Abstract
The nuclear factor κB (NF-κB) transcription factors coordinate the inflammatory immune response during microbial infection. Pathogenic substances engage canonical NF-κB signaling through the heterodimer RelA:p50, which is subjected to rapid negative feedback by inhibitor of κBα (IκBα). The noncanonical NF-κB pathway is required for the differentiation of immune cells; however, crosstalk between both pathways can occur. Concomitantly activated noncanonical signaling generates p52 from the p100 precursor. The synthesis of p100 is induced by canonical signaling, leading to formation of the late-acting RelA:p52 heterodimer. This crosstalk prolongs inflammatory RelA activity in epithelial cells to ensure pathogen clearance. We found that the Toll-like receptor 4 (TLR4)–activated canonical NF-κB signaling pathway is insulated from lymphotoxin β receptor (LTβR)–induced noncanonical signaling in mouse macrophage cell lines. Combined computational and biochemical studies indicated that the extent of NF-κB–responsive expression of Nfkbia, which encodes IκBα, inversely correlated with crosstalk. The Nfkbia promoter showed enhanced responsiveness to NF-κB activation in macrophages compared to that in fibroblasts. We found that this hyperresponsive promoter engaged the RelA:p52 dimer generated during costimulation of macrophages through TLR4 and LTβR to trigger synthesis of IκBα at late time points, which prevented the late-acting RelA crosstalk response. Together, these data suggest that despite the presence of identical signaling networks in cells of diverse lineages, emergent crosstalk between signaling pathways is subject to cell type–specific regulation. We propose that the insulation of canonical and noncanonical NF-κB pathways limits the deleterious effects of macrophage-mediated inflammation.
Introduction
The nuclear factor κB (NF-κB) family of transcription factors coordinates innate immune responses to various microbial agents. Pathogen recognition through Toll-like receptors (TLRs) activates the RelA NF-κB subunits, which induce the expression of genes encoding mediators of inflammation and pro-survival factors in tissue-resident cells. In turn, NF-κB–induced cytokines and chemokines propagate inflammation through paracrine mechanisms that involve other immune cells for pathogen clearance. Insufficient NF-κB activation dampens the inflammatory response and is associated with immune deficiencies. Conversely, persistently increased NF-κB activity is implicated in chronic inflammatory disorders, as well as in neoplastic diseases (1). Therefore, it is important to understand fully the molecular mechanism controlling the inflammatory RelA-dependent response in various innate immune cells.
In unstimulated cells, RelA dimers are held inactive in the cytoplasm by the inhibitor of κB (IκB) α, β, and ε proteins, and activation of these dimers are mediated by the canonical NF-κB pathway in inflammatory settings. In this pathway, activation of the IκB kinase (IKK) complex consisting of NEMO-IKK2 (NEMO-IKKβ) promotes the phosphorylation and subsequent proteasomal degradation of IκBs, thereby liberating RelA dimers so that they can translocate to the nucleus. It is thought that inflammation-induced NF-κB activity is mostly mediated by RelA:p50 heterodimers. Furthermore, the RelA:p50 heterodimer rapidly induces synthesis of Nfkbia mRNA, which encodes IκBα, thus attenuating this early RelA activity in a negative feedback loop (2). Indeed, stringent dynamic controls ensure transient NF-κB activity in the canonical pathway (3).
The noncanonical NF-κB pathway is activated during immune cell differentiation and immune organ development (4). In this pathway, the kinases NF-κB–inducing kinase (NIK) and IKK1 (also known as IKKα) phosphorylate the Nfkb2-encoded precursor p100, which is bound to RelB. Subsequent proteasomal processing removes the C-terminal inhibitory domain from p100 to produce the mature p52 subunit, which gives rise to RelB:p52 activity in the nucleus. The lymphotoxin-β receptor (LTβR) induces noncanonical NF-κB signaling in lymph node stromal cells, and RelB:p52 dimer activity induced through the non-canonical NF-κB pathway is implicated in secondary lymph node development (5, 6).
The non-canonical NF-κB pathway also modulates canonical NF-κB activity induced during the immune response (7–9). In particular, LTβR-activated non-canonical signaling extends the duration of the canonical, RelA-mediated response to TLR4 in fibroblasts and intestinal epithelial cells (8). Mechanistic investigation revealed that canonical signaling induces the synthesis of p100, whereas concomitant noncanonical signaling by NIK converts this newly synthesized p100 into p52 to generate an alternate RelA:p52 dimer. Crosstalk between these NF-κB activating pathways promotes the progressive nuclear accumulation of the RelA:p52 dimer in cells costimulated through TLR4 and LTβR (8). Furthermore, RelA:p50 and RelA:p52 dimers exhibit largely overlapping DNA-binding (10) and gene-expression specificities (11). Accordingly, it was suggested that the noncanonical pathway alleviates infection of the intestine by Citrobacter rodentium by prolonging the RelA-dependent response in epithelial cells through the generation of RelA:p52 (8, 9).
Macrophages play a critical role in the inflammatory immune response; however, excessive RelA activation in macrophages during inflammation results in severe tissue damage and is considered detrimental to health (12). Persistent canonical signaling in myeloid cells exacerbates chronic colitis in an experimental animal model of inflammatory bowel disease (13). Macrophage-derived proinflammatory cytokines, whose generation relies on RelA signaling, stimulate tumor growth in colitis-associated cancer (14). A previous investigation suggested that in addition to IκBα-mediated negative feedback, proteasomal degradation of nuclear RelA confers dynamic control over canonical RelA activity in macrophages (15). Indeed, macrophages express LTβR and transduce noncanonical NF-κB signal (16, 17). Noncanonical signaling prolongs the canonical NF-κB response in fibroblasts, epithelial cells, and B cells (7, 8). We asked whether such a cross-regulatory mechanism perpetuated canonical RelA activity in macrophages, and thereby exacerbated inflammation.
Here, we report that macrophages instead use a distinct mechanism to insulate the TLR4-induced, canonical, RelA NF-κB pathway from LTβR-induced non-canonical signaling. In an iterative systems-modeling approach, we characterized the macrophage-associated biochemical parameters that indicated the presence of an Nfkbia promoter with enhanced responsiveness to RelA. Our mechanistic study demonstrated that this hyperactive promoter engaged the RelA:p52 dimer, which was produced during costimulation of macrophages through TLR4 and LTβR, to induce the synthesis of IκBα additionally at late time points. The production of IκBα at late time points prevented the progressive nuclear accumulation of RelA upon costimulation of macrophages. Together, these data suggest that despite the identical signaling network being present in different cell types, pathway crosstalk is subjected to cell type–specific control.
Results
The TLR4-activated canonical RelA pathway is insulated from LTβR-induced noncanonical NF-κB signaling in macrophages
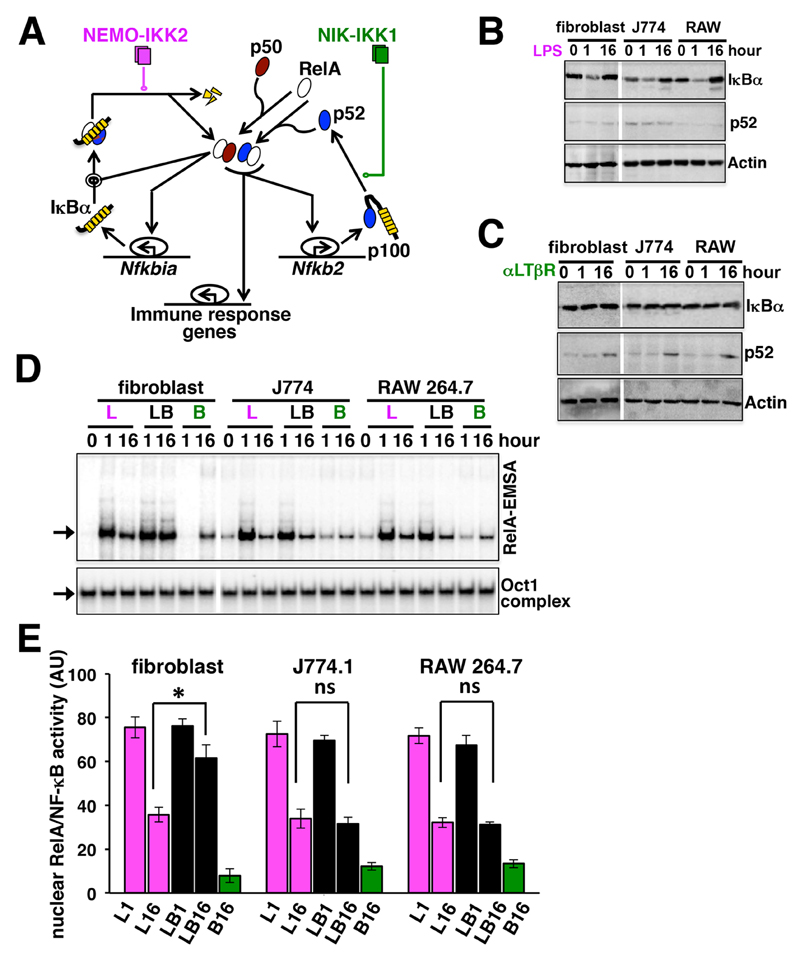
LTβR-stimulated noncanonical signaling prolongs TLR4-induced canonical RelA activity through RelA:p52 dimer generation in fibroblasts (Fig. 1A) (8). Considering the importance of macrophages in the inflammatory response, we sought to examine whether similar signaling crosstalk modulated RelA activity in these cells. We stimulated cells with the TLR4 ligand lipopolysaccharide (LPS), an agonistic anti-LTβR antibody (αLTβR), or both. Our Western blotting analyses revealed that LPS stimulated the degradation of IκBα after 1 hour during canonical signaling in immortalized mouse embryonic fibroblasts (MEFs) (8) as well as in the J774.1 monocyte and RAW 264.7 macrophage mouse cell lines (Fig. 1B). Treatment with LPS alone did not alter p52 abundance in these cell-lines. We previously showed in fibroblasts that a sub-saturating concentration of αLTβR (0.3 µg/ml) fails to induce IκBα degradation in subtly stimulating RelA:p50 activity, and this activity is liberated from an oligomeric p100 complex, termed IκBδ, which sequesters a subpopulation of RelA:p50 dimers in resting cells (18). Stimulation of J774.1 or RAW 264.7 cells with this low concentration of αLTβR did not induce IκBα degradation in our experiments (Fig. 1C). However, comparable signal-induced p52 generation after 16 hours of stimulation of fibroblasts, J774.1 cells, and RAW 264.7 cells with αLTβR confirmed that intact noncanonical signaling functioned in macrophages (Fig. 1C) (17).
