Abstract
Regeneration and tumorigenesis share common molecular pathways, nevertheless the outcome of regeneration is life, whereas tumorigenesis leads to death. Although the process of regeneration is strictly controlled, malignant transformation is unrestrained. In this review, we discuss the involvement of TP53, the major tumor-suppressor gene, in the regeneration process. We point to the role of p53 as coordinator assuring that regeneration will not shift to carcinogenesis. The fluctuation in p53 activity during the regeneration process permits a tight control. On one hand, its inhibition at the initial stages allows massive proliferation, on the other its induction at advanced steps of regeneration is essential for preservation of robustness and fidelity of the regeneration process. A better understanding of the role of p53 in regulation of regeneration may open new opportunities for implementation of TP53-based therapies, currently available for cancer patients, in regenerative medicine.
Facts
Molecular pathways and gene expression patterns underlying regeneration and tumorigenesis are akin.
Fluctuations in p53 levels in the regeneration process were observed during salamander limb regeneration, as well as during liver and nerve regeneration in mice.
Following liver regeneration, TP53-deficient mice acquire more chromosomal segregation errors than their p53 wild-type counterparts.
p53 serves as a regeneration coordinator that blocks the shift from regeneration to carcinogenesis.
Open questions
Does proper regeneration restrain malignant transformation or may impaired regeneration lead to carcinogenesis?
What are the roles of tumor suppressors during the regeneration process?
What is the contribution of liver stem/progenitor cells to liver regeneration process? What is the role of TP53 in these cells?
The link between regeneration and cancer
Regeneration is a homeostatic process of renewal that comprises well-coordinated restoration of cells, tissues and organs that have been damaged or lost. Hence, regeneration enables maintenance of structural and functional integrity of the tissue/organ. This is manifested by restoration in states of injury and pathology, as well as by cells turnover under normal physiological conditions. At large, tissue regeneration is characterized by three different overlapping stages; inflammation, tissue reconstruction and remodeling.1 The process of regeneration entails extensive cellular proliferation that is tightly controlled by specific signals, eventually resulting in a finite number of cells. Cellular processes, such as senescence,1 apoptosis2 and differentiation,3 are evoked at different stages of regeneration to ensure controlled expansion. Apoptosis-induced proliferation was also documented in regeneration processes. In this case, damaged or faulty cells undergoing apoptosis are signaling their healthy neighboring cells to proliferate.4 Finally, once the regeneration process is completed, specific signals are released for the termination of the cell proliferation.
Regeneration may be accomplished by several different mechanisms that vary depending on the given organism species, organ type or cell fate (Table 1). For example, in amphibians such as adult newts, regeneration may be mediated by differentiated post-mitotic cells that re-enter the S-phase of the cell cycle and undergo dedifferentiation.5, 6 In planarians, flatworms, the main regeneration mechanism involves proliferation of resident adult somatic stem cells (SCs).7, 8 Similarly, utilization of dedicated SCs to sustain normal cell turnover is evident in mammalian organs such as skin and intestine, which consist of highly proliferative tissues.9 Conversely, it was suggested that quiescent tissues such as liver or pancreas, display alternative regenerative mechanisms involving dormant SCs activation, trans-differentiation, metaplasia and compensatory proliferation of mature cells.10, 11 Importantly, one should bear in mind that activation or formation of SCs should be tightly controlled in order to prevent the acquisition of cancer SCs (CSCs) phenotype (Box 1). SCs and CSCs often share similar regulatory factors that modulate their biological functions.12 Although the regulation of normal SCs division and differentiation remains under physiological control, in CSCs these processes are unleashed.13, 14 The absence of proper regulation leads to asymmetric and uncontrolled divisions, which give rise to a bulk of tumor cells and a CSC with the capability to initiate new tumors.15
Table 1. Cellular sources tangled in regeneration processes of different tissues and organisms.
| Cell type | Process | Regenerating tissue/organism | Reference |
|---|---|---|---|
| Interstitial stem cells | Differentiation to zymogen gland cells | Hydra head | 107 |
| Zymogen gland cells | Trans-differentiation to granular mucous cells | Hydra head | 108 |
| Mesenchymal stem cells/neoblasts | Self-renewal and pluripotent differentiation potential | Lethally irradiated planarians | 109, 110 |
| Liver progenitor cells | Differentiation to hepatocytes | Chronic liver injury in mice | 77, 78 111 |
| Hepatocytes | Proliferation | Partial hepatectomy in mice | 88, 89, 90 111 |
| Cardiomyocytes | Proliferation and differentiation | Damaged heart in zebrafish | 112, 113 |
| Pigmented epithelial cells | Dedifferentiation, proliferation and differentiation to lens cells | Lens regeneration in newt | 114 |
| Syncytial skeletal myotubes | Dedifferentiation to mononucleate cells that are able to proliferate | Appendage regeneration in urodele | 25, 115 |
| Skeletal muscle satellite cells | Activation | Limb regeneration in salamander | 116 |
One of the essential processes underlying tissue regeneration is production of new cells. These new cells can be derived from distinct origins such as amplification and differentiation of resident stem and progenitor cells, proliferation of mature cells, dedifferentiation of cells to a more stem state or trans-differentiation of one cell type to another cell type.106 In the table above, the different cell types involved in specific regeneration processes are listed
Cancer stem cells.
