Nestin defines a novel tendon stem/progenitor cell subpopulation with high tenogenesis potential.
Keywords: Tendons stem/progenitor cells, tenogenesis, nestin, single-cell gene analysis
Abstract
The repair of injured tendons remains a formidable clinical challenge because of our limited understanding of tendon stem cells and the regulation of tenogenesis. With single-cell analysis to characterize the gene expression profiles of individual cells isolated from tendon tissue, a subpopulation of nestin+ tendon stem/progenitor cells (TSPCs) was identified within the tendon cell population. Using Gene Expression Omnibus datasets and immunofluorescence assays, we found that nestin expression was activated at specific stages of tendon development. Moreover, isolated nestin+ TSPCs exhibited superior tenogenic capacity compared to nestin− TSPCs. Knockdown of nestin expression in TSPCs suppressed their clonogenic capacity and reduced their tenogenic potential significantly both in vitro and in vivo. Hence, these findings provide new insights into the identification of subpopulations of TSPCs and illustrate the crucial roles of nestin in TSPC fate decisions and phenotype maintenance, which may assist in future therapeutic strategies to treat tendon disease.
INTRODUCTION
Tendons are specialized tissues that connect bone to muscle, transmitting the forces generated by these musculoskeletal tissues to enable body movement. Tendon injuries due to overuse or age-related degeneration are a common clinical problem. However, the hypocellularity and hypovascularity of tendon tissue make its natural healing extremely slow and inefficient, and the structural integrity and mechanical strength of normal undamaged tendons are rarely attained after healing. The repair of injured tendons remains a formidable challenge, largely due to our limited understanding of the basic biology of tendon stem cells and the regulation of tenogenesis (1, 2).
Tendon stem/progenitor cells (TSPCs) were first identified by Bi et al. (3) and exhibited the various common properties of stem cells. Since then, stem cells from tendon tissues were isolated from various species and were shown to display clonogenicity, multipotency, and self-renewal (4–7). Despite these findings and extensive studies of TSPCs in tissue engineering application, there is a huge need for reliable and specific markers to characterize and define the biological characteristics of TSPCs and their subpopulations in vitro and to study their identity and functions in vivo. To date, a broad panel of several stem cell markers is used to define TSPCs, which meet the marker criteria for mesenchymal stem cells (MSCs). The lack of a specific marker and a subpopulation of TSPCs is in part due to the limited number of lineage markers and the effects of ensemble averaging in conventional transcriptome analysis on cell populations. During embryonic development, expression of Scleraxis (Scx), the earliest known marker of tendon progenitor cells and tenocytes, is highly specific. Because Scx-null mouse showed severe tendon defects, Scx has been described to play a vital role in tendon development (8). More recently, the transcription factors Mohawk (Mkx) and Egr1 were found to be essential for tendon development during embryogenesis, through the transforming growth factor–β (TGF-β) pathway (9–13), but these two transcription factors are not specific to tendons (14). Scx can also be classified as the first signal for tendon progenitor generation. Moreover, Scx+/Sox9+ progenitors contribute to the establishment of the chondrotendinous/chondroligamentous junction, which indicates that tendon progenitor cells may be divided into different stages and subpopulations. Although some tendon-related genes are highly expressed in TSPCs, such as the genes for the matrix proteins collagen type I (Col I), collagen type III (Col III), and tenascin C (TnC) and the transcription factors Scx and tenomodulin (Tnmd), these genes alone are not useful for distinguishing TSPCs from tendon progenitor cells or tenocytes. Isolated TSPCs are a heterogeneous population, potentially reflecting different propensity in multilineage differentiation (3), which raises questions about the identity of the TSPC subpopulation that can contribute to tenogenesis. The ability to specifically identify and isolate the TSPC subpopulation will not only facilitate their clinical applications but also provide greater understanding of tendon development. Advancement in single-cell quantitative gene analysis allows unbiased and high-throughput analysis of gene expression profiles of individual cells for investigating the heterogeneity of cell population (15, 16). This approach overcomes the limitations involving heterogeneous cell populations and sample amounts and may shed light on the subpopulation and unique markers in TSPCs.