Fig. 1. Costimulatory LTβR signaling prolongs nuclear RelA activity in response to TLR4 stimulation in fibroblasts, but not macrophages.
(A) Graphical depiction of the signaling circuitry underlying the crosstalk control of RelA activity. Magenta and green lines represent canonical NEMO-IKK2 and noncanonical NIK-IKK1 signals, respectively. (B and C) Immortalized mouse embryonic fibroblasts (MEFs) as well as J774.1 cells and RAW 264.7 cells were treated with LPS (B) or αLTβR (C) for the indicated times before the cells were lysed and analyzed by Western blotting with antibodies against the indicated proteins. Western blots are representative of three experiments. (D) Top: Fibroblasts, J774.1 cells, and RAW 264.7 cells were treated with LPS (L), αLTβR (B), or a combination of both stimuli (LB) for the indicated times. Supershifting RelB with an anti-RelB antibody residual RelA activities were revealed by RelA-EMSA. The arrow indicates the NF-κB DNA-binding complex composed of RelA heterodimers (also see fig. S1). Bottom: The DNA-binding activity of Oct1 served as a loading control. Data are representative of three experiments. (E) The signals corresponding to the DNA-binding activities of RelA were quantified from three independent experiments. Data are means ± SEM. Statistical significance was evaluated with the Student’s t test. *P < 0.05; ns, not significant.
Our electrophoretic mobility shift assay (EMSA) demonstrated that stimulation with LPS (1 µg/ml) alone induced similar DNA-binding activity by RelA in these cell lines at 1 hour, which was subsequently diminished after 16 hours (Fig. 1D, and fig. S1, A and B) as a result of negative feedback. Treatment of cells with αLTβR weakly induced the formation of nuclear RelA dimers at 16 hours (Fig. 1D). Consistent with our previous report (8), costimulation of fibroblasts with αLTβR and LPS synergistically enhanced the activity of RelA at 16 hours (Fig. 1, D and E). This crosstalk effect involved the progressive nuclear accumulation of the RelA:p52 dimer (fig. S1C). Unlike its effects in fibroblasts, however, αLTβR failed to prolong TLR4-stimulated RelA activity in J774.1 and RAW 264.7 cells (Fig. 1, D and E). Our analyses of primary MEFs and bone marrow–derived macrophages (BMDMs) substantiated that macrophages failed to exhibit pathway crosstalk and that fibroblasts were permissive for pathway crosstalk at different concentrations of LPS (fig. S1, D and E). Based on these analyses, we conclude that macrophages do not exhibit pathway crosstalk, despite having functional canonical and noncanonical NF-κB signaling.
The RelA-induced expression of Nfkbia mRNA inhibits pathway crosstalk
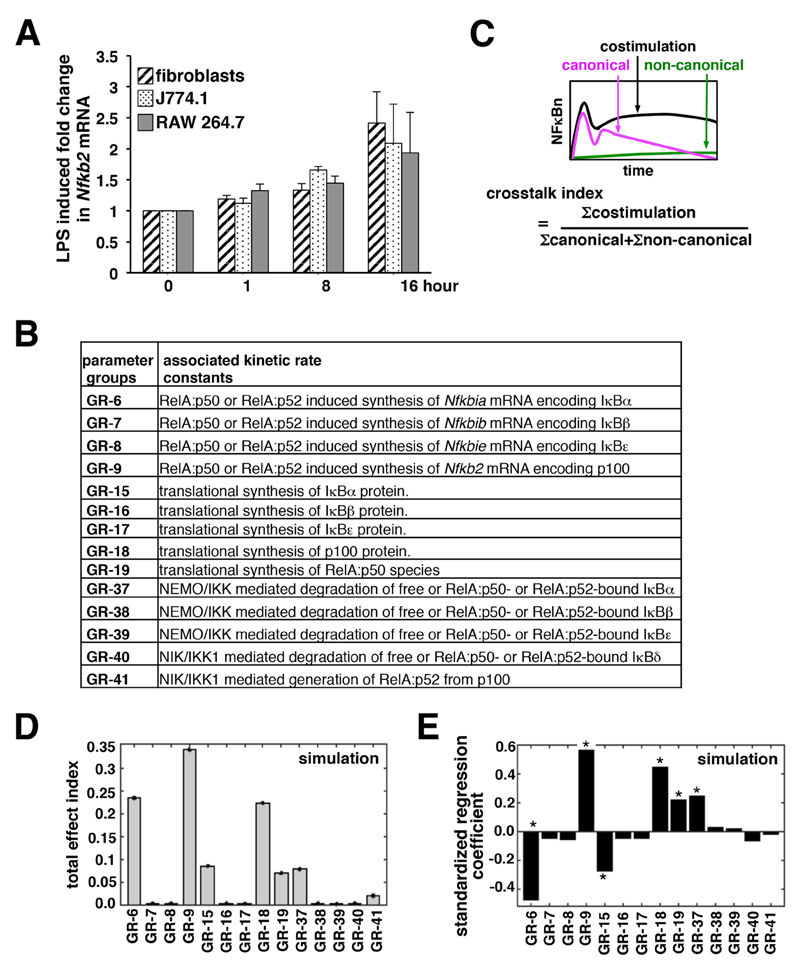
Mathematical reconstructions of a dynamical signaling network may offer insight into its emergent properties, such as crosstalk (19, 20). We previously rewired the underlying signaling network of the NF-κB system in a mass-action kinetics–based mathematical model [NF-κB systems model v1.0, (8)] and showed in silico how pathway crosstalk occurs. With our mathematical model, we predicted, and then experimentally confirmed, that the RelA-dependent expression of Nfkb2 mRNA (which encodes p100) by canonical signaling ensures an abundant supply of p100 for the concomitantly activated noncanonical module to generate RelA:p52 dimers in crosstalk-proficient fibroblasts. Because we observed a lack of crosstalk in macrophages, we asked whether these cells were defective in the LPS-induced, NF-κB-mediated expression of Nfkb2 mRNA. Our quantitative reverse transcription polymerase chain reaction (RT-PCR) analyses instead revealed a similar onset of Nfkb2 mRNA expression in fibroblasts, J774.1 cells, and RAW 264.7 cells (Fig. 2A) in response to LPS, with almost equivalent mRNA abundance at 16 hours after stimulation. Moreover, the basal amounts of Nfkb2 mRNA were not discernibly different between these cell lines (fig. S2). We reasoned that pathway crosstalk was regulated by additional Nfkb2-independent mechanisms, which provide for cell-type specific control.
Fig. 2. IκBα controls crosstalk between the NF-κB pathways.
(A) Fibroblasts, J774.1 cells, and RAW 264.7 cells were treated with LPS for the indicated times before being subjected to quantitative RT-PCR analysis of Nfkb2 mRNA abundance normalized to that of Actb mRNA. Data are means ± SEM of three biological replicates. (B) The parameter groups analyzed in the variance-based, multi-parametric perturbation analysis. For the individual parameter groups, incorporated kinetic rate constants and associated biochemical reactions are indicated. (C) A schematic defining the crosstalk index. Nuclear RelA activity induced by the indicated type of signal was measured as the area under the curve corresponding to the 24-hour time course from which the basal activity was subtracted, and this value was used to estimate the crosstalk index. (D) The total effect index was derived from Monte Carlo simulations identifying biochemical reactions critical for synergistic crosstalk. A standard bootstrapping technique was used to calculate the error ranges of the total effect indexes. (E) Bar diagram depicting the standardized regression coefficients of the parameter groups. The asterisk indicates statistically significant deviation (P < 0.005) from the null sensitivity for certain parameter groups as determined by two-tailed student’s t test.
We looked for the sensitive parameters that regulated crosstalk with a variance-based global sensitivity analysis (21). To implement our global analyses, we first catalogued 105 kinetic rate parameters into 43 parameter groups (Fig. 2B and table S1). In general, we grouped together parameters corresponding to potentially co-regulated biochemical processes associated with a given molecular species. It was previously suggested that kinetic parameters associated with signal-responsive processes, such as stimulus-induced degradation or transcription events, are divergent among different cell types, whereas those associated with signal-independent processes, such as molecular association and dissociation or nuclear import and export, are mostly invariant (19). To identify the molecular mechanism governing the cell type–specific control of pathway crosstalk, therefore, we focused on the signal-induced degradation of various IκBs, the stimulus-responsive processing of p100 to generate p52, as well as the RelA-induced synthesis of mRNAs encoding various IκBs and p100 (Fig. 2B). Although signal-independent protein translation rates vary between different cell types (22) and was included in our sensitivity analysis. Conversely, rate parameters corresponding to the constitutive synthesis and degradation of mRNAs, the signal-independent degradation of proteins, and association and dissociation between NF-κB and IκB species, as well as the nuclear import and export of complexes were excluded (see table S1 for detailed justifications for these decisions).
Next, we simulated our mathematical model and computed the nuclear RelA activity in response to the TLR4-induced activity of NEMO-IKK2 (canonical), the LTβR-induced activity of NIK-IKK1 (noncanonical), or both, which were measured experimentally (8). The crosstalk index, which signifies a synergistic response with a score >1, was estimated as the ratio of the total nuclear activity of RelA induced in a 24-hour time period upon costimulation to the sum of the nuclear activities of RelA induced by individual stimuli (Fig. 2C). We simultaneously explored a predefined range (± 25%) of parameter space around the initial values for multiple parameter groups with iterative Monte Carlo sampling (N = 1000 simulations) (see the Materials and Methods, and Supplementary Materials for details). The parameters in a given group were altered by the same factor. We determined the effect of parameter uncertainty on the crosstalk index for the individual parameter groups by summarizing these results into the total effect index (Fig. 2D and fig. S3A). In addition to Nfkb2 transcription (Group 9, GR-9), our variance-based, multiparametric sensitivity analysis identified the kinetic rate parameter associated with the RelA-induced synthesis of Nfkbia mRNA (which encodes IκBα) as being critical in controlling crosstalk (Group 6, GR-6). Consistent with a regulatory role for p100, crosstalk was sensitive to perturbations in the kinetic parameter corresponding to the production of p100 from Nfkb2 mRNA (Group 18, GR-18). Similarly, crosstalk was sensitive, albeit to a lesser extent, to the variation in the rate parameters associated with the NEMO-IKK2–mediated degradation of IκBα (Group 37, GR-37) or the translational synthesis of IκBα (Group 15, GR-15). In addition to IκBα, the IκBβ and IκBε isoforms also inhibit the activity of RelA during inflammatory signaling. Our mathematical analyses, however, indicated that biochemical processes associated with the synthesis and degradation of IκBβ (Groups 7, 16, and 38) and IκBε (Groups 8, 17, and 39) were less likely to contribute to crosstalk control. Finally, we calculated standardized regression coefficients from these randomly sampled values for a given parameter group as independent variables and crosstalk index output as the dependent variable. Our regression analyses confirmed that these sensitive parameter groups deviated from null sensitivity, and that the rate of RelA-induced IκBα synthesis was inversely related with crosstalk (Fig. 2E and fig. S3, B and C). These mathematical studies predicted that RelA-induced IκBα synthesis was an important determinant of crosstalk between the two NF-κB–activating pathways.
Enhancing the Nfkbia promoter activity in fibroblasts abrogates crosstalk by limiting the expression of Nfkb2 mRNA
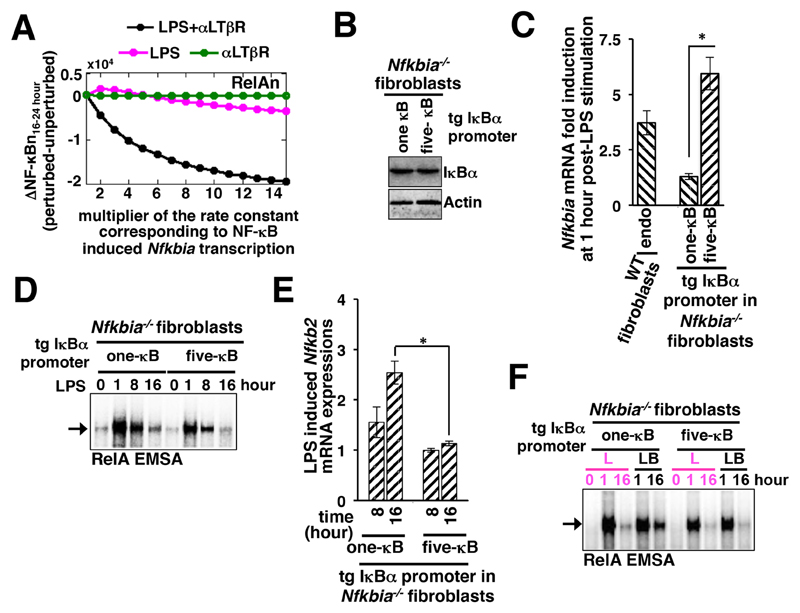
Next, we investigated how the RelA-induced synthesis of IκBα affected pathway crosstalk. We simulated individual canonical TLR4-dependent or noncanonical LTβR-dependent regimes in our mathematical model, as well as the TLR4 and LTβR costimulation regime. We computed late RelA activities induced between 16 and 24 hours after stimulation in these settings for different values of the kinetic rate parameter describing NF-κB–dependent Nfkbia transcription (see the Materials and Methods for details). Then we plotted the differential between the RelA activity induced for a given value of the rate constant and that for the nominal value against increasing values of the rate parameter of Nfkbia transcription (Fig. 3A). Our mathematical analyses suggested that an increased rate of Nfkbia transcription substantially dampened the activation of RelA at late time points in response to costimulation, with only a subtle effect on the activities of RelA induced by individual stimuli, and abrogated crosstalk (Fig. 3A and fig. S4A).
Fig. 3. Enhancing the NF-κB responsiveness of the Nfkbia promoter in fibroblasts abrogates crosstalk by reducing the expression of Nfkb2 mRNA.
(A) Computational simulations showing changes in the nuclear activity of RelA (RelAn) at late times in response to the indicated stimuli as a function of the rate constant corresponding to the NF-κB–induced expression of Nfkbia mRNA (which encodes IκBα). The activity of RelA at late time points was determined as the area under the respective time-course curve measured between 16 and 24 hours. The differences in the RelA activity induced for a given value of the rate constant and the nominal value were computed and plotted along the y-axis. (B) Western blotting analysis of IκBα proteins in Nfkbia–/– fibroblasts expressing transgenic (tg) constructs containing promoters with either one or five κB sites. Actin was used as a loading control. Blots are representative of two technical replicates. (C) WT and Nfkbia–/– fibroblasts expressing the indicated constructs were treated with LPS for 1 hour before being subjected to RT-PCR analysis of Nfkbia mRNA abundance normalized to that of Actb mRNA. Data are means ± SEM of three biological replicates. *P < 0.05. LPS-induced changes in Nfkbia mRNA abundance expressed by the endogenous (endo) promoter in WT fibroblasts were measured as a control. (D) Nfkbia–/– fibroblasts expressing the indicated constructs were treated with LPS for the indicated times before being subjected to RelA-EMSA. The arrow indicates the NF-κB DNA-binding complex composed of RelA heterodimers. Data are representative of three independent experiments. (E) Nfkbia–/– fibroblasts expressing the indicated constructs were treated with LPS for the indicated times before being subjected to RT-PCR analysis of Nfkb2 mRNA abundance. Data are means ± SEM of three biological replicates. *P < 0.05. (F) Nfkbia–/– fibroblasts expressing the indicated constructs were treated with LPS alone (L) or were costimulated with LPS and αLTβR (LB) for the indicated times before being subjected to RelA-EMSA. The arrow indicates the NF-κB DNA-binding complex composed of RelA heterodimers. Data are representative of three experiments.
It is thought that cooperative binding of the RelA:p50 heterodimers to the adjacent κB sites present in the endogenous promoter provides for the rapid, but transient, induction of Nfkbia mRNA synthesis (23). To validate our computational predictions, we engineered Nfkbia-/- fibroblasts to stably express IκBα from a transgenic, artificial promoter consisting of either one or five tandem κB sites (Fig. 3B and fig. S4B). Stimulation of these cells with LPS confirmed the different strengths of the promoters (Fig. 3C). Whereas we observed a six-fold increase in the amount of Nfkbia mRNA 1 hour after stimulation in cells expressing the five-κB promoter, only minor increases in Nfkbia mRNA abundance were observed in cells expressing the one-κB promoter (Fig. 3C). Note that the abundance of Nfkbia mRNA expressed from the five-κB promoter was greater than that induced by RelA in crosstalk-proficient wild-type fibroblasts containing the endogenous promoter (Fig. 3C), which contains three κB sites (23).
Nevertheless, we used this pair of engineered cell lines to examine the role of RelA-responsive Nfkbia mRNA expression in pathway crosstalk. Our time-course EMSA analyses indicated that strong IκBα-mediated negative feedback generated from the five-κB promoter rapidly terminated the LPS-induced early (1 hour) activation of RelA, with only minor RelA activity observable in the nucleus at 8 hours, as well as at 16 hours (Fig. 3D). On the other hand, weakened IκBα-dependent feedback from the one-κB promoter led to somewhat augmented RelA activity at 8 hours after treatment with LPS, which was subsequently attenuated by 16 hours (Fig. 3D). Consistent with the reduced activity of RelA at late time points after stimulation, LPS failed to induce the late expression of Nfkb2 mRNA in cells containing the five-κB Nfkbia promoter (Fig. 3E). However, LPS efficiently induced Nfkb2 mRNA expression in cells containing the one-κB Nfkbia promoter within 8 hours of stimulation, with increased mRNA amounts observed at 16 hours (Fig. 3E). The NF-κB–driven expression of Nfkb2 mRNA produces p100, which was previously shown to form an oligomeric complex, termed IκBδ, and inhibit RelA in both unstimulated MEFs as well as MEFs treated with LPS alone (24–26). Efficient attenuation of the NF-κB response to LPS at 16 hours (Fig. 3D) despite the weakened IκBα-mediated feedback in cells expressing the one-κB promoter therefore corroborated the previous finding that IκBα- and IκBδ/Nfkb2-mediated negative feedback collaborate during responses to LPS alone (24).
IκBα generated from the five-κB promoter abrogated the LTβR-mediated amplification of the late NF-κB response to LPS (Fig. 3F). However, αLTβR costimulation extended the duration of LPS-induced RelA activity in cells producing IκBα from the one-κB promoter, with robust late activity observable at 16 hours (Fig. 3F). In costimulated cells, NIK acts on LPS-induced, newly synthesized p100 to enable generation of the RelA:p52 dimer, which progressively accumulates in the nucleus to cause pathway crosstalk at late times (Fig. 3F). Our analyses suggest that an enhanced Nfkbia promoter, which determines the rate of NF-κB–dependent synthesis of IκBα, is indeed capable of abrogating crosstalk between the two pathways. However, in the engineered fibroblast cell line, enhanced IκBα synthesis disrupted crosstalk between the pathways by limiting the LPS-induced, NF-κB–responsive expression of Nfkb2 (which encodes p100). Note that our biochemical analyses suggest that the inability of macrophages to exhibit pathway crosstalk involves Nfkb2-independent mechanisms (Fig. 2A).
A hyperresponsive Nfkbia promoter functions independently of Nfkb2 in crosstalk-deficient macrophages
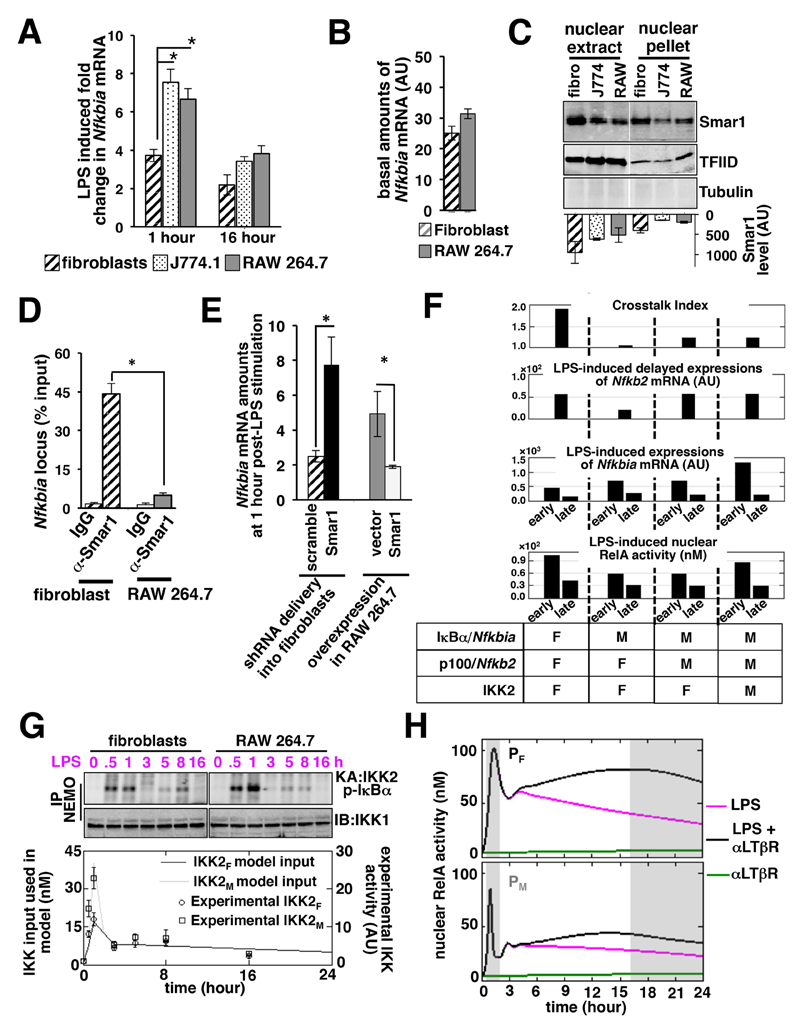
Next, we compared the Nfkbia promoter activity between the crosstalk-proficient fibroblast cell line and the crosstalk-deficient J774.1 and RAW 264.7 cell lines. Our quantitative RT-PCR analyses revealed that the abundance of Nfkbia mRNA after 1 hour of stimulation with LPS (1 µg/ml) was substantially greater in J774.1 and RAW 264.7 cells than in fibroblasts, suggesting that the Nfkbia promoter in macrophages was more responsive (Fig. 4A). The residual amounts of Nfkbia mRNA at 16 hours after treatment with LPS were not discernibly different between the different cell types. Similarly, the basal amounts of Nfkbia mRNA in fibroblasts and RAW 264.7 cells were comparable (Fig. 4B). Our analyses involving primary cells (fig. S5A) further validated our cell line–based finding that LPS stimulation led to the increased early induction of Nfkbia mRNA expression in macrophages as compared to fibroblasts. Moreover, TNF-stimulated canonical RelA signaling also led to augmented early expression of Nfkbia mRNA in macrophages (fig. S5B). These studies suggest that the Nfkbia promoter in macrophages is hyperresponsive to RelA activated by multiple proinflammatory stimuli.
Fig. 4. A hyperactive Nfkbia promoter abrogates crosstalk independently of Nfkb2 in macrophages.
(A) Fibroblasts, J774.1 cells, and RAW 264.7 cells were stimulated with LPS for the indicated times before being subjected to RT-PCR analysis of Nfkbia mRNA abundance normalized to that of Actb mRNA. Data are means ± SEM of three independent experiments. *P < 0.05 by student’s t test. (B) Quantitative RT-PCR analysis of the basal amounts of Nfkbia mRNA in unstimulated fibroblast and RAW 264.7 cell lines. Data are means ± SEM of three independent experiments. (C) Fibroblasts, J774.1 cells, and RAW 264.7 cells were processed to generate nuclear extracts and nuclear pellets, which were then subjected to Western blotting analysis of the relative amounts of Smar1. TFIID served as a loading control. Tubulin was analyzed to examine the purity of the nuclear fractions. Blots are representative of three independent experiments. Average amounts of Smar1 in the individual lanes were quantified from three independent experiments and indicated at the bottom of the blot. (D) Fibroblasts and RAW 264.7 cells were subjected to ChIP analysis with anti-Smar1 or IgG control antibodies. Quantification of the abundance of Nfkbia promoter DNA in the immunopellet was performed by semi-quantitative PCR analysis and plotted relative to the respective inputs. Data are means ± SEM of three biological replicates. *P < 0.05. (E) Fibroblasts transduced with lentiviruses expressing either scrambled shRNA or Smar1-specific shRNA (left) and RAW 264.7 transfected with either p3XFLAG-CMV or p3XFLAG-CMV-Smar1 (right) were treated with LPS for 1 hour before being subjected to RT-PCR analysis of Nfkbia mRNA abundance normalized to that of Actb mRNA. Data are means ± SEM of three biological replicates. *P < 0.05. (F) Simulations revealing the relative contributions of macrophage rate parameters and IKK activity in capturing various characteristics of NF-κB signaling in macrophages. Kinetic rate parameters corresponding to the NF-κB–responsive synthesis of Nfkbia and Nfkb2 mRNAs, as well as macrophage IKK2 activities, are indicated with “M,” and those related to the fibroblastic parameter ensemble are represented with “F.” Early and late times of gene expression reflect the average amounts of mRNA synthesized between 1 to 2 and 16 to 24 hours, respectively. Similarly, simulation data corresponding to the early peak of RelA activity and the late (16 hour) time point of activity are presented. (G) Kinase assay comparing the LPS-induced activity of NEMO-IKK2 over time. Fibroblasts and RAW 264.7 cells were treated with LPS for the indicated times before being subjected to immunoprecipitation with anti-NEMO antibody. The immunoprecipitate was subsequently examined for the presence of NEMO-IKK activity using recombinant GST-IκBα as a substrate (Top) and was subjected to Western blotting (IB) analysis, where IKK1 served as a loading control (Middle). Bottom: Quantified data points, representing means ± SEM from three independent experiments, were used to derive fibroblast-associated (IKKF) and macrophage-related (IKKM) IKK input profiles. (H) Computational simulations comparing RelA activities in the PF and PM systems in response to stimulation with LPS, αLTβR, or both. The early and late phases are indicated by gray boxes.
In addition to regulation by RelA-containing dimers, IκBα synthesis is suppressed by Smar1 (27, 28), a protein that was originally shown to bind to the matrix attachment region (MAR) of the Trb locus (which encodes T cell receptor β) (29). In cancer cells, Smar1 binds to an MAR that is proximal to the κB sites in the Nfkbia promoter (27, 28). Furthermore, Smar1 mediates recruitment of the histone deacetylase 1 (HDAC1) repressor complex to the Nfkbia promoter in unstimulated cells and thereby restricts Nfkbia mRNA synthesis upon subsequent activation of RelA (28). Given that LPS-induced RelA activity was comparable between fibroblasts and macrophages, we asked whether Smar1 accounted for the cell-type specific differences in the RelA-dependent expression of Nfkbia. Western blotting analysis showed that nuclear extracts of J774.1 and RAW 264.7 cells had less Smar1 protein than did nuclear extracts of fibroblasts (Fig. 4C), and the difference was even more substantial when nuclear pellets were compared. Similar differences in the amounts of Smar1 were also noted between primary BMDMs and MEFs (fig. S5C). Consistent with these findings, chromatin immunoprecipitation analyses revealed reduced binding of Smar1 to the Nfkbia locus in RAW 264.7 cells compared to that in fibroblasts (Fig. 4D). We then depleted Smar1 in fibroblasts with a lentivirus expressing Smar1-specific shRNA (fig. S5D). Indeed, compared to Smar1-suffcient fibroblasts, Smar1-depleted fibroblasts had increased amounts of Nfkbia mRNA early after stimulation with LPS (Fig. 4E). Conversely, overexpression of Smar1 in RAW 264.7 cells led to inhibition of LPS-induced Nfkbia mRNA expression 1 hour after treatment (Fig. 4E and fig. S5E). Our results suggest that the relatively reduced amount of Smar1 in macrophages maintains the Nfkbia promoter in a poised state, which results in the heightened early expression of Nfkbia mRNA upon canonical RelA signaling stimulated by LPS and other proinflammatory mediators.
The nominal kinetic parameter ensemble (PF) and the IKK2 input (IKKF) that revealed the pathway crosstalk in our model were mostly derived from fibroblast-based experiments (8). Through our computational model, we then investigated whether the hyperactive Nfkbia promoter explained the defect in Nfkb2-independent crosstalk in macrophages. To capture the hyperresponsive Nfkbia promoter in our model, we fitted a 15-fold increased value for the kinetic rate parameter corresponding to RelA-induced IκBα synthesis. Our analysis suggested that the increased rate of IκBα synthesis was sufficient to abrogate pathway crosstalk (Fig. 4F, compare the first and second columns from the left). However, IκBα synthesis from the hyperactive promoter restricted the LPS-induced synthesis of Nfkb2 mRNA at late time points (Fig. 4F), as was also experimentally observed in fibroblasts expressing Nfkbia mRNA from the five-κB-containing promoter (Fig. 3E). Note that the macrophage cell lines did not show any defect in the LPS-induced expression of Nfkb2 in our experiments (Fig. 2A). To adopt the computational model for macrophages, a six-fold increase in the parameter value corresponding to RelA-induced Nfkb2 transcription was implemented. Although this change improved the model performance with respect to Nfkb2 expression without having any effect on the crosstalk index (Fig. 4F, compare first and third columns), we noted a deviation from the experimental observations (Fig. 1D), with reduced early RelA signaling in response to LPS in silico.
Next, we experimentally measured LPS-induced IKK2 activity in macrophages over time. Our biochemical analyses revealed an enhanced IKK2 activity at 30 min and 1 hour in LPS-stimulated RAW 264.7 cells compared to that in fibroblasts, although IKK2 activities at later time points were similar (Fig. 4G). We simulated LPS signaling in our mathematical model involving the altered parameter ensemble, denoted as PM, using the macrophage IKK2 profile (IKKM) as input. Model simulations indeed captured various characteristics of macrophage NF-κB signaling (Fig. 4F, compare first and fourth columns). For example, it mirrored the experimental observation that treatment with LPS alone induced comparable DNA-binding activities of RelA in macrophages and fibroblasts. Our mathematical analysis recapitulated the enhanced expression of Nfkbia mRNA in macrophages as compared to fibroblasts at early times after treatment with LPS and the similarly reduced expression at later times. In addition, the LPS-induced synthesis of Nfkb2 mRNA at late time points was similar between the PF and PM ensembles. Corroborating our experimental data, our model showed that noncanonical LTβR-dependent signaling only weakly induced RelA activity (Fig. 4H). Furthermore, simulations involving the PM revealed diminished NF-κB crosstalk in macrophages with the residual late RelA activity during LPS signaling that was not substantially enhanced upon costimulation of the LTβR (Fig. 4, F and H).