Cancer stem cells (CSCs) are rare quiescent cells within the tumor that possess augmented tumorigenic potential and drug resistance. Alike normal SCs, CSCs are able to self-renew and differentiate. CSCs account for tumor heterogeneity and are able to give rise to a complex tumor bulk following injection into immune-compromised mice. CSCs were found to contribute to various aspects of tumorigenic process including tumor initiation, progression, invasiveness and metastasis. 99
Accumulated data suggested that CSCs may originate from normal SCs that underwent genetic and epigenetic alterations, or alternatively by dedifferentiation of progenitor or mature cells induced by specific signals from the microenvironment. 100
Although wild-type p53 serves as a barrier to CSCs formation regardless of their origin, mutant p53 proteins exhibit their oncogenic gain-of-function by facilitating the acquisition of CSCs phenotype. 44, 101
Moreover, growing experimental evidence indicates that regeneration and tumorigenesis are related processes, whereby dysregulated regeneration process may lead to tumor development.16, 17 It is well known that chronic inflammation and preceding injuries serve as a precondition for tumorigenesis.17 This notion was initially postulated in 1863 by Rudolf Virehow.18 Later, in 1972, Sir Alexander Haddow19 suggested that ‘tumor production is a possible of over healing'. Finally, the association between the term ‘wound' and cancer was proposed by Dvorak20 who stated that tumors are wounds that do not heal. The basis suggested for this statement is that both tissue regeneration restoring wounded organ, as well as carcinogenesis encompass cell proliferation, survival and migration that are regulated by growth factors and cytokines, as well as inflammatory and angiogenic signals. However, in contrast to wound healing, cancer is not self-limiting resulting in uncontrolled cell proliferation, invasion and metastasis.21 Twenty years later, the statement that regeneration and cancer share common features was corroborated by the study of Riss et al.,22 who compared microarray data from a model of renal regeneration and repair (RRR) with gene expression of renal cell carcinoma (RCC). The data analysis suggested that the majority (77%) of the genes expressed in RRR and RCC were concordantly regulated, whereas only 23% were discordant (i.e., changed in opposite directions), thus supporting the hypothesis that cancer is an aberrancy of the physiologic processes of wound healing.
Interestingly, it was suggested that the regeneration process may restrain transformation.23 For example, simultaneous exposure of dorsal and ventral iris of newt to carcinogen showed that the regenerating dorsal iris was persistently resistant to carcinogen, whereas the ventral iris, which cannot regenerate, was more susceptible to tumor induction.24 Furthermore, it was shown that blastemal cells, which are a mass of cells capable of growth and regeneration into organs, are resistant to tumor formation.25 Therefore, it is intriguing to understand the mechanisms that assure the high fidelity of regeneration progression.
Cancer is known to be restricted by the action of tumor suppressors.26 Interestingly, tumor suppressors were proposed to possess ‘regeneration suppressor' activities by which they orchestrate major aspects of the regeneration process and ensure its robustness.21 In this review, we will focus on the role of the pivotal tumor-suppressor TP53 in the regeneration process at large, and on its contribution to the fidelity of the regeneration process, in particular.
The transcription factor p53 – more than a tumor suppressor
TP53 is one of the most important tumor-suppressor genes that is activated via different stress signals and functions to determine cell fate. TP53 is designated as the ‘guardian of the genome' because of its ability to protect cells from DNA damage and thus to prevent tumor development.27 Therefore, it is not surprising that TP53 is mutated in >50% of human tumors. The majority of TP53 mutations rise because of missense substitutions.28 Importantly, most common TP53 mutations do not only abrogate its tumor-suppressor function, rather they confer it with new oncogenic functions.29, 30
It is well known that cellular stress such as DNA damage, oncogene activation, hypoxia and telomere shortening can activate p53 and stabilize its protein levels.31 When activated, the p53 protein functions as a transcriptional regulator, hence initiating a cascade of events that determines the cellular outcome including cell cycle arrest, apoptosis, senescence, DNA repair, development, differentiation and tissue homeostasis.32 Interestingly, all these cellular activities are part of the regeneration process, pointing to p53 as a potential regeneration coordinator. Notably, besides the full-length p53 protein, different p53 isoforms were identified in multiple human tissues and in various animal models such as Drosophila, zebrafish and mouse.33, 34, 35, 36, 37 These p53 isoforms can be generated as a consequence of either alternative splicing, alternative translation initiation or transcription from an alternate promoter.38 Apparently, p53 isoforms are conserved during evolution and involved in various aspects of cell fate decisions.36, 37, 39
Indeed, full-length p53 and its isoforms often have a regulatory role during normal tissue regeneration following injury. Modulation in p53 levels was observed in different stages of the regeneration process, in a context-dependent manner. For example, in the initial stages of the regeneration process, while mitogenic growth factors promoting cell proliferation were highly expressed, a concomitant suppression of p53 was evident. Whereas, at the late stages of healing, at the remission of the regeneration process, the suppression of mitogenic growth factors was accompanied by strong induction of p53 expression, which served to downregulate cellular growth.40 Variations in p53 levels were also found during salamander limb regeneration. It was shown that at first p53 activity decreased, thus allowing both the formation of the blastema and the cell cycle reentry of post-mitotic differentiated cells. Then, p53 level returned to baseline allowing the re-differentiation to muscle. The authors suggested that the regulation of p53 activity is a pivotal mechanism that controls the plasticity of the differentiated state during regeneration.41 The role of TP53 in regeneration-related processes such as proliferation and differentiation is conserved through evolution in additional multicellular organisms such as planarians and Drosophila42, 43 (Box 2).
The p53 duality: p53 can differentially regulate numerous molecular pathways dependent on the cell fate and cellular surroundings.
p53 is termed the ‘guardian of the genome' 27 because of its profound role in preservation of cell genomic fidelity. Following exposure to cellular insults, p53 activity may lead to dual outcomes, such as cell cycle arrest or cell death dependent on various circumstances.
p53 has a dual role in cancer and aging. Although p53 activation blocks cancer development, it promotes aging by and may restrict normal tissue turnover and regeneration. 102
p53 was found to control autophagy by two opposing mechanisms, depending on its cellular localization: the nuclear p53 induces autophagy, whereas, the cytoplasmic p53 may repress it. 103
p53 in the liver is activated following tissue damage and acts as two-edged sword. In the short term, p53 activity prevents carcinogenesis, however, in the long term, same p53 activities may contribute to progress of liver disease, which may eventually lead to cancer development. 104
Drosophila p53 exhibits dual roles in cells death and cell differentiation. On one hand, p53 induces apoptosis via the hid gene, on the other it attenuates the differentiation of the photoreceptor neurons and cone cells in the eye, independently of cell death induction. 105
Taken together, it is conceivable to assume that p53 may function as a coordinator of the regeneration process assuring the quality and quantity of the SCs nourishing the regenerative site, as well as the differentiated cells, at the end of the process. This is in line with the notion that p53 functions as the barrier to CSCs formation,44 by governing the quantity and quality of various SCs, by restricting either expansion or dedifferentiation processes of SCs.
The role of p53 in nerve regeneration
Accomplishing successful regeneration and functional recovery following neural damage, because of either physical injury or pathological conditions, is one of the biggest challenges of neuroscientists and clinicians. Identification of the molecular factors and understanding the molecular pathways is a major step toward achievement of such a goal. The nervous system comprises the peripheral nervous system (PNS) and the central nervous system (CNS) possessing different regenerative capacities. Although the PNS neurons successfully regenerate after damage, the CNS has limited regeneration potential because of the presence of glial inhibitory environment and suppressive intrinsic molecular networks.45, 46, 47
It should be emphasized that impaired tissue regeneration is often associated with aging, carcinogenesis and augmented degenerative disease.48 p53 and its isoforms are associated with development of neurodegenerative diseases such as Parkinson's and Alzheimer's because of its induction of cell death in response to stress and interaction with distinct cellular factors that have the ability to facilitate the development of these diseases.49, 50, 51, 52
Recent accumulating data suggest that p53 has a role in regulation of PNS regeneration. Di Giovanni et al.53 have shown that TP53 null mice exhibited limited nerve regeneration and suggested that this is due to the transcriptional activity mediated by p53, which activates the axon regenerating genes Coronin 1b and Rab13. p53 exhibited dual activity that may lead to diverse outcomes depending on the different stages of the regeneration process. Indeed, it was demonstrated that following neurons injury p53 exerted its pro-apoptotic effect via transactivation of noxa, 54, 55 as well as promoted cell cycle arrest by the MAPK, JNK and p38 signaling pathways.56, 57, 58 However, it was found that p53 enhanced proliferation and axonal outgrowth that are required for axonal regeneration following injury, through other mechanisms.59, 60 Furthermore, ample data indicate that post-translational modifications affect p53 pro-regenerative activity. For instance, acetylation of p53 by CBP/p300 enables its regulation of GAP-43, which expression is essential for axonal outgrowth and regeneration.61, 62, 63
Although the regeneration capacity of the injured CNS is restricted, in some cases it may be mediated via modulation of the neuronal intrinsic potential of neuronal stem/progenitor cell (NSCs/NPCs) populations.64 p53 was suggested to have a role in maintaining self-renewal and differentiation of NSCs under homeostatic conditions. In the absence of p53, the number of differentiated neurons increases.60 Moreover, p53 was also suggested to be critical for the CNS regeneration upon damage.65 Following brain injury, p53 was shown to inhibit Rho kinase activity leading to axonal growth and motility, eventually resulting in axonal regeneration.66 In addition, it was reported that regeneration and axonal sprouting can be attenuated by the MDM4/2-p53-IGF1 signaling complex. Accordingly, inhibition of the triad or one of its components promoted functional recovery after spinal cord injury.67
Taking together, it seems that p53 has a role in both PNS and CNS regeneration upon nerve damage. p53 acts beyond its pro-apoptotic regulation activity and facilitates axonal proliferation and regeneration. Yet, to date, the underlying mechanisms are not fully deciphered and more research is needed to uncover the specific molecular networks. With our growing understanding and the emergence of novel findings, it is conceivable to hope that in the future pharmacological reagents modulating p53 activity, which are currently applied for cancer treatment, may be also implemented in neurodegenerative therapy.
The role of p53 in liver regeneration
One of the primary characteristics of the liver is its ability to regenerate.68 As the liver is the main site of drug detoxification, being constantly subjected to a myriad of toxic chemicals that may induce injury, its remarkable regeneration capacity is of great importance. Moreover, high hepatic regeneration potential permits the use of surgery as a major strategy for treatment of liver diseases including hepatocellular carcinoma (HCC) and liver fibrosis.69 The liver structure is composed of many sub-units named the ‘hepatic lobules'. These well-organized structures contain diverse cell types that reside in different compartments of the lobules. The central part comprises the central veins and contains mainly hepatocytes. The peripheral part named ‘canal of Hering' contains the portal tracts and is primarily populated with the biliary epithelial cells (BECs). In addition, other cells types found in the lobules are fibroblasts, endothelial cells and the macrophage-like cells (Kupper cells).70, 71
Apparently, mechanisms underlying the liver regeneration process vary according to different conditions: the regeneration mechanisms upon normal homeostasis are different from regeneration upon chronic or acute injury.71
The liver is characterized by slow turnover rate and the mechanisms underlying its normal homeostasis maintenance are still debatable. Until recently, the accepted hypothesis has been the ‘streaming liver', proposing that the entire lobule is eventually replaced by sub-population of hepatocytes, which reside near the portal tracts and possess the ability to regenerate and to stream along the lobule.72 However, many studies have refuted this hypothesis as lineage tracing methods have failed to prove it.73, 74 Currently, the prevalent assumption is that normal liver turnover is maintained by pre-existing hepatocytes cells.70 Interestingly, recently Wang et al.75 have identified a small population of cells expressing Axin2+ that possess self-renewal ability as well as the capacity to differentiate into hepatocytes.75 Therefore, this population of cells was suggested to be referred as hepatocyte SCs, which participate in maintaining the normal liver homeostasis.
Importantly, in addition to preservation of normal homeostasis, the liver has the capacity to regenerate following damage. Different regeneration mechanisms are executed upon acute or chronic injury of the liver. Chronic disease of the liver can be triggered by several agents including viral infections, alcohol abuse and nonalcoholic steatohepatitis (NASH). These agents can cause long-term damage that may lead to liver fibrosis and even to liver cirrhosis, which eventually may result in HCC.76 Chronic injuries lead to reduction of hepatocytes proliferative capacity, inducing them to undergo senescence, mediated by p53. Under these conditions, the vast majority of the liver is replaced by hepatic progenitor cells.77, 78 For example, it was demonstrated that upon chronic injury induced by CCl4 treatment, the quiescent hepatocyte stellate cells (HSCs) become activated and regenerate the fibrotic scar. p53 was found to attenuate fibrosis by inducing HSC senescence in non-cell autonomous mechanism. Thus, p53 regulates the fibrosis response and may prevent the deterioration to HCC.77, 79
To date, many studies have been addressing the question whether there are specific SCs in the liver.80 The most prevalent hypothesis refers to the ovals cells as the facultative SCs of the liver.71 It was suggested that these cells arise upon injury and have common characteristics of both hepatocytes and BECs.81 The oval cells reside in a niche inside the canal of Hering in the liver lobule.82 Nevertheless, several reports claimed that the oval cells cannot be regarded as liver SCs as they are incapable of undergoing terminal differentiation into hepatocytes.71, 83 Moreover, it was shown that the oval cells may promote the progression of HCC.84 Considering the profound role of p53 in the life of normal SCs and CSC, 44 it is not surprising that oval cells that were isolated from TP53 null mice and maintained in culture, gave rise to HCC in vivo.85
Accumulating experimental evidence indicate that besides oval cells, other cellular sub-population may also contribute to liver regeneration upon chronic injury. One example is a sub-population of liver cells expressing the Lgr5 marker. These cells, unlike the oval cells, can differentiate to hepatocytes or BECs after in vitro cultivation.86 Another example is the hybrid hepatocytes residing at the periportal region of the lobule that are capable to regenerate after chronic injury without promoting HCC.87 However, the role of p53 in these diverged sub-populations of cells is still unknown and requires further investigation.
Following acute damage such as hepatectomy, the liver is able to restore up to 70% of the tissue resection. The main source of cells that renovate the liver are mature hepatocytes. This process comprises three major phases. Priming – adult hepatocytes re-enter the cell cycle and undergo transition from G0 to the G1 phase. Progression – the cells complete the mitosis process. Termination – the cells return to the G0 phase and the liver retains its original size.88, 89, 90
Numerous proteins are implicated in the regulation of these phases, among them is p53. Of note, at first glance it seemed as if p53 activity is not crucial for liver regeneration following partial hepatectomy, as TP53 null mice exhibited complete liver regeneration.91, 92 Strikingly, it was later discovered that p53 function is essential for controlling the robustness of the regeneration process and its fidelity.93 Similar to the regeneration process in the nerves, modulation in p53 levels was also documented in liver regeneration. In the priming phase, p53 activity is repressed by c-JUN to enable the hepatocytes re-enter the cell cycle.94 Ample data suggest that following liver regeneration hepatocytes are tolerant to nuclear ploidy without gaining tumorigenic potential.95, 96 This phenomenon may be attributed to the presence of functional p53, which is known to protect genome stability.97 Indeed, it was recently demonstrated that TP53-deficient mice acquire more chromosome segregation errors following liver regeneration than their TP53-expressing counterparts. Moreover, it was reported that p53 is involved in controlling the levels of hepatic ploidy by direct regulation of specific target genes, such as Foxm1, Plk2/4, Lats2 and Aurka, at the different phases of the cell cycle.93, 98 Thus, the activity of p53 during the regeneration process following acute damage of the liver is context and time dependent. Despite the inhibition of p53 activity in the initial stage of the regeneration, its function in more advanced steps is essential for keeping the robustness and assuring the fidelity of the process.
All in all, liver regeneration is a complex process that is so far not completely elucidated. It may involve mature hepatocytes as suggested for regeneration upon partial hepatectomy as well as vast sub-population of SCs following other injuries and normal homeostasis turnover. Collectively, it seems that preservation of DNA fidelity and tumor-suppressor activities are crucial to ensure proper regeneration and prevent HCC development.
Concluding remarks
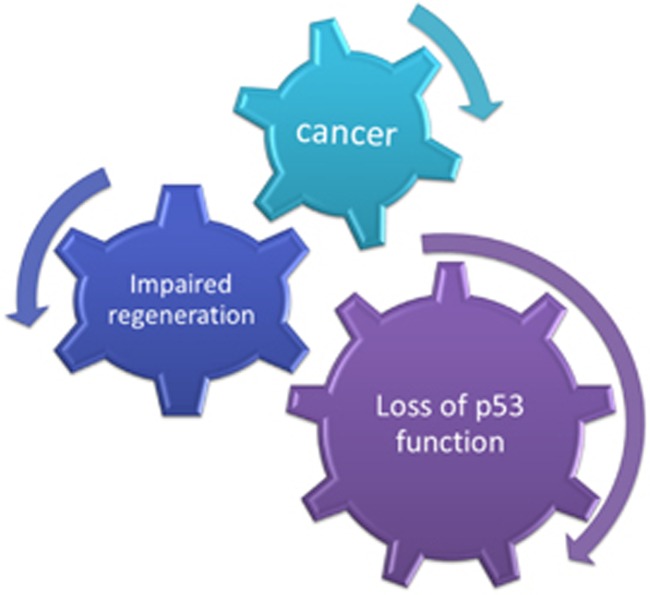
Regeneration and tumorigenesis have been proposed to be related processes and yet the former is a well-orchestrated and controlled process, while the latter is an unrestrained one. As p53 is a major tumor suppressor, it is tempting to speculate that p53 has a key role in regulation of the regeneration process, blocking the shift toward tumorigenesis. p53 activity is fluctuated during regeneration. Although p53 is inhibited during the initial proliferative steps, it is upregulated toward the final stage, when preservation of fidelity and integrity of the regeneration process is of great importance. Activated p53 may induce a variety of signaling pathways such as DNA repair, apoptosis, senescence and others that contribute to the elimination of faulty cells in order to prevent the drifting from regeneration to malignancy. Thus, induction of p53 activity may serve as the quality control checkpoint in the regeneration process, thereby preventing carcinogenesis (Figure 1).
Figure 1.
p53 coordinates proper regeneration process to prevent cancer development. p53 serves as a quality control protein that assures the integrity of the regeneration process. When p53 function is lost, the regeneration process is impaired leading to cancer development
Bearing in mind the great opportunities offered by regenerative medicine to repair pathologic tissues, a better understanding of the regulatory landscape fundamental for p53 function as a coordinator of regeneration may pave the way for overcoming the current challenges.
Acknowledgments
Research in the laboratory of VR is supported by a center of excellence grant from the Israel Science Foundation (ISF) center from the Israeli Academy of Science and a Center of Excellence grant from the Flight Attendant Medical Research Institute (FAMRI). VR is the incumbent of the Norman and Helen Asher Professorial Chair Cancer Research at The Weizmann Institute.
Glossary
- BECs
biliary epithelial cells
- CNS
central nervous system
- CSC
cancer stem cells
- HCC
hepatocellular carcinoma
- HSCs
hepatocyte stellate cells
- NASH
nonalcoholic steatohepatitis
- NPCs
neuronal progenitor cells
- NSCs
neuronal stem cells
- PNS
peripheral nervous system
- RCC
renal cell carcinoma
- RRR
renal regeneration and repair
- SCs
stem cells
The authors declare no conflict of interest.
Footnotes
Edited by RA Knight
References
- Roux I, Wu JS, McIntosh JM, Glowatzki E. Assessment of the expression and role of the alpha1 nAChR subunit in efferent cholinergic function during the development of the mammalian cochlea. J Neurophysiol 2016; 116: 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng AS, Adams DS, Qiu D, Koustubhan P, Levin M. Apoptosis is required during early stages of tail regeneration in Xenopus laevis. Dev Biol 2007; 301: 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Gordon SP, Meyerowitz EM. Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol 2011; 21: 212–218. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A. The role of apoptosis-induced proliferation for regeneration and cancer. Cold Spring Harb Perspect Biol 2012; 4: a008797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odelberg SJ. Inducing cellular dedifferentiation: a potential method for enhancing endogenous regeneration in mammals. Semin Cell Dev Biol 2002; 13: 335–343. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Tanaka EM. Mechanisms of muscle dedifferentiation during regeneration. Semin Cell Dev Biol 2002; 13: 353–360. [DOI] [PubMed] [Google Scholar]
- Agata K, Watanabe K. Molecular and cellular aspects of planarian regeneration. Semin Cell Dev Biol 1999; 10: 377–383. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sanchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol 2004; 20: 725–757. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science 2014; 344: 1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 2013; 14: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JM. Metaplasia and transdifferentiation: from pure biology to the clinic. Nat Rev Mol Cell Biol 2007; 8: 369–378. [DOI] [PubMed] [Google Scholar]
- Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res 2005; 65: 6207–6219. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Shen H, Franklin DS, Scadden DT, Cheng T. In vivo self-renewing divisions of haematopoietic stem cells are increased in the absence of the early G1-phase inhibitor, p18INK4C. Nat Cell Biol 2004; 6: 436–442. [DOI] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 2000; 287: 1804–1808. [DOI] [PubMed] [Google Scholar]
- Alison MR, Islam S, Wright NA. Stem cells in cancer: instigators and propagators? J Cell Sci 2010; 123(Pt 14): 2357–2368. [DOI] [PubMed] [Google Scholar]
- Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature 2004; 432: 324–331. [DOI] [PubMed] [Google Scholar]
- Schafer M, Werner S. Cancer as an overhealing wound: an old hypothesis revisited. Nat Rev Mol Cell Biol 2008; 9: 628–638. [DOI] [PubMed] [Google Scholar]
- Virehow R. Aetiologie der neoplastischen Geschwulste/Pathogenie der neoplastischen Geschwulste. Verlag von August Hirschwald Berlin, Germany 1863.
- Haddow A. Molecular repair, wound healing, and carcinogenesis: tumor production a possible overhealing? Adv Cancer Res 1972; 16: 181–234. [DOI] [PubMed] [Google Scholar]
- Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med 1986; 315: 1650–1659. [DOI] [PubMed] [Google Scholar]
- Pomerantz JH, Blau HM. Tumor suppressors: enhancers or suppressors of regeneration? Development 2013; 140: 2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riss J, Khanna C, Koo S, Chandramouli GV, Yang HH, Hu Y et al. Cancers as wounds that do not heal: differences and similarities between renal regeneration/repair and renal cell carcinoma. Cancer Res 2006; 66: 7216–7224. [DOI] [PubMed] [Google Scholar]
- Brockes JP. Regeneration and cancer. Biochim Biophys Acta 1998; 1377: M1–M11. [DOI] [PubMed] [Google Scholar]
- Okamoto M. Simultaneous demonstration of lens regeneration from dorsal iris and tumour production from ventral iris in the same newt eye after carcinogen administration. Differentiation 1997; 61: 285–292. [DOI] [PubMed] [Google Scholar]
- Lo DC, Allen F, Brockes JP. Reversal of muscle differentiation during urodele limb regeneration. Proc Natl Acad Sci USA 1993; 90: 7230–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Principles of tumor suppression. Cell 2004; 116: 235–246. [DOI] [PubMed] [Google Scholar]
- Lane DP. Cancer. p53, guardian of the genome. Nature 1992; 358: 15–16. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science 1991; 253: 49–53. [DOI] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009; 9: 701–713. [DOI] [PubMed] [Google Scholar]
- Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2010; 2: a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000; 408: 307–310. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer 2002; 2: 594–604. [DOI] [PubMed] [Google Scholar]
- Marcel V, Fernandes K, Terrier O, Lane DP, Bourdon JC. Modulation of p53beta and p53gamma expression by regulating the alternative splicing of TP53 gene modifies cellular response. Cell Death Differ 2014; 21: 1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtel-Danjoy ML, Ma D, Dourlen P, Chatelain G, Napoletano F, Robin M et al. Drosophila p53 isoforms differentially regulate apoptosis and apoptosis-induced proliferation. Cell Death Differ 2013; 20: 108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ng SM, Chang C, Zhang Z, Bourdon JC, Lane DP et al. p53 isoform delta113p53 is a p53 target gene that antagonizes p53 apoptotic activity via BclxL activation in zebrafish. Genes Dev 2009; 23: 278–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon JC. p53 isoforms change p53 paradigm. Mol Cell Oncol 2014; 1: e969136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joruiz SM, Bourdon JC. p53 isoforms: key regulators of the cell fate decision. Cold Spring Harb Perspect Med 2016; 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev 2005; 19: 2122–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewitter E, Scrable H. Delta40p53 controls the switch from pluripotency to differentiation by regulating IGF signaling in ESCs. Genes Dev 2010; 24: 2408–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniades HN, Galanopoulos T, Neville-Golden J, Kiritsy CP, Lynch SE. p53 expression during normal tissue regeneration in response to acute cutaneous injury in swine. J Clin Invest 1994; 93: 2206–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH, Gates PB, Brockes JP. Regulation of p53 is critical for vertebrate limb regeneration. Proc Natl Acad Sci USA 2013; 110: 17392–17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells BS, Yoshida E, Johnston LA. Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr Biol 2006; 16: 1606–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Sanchez Alvarado A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development 2010; 137: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni-Grinstein R, Shetzer Y, Kaufman T, Rotter V. p53: the barrier to cancer stem cell formation. FEBS Lett 2014; 588: 2580–2589. [DOI] [PubMed] [Google Scholar]
- Filbin MT. Myelin-associated inhibitors of axonal regeneration in the adult mammalian CNS. Nat Rev Neurosci 2003; 4: 703–713. [DOI] [PubMed] [Google Scholar]
- Huebner EA, Strittmatter SM. Axon regeneration in the peripheral and central nervous systems. Results Probl Cell Differ 2009; 48: 339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson TA, Son YJ. Extrinsic and intrinsic determinants of nerve regeneration. J Tissue Eng 2011; 2: 2041731411418392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Morrison SJ. Mechanisms that regulate stem cell aging and life span. Cell Stem Cell 2013; 12: 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JR, Ghafouri M, Mukerjee R, Bagashev A, Chabrashvili T, Sawaya BE. Role of p53 in neurodegenerative diseases. Neurodegener Dis 2012; 9: 68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royds JA, Iacopetta B. p53 and disease: when the guardian angel fails. Cell Death Differ 2006; 13: 1017–1026. [DOI] [PubMed] [Google Scholar]