Nestin, a type IV intermediate filament (IF) protein, is also expressed in tissues outside the central nervous system, particularly in proliferating cells during the developmental stages of a variety of tissues (17). Nestin was also expressed in a variety of adult stem/progenitor cell populations (18–23) and was found to be important for the proper self-renewal of stem cells (24–26). Upon analyzing Gene Expression Omnibus (GEO) datasets, we noticed that nestin expression was elevated during initiation of tenogenesis and at specific stages of differentiation during development. It was reported that nestin was expressed by perivascular cells and some spindle-shaped cells in adult tendon tissue (27), as well as at myotendinous junctions (28, 29). Additionally, it was noticed that the TSPCs isolated from human Achilles tendon expressed a high level of nestin (30), which indicates that nestin might be a candidate marker for TSPCs. Moreover, the single-cell gene analysis of tendon-derived cells revealed the enrichment of nestin in TSPCs, which inspired us to investigate the expression and functional roles of nestin in tendon stem cells during tendon differentiation and injury repair. This offers new insights into the biology of tendon stem cells and may assist in future cell-based therapy of tendon injury.
RESULTS
Gene expression profiling distinguishes three major subpopulations of tendon-derived cells
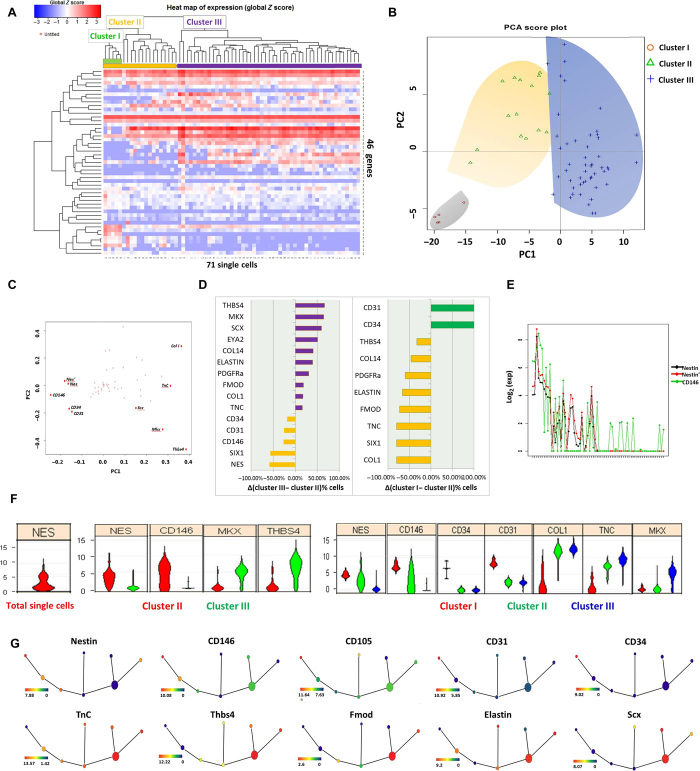
For molecular profiling of individual cells derived from adult tendon, we used highly parallel single-cell quantitative reverse transcription polymerase chain reaction (qRT-PCR) on the Biomark (Fluidigm) platform, analyzing a total of 46 genes from each cell. We used the C1 Single-Cell AutoPrep System to isolate individual cells from adult tendons for single-cell transcriptional analysis. Hierarchical clustering (HC) of the cells based on their gene expression profiles revealed that cells derived from tendons comprised two major clusters (Fig. 1A). One population (cluster II), marked in orange, displayed relatively high expression levels of nestin, whereas the other population (cluster III), marked in purple, displayed low expression levels of nestin but had elevated expression of Scx, Col I, TnC, Thbs4, Mkx, Comp, and Eln (Fig. 1A). Within these two major populations, at least one subpopulation was further identified: cluster I, marked in green, was readily discerned as a subpopulation within cluster II and expressed high levels of CD31 and CD34. To reduce data complexity, we used principal components analysis (PCA). A projection of the cells’ expression patterns onto PC1 and PC2 could differentiate individual cells into three distinct subpopulations (Fig. 1B). PC1 separates the two clusters, indicating that this is the primary source of variation in the dataset. PC2 largely separates a further subcluster from the remainder of cluster II cells. When we projected the first two PC loadings for all 46 transcripts, we could categorize two distinct cohorts of genes based on high-differential loadings between PC1 and PC2 (Fig. 1C). In addition, comparison of the relative proportion of cells expressing individual genes and the expression levels of individual genes reveals that teno-lineage–related transcripts are linked to cells belonging to cluster