Cell type–specific variations in the abundances of NF-κB regulators or IKK2 activities have been noted previously. In particular, Nfkbia promoter activity is reduced in colonic epithelial cells because of repressive DNA methylation (30). Differences in the abundance of Nfkb2 mRNAs between fibroblasts and dendritic cells have also been observed (31). Our computational analyses indicated that IKKM and the macrophage-specific Nfkb2 transcription rate do not account for the observed defect in crosstalk, and that the hyperactive Nfkbia promoter acts independently of Nfkb2 in abrogating crosstalk. Macrophages are likely to be divergent from fibroblasts in several NF-κB-dependent and - independent mechanisms. Our in silico analyses involving the PM parameter ensemble suggest that consideration of an IκBα-dependent mechanism is sufficient to explain the observed crosstalk defect in macrophages.
Increased RelA-IκBα complex formation decreases nuclear RelA activity in response to costimulation at late times in macrophages
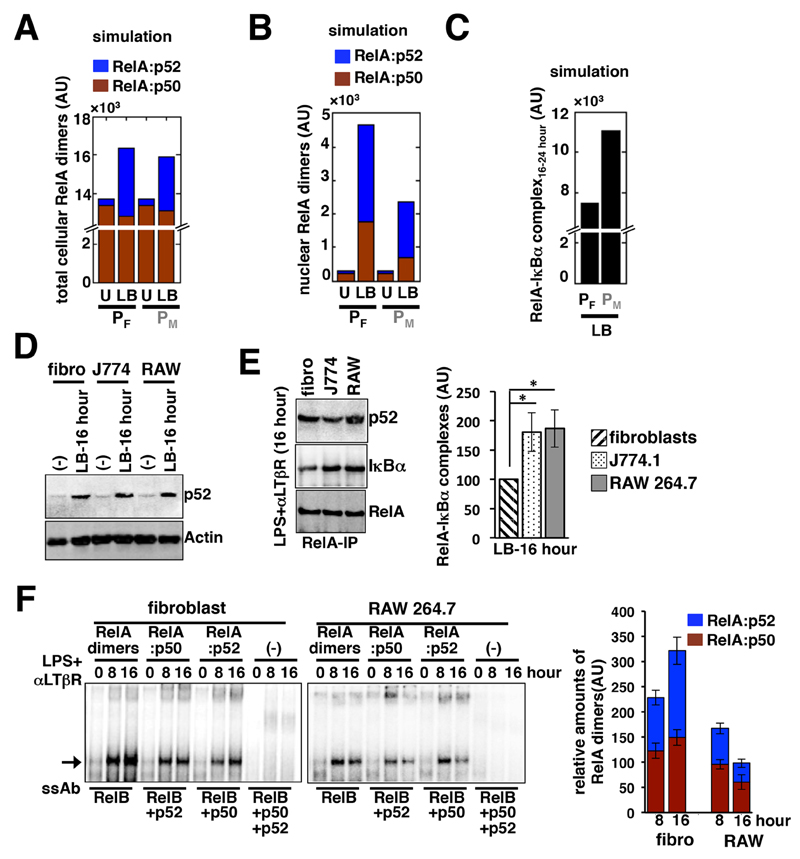
Our experimental and computational studies confirmed that costimulation of fibroblasts with LPS and αLTβR induced late-acting RelA signaling, which was rather diminished in macrophages. Given that the RelA:p52 dimer reinforced late RelA activity in costimulated fibroblasts, we asked whether the hyperactive Nfkbia promoter disrupted pathway-crosstalk in macrophages by preventing the generation of RelA:p52 dimers despite the fact that Nfkb2 mRNA synthesis was unperturbed. Computational simulations involving the PM parameter ensemble predicted that costimulation of macrophages would efficiently augment the total cellular amount of the RelA:p52 dimer (Fig. 5A). Computational studies further revealed a marked reduction in the nuclear activity of both the RelA:p50 and RelA:p52 dimers at late time points (between 16 and 24 hours) in costimulated macrophages as compared to costimulated fibroblasts (Fig. 5B). Finally, modeling studies predicted the enhanced accumulation of the RelA-IκBα complex, which restricts nuclear RelA activity, specifically in macrophages between 16 and 24 hours after costimulation (Fig. 5C).
Fig. 5. Compared to fibroblasts, costimulated macrophages have similar RelA:p52 amounts, but increased amounts of RelA-IκBα complex.
(A and B) Simulations comparing the average values for the abundances of total (A) and nuclear (B) RelA:p50 and RelA:p52 dimers that were estimated to occur between 16 and 24 hours after costimulation (LB) in the PF and PM systems. U, untreated cells. (C) Simulation charting the average abundance of the RelA-IκBα complex between 16 and 24 hours after costimulation in the PF and PM systems. (D) Fibroblasts, J774.1 cells, and RAW 264.7 cells were left untreated or were costimulated with LPS and αLTβR for 16 hours before being subjected to Western blotting analysis with antibodies against the indicated proteins. Blots are representative of three experiments. (E) Left: Fibroblasts, J774.1 cells, and RAW 264.7 cells were costimulated with LPS and αLTβR for 16 hours before being subjected to immunoprecipitation (IP) with anti-RelA antibody and Western blotting analysis with antibodies against the indicated proteins. Right: Densitometric analysis of the relative abundances of RelA-IκBα complexes in the indicated cells. Data are means ± SEM of three biological replicates. (F) Left: Fibroblasts and RAW 264.7 cells were costimulated with LPS and αLTβR for the indicated times before being subjected to Supershift analyses to distinguish between nuclear RelA:p50 and RelA:p52 dimers. Right: Quantitative analysis of the relative abundances of the different NF-κB dimers in the indicated cells. Data are means ± SEM of three biological replicates.
We next tested these predictions in our biochemical assays. Western blotting analysis of whole-cell extracts (Fig. 5D) and immunoprecipitated RelA samples (Fig. 5E) confirmed the presence of comparable amounts of p52 and RelA:p52 dimer, respectively, in fibroblasts, RAW 264.7 cells, and J774.1 cells after 16 hours of costimulation with LPS and αLTβR. Ablating the DNA-binding activity of one or more NF-κB subunits resulted in the identification by EMSA supershift analyses of only brief nuclear RelA:p52 activity in RAW 264.7 cells at 8 hours after costimulation, with a subsequent decline at 16 hours (Fig. 5F). Similarly late RelA:p50 activity was also diminished in costimulated RAW 264.7 cells. In contrast, costimulation of fibroblasts led to progressive nuclear accumulation of the RelA:p52 dimer (Fig. 5F). Our immunoprecipitation-based analyses and corresponding quantification of the data further confirmed that, as compared to fibroblasts, RAW 264.7 and J774.1 cells had increased amounts of RelA-IκBα complex after 16 hours of costimulation (Fig. 5E). These results suggest that the noncanonical pathway continuously generates RelA:p52 dimers during the costimulation of macrophages, but that IκBα (which is synthesized due to expression driven from a hyperresponsive promoter) directly sequesters late-acting RelA-containing dimers to abrogate crosstalk between the two pathways.
The RelA:p52 dimer induces late-phase IκBα synthesis during the costimulation of macrophages
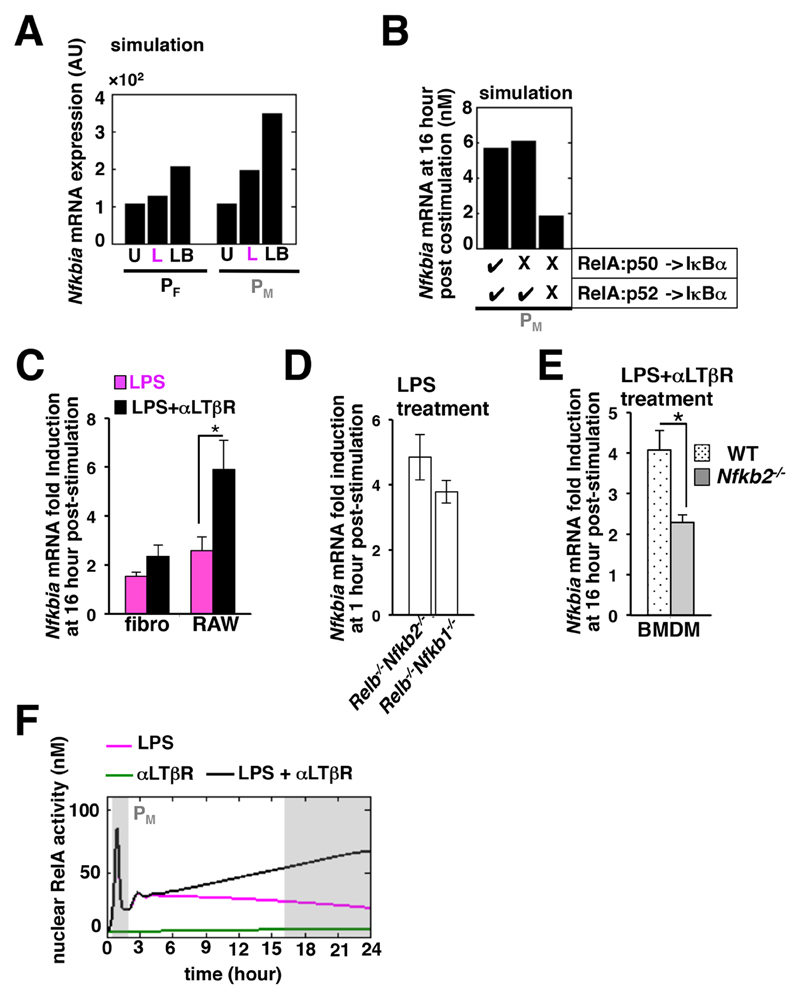
In our experiments, the LPS-induced expression of Nfkbia mRNA at late time points was similarly reduced between the fibroblast and the macrophage cell lines, despite their having differences in mRNA synthesis at earlier times (Fig. 4, A and F). We investigated whether costimulation of macrophages with LPS and αLTβR would affect the activity of the hyperactive promoter in such a way as to cause delayed synthesis of IκBα, which then would sequester RelA at late time points to inhibit its activity. Computational studies involving the macrophage parameter ensemble PM indicated that there would be a considerable increase in the amount of Nfkbia mRNA after 16 to 24 hours of costimulation as compared to that in response to treatment with LPS alone (Fig. 6A). The difference in Nfkbia mRNA abundance was less obvious for the fibroblast-associated parameter space, PF. Our simulation results predicted that the RelA:p50–induced expression of Nfkbia mRNA was not required for late-phase IκBα synthesis in the costimulated macrophage system (Fig. 6B), but that RelA:p52–mediated transcription was necessary.
Fig. 6. Costimulation of macrophages induces the synthesis of IκBα at late time points mediated by the RelA:p52 dimer.
(A) In silico analysis predicting the averaged values for the expression of Nfkbia mRNA measured between 16 and 24 hours after costimulation (LB) in the fibroblast and macrophage cell systems. U, unstimulated; L, LPS-stimulated. (B) Computational simulation charting the expression of Nfkbia mRNA at 16 hours after costimulation in the macrophage PM parameter ensemble in the absence of either RelA:p50–mediated Nfkbia transcription or of both RelA:p50- and RelA:p52-mediated Nfkbia transcription. (C) Fibroblast and RAW 264.7 cell lines were stimulated for 16 hours with LPS alone or together with αLTβR before being subjected to quantitative RT-PCR analysis of Nfkbia mRNA abundance normalized to that of Actb mRNA. Data are means ± SEM of three independent experiments. *P < 0.05 by two-tailed student’s t test. (D) Quantitative RT-PCR analysis of Nfkbia mRNA abundance in Relb-/-Nfkb1-/- fibroblasts (which exhibit RelA:p52 dimer activity) and in Relb-/-Nfkb2-/- cells (which exhibit RelA:p50 dimer activity) 1 hour after stimulation with LPS. Data are means ± SEM of three independent experiments. (E) Quantitative RT-PCR analysis of Nfkbia mRNA abundance in WT and Nfkb2-/- BMDMs in response to costimulation with LPS and αLTβR. Data are means ± SEM of three independent experiments. *P < 0.05. (F) Time-course analysis demonstrating the rescue of pathway crosstalk in silico in the PM parameter space in the absence of RelA:p52–mediated Nfkbia transcription.
Consistent with our simulation results, experimental analyses revealed a substantial increase in the LPS-induced expression of Nfkbia mRNA at late time points in costimulated RAW 264.7 cells, with only a moderate effect apparent in fibroblasts (Fig. 6C). The canonical pathway predominantly activates the RelA:p50 dimer, but not the RelA:p52 dimer, which is abundantly produced only upon costimulation through TLR4 and LTβR (8). However, a study showed that a lack of RelA:p50 in Nfkb1-/- fibroblasts (Nfkb1 encodes p105, which produces the mature p50 subunit) was compensated for by the RelA:p52 dimer, which was generated constitutively in these cells and was activated upon canonical signaling (11, 32). In our experiments, LPS induced the expression of Nfkbia mRNA in Nfkb1-/- fibroblasts, which suggested that the RelA:p52 dimer was capable of activating the Nfkbia promoter (Fig. 6D). Furthermore, a comparison with wild-type BMDMs revealed a substantial decrease in the abundance of Nfkbia mRNA at late time points during the costimulation of Nfkb2-/- macrophages, which were devoid of the RelA:p52 dimer (Fig. 6E).
Finally, computational analyses demonstrated that disruption of the RelA:p52–mediated synthesis of IκBα almost completely restored RelA activity at late time points in response to costimulation in the PM parameter space (Fig. 6F). Because of the lack of a RelA:p52 mutant that is specifically defective in activating Nfkbia transcription, we were unable to further test pathway crosstalk in the absence of RelA:p52–mediated IκBα synthesis experimentally. Together, our results support a mechanistic model wherein the hyperresponsive Nfkbia promoter in macrophages engages the RelA:p52 dimer produced upon costimulation to induce synthesis of IκBα at late time points, which in turn prevents the progressive nuclear accumulation of both the RelA:p50 and RelA:p52 NF-κB dimers to abrogate crosstalk.
Discussion
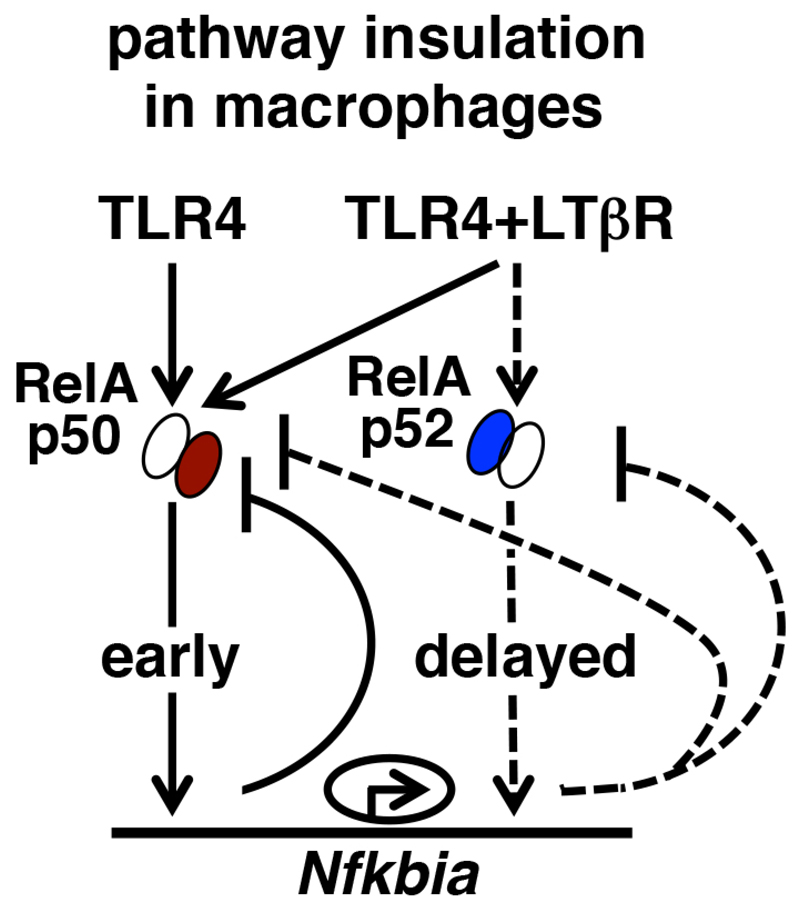
In addition to its amplitude, the duration of NF-κB signaling also plays an important role in immune regulation (33). TLR4 induces early RelA:p50 activity, which is rapidly terminated by IκBα-mediated negative feedback. We previously showed that crosstalk between TLR4-activated canonical NF-κB signaling and LTβR-induced noncanonical signaling produces an alternate RelA:p52 dimer, which sustains RelA activity in fibroblasts and epithelial cells (8). Here, we found that TLR4 signaling was insulated from LTβR pathway in macrophages despite the fact that the RelA:p52 dimer was generated upon costimulation. As such, the RelA:p52 dimer briefly accumulated in the nucleus, which was followed by a decline in the nuclear activities of both RelA:p50 and RelA:p52 (Fig. 5F). The reduced amount of Smar1 in macrophages resulted in the Nfkbia promoter becoming hyperresponsive to activation by NF-κB. Our mechanistic study suggested that the RelA:p52 dimer that was generated upon costimulation activated this hyperactive promoter leading to IκBα synthesis at late time points which prevented the late-acting RelA crosstalk response in macrophages (Fig. 7).
Fig. 7. The role of the RelA:p52–induced, late-phase synthesis of IκBα in costimulated macrophages in insulating canonical and noncanonical NF-κB signalingpathways.
TLR4 stimulation induces the nuclear activity of the RelA:p50 dimer, which is subjected to rapid negative feedback by IκBα. Costimulation through LTβR generates a late RelA:p52–containing NF-κB activity in fibroblasts. In costimulated macrophages, however, the RelA:p52 dimer triggersed the hyperactive Nfkbia promoter, which leads to the production of IκBα additionally at late time points. This late-phase IκBα synthesis attenuatesd the late-acting RelA:p50 and RelA:p52 activities in costimulated macrophages and abrogatesd crosstalk between the canonical and noncanonical NF-κB pathways. Early and late signaling events have been indicated with solid and dashed lines, respectively.
Smar1 binds to the Nfkbia promoter and recruits HDAC1, which then deacetylates histone 3 at Lys9 to maintain the Nfkbia promoter in a repressed state (28). Although Smar1 binding did not impede the recruitment of RelA to the Nfkbia promoter, it prevented the RelA-dependent expression of Nfkbia. We found that the extent of binding of Smar1 to the Nfkbia locus in macrophages was reduced compared to that in fibroblasts. Our study suggested that this pre-existing difference (under basal conditions) led to a hyperactive promoter, which resulted in enhanced early expression of Nfkbia mRNA upon RelA activation in macrophages. Furthermore, the basal amount of Nfkbia mRNA was not substantially different between macrophages and fibroblasts (Fig. 4B). As such, unstimulated cells maintained only minimal constitutive RelA activity in the nucleus (Figs. 1D, 4H, and 5B). Our results suggest that this limited RelA activity, despite the reduction in Smar1 abundance, resulted in restricted basal Nfkbia mRNA synthesis from the hyperactive promoter in unstimulated macrophages. Note that our computational model also recapitulated the comparable basal amounts of Nfkbia mRNA in unstimulated fibroblasts and macrophages (Fig. 6A). Unlike in macrophages, the amount of Smar1 is substantially increased during T cell differentiation and maturation (29). Whereas the reduced amount of Smar1 in cancer cells is attributed to modulation of the p53-mediated transcription of the gene encoding Smar1 (34), the molecular mechanism underlying the reduced nuclear abundance of Smar1 in macrophages remains elusive. Given the profound contrast between lymphocytes and macrophages, it is tempting to speculate that hematopoietic cell differentiation differentially regulates Smar1 abundance and function. Considering the abundance of Smar1-binding sites in the genome, it is also likely that the reduced abundance of Smar1 affects many facets of inflammatory signaling in macrophages in addition to the control of NF-κB crosstalk.
Crosstalk between NF-κB signaling pathways reflects the RelA response to costimulation relative to its response to stimulation by LPS or αLTβR individually (Fig. 2C). To tune crosstalk, therefore, a molecule should be capable of differentially modulating RelA response to costimulation as opposed to responses induced by individual stimuli. IκBα indiscriminately attenuates RelA activity that is induced by a wide spectrum of cell-activating stimuli. Here, we identified that IκBα, despite being a generic inhibitor, modulated NF-κB crosstalk and imparted cell type–specific control. Our mechanistic study revealed that a hyperactive Nfkbia promoter might abrogate crosstalk in both an Nfkb2-dependent or -independent manner. In the engineered fibroblast cell line, the hyperactive Nfkbia promoter rapidly attenuated LPS-induced RelA signaling (Fig. 3D) and thereby restricted the synthesis of p100 (encoded by Nfkb2) at late time points (Fig. 3E), which potentiates the crosstalk between these pathways. In macrophages, however, the hyperactive Nfkbia promoter acted independently of Nfkb2 (Figs. 4 to 6), whose LPS-induced expression was unaffected (Fig. 2A and Fig. 4F). Instead, the RelA:p52 dimer that accumulated upon costimulation of macrophages activated this hyperactive Nfkbia promoter to induce the synthesis of IκBα at late time points. In contrast, delayed IκBα synthesis in macrophages treated with LPS alone was rather subtle (Fig. 4A). Therefore, the hyperactive Nfkbia promoter in macrophages differentially affected the activity of RelA at late time points depending on whether the cells were stimulated with LPS alone or with both LPS and αLTβR (Fig. 1D and Fig. 4, F and H). Together, these data suggest that IκBα enforces cell type–specific crosstalk control by distinguishing between stimulation through TLR4 alone and costimulation through both TLR4 and LTβR.
Regulatory mechanisms governing the cell type–specificity of signaling have remained an active area of research (35). It is thought that stimulus-induced activation of ubiquitous transcription factors is mostly conserved among different cell types, and that the collaboration of these common transcriptional activators with cell lineage–restricted transcription factors generates cell type–specific responses (36, 37). For example, the lineage-restricted transcription factor PU.1 directs the activity of stress-inducible transcription factors, such as RelA, to a subset of enhancers to tune macrophage-specific responses to endotoxins (38). Similarly, Oct4 in stem cells, MyoD1 in myotubes, and PU.1 in pro-B cells determine the genome occupancy of Smad3 to orchestrate cell type–specific controls over the transforming growth factor–β (TGF-β) pathway (39). Here, we illustrated that the quantitative differences in biochemical rate parameters associated with a stress-signaling pathway imparted cell type–specific control over the activity of ubiquitous transcription factors. In particular, the rate of IκBα synthesis, which characterizes promoter strength, appeared to specifically determine the magnitude of RelA activity at late time points in response to the costimulation of TLR4 and LTβR in different cell types.
In a previous study, an NF-κB network model was constructed from fibroblast-based experimental data, and it was further refined to capture the complexity of NF-κB signaling in dendritic cells (31). Our investigation reiterates the suitability of this method by recapitulating macrophage RelA signaling, particularly its regulation by crosstalk. In general, we argue that a core network model can be conveniently adapted for various cell types, and that such an approach may offer mechanistic insights into cell type–specific regulations. The LPS-induced activation of RelA exhibits cell-to-cell variations within a population in single-cell studies (40, 41). However, our mathematical model assumed deterministic cell behavior, and our experimental system relied on bulk measurements of biochemical entities. A theoretical study suggested that stochastic variations in the abundance of IκBα underlie the differences in the NF-κB activity at the single-cell level and may provide for distinct types of crosstalk regulation (42). In future, the possible contributions of cell-to-cell variations in crosstalk between NF-κB pathways should be examined.
Crosstalk between the NF-κB pathways prolongs RelA activity in epithelial cells during bacterial infections and is important for the protective innate immune response (8). Similarly, BAFFR-dependent noncanonical NF-κB signaling sustains BCR-stimulated canonical NF-κB signaling through crosstalk to potentiate the proliferation of B cells (7). In physiological settings, macrophages receive signals from myriad cell-activating cues that likely result in the concomitant activation of both canonical and noncanonical NF-κB pathways (17). Persistent NF-κB activity in macrophages, however, leads to autoimmune and inflammatory disorders, as well as to neoplastic diseases (43). Through our investigation of the cell type–specific control of crosstalk, we would suggest that macrophages use a distinct mechanism to insulate canonical NF-κB signaling from the noncanonical pathway. Homodimeric p52 complexes restrict the expression of RelA target genes in myeloid cells (16). Our data suggest that by disrupting crosstalk between the NF-κB–activating pathways, macrophages additionally ensure the transient activation of nuclear RelA during canonical signaling. We hypothesize that this insulation of NF-κB– activating pathways in macrophages avoids the deleterious effects of inflammation during they physiological immune response. Future studies should examine potential aberrations in the cell type–specific control of crosstalk in inflammatory and autoimmune diseases. NF-κB activity is mediated by multiple dimeric transcription factors. Canonical signaling activates cRel activity in certain cell types, and RelB-containing NF-κB dimers limit the expression of inflammation-associated genes (8). In this context, it is important to investigate potential crosstalk-mediated regulation of other NF-κB dimers in physiological settings.
Materials and Methods
Cell culture and reagents
Wild-type (WT) or specific gene-deficient C57BL/6 mice were housed in the small animal facility at NII and used in accordance with the guidelines of the Institutional Animal Ethics Committee. MEFs, used either as primary cells or subsequent to their immortalization by the 3T3 protocol, were generated from E13.5 embryos as described previously (8, 18). J774.1 and RAW 264.7 cells were obtained from the ATCC. Bone marrow cells derived from WT or Nfkb2-/- mice were differentiated into macrophages in vitro as described previously (44). The HRS.puro retroviral construct expressing mouse IκBα from an engineered transgenic promoter containing five tandem κB sites was a gift from Riku Fagerlund, UCSD (45). Briefly, the tetO7 promoter present in the HRSp-puro retroviral plasmid was replaced with the cDNA sequence derived from a five-κB site–driven luciferase construct (pOSA) to generate the five-κB promoter construct. Through site-directed mutagenesis, we individually mutated four of the five κB sites to generate the one-κB promoter construct (see fig. S4B for sequences). For knockdown studies, we used the previously described pSIREN-RetroQ-ZsGreen1 (Clonetech) based retrovirus particles expressing an shRNA specific for Smar1 (shRNA sequence: 5’-GGATCAAGC AGAGCATTGA-3’) or a scrambled shRNA (shRNA sequence: 5’-ACCGAAGGC AAGCAAAGCTT-3’) to transduce fibroblasts (34). For overexpression studies, we transfected RAW 264.7 cells with either p3XFLAG-CMV or p3XFLAG-CMV-Smar1 with Lipofectamine2000 (Invitrogen), as described previously (34). Cells were stimulated 40 hours after transfection.
Biochemical analyses
Cells were stimulated with αLTβR (0.3 µg/ml, a gift from J. Browning and A. Papandile, Biogen, Cambridge, USA), LPS (1 μg/ml, Enzo, USA), or a combination thereof. Subsequently cells were harvested, and nuclear, cytoplasmic, or whole-cell extracts were prepared and analyzed by EMSA, super-shift analysis, or Western blotting or were subjected to immunoprecipitation as described previously (8). The gel images were acquired with a PhosphorImager (GE, Amersham, UK) and quantified with ImageQuant 5.2 software. Western blotting analysis of immunoprecipitates was performed with TrueBlot (eBioscience, San Diego, USA). The IKK2 kinase assay was performed as described previously (8).
Gene-expression analyses
Total RNA was isolated from cells treated with stimuli as indicated in the figure legends, and quantitative RT-PCR analysis was performed with the cDNA synthesis kit from BioBharati Life Sciences Ltd, as described previously (8). For ChIP studies, 3 x 106 cells were fixed with 2% formaldehyde, and the nuclei were isolated, sonicated, and lysates were subjected to immunoprecipitation with an anti-Smar1 antibody (Bathyl Laboratories). Subsequent to reversal of crosslinking, genomic DNA fragments were extracted from the pellet, and the presence of the Nfkbia locus was determined by semi-quantitative PCR, as described previously (28).
Computational modeling
NF-κB Systems Model v. 1.0 was published previously (8). In brief, biochemical reactions involving the association or dissociation, synthesis or degradation, and nuclear import or export of various NF-κB and IκB species were described with mass action kinetics. The transcription of genes encoding Nfkbia or Nfkb2 was computed as a sum of constitutive and NF-κB-dependent transcriptions. A Monte Carlo simulation was designed for variance-based global sensitivity analyses as described previously (21), and was implemented in MATLAB (version 2012a). Total effect index, which captured the importance of a parameter group in regulating crosstalk, was determined based on algorithms described by Saltelli et al. (21). A detailed description of Monte Carlo sampling and related methods can be found in the Supplementary Materials. Nominal values of the individual parameters and various parameter groups are described in table S1. Notations used in the computational model are listed in table S2. Computation simulations were performed involving the PF or PM parameter ensemble to predict the abundance of various molecular species in fibroblasts and macrophages, respectively. The refinement of parameters and input values to capture macrophage NF-κB signaling was described in the text. The abundance of molecular species during early signaling was determined as the area under the respective time-course curve between 1 and 2 hours after the stimulation and averaged over time in hours. Similarly, the abundance of various molecular species during late signaling was estimated as an average between 16 and 24 hours after the stimulation time period.
Statistical analysis
Error bars in the figures show the SEM of at least three biological replicates. Statistical significance was calculated by two-tailed Student’s t test.
Supplementary Materials
Acknowledgements
We thank N. Chatterjee, IIT-Delhi, for guidance with statistical analyses, R. Fagerlund, UCSD, for the five-κB promoter plasmid, J. Browning and A. Papandile, Biogen, for the αLTβR antibody. We also thank V. Kumar for technical help and M.V. R. Prasad for help with computational studies. Funding: This work was supported by an intermediate fellowship from Wellcome Trust DBT India Alliance (grant no: 500094/Z/09/Z) to S.B. and National Institute of Immunology-core funding. B.B. thanks the Council for Scientific and Industrial Research, India, and B.C. and T.M. thank the University Grant Commission, India for research fellowships.
Footnotes
Author contributions: B.B. performed wet laboratory experiments with assistance from T.M., N.T., and B.V. and guidance from S.C. and S.B.; B.C. performed in silico analyses under the supervision of J.G. and S.B.; and S.B. wrote the manuscript with help from B.C. and B.B.
Competing interests: The authors declare that they have no competing interests.
References and Notes
- 1.Vlahopoulos SA, Cen O, Hengen N, Agan J, Moschovi M, Critselis E, Adamaki M, Bacopoulou F, Copland JA, Boldogh I, Karin M, et al. Dynamic aberrant NF-kappaB spurs tumorigenesis: a new model encompassing the microenvironment. Cytokine & growth factor reviews. 2015;26:389–403. doi: 10.1016/j.cytogfr.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayden MS, Ghosh S. NF-kappaB in immunobiology. Cell research. 2011;21:223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunological reviews. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 4.Sun SC. The noncanonical NF-kappaB pathway. Immunological reviews. 2012;246:125–140. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yilmaz ZB, Weih DS, Sivakumar V, Weih F. RelB is required for Peyer's patch development: differential regulation of p52-RelB by lymphotoxin and TNF. The EMBO journal. 2003;22:121–130. doi: 10.1093/emboj/cdg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shinkura R, Kitada K, Matsuda F, Tashiro K, Ikuta K, Suzuki M, Kogishi K, Serikawa T, Honjo T. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-kappa b-inducing kinase. Nature genetics. 1999;22:74–77. doi: 10.1038/8780. [DOI] [PubMed] [Google Scholar]
- 7.Almaden JV, Tsui R, Liu YC, Birnbaum H, Shokhirev MN, Ngo KA, Davis-Turak JC, Otero D, Basak S, Rickert RC, Hoffmann A. A pathway switch directs BAFF signaling to distinct NFkappaB transcription factors in maturing and proliferating B cells. Cell reports. 2014;9:2098–2111. doi: 10.1016/j.celrep.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banoth B, Chatterjee B, Vijayaragavan B, Prasad M, Roy P, Basak S. Stimulus-selective crosstalk via the NF-kappaB signaling system reinforces innate immune response to alleviate gut infection. eLife. 2015;4 doi: 10.7554/eLife.05648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giacomin PR, Moy RH, Noti M, Osborne LC, Siracusa MC, Alenghat T, Liu B, McCorkell KA, Troy AE, Rak GD, Hu Y, et al. Epithelial-intrinsic IKKalpha expression regulates group 3 innate lymphoid cell responses and antibacterial immunity. The Journal of experimental medicine. 2015;212:1513–1528. doi: 10.1084/jem.20141831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siggers T, Chang AB, Teixeira A, Wong D, Williams KJ, Ahmed B, Ragoussis J, Udalova IA, Smale ST, Bulyk ML. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-kappaB family DNA binding. Nature immunology. 2012;13:95–102. doi: 10.1038/ni.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann A, Leung TH, Baltimore D. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. The EMBO journal. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harbor perspectives in biology. 2012;4 doi: 10.1101/cshperspect.a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckmann L, Nebelsiek T, Fingerle AA, Dann SM, Mages J, Lang R, Robine S, Kagnoff MF, Schmid RM, Karin M, Arkan MC, et al. Opposing functions of IKKbeta during acute and chronic intestinal inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15058–15063. doi: 10.1073/pnas.0808216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- 16.Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu ZG. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nature immunology. 2010;11:799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wimmer N, Huber B, Barabas N, Rohrl J, Pfeffer K, Hehlgans T. Lymphotoxin beta receptor activation on macrophages induces cross-tolerance to TLR4 and TLR9 ligands. Journal of immunology. 2012;188:3426–3433. doi: 10.4049/jimmunol.1103324. [DOI] [PubMed] [Google Scholar]
- 18.Basak S, Kim H, Kearns JD, Tergaonkar V, O'Dea E, Werner SL, Benedict CA, Ware CF, Ghosh G, Verma IM, Hoffmann A. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basak S, Behar M, Hoffmann A. Lessons from mathematically modeling the NF-kappaB pathway. Immunological reviews. 2012;246:221–238. doi: 10.1111/j.1600-065X.2011.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughey JJ, Lee TK, Covert MW. Computational modeling of mammalian signaling networks. Wiley interdisciplinary reviews. Systems biology and medicine. 2010;2:194–209. doi: 10.1002/wsbm.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saltelli A, Annoni P, Azzini I, Campolongo F, Ratto M, Tarantola S. Variance based sensitivity analysis of model output. Design and estimator for the total sensitivity index. Computer Physics Communications. 2010;181:259–270. [Google Scholar]
- 22.Kristensen AR, Gsponer J, Foster LJ. Protein synthesis rate is the predominant regulator of protein expression during differentiation. Molecular systems biology. 2013;9:689. doi: 10.1038/msb.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito CY, Kazantsev AG, Baldwin AS., Jr Three NF-kappa B sites in the I kappa B-alpha promoter are required for induction of gene expression by TNF alpha. Nucleic acids research. 1994;22:3787–3792. doi: 10.1093/nar/22.18.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shih VF, Kearns JD, Basak S, Savinova OV, Ghosh G, Hoffmann A. Kinetic control of negative feedback regulators of NF-kappaB/RelA determines their pathogen- and cytokine-receptor signaling specificity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9619–9624. doi: 10.1073/pnas.0812367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savinova OV, Hoffmann A, Ghosh G. The Nfkb1 and Nfkb2 proteins p105 and p100 function as the core of high-molecular-weight heterogeneous complexes. Molecular cell. 2009;34:591–602. doi: 10.1016/j.molcel.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao Z, Fusco A, Huang DB, Gupta K, Young Kim D, Ware CF, Van Duyne GD, Ghosh G. p100/IkappaBdelta sequesters and inhibits NF-kappaB through kappaBsome formation. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1408552111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malonia SK, Sinha S, Lakshminarasimhan P, Singh K, Jalota-Badhwar A, Rampalli S, Kaul-Ghanekar R, Chattopadhyay S. Gene regulation by SMAR1: Role in cellular homeostasis and cancer. Biochimica et biophysica acta. 2011;1815:1–12. doi: 10.1016/j.bbcan.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Singh K, Sinha S, Malonia SK, Bist P, Tergaonkar V, Chattopadhyay S. Tumor suppressor SMAR1 represses IkappaBalpha expression and inhibits p65 transactivation through matrix attachment regions. The Journal of biological chemistry. 2009;284:1267–1278. doi: 10.1074/jbc.M801088200. [DOI] [PubMed] [Google Scholar]
- 29.Chattopadhyay S, Kaul R, Charest A, Housman D, Chen J. SMAR1, a novel, alternatively spliced gene product, binds the Scaffold/Matrix-associated region at the T cell receptor beta locus. Genomics. 2000;68:93–96. doi: 10.1006/geno.2000.6279. [DOI] [PubMed] [Google Scholar]
- 30.O'Gorman A, Colleran A, Ryan A, Mann J, Egan LJ. Regulation of NF-kappaB responses by epigenetic suppression of IkappaBalpha expression in HCT116 intestinal epithelial cells. American journal of physiology. Gastrointestinal and liver physiology. 2010;299:G96–G105. doi: 10.1152/ajpgi.00460.2009. [DOI] [PubMed] [Google Scholar]
- 31.Shih VF, Davis-Turak J, Macal M, Huang JQ, Ponomarenko J, Kearns JD, Yu T, Fagerlund R, Asagiri M, Zuniga EI, Hoffmann A. Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-kappaB pathways. Nature immunology. 2012;13:1162–1170. doi: 10.1038/ni.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basak S, Shih VF, Hoffmann A. Generation and activation of multiple dimeric transcription factors within the NF-kappaB signaling system. Molecular and cellular biology. 2008;28:3139–3150. doi: 10.1128/MCB.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behar M, Hoffmann A. Understanding the temporal codes of intra-cellular signals. Current opinion in genetics & development. 2010;20:684–693. doi: 10.1016/j.gde.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha S, Malonia SK, Mittal SP, Singh K, Kadreppa S, Kamat R, Mukhopadhyaya R, Pal JK, Chattopadhyay S. Coordinated regulation of p53 apoptotic targets BAX and PUMA by SMAR1 through an identical MAR element. The EMBO journal. 2010;29:830–842. doi: 10.1038/emboj.2009.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bajikar SS, Janes KA. Multiscale models of cell signaling. Annals of biomedical engineering. 2012;40:2319–2327. doi: 10.1007/s10439-012-0560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nature reviews. Molecular cell biology. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nature reviews. Immunology. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 38.Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, Ragoussis J, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 39.Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee TK, Denny EM, Sanghvi JC, Gaston JE, Maynard ND, Hughey JJ, Covert MW. A noisy paracrine signal determines the cellular NF-kappaB response to lipopolysaccharide. Science signaling. 2009;2:ra65. doi: 10.1126/scisignal.2000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sung MH, Li N, Lao Q, Gottschalk RA, Hager GL, Fraser ID. Switching of the relative dominance between feedback mechanisms in lipopolysaccharide-induced NF-kappaB signaling. Science signaling. 2014;7:ra6. doi: 10.1126/scisignal.2004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mothes J, Busse D, Kofahl B, Wolf J. Sources of dynamic variability in NF-kappaB signal transduction: a mechanistic model. BioEssays : news and reviews in molecular, cellular and developmental biology. 2015;37:452–462. doi: 10.1002/bies.201400113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Molecular cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weischenfeldt J, Porse B. Bone Marrow-Derived Macrophages (BMM): Isolation and Applications. CSH protocols. 2008;2008 doi: 10.1101/pdb.prot5080. pdb prot5080. [DOI] [PubMed] [Google Scholar]
- 45.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nature immunology. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 46.Sobol’ IM. Sensitivity estimates for nonlinear mathematical models. Mathematical Modelling and Computational Experiment. 1993;1:407–414. [Google Scholar]
- 47.Homma T, Saltelli A. Importance measures in global sensitivity analysis of nonlinear models. Reliability Engineering & System Safety. 1996;52:1–17. [Google Scholar]
- 48.Saltelli A, Ratto M, Andres T, Campolongo F, Cariboni J, Gatelli D, Saisana M, Tarantola S. Global sensitivity analysis: the primer. Wiley; 2008. [Google Scholar]
- 49.Ori A, Banterle N, Iskar M, Andrés-Pons A, Escher C, Khanh Bui H, Sparks L, Solis-Mezarino V, Rinner O, Bork P, Lemke EA, et al. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Molecular Systems Biology. 2013;9:648. doi: 10.1038/msb.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.