- Pehar M, O'Riordan KJ, Burns-Cusato M, Andrzejewski ME, del Alcazar CG, Burger C et al. Altered longevity-assurance activity of p53:p44 in the mouse causes memory loss, neurodegeneration and premature death. Aging Cell 2010; 9: 174–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnquist C, Horikawa I, Foran E, Major EO, Vojtesek B, Lane DP et al. p53 isoforms regulate astrocyte-mediated neuroprotection and neurodegeneration. Cell Death Differ 2016; 23: 1515–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giovanni S, Knights CD, Rao M, Yakovlev A, Beers J, Catania J et al. The tumor suppressor protein p53 is required for neurite outgrowth and axon regeneration. EMBO J 2006; 25: 4084–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiryu-Seo S, Hirayama T, Kato R, Kiyama H. Noxa is a critical mediator of p53-dependent motor neuron death after nerve injury in adult mouse. J Neurosci 2005; 25: 1442–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs WB, Kaplan DR, Miller FD. The p53 family in nervous system development and disease. J Neurochem 2006; 97: 1571–1584. [DOI] [PubMed] [Google Scholar]
- Song J, Chao C, Xu Y. Ser18 and Ser23 phosphorylation plays synergistic roles in activating p53-dependent neuronal apoptosis. Cell Cycle 2007; 6: 1412–1414. [PubMed] [Google Scholar]
- Hughes AL, Gollapudi L, Sladek TL, Neet KE. Mediation of nerve growth factor-driven cell cycle arrest in PC12 cells by p53. Simultaneous differentiation and proliferation subsequent to p53 functional inactivation. J Biol Chem 2000; 275: 37829–37837. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Mao XO, Sun Y, Xia Z, Greenberg DA. p38 Mitogen-activated protein kinase mediates hypoxic regulation of Mdm2 and p53 in neurons. J Biol Chem 2002; 277: 22909–22914. [DOI] [PubMed] [Google Scholar]
- Kiryu-Seo S, Kiyama H. The nuclear events guiding successful nerve regeneration. Front Mol Neurosci 2011; 4: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A, Di Giovanni S. The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO Rep 2009; 10: 576–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A, Nguyen T, Puttagunta R, Gaub P, Di Giovanni S. A p53-CBP/p300 transcription module is required for GAP-43 expression, axon outgrowth, and regeneration. Cell Death Differ 2009; 16: 543–554. [DOI] [PubMed] [Google Scholar]
- Di Giovanni S, Rathore K. p53-Dependent pathways in neurite outgrowth and axonal regeneration. Cell Tissue Res 2012; 349: 87–95. [DOI] [PubMed] [Google Scholar]
- Gaub P, Joshi Y, Wuttke A, Naumann U, Schnichels S, Heiduschka P et al. The histone acetyltransferase p300 promotes intrinsic axonal regeneration. Brain 2011; 134(Pt 7): 2134–2148. [DOI] [PubMed] [Google Scholar]
- Martino G, Pluchino S, Bonfanti L, Schwartz M. Brain regeneration in physiology and pathology: the immune signature driving therapeutic plasticity of neural stem cells. Physiol Rev 2011; 91: 1281–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano S, Burns-Cusato M, Atienza MB, Rahimi D, Scrable H. Regenerative capacity of neural precursors in the adult mammalian brain is under the control of p53. Neurobiol Aging 2009; 30: 483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q, Baudry M, Liao G, Noniyev A, Galeano J, Bi X. A novel function for p53: regulation of growth cone motility through interaction with Rho kinase. J Neurosci 2009; 29: 5183–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi Y, Soria MG, Quadrato G, Inak G, Zhou L, Hervera A et al. The MDM4/MDM2-p53-IGF1 axis controls axonal regeneration, sprouting and functional recovery after CNS injury. Brain 2015; 138(Pt 7): 1843–1862. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science 1997; 276: 60–66. [DOI] [PubMed] [Google Scholar]
- Mehendale HM. Tissue repair: an important determinant of final outcome of toxicant-induced injury. Toxicol Pathol 2005; 33: 41–51. [DOI] [PubMed] [Google Scholar]
- Stanger BZ. Cellular homeostasis and repair in the mammalian liver. Annu Rev Physiol 2015; 77: 179–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K, Stanger BZ. Facultative stem cells in liver and pancreas: fact and fancy. Dev Dyn 2011; 240: 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajicek G, Oren R, Weinreb M Jr. The streaming liver. Liver 1985; 5: 293–300. [DOI] [PubMed] [Google Scholar]
- Magami Y, Azuma T, Inokuchi H, Kokuno S, Moriyasu F, Kawai K et al. Cell proliferation and renewal of normal hepatocytes and bile duct cells in adult mouse liver. Liver 2002; 22: 419–425. [DOI] [PubMed] [Google Scholar]
- Malato Y, Naqvi S, Schurmann N, Ng R, Wang B, Zape J et al. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J Clin Invest 2011; 121: 4850–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature 2015; 524: 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataller R, Brenner DA. Liver fibrosis. J Clin Invest 2005; 115: 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C et al. Senescence of activated stellate cells limits liver fibrosis. Cell 2008; 134: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu WY, Bird TG, Boulter L, Tsuchiya A, Cole AM, Hay T et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol 2015; 17: 971–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Akkari L, Simon J, Grace D, Tschaharganeh DF, Bolden JE et al. Non-cell-autonomous tumor suppression by p53. Cell 2013; 153: 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell C, Erker L, Schug J, Kopp JL, Canaday PS, Fox AJ et al. Prospective isolation of a bipotential clonogenic liver progenitor cell in adult mice. Genes Dev 2011; 25: 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Rogler LE, Teperman L, Morgan G, Rogler CE. Identification of hepatocytic and bile ductular cell lineages and candidate stem cells in bipolar ductular reactions in cirrhotic human liver. Hepatology 2007; 45: 716–724. [DOI] [PubMed] [Google Scholar]
- Miyajima A, Tanaka M, Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 2014; 14: 561–574. [DOI] [PubMed] [Google Scholar]