III, distinguishing these cells from cluster II and indicating that cluster III may be differentiated tenocytes (Fig. 1D). Comparison of the relative proportions of cells between cluster I and the remainder of cluster II showed that all the cells in cluster I were CD31+ and CD34+, but a much larger number of cells express teno-lineage–related genes in the remainder of cluster II, which are likely to be TSPCs (Fig. 1D). Correlation analysis was conducted on the basis of nestin expression, and CD146 turned out to have the strongest positive correlation (R = 0.753). Meanwhile, the two primers of nestin showed perfect consistency (R = 0.991) (Fig. 1E). Violin plots, which depict the probability density of the data at different values, showed bimodal distributions, indicating that the nestin gene was differentially expressed by at least two subpopulations among these single cells isolated from the tendon (Fig. 1F). Furthermore, feature reduction by analysis of variance (ANOVA) revealed a reduced set of markers with high differential expression between the clusters. Upon comparing cluster II with cluster III, we found that the multipotent stem cell marker nestin, together with CD146, was enriched in cluster II, whereas the tendon-related markers Mkx and Thbs4 were highly expressed in cluster III (Fig. 1F). Upon separating cluster I from cluster II, we found that Nes, CD146, CD34, CD31, Col I, TnC, and Mkx were significantly differentially expressed and were among the top 10 differentially expressed genes ranked by ANOVA P values (Fig. 1F), thus suggesting that there are two subpopulations of nestin+ cells. The tendon-derived cells in cluster I were CD31+ and C34+, indicating their endothelial or hematopoietic origin, whereas the remaining cells in cluster II that expressed both intermediate levels of stem cell markers and teno-lineage markers are likely to be TSPCs. We used spanning-tree progression analysis of density-normalized events (SPADE) to distill multidimensional single-cell data down to a single interconnected cluster of transitional cell populations. In this tree plot, each node of cells is connected to its “most related” node of cells (16). This unsupervised computationally constructed hierarchy showed the interconnected clusters of rare, transitional, and abundant cell subpopulations in tendon-derived cells. The overlaid expression of different genes revealed the different cell clusters with distinct, regimented profiles (Fig. 1G). In Fig. 1G, red to yellow suggests high to middle expression, whereas green to blue suggests low to no expression. On the basis of the teno-lineage–related TnC, Thbs4, Fmod, Elastin, and Scx gene expression, the biggest node represented the tenocyte population that expressed the highest level of teno-lineage markers and a relatively low level of stem cell markers. On the basis of stem cell marker (Nes, CD146, CD31, CD34, and CD105) expression on different nodes, the nodes at the left sides were likely TSPCs at different stages, whereas some nodes and branches encompassed transitional and less-understood cell types. Collectively, single-cell transcriptional profiling showed the heterogeneity of tendon-derived cells and the existence of nestin+ TSPCs.
Fig. 1. Single-cell transcriptional profiling reveals distinct subpopulations of tendon cells and their corