulfide isomerase, at Cys148, which is oxidized when IRE1α is activated. Furthermore, in the cytoplasmic domains of IRE1α, IRE1α activity is limited cytoplasmically by interaction with BCL-2-related proteins and Bax inhibitor-1 (BI-1), BCL2 binding component 3 (PUMA), or Bcl-2-interacting mediator of cell death (BIM) (Eletto et al., 2014). The RNase activity of IRE1α can be modulated by unique amino acid sequences in subdomains V and VIA within the adenosine diphosphate (ADP)/adenosine triphosphate (ATP) binding domain (Poothong et al., 2010). IRE1α activity can also be regulated by the scaffold protein receptor for activated C-kinase 1 (RCK1), which binds to IRE1α and PP2A and forms the glucose-inducible ternary IRE1α-RCK1-PP2A complex (Fig. 1: F3-1-1–F3-1-3). This association forms the ternary IRE1α-RCK1-PP2A complex, promotes the dephosphorylation of IRE1α by PP2A, and attenuates IRE1α-dependent increases in insulin production (Qiu et al., 2010).
4.3.4 Downstream targets and their functions regulated by the IRE1α/XBP1 signal cascade
The primary function of IRE1α/XBP1 is to promote cell survival by up-regulating genes encoding proteins involved in protein folding, quality control, and ERAD by both transcription factor activity of XBP1 and RNase activity of IRE1α. The cleaved IRE1α also has been reported to function as a transcription factor in mammals when increased presenilin-1 under UPR controls IRE1α proteolysis (Niwa et al., 1999). IRE1β can interact with the transcriptional coactivator, Gcn5p, and recruit a transcription coactivator complex to a specific chromosomal locus to mediate localized histone acetylation, thus making specific gene sequences accessible for transcription (Welihinda et al., 1997).
The IRE1α/XBP1 pathway simultaneously induces the expression of all four mammalian translocon-associated protein (TRAP) complex subunits, which accelerates misfolded protein ERAD by distinguishing them from correctly folded proteins (Nagasawa et al., 2007). Furthermore, the actions of IRE1α can vary depending on the tissue and cell type. In chondrocytes, IRE1α expression is induced by the granulin-epithelin precursor (GEP), a growth factor known to stimulate chondrogenesis. It inhibits GEP-mediated differentiation by reducing collagen II (ColII), sex determining region Y-box 9 (Sox9), collagen X (ColX), matrix metalloproteinase 13 (MMP-13), Indian hedgehog (IHH), and Runx2 expressions, while enhancing parathyroid hormone-related peptide (PTHrP) expression (Guo et al., 2014a). IRE1α-XBP1-PDI signaling also regulates hepatic triglyceride export and the assembly of lipid-rich very low-density lipoprotein (VLDL) particles (Wang et al., 2012). Moreover, in solid tumors, IRE1α up-regulates the expression of VEGF-A (Drogat et al., 2007) and the expression of valosin containing protein (VCP). VCP is involved in protein folding, cell cycle control, and apoptosis as part of ERAD, and regulates ubiquitin-mediated degradation of misfolded proteins (Wang et al., 2014). More significantly, the IRE1 target XBP1 acts as a negative regulator of apoptosis in osteoarthritis by affecting caspase 3, caspase 9, caspase 12, p-JNK1, and CHOP (Guo et al., 2014b).
Apart from cleavage of XBP1 mRNA, some microRNAs (miRs) that repress translation are also cleaved by IRE1α and release translational blocks. For example, they are involved in regulating the expression of caspase 2 (Hassler et al., 2012).
Furthermore, some genes are regulated synergistically by several pathways. For example, PERK signaling induces ATF5 to activate pro-oxidant thioredoxin-interacting protein (Txnip) transcription while IRE1α cleaves miR-17 to stabilize Txnip mRNA (Hassler et al., 2012; Oslowski et al., 2012). A specificity protein 1 (Sp1)-binding site within a GC-rich region of the cationic amino acid transporter-1 (Cat-1) gene controls its basal expression, while ATF4 and XBP1 are required for sustained transcriptional induction of the Cat-1 gene during the UPR (Huang et al., 2010). All these positive and negative feedback mechanisms provide survival signals and inhibit cell death.
4.3.5 Other signal pathways related to IRE1α and IRE1β
In addition to the traditional IRE1α/XBP1 signal cascade, several other cascades can be activated by IRE1α oligomerization, including: (1) ER stress-induced IRE1α phosphorylation leading to tumor necrosis factor receptor-associated factor 2 (TRAF2) and apoptosis signal-regulating kinase 1 (ASK1) recruitment to the cytosolic leaflet of the ER membrane. ASK1 then stimulates JNK phosphorylation, subsequently activating c-Jun and inducing apoptosis (Chen et al., 2014a; Nagelkerke et al., 2014) (Fig. 1: II-1, II-2, II-2-4). (2) Activated JNK interacts with Sab, a mitochondrial JNK binding protein, and consequently impairs respiration, which increases mitochondrial reactive oxygen species (ROS) culminating in apoptosis (Win et al., 2014) (Fig. 1: II-2-1). (3) The activated JNK also phosphorylates the transcription factor activator protein 1 (AP-1) and induces the expressions of inflammatory genes (Chaudhari et al., 2014) (Fig. 1: II-2-2). (4) The TRAF2 associates with IκB kinase (IKK) and activates NF-κB by promoting degradation of IκBα, resulting in NF-κB nuclear translocation (Chaudhari et al., 2014) (Fig. 1: I-1–I-5). (5) IRE1β-mediated ASK1/JNK activation leads to the cleavage and activation of procaspase 12 (in mouse) or procaspase 4 (in humans) to promote cell death (Fabrizio et al., 2014; Wang et al., 2014); furthermore, the IRE1α-TRAF2-ASK1 complex inhibits the Ca2+-binding inositol 1,4,5-triphosphate receptor (InsP3R) from binding its inhibitor CIB1, thus inhibiting ER-to-cytosolic efflux of Ca2+. The severe UPR induces the dissociation of ASK1 from the IRE1α-TRAF2-ASK1 complex and increases the assembly of ASK1/calcium and integrin binding 1 (CIB1) complex. CIB1 dissociates from CIB1/InsP3R and increases the cytosolic concentration of Ca2+, which induces mitochondrial production of ROS and cell apoptosis (Son et al., 2014) (Fig. 1: III-1–III-6). A region of mitochondria-associated membranes (MAMs) connects the ER membrane and mitochondrial membrane by some proteins interacting within the two organelle membranes. Interaction of the sigma-1 receptor with IRE1α and BiP allows the receptors to efficiently modulate cell survival signals from the ER to the mitochondria. The sigma-1 receptor interacts with and stabilizes IRE1α to enhance a prolonged activation of the IRE1α-XBP1 pathway, thus promoting cell survival. When ER calcium is depleted, BiP dissociates from the sigma-1 receptor, enabling it to stabilize InsP3Rs at the MAMs and facilitate calcium signaling to the mitochondria during ER stress conditions (Wang et al., 2014). Progranulin (PGRN) induced by XBP1 binds directly to the 2nd and 3rd cysteine-rich domains (CRDs) in the extracellular portion of TNFR2 and activates ERK1/2 as well as the AKT signaling pathway (Li et al., 2014a).
5. Concluding remarks
5.1. ER stress and apoptosis
Cellular homeostasis acts in opposition to both internal and external stimuli to maintain optimal metabolic and proliferative levels via a series of positive and negative feedback networks. Despite recent improvements in our knowledge of the UPR, many of its underlying mechanisms remain poorly understood. In addition to the conventional UPR activation mechanism involving BiP/ER stress sensors, PERK and ATF6 can also be activated by VEGF-PLCγ-mTORC1 pathway in tumors (Urra and Hetz, 2014), and IRE1α can be activated by activated AKT (Karali et al., 2014). In normal cells, moderate ER stress, functioning in re-establishing cellular homeostasis, is strictly regulated and favors survival, while prolonged UPR activation induces apoptosis (Rutkowski et al., 2006). CHOP increases folding ability as well as the burden of misfolding in the ER by re-establishing protein folding (Malhotra and Kaufman, 2011; Wang et al., 2014). IRE1α also functions as a double-edged sword: on the one hand, protein synthesis is decreased by RIDD and successively protects cells by reducing the protein-folding burden, but on the other hand, the mRNAs that encode pro-survival proteins are also degraded (Hassler et al., 2012).
Apoptosis induced by ER stress is achieved through the expressions of pro-apoptotic proteins (Hassler et al., 2012; Chen et al., 2014a; Lu et al., 2014; Nagelkerke et al., 2014; Wang et al., 2014) and the stabilities of these proteins and their mRNAs, as previously discussed (Häcker, 2014). An additional apoptotic mechanism includes decreasing the translational efficiency of anti-apoptotic proteins, such as hypertonicity-induced eIF2α phosphorylation inducing cytoplasmic hnRNP A1 accumulation, which attenuates IRES-mediated translation of anti-apoptotic mRNAs, including Bcl-xL (Bevilacqua et al., 2010). Another apoptotic mechanism involves increasing the expression of tumor necrosis factor α (TNF-α) via IRE1α-and NF-κB-dependent activation, which is mediated through TNF receptor 1/caspase 8 (Hu et al., 2006). Lastly, apoptosis can occur by augmenting the expression or proteolytic cleavage of caspases. In force-induced apoptosis, PERK, not PKR, GCN2, eIF2α, CHOP, IRE1, or ATF6, activates caspase 3 in a caspase 9-dependent and mitochondria-independent fashion, implying a novel function of PERK that occurs in addition to its canonical UPR role (Mak et al., 2008). Expressions of caspase 2 and subsequent Bid proteolytic cleavage increase, contributing to apoptotic cell death (Hassler et al., 2012). Furthermore, IRE1-mediated ASK1/JNK activation leads to the cleavage and activation of procaspase 12 (in mouse) or procaspase 4 (in humans) to promote cell death (Fabrizio et al., 2014; Wang et al., 2014).
A systemic inflammatory response is another important mechanism resulting in cell death apart from the activation of AP-1 and NF-κB, as described previously (Malhotra and Kaufman, 2011; Chaudhari et al., 2014; Chen et al., 2014a; Nagelkerke et al., 2014). ER stress-induced activation of the JAK1/STAT3 axis leads to the expressions of IL-6 and several chemokines to drive inflammation (Meares et al., 2014). Txnip, induced by ER stress through the PERK and IRE1 pathways, induces IL-1β mRNA transcription, activates IL-1β production by the NLRP3 inflammasome, and mediates ER stress-mediated β cell death (Hassler et al., 2012; Oslowski et al., 2012).
ROS and Ca2+ are two important apoptosis executors. ROS, harmful at high concentrations while beneficial at moderate/low concentration, increase in response to ER stresses (Chaudhari et al., 2014). The formation of disulfide in the ER is believed to contribute to 25% of the ROS generated by the cell (Chaudhari et al., 2014). Furthermore, mitochondrial ROS are increased by JNK/Sab complex activation when ER stress activates the JNK (Win et al., 2014). CHOP induces endoplasmic reticulum oxidoreductase 1α (ERO1α), which hyperoxidizes the ER environment and further commits the cell to apoptosis (Wang et al., 2014).
The ER, mitochondria and nucleus are the main intracellular Ca2+ stores, with the ER being able to store up to 10–100 mmol/L Ca2+ (versus 100–300 nmol/L in the cytoplasm). InsP3Rs and ryanodine receptors on the ER membrane facilitate the transfer of Ca2+ from the lumen of the ER to the cytoplasm, and Ca2+ is taken up again by the sarco-endoplasmic reticulum Ca2+-ATPases (SERCAs) also located in the ER membrane. However, Ca2+ uptake into the mitochondria is controlled by the voltage-dependent anion channel (VDAC) residing at the outer mitochondrial membrane interface and the Ca2+ uniporter residing at the inner mitochondrial membrane interface, while mitochondrial Ca2+ is expelled by antiporters in an exchange process for either Na+ or H+. Thus, the antiporter and the exchanger maintain mitochondrial membrane potential and optimal Ca2+ concentrations in the mitochondria (Malhotra and Kaufman, 2011). Furthermore, InsP3Rs are primarily clustered in the MAM regions and serve as primary subcellular microdomains of Ca2+ transfer from the ER to the mitochondria (Son et al., 2014). ER stress promotes the association of the CIB1/InsP3R complex, which contributes to the increase in cytosolic Ca2+ concentration (Hu et al., 2006; Son et al., 2014). Bak and Bax, which localize to both the ER and mitochondria, have been shown to be associated with ER calcium release concomitant with increased mitochondrial calcium (Malhotra and Kaufman, 2011). ER stress also up-regulates FBXO6, which inhibits cadmium-induced ER stress and JNK1 activation (Du et al., 2014). Pathological conditions accompanied by a prolonged increase in calcium levels can overwhelm the calcium regulatory network, making it difficult for cells to recuperate. A disruption in Ca2+ homeostasis can trigger apoptosis through the activation of ER resident caspases or mitochondrial dysfunction due to Ca2+ overload (Son et al., 2014).
5.2. Persistent ER stress and cancer
To re-establish cellular homeostasis, UPRs are activated at different levels. They also serve as apoptotic inducers that denote normal cells destined for death during persistent ER stress. However, the precise regulatory mechanisms remain unclear. Surprisingly, cancer cells have evolved ways to adapt to unfavorable microenvironments consisting of low pH, oxygen, glucose, or other nutrient levels, in which a persistent UPR is necessary for tumor survival. These cell fate differences between normal cells and cancer cells in response to a persistent UPR can be attributed to differing UPR molecular mechanisms in tumor versus normal cells. For example, in normal hypoxic/ischemic retinal ganglion neurons, activated IRE1α degrades the classical guidance cue netrin-1, subsequently hindering vascular regeneration (Binet et al., 2013), while in paralleled tumors IRE1α induces HIF-1α expression, which induces VEGF expression and vascular regeneration (Urra and Hetz, 2014). Several reviews have concluded that ER stress and UPR activation stimulate tumor angiogenesis (Nagelkerke et al., 2014; Wang and Kaufman, 2014; Wang et al., 2014; Yadav et al., 2014). The relation between tumorigenesis and persistent ER stress still remains unclear. However, it can be reasonably hypothesized that cells have a higher probability of cancerization due to abnormal activation of signal cascades, mutations in vital signaling proteins, a persistent inflammation response, or altered gene expression under persistent ER stress. For instance, chaperones and oxidoreductases promote tumorigenesis when they are abnormally translocated on the plasma membrane during a persistent stress response (Malhotra and Kaufman, 2007). As a consequence of disrupted homeostasis, the probability of cancerization and disease is increased.
Footnotes
Project supported by the National Basic Research Program (973) of China (No. 2012CB518900), the National Natural Science Foundation of China (Nos. 31160240 and 31260621), the National Major Scientific and Technological Special Project during the Twelfth Five-year Plan Period of China (No. 2012ZX10002006), the Hangzhou Normal University Supporting Project (No. PE13002004042), and the Natural Science Foundation of Jiangxi Province (No. 20114BAB204016), China
Compliance with ethics guidelines: Jing GONG, Xing-zhi WANG, Tao WANG, Jiao-jiao CHEN, Xiao-yuan XIE, Hui HU, Fang YU, Hui-lin LIU, Xing-yan JIANG, and Han-dong FAN declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Awad W, Estrada I, Shen Y, et al. BiP mutants that are unable to interact with endoplasmic reticulum Dnaj proteins provide insights into interdomain interactions in BiP. PNAS. 2008;105(4):1164–1169. doi: 10.1073/pnas.0702132105. (Available from: http://dx.doi.org/10.1073/pnas.0702132105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bevilacqua E, Wang X, Majumder M, et al. eIF2α phosphorylation tips the balance to apoptosis during osmotic stress. J Biol Chem. 2010;285(22):17098–17111. doi: 10.1074/jbc.M110.109439. (Available from: http://dx.doi.org/10.1074/jbc.M110.109439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binet F, Mawambo G, Sitaras N, et al. Neuronal ER stress impedes myeloid-cell-induced vascular regeneration through IRE1α degradation of netrin-1. Cell Metab. 2013;17(3):353–371. doi: 10.1016/j.cmet.2013.02.003. (Available from: http://dx.doi.org/10.1016/j.cmet.2013.02.003) [DOI] [PubMed] [Google Scholar]
- 4.Bommiasamy H, Back SH, Fagone P, et al. ATF6α induces XBP1-independent expansion of the endoplasmic reticulum. J Cell Sci. 2009;122(10):1626–1636. doi: 10.1242/jcs.045625. (Available from: http://dx.doi.org/10.1242/jcs.045625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braakman I, Bulleid NJ. Protein folding and modification in the mammalian endoplasmic reticulum. Annu Rev Biochem. 2011;80(1):71–99. doi: 10.1146/annurev-biochem-062209-093836. (Available from: http://dx.doi.org/10.1146/annurev-biochem-062209-093836) [DOI] [PubMed] [Google Scholar]
- 6.Cao SS, Zimmermann EM, Chuang BM, et al. The unfolded protein response and chemical chaperones reduce protein misfolding and colitis in mice. Gastroenterology. 2013;144(5):S–989. doi: 10.1053/j.gastro.2013.01.023. (Available from: http://dx.doi.org/10.1053/j.gastro.2013.01.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudhari N, Talwar P, Parimisetty A, et al. A molecular web: endoplasmic reticulum stress, inflammation, and oxidative stress. Front Cell Neurosci. 2014;8:213. doi: 10.3389/fncel.2014.00213. (Available from: http://dx.doi.org/10.3389/fncel.2014.00213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Xu S, Liu L, et al. Cab45S inhibits the ER stress-induced IRE1-JNK pathway and apoptosis via GRP78/BiP. Cell Death Dis. 2014;5(5):e1219. doi: 10.1038/cddis.2014.193. (Available from: http://dx.doi.org/10.1038/cddis.2014.193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Iliopoulos D, Zhang Q, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 2014;508(7494):103–107. doi: 10.1038/nature13119. (Available from: http://dx.doi.org/10.1038/nature13119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung KT, Shen Y, Hendershot LM. Bap, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J Biol Chem. 2002;277(49):47557–47563. doi: 10.1074/jbc.M208377200. (Available from: http://dx.doi.org/10.1074/jbc.M208377200) [DOI] [PubMed] [Google Scholar]
- 11.Cullinan SB, Zhang D, Hannink M, et al. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol. 2003;23(20):7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. (Available from: http://dx.doi.org/10.1128/MCB.23.20.7198-7209.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng J, Lu PD, Zhang Y, et al. Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2. Mol Cell Biol. 2004;24(23):10161–10168. doi: 10.1128/MCB.24.23.10161-10168.2004. (Available from: http://dx.doi.org/10.1128/MCB.24.23.10161-10168.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drogat B, Auguste P, Nguyen DT, et al. IRE1 signaling is essential for ischemia-induced vascular endothelial growth factor-A expression and contributes to angiogenesis and tumor growth in vivo. Cancer Res. 2007;67(14):6700–6707. doi: 10.1158/0008-5472.CAN-06-3235. (Available from: http://dx.doi.org/10.1158/0008-5472.CAN-06-3235) [DOI] [PubMed] [Google Scholar]
- 14.Du K, Takahashi T, Kuge S, et al. FBXO6 attenuates cadmium toxicity in HEK293 cells by inhibiting ER stress and JNK activation. J Toxicol Sci. 2014;39(6):861–866. doi: 10.2131/jts.39.861. (Available from: http://dx.doi.org/10.2131/jts.39.861) [DOI] [PubMed] [Google Scholar]
- 15.DuRose JB, Scheuner D, Kaufman RJ, et al. Phosphorylation of eukaryotic translation initiation factor 2α coordinates rRNA transcription and translation inhibition during endoplasmic reticulum stress. Mol Cell Biol. 2009;29(15):4295–4307. doi: 10.1128/MCB.00260-09. (Available from: http://dx.doi.org/10.1128/MCB.00260-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberle AB, Lykke-Andersen S, Mühlemann O, et al. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16(1):49–55. doi: 10.1038/nsmb.1530. (Available from: http://dx.doi.org/10.1038/nsmb.1530) [DOI] [PubMed] [Google Scholar]
- 17.Eletto D, Eletto D, Dersh D, et al. Protein disulfide isomerase A6 controls the decay of IRE1α signaling via disulfide-dependent association. Mol Cell. 2014;53(4):562–576. doi: 10.1016/j.molcel.2014.01.004. (Available from: http://dx.doi.org/10.1016/j.molcel.2014.01.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabrizio G, Di Paola S, Stilla A, et al. ARTC1-mediated ADP-ribosylation of GRP78/BiP: a new player in endoplasmic-reticulum stress responses. Cell Mol Life Sci. 2014;72(6):1209–1225. doi: 10.1007/s00018-014-1745-6. (Available from: http://dx.doi.org/10.1007/s00018-014-1745-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farhan H, Weiss M, Tani K, et al. Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. EMBO J. 2008;27(15):2043–2054. doi: 10.1038/emboj.2008.136. (Available from: http://dx.doi.org/10.1038/emboj.2008.136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo FJ, Jiang R, Li X, et al. Regulation of chondrocyte differentiation by IRE1α depends on its enzymatic activity. Cell Signal. 2014;26(9):1998–2007. doi: 10.1016/j.cellsig.2014.05.008. (Available from: http://dx.doi.org/10.1016/j.cellsig.2014.05.008) [DOI] [PubMed] [Google Scholar]
- 21.Guo FJ, Xiong Z, Lu X, et al. ATF6 upregulates XBP1s and inhibits ER stress-mediated apoptosis in osteoarthritis cartilage. Cell Signal. 2014;26(2):332–342. doi: 10.1016/j.cellsig.2013.11.018. (Available from: http://dx.doi.org/10.1016/j.cellsig.2013.11.018) [DOI] [PubMed] [Google Scholar]
- 22.Häcker G. ER-stress and apoptosis: molecular mechanisms and potential relevance in infection. Microbes Infect. 2014;16(10):805–810. doi: 10.1016/j.micinf.2014.08.009. (Available from: http://dx.doi.org/10.1016/j.micinf.2014.08.009) [DOI] [PubMed] [Google Scholar]
- 23.Harding HP, Zhang Y, Scheuner D, et al. Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 alpha (eIF2α) dephosphorylation in mammalian development. PNAS. 2009;106(6):1832–1837. doi: 10.1073/pnas.0809632106. (Available from: http://dx.doi.org/10.1073/pnas.0809632106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassler J, Cao SS, Kaufman RJ. IRE1, a double-edged sword in pre-miRNA slicing and cell death. Dev Cell. 2012;23(5):921–923. doi: 10.1016/j.devcel.2012.10.025. (Available from: http://dx.doi.org/10.1016/j.devcel.2012.10.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiramatsu N, Messah C, Han J, et al. Translational and posttranslational regulation of XIAP by eIF2α and ATF4 promotes ER stress-induced cell death during the unfolded protein response. Mol Biol Cell. 2014;25(9):1411–1420. doi: 10.1091/mbc.E13-11-0664. (Available from: http://dx.doi.org/10.1091/mbc.E13-11-0664) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirsch I, Weiwad M, Prell E, et al. ERp29 deficiency affects sensitivity to apoptosis via impairment of the ATF6-CHOP pathway of stress response. Apoptosis. 2014;19(5):801–815. doi: 10.1007/s10495-013-0961-0. (Available from: http://dx.doi.org/10.1007/s10495-013-0961-0) [DOI] [PubMed] [Google Scholar]
- 27.Hu P, Han Z, Couvillon AD, et al. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1α-mediated NF-κB activation and down-regulation of TRAF2 expression. Mol Cell Biol. 2006;26(8):3071–3084. doi: 10.1128/MCB.26.8.3071-3084.2006. (Available from: http://dx.doi.org/10.1128/MCB.26.8.3071-3084.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang CC, Li Y, Lopez AB, et al. Temporal regulation of Cat-1 (cationic amino acid transporter-1) gene transcription during endoplasmic reticulum stress. Biochem J. 2010;429(1):215–224. doi: 10.1042/BJ20100286. (Available from: http://dx.doi.org/10.1042/BJ20100286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang J, Sato H, Tang Y, et al. UPF1 association with the CAP-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell. 2010;39(3):396–409. doi: 10.1016/j.molcel.2010.07.004. (Available from: http://dx.doi.org/10.1016/j.molcel.2010.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karali E, Bellou S, Stellas D, et al. VEGF signals through ATF6 and PERK to promote endothelial cell survival and angiogenesis in the absence of ER stress. Mol Cell. 2014;54(4):559–572. doi: 10.1016/j.molcel.2014.03.022. (Available from: http://dx.doi.org/10.1016/j.molcel.2014.03.022) [DOI] [PubMed] [Google Scholar]
- 31.Kaufman RJ. Regulation of mRNA translation by protein folding in the endoplasmic reticulum. Trends Biochem Sci. 2004;29(3):152–158. doi: 10.1016/j.tibs.2004.01.004. (Available from: http://dx.doi.org/10.1016/j.tibs.2004.01.004) [DOI] [PubMed] [Google Scholar]
- 32.Kaufman RJ, Cao S. Inositol-requiring 1/X-box-binding protein 1 is a regulatory hub that links endoplasmic reticulum homeostasis with innate immunity and metabolism. EMBO Mol Med. 2010;2(6):189–192. doi: 10.1002/emmm.201000076. (Available from: http://dx.doi.org/10.1002/emmm.201000076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Choi TG, Ding Y, et al. Overexpressed cyclophilin B suppresses apoptosis associated with ROS and Ca2+ homeostasis after ER stress. J Cell Sci. 2008;121(21):3636–3648. doi: 10.1242/jcs.028654. (Available from: http://dx.doi.org/10.1242/jcs.028654) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouroku Y, Fujita E, Tanida I, et al. ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14(2):230–239. doi: 10.1038/sj.cdd.4401984. (Available from: http://dx.doi.org/10.1038/sj.cdd.4401984) [DOI] [PubMed] [Google Scholar]
- 35.Li M, Liu Y, Xia F, et al. Progranulin is required for proper ER stress response and inhibits ER stress-mediated apoptosis through TNFR2. Cell Signal. 2014;26(7):1539–1548. doi: 10.1016/j.cellsig.2014.03.026. (Available from: http://dx.doi.org/10.1016/j.cellsig.2014.03.026) [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Guo Y, Tang J, et al. New insights into the roles of CHOP-induced apoptosis in ER stress. Acta Biochim Biophys Sin (Shanghai) 2014;46(8):629–640. doi: 10.1093/abbs/gmu048. (Available from: http://dx.doi.org/10.1093/abbs/gmu048) [DOI] [PubMed] [Google Scholar]
- 37.Lu M, Lawrence DA, Marsters S, et al. Cell death. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345(6192):98–101. doi: 10.1126/science.1254312. (Available from: http://dx.doi.org/10.1126/science.1254312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majumder M, Huang C, Snider MD, et al. A novel feedback loop regulates the response to endoplasmic reticulum stress via the cooperation of cytoplasmic splicing and mRNA translation. Mol Cell Biol. 2012;32(5):992–1003. doi: 10.1128/MCB.06665-11. (Available from: http://dx.doi.org/10.1128/MCB.06665-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mak BC, Wang Q, Laschinger C, et al. Novel function of PERK as a mediator of force-induced apoptosis. J Biol Chem. 2008;283(34):23462–23472. doi: 10.1074/jbc.M803194200. (Available from: http://dx.doi.org/10.1074/jbc.M803194200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malhotra JD, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid Redox Signal. 2007;9(12):2277–2293. doi: 10.1089/ars.2007.1782. (Available from: http://dx.doi.org/10.1089/ars.2007.1782) [DOI] [PubMed] [Google Scholar]
- 41.Malhotra JD, Kaufman RJ. ER stress and its functional link to mitochondria: role in cell survival and death. Cold Spring Harb Perspect Biol. 2011;3(9):a004424. doi: 10.1101/cshperspect.a004424. (Available from: http://dx.doi.org/10.1101/cshperspect.a004424) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao T, Shao M, Qiu Y, et al. PKA phosphorylation couples hepatic inositol-requiring enzyme 1α to glucagon signaling in glucose metabolism. PNAS. 2011;108(38):15852–15857. doi: 10.1073/pnas.1107394108. (Available from: http://dx.doi.org/10.1073/pnas.1107394108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurel M, Chevet E, Tavernier J, et al. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci. 2014;39(5):245–254. doi: 10.1016/j.tibs.2014.02.008. (Available from: http://dx.doi.org/10.1016/j.tibs.2014.02.008) [DOI] [PubMed] [Google Scholar]
- 44.Meares GP, Liu Y, Rajbhandari R, et al. PERK-dependent activation of JAK1 and STAT3 contributes to endoplasmic reticulum stress-induced inflammation. Mol Cell Biol. 2014;34(20):3911–3925. doi: 10.1128/MCB.00980-14. (Available from: http://dx.doi.org/10.1128/MCB.00980-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muaddi H, Majumder M, Peidis P, et al. Phosphorylation of eIF2α at serine 51 is an important determinant of cell survival and adaptation to glucose deficiency. Mol Biol Cell. 2010;21(18):3220–3231. doi: 10.1091/mbc.E10-01-0023. (Available from: http://dx.doi.org/10.1091/mbc.E10-01-0023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mühlemann O, Lykke-Andersen J. How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol. 2010;7(1):28–32. doi: 10.4161/rna.7.1.10578. (Available from: http://dx.doi.org/10.4161/rna.7.1.10578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagasawa K, Higashi T, Hosokawa N, et al. Simultaneous induction of the four subunits of the TRAP complex by ER stress accelerates ER degradation. EMBO Rep. 2007;8(5):483–489. doi: 10.1038/sj.embor.7400933. (Available from: http://dx.doi.org/10.1038/sj.embor.7400933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagelkerke A, Bussink J, Sweep FC, et al. The unfolded protein response as a target for cancer therapy. Biochim Biophys Acta. 2014;1846(2):277–284. doi: 10.1016/j.bbcan.2014.07.006. (Available from: http://dx.doi.org/10.1016/j.bbcan.2014.07.006) [DOI] [PubMed] [Google Scholar]
- 49.Niwa M, Sidrauski C, Kaufman RJ, et al. A role for presenilin-1 in nuclear accumulation of IRE1 fragments and induction of the mammalian unfolded protein response. Cell. 1999;99(7):691–702. doi: 10.1016/s0092-8674(00)81667-0. (Available from: http://dx.doi.org/10.1016/S0092-8674(00)81667-0) [DOI] [PubMed] [Google Scholar]
- 50.Oslowski CM, Hara T, O'sullivan-Murphy B, et al. Thioredoxin-interacting protein mediates ER stress-induced β cell death through initiation of the inflammasome. Cell Metab. 2012;16(2):265–273. doi: 10.1016/j.cmet.2012.07.005. (Available from: http://dx.doi.org/10.1016/j.cmet.2012.07.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poothong J, Sopha P, Kaufman RJ, et al. Domain compatibility in IRE1 kinase is critical for the unfolded protein response. FEBS Lett. 2010;584(14):3203–3208. doi: 10.1016/j.febslet.2010.06.003. (Available from: http://dx.doi.org/10.1016/j.febslet.2010.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu Y, Mao T, Zhang Y, et al. A crucial role for RACK1 in the regulation of glucose-stimulated IRE1α activation in pancreatic β cells. Sci Signal. 2010;3(106):ra7. doi: 10.1126/scisignal.2000514. (Available from: http://dx.doi.org/10.1126/scisignal.2000514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Renna M, Caporaso MG, Bonatti S, et al. Regulation of ERGIC-53 gene transcription in response to endoplasmic reticulum stress. J Biol Chem. 2007;282(31):22499–22512. doi: 10.1074/jbc.M703778200. (Available from: http://dx.doi.org/10.1074/jbc.M703778200) [DOI] [PubMed] [Google Scholar]
- 54.Rutkowski DT, Kaufman RJ. That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem Sci. 2007;32(10):469–476. doi: 10.1016/j.tibs.2007.09.003. (Available from: http://dx.doi.org/10.1016/j.tibs.2007.09.003) [DOI] [PubMed] [Google Scholar]
- 55.Rutkowski DT, Arnold SM, Miller CN, et al. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4(11):e374. doi: 10.1371/journal.pbio.0040374. (Available from: http://dx.doi.org/10.1371/journal.pbio.0040374) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakaki K, Yoshina S, Shen X, et al. RNA surveillance is required for endoplasmic reticulum homeostasis. PNAS. 2012;109(21):8079–8084. doi: 10.1073/pnas.1110589109. (Available from: http://dx.doi.org/10.1073/pnas.1110589109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen X, Zhang K, Kaufman RJ. The unfolded protein response–a stress signaling pathway of the endoplasmic reticulum. J Chem Neuroanat. 2004;28(1-2):79–92. doi: 10.1016/j.jchemneu.2004.02.006. (Available from: http://dx.doi.org/10.1016/j.jchemneu.2004.02.006) [DOI] [PubMed] [Google Scholar]
- 58.Shen Y, Meunier L, Hendershot LM. Identification and characterization of a novel endoplasmic reticulum (ER) Dnaj homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J Biol Chem. 2002;277(18):15947–15956. doi: 10.1074/jbc.M112214200. (Available from: http://dx.doi.org/10.1074/jbc.M112214200) [DOI] [PubMed] [Google Scholar]
- 59.Son SM, Byun J, Roh SE, et al. Reduced IRE1α mediates apoptotic cell death by disrupting calcium homeostasis via the INSP3 receptor. Cell Death Dis. 2014;5(4):e1188. doi: 10.1038/cddis.2014.129. (Available from: http://dx.doi.org/10.1038/cddis.2014.129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. (Available from: http://dx.doi.org/10.1016/j.cell.2009.01.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsukumo Y, Tsukahara S, Furuno A, et al. TBL2 is a novel PERK-binding protein that modulates stress-signaling and cell survival during endoplasmic reticulum stress. PLoS ONE. 2014;9(11):e112761. doi: 10.1371/journal.pone.0112761. (Available from: http://dx.doi.org/10.1371/journal.pone.0112761) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Urra H, Hetz C. A novel ER stress-independent function of the UPR in angiogenesis. Mol Cell. 2014;54(4):542–544. doi: 10.1016/j.molcel.2014.05.013. (Available from: http://dx.doi.org/10.1016/j.molcel.2014.05.013) [DOI] [PubMed] [Google Scholar]
- 63.Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer. 2014;14(9):581–597. doi: 10.1038/nrc3800. (Available from: http://dx.doi.org/10.1038/nrc3800) [DOI] [PubMed] [Google Scholar]
- 64.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J Cell Biol. 2012;197(7):857–867. doi: 10.1083/jcb.201110131. (Available from: http://dx.doi.org/10.1083/jcb.201110131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang S, Chen Z, Lam V, et al. IRE1α-XBP1s induces PDI expression to increase MTP activity for hepatic VLDL assembly and lipid homeostasis. Cell Metab. 2012;16(4):473–486. doi: 10.1016/j.cmet.2012.09.003. (Available from: http://dx.doi.org/10.1016/j.cmet.2012.09.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang WA, Groenendyk J, Michalak M. Endoplasmic reticulum stress associated responses in cancer. BBA-Mol Cell Res. 2014;1843(10):2143–2149. doi: 10.1016/j.bbamcr.2014.01.012. (Available from: http://dx.doi.org/10.1016/j.bbamcr.2014.01.012) [DOI] [PubMed] [Google Scholar]
- 67.Welihinda AA, Tirasophon W, Green SR, et al. Gene induction in response to unfolded protein in the endoplasmic reticulum is mediated through Ire1p kinase interaction with a transcriptional coactivator complex containing Ada5p. PNAS. 1997;94(9):4289–4294. doi: 10.1073/pnas.94.9.4289. (Available from: http://dx.doi.org/10.1073/pnas.94.9.4289) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Win S, Than TA, Fernandez-Checa JC, et al. JNK interaction with Sab mediates ER stress induced inhibition of mitochondrial respiration and cell death. Cell Death Dis. 2014;5(1):e989. doi: 10.1038/cddis.2013.522. (Available from: http://dx.doi.org/10.1038/cddis.2013.522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu J, Ruas JL, Estall JL, et al. The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1α/ATF6α complex. Cell Metab. 2011;13(2):160–169. doi: 10.1016/j.cmet.2011.01.003. (Available from: http://dx.doi.org/10.1016/j.cmet.2011.01.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yadav RK, Chae SW, Kim HR, et al. Endoplasmic reticulum stress and cancer. J Cancer Prev. 2014;19(2):75–88. doi: 10.15430/JCP.2014.19.2.75. (Available from: http://dx.doi.org/10.15430/JCP.2014.19.2.75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamamoto K, Yoshida H, Kokame K, et al. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem. 2004;136(3):343–350. doi: 10.1093/jb/mvh122. (Available from: http://dx.doi.org/10.1093/jb/mvh122) [DOI] [PubMed] [Google Scholar]
- 72.Zhang K, Shen X, Wu J, et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124(3):587–599. doi: 10.1016/j.cell.2005.11.040. (Available from: http://dx.doi.org/10.1016/j.cell.2005.11.040) [DOI] [PubMed] [Google Scholar]
- 73.Zhang P, Sun Q, Zhao C, et al. 2014. [Google Scholar]
- 74.Zhu JJ, Chai XL, Zhang YS. Endoplasmic reticulum stress and vascular endothelial injury in type 2 diabetes mellitus. Progress Physiol Sci. 2014;45(1):72–74. (in Chinese) [PubMed] [Google Scholar]