- Yanger K, Knigin D, Zong Y, Maggs L, Gu G, Akiyama H et al. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 2014; 15: 340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell S, Leffert HL. Liver cancer stem cells. J Clin Oncol 2008; 26: 2800–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumble ML, Croager EJ, Yeoh GC, Quail EA. Generation and characterization of p53 null transformed hepatic progenitor cells: oval cells give rise to hepatocellular carcinoma. Carcinogenesis 2002; 23: 435–445. [DOI] [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M et al In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 2013; 494: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font-Burgada J, Shalapour S, Ramaswamy S, Hsueh B, Rossell D, Umemura A et al. Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell 2015; 162: 766–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK. Liver regeneration. J Cell Physiol 2007; 213: 286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology 2006; 43(2 Suppl 1): S45–S53. [DOI] [PubMed] [Google Scholar]
- Cienfuegos JA, Rotellar F, Baixauli J, Martinez-Regueira F, Pardo F, Hernandez-Lizoain JL. Liver regeneration—the best kept secret. A model of tissue injury response. Rev Esp Enferm Dig 2014; 106: 171–194. [PubMed] [Google Scholar]
- Albrecht JH, Meyer AH, Hu MY. Regulation of cyclin-dependent kinase inhibitor p21(WAF1/Cip1/Sdi1) gene expression in hepatic regeneration. Hepatology 1997; 25: 557–563. [DOI] [PubMed] [Google Scholar]
- Kurinna S, Stratton SA, Tsai WW, Akdemir KC, Gu W, Singh P et al. Direct activation of forkhead box O3 by tumor suppressors p53 and p73 is disrupted during liver regeneration in mice. Hepatology 2010; 52: 1023–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurinna S, Stratton SA, Coban Z, Schumacher JM, Grompe M, Duncan AW et al. p53 regulates a mitotic transcription program and determines ploidy in normal mouse liver. Hepatology 2013; 57: 2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepniak E, Ricci R, Eferl R, Sumara G, Sumara I, Rath M et al. c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev 2006; 20: 2306–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 2010; 467: 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature 2004; 432: 338–341. [DOI] [PubMed] [Google Scholar]
- Aylon Y, Oren M. p53: guardian of ploidy. Mol Oncol 2011; 5: 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CC, Chu HY, Yang CW, Chou CK, Tsai TF. Aurora-A overexpression in mouse liver causes p53-dependent premitotic arrest during liver regeneration. Mol Cancer Res 2009; 7: 678–688. [DOI] [PubMed] [Google Scholar]
- Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer 2012; 12: 133–143. [DOI] [PubMed] [Google Scholar]
- Sugihara E, Saya H. Complexity of cancer stem cells. Int J Cancer 2013; 132: 1249–1259. [DOI] [PubMed] [Google Scholar]
- Shetzer Y, Molchadsky A, Rotter V. Oncogenic mutant p53 gain of function nourishes the vicious cycle of tumor development and cancer stem-cell formation. Cold Spring Harb Perspect Med 2016; 6: a026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. p53: good cop/bad cop. Cell 2002; 110: 9–12. [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Chiara Maiuri M, Morselli E, Criollo A, D'Amelio M, Djavaheri-Mergny M et al. A dual role of p53 in the control of autophagy. Autophagy 2008; 4: 810–814. [DOI] [PubMed] [Google Scholar]
- Charni M, Rivlin N, Molchadsky A, Aloni-Grinstein R, Rotter V. p53 in liver pathologies-taking the good with the bad. J Mol Med (Berl) 2014; 92: 1229–1234. [DOI] [PubMed] [Google Scholar]
- Fan Y, Lee TV, Xu D, Chen Z, Lamblin AF, Steller H et al. Dual roles of Drosophila p53 in cell death and cell differentiation. Cell Death Differ 2010; 17: 912–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RS, Newmark PA. The cell biology of regeneration. J Cell Biol 2012; 196: 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, Anton-Erxleben F, Milde S, Plotz C, Wittlieb J, Hemmrich G et al. Transgenic stem cells in Hydra reveal an early evolutionary origin for key elements controlling self-renewal and differentiation. Dev Biol 2007; 309: 32–44. [DOI] [PubMed] [Google Scholar]
- Siebert S, Anton-Erxleben F, Bosch TC. Cell type complexity in the basal metazoan Hydra is maintained by both stem cell based mechanisms and transdifferentiation. Dev Biol 2008; 313: 13–24. [DOI] [PubMed] [Google Scholar]
- Baguna J, Salo E, Romero R. Effects of activators and antagonists of the neuropeptides substance P and substance K on cell proliferation in planarians. Int J Dev Biol 1989; 33: 261–266. [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 2011; 332: 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehle KJ, Dan YY, Campbell JS, Fausto N. New concepts in liver regeneration. J Gastroenterol Hepatol 2011; 26: 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010; 464: 606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 2010; 464: 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry JJ, Tsonis PA. Molecular and cellular aspects of amphibian lens regeneration. Prog Retin Eye Res 2010; 29: 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Velloso CP, Imokawa Y, Brockes JP. The regenerative plasticity of isolated urodele myofibers and its dependence on MSX1. PLoS Biol 2004; 2: E218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JI, Loof S, He P, Simon A. Salamander limb regeneration involves the activation of a multipotent skeletal muscle satellite cell population. J Cell Biol 2006; 172: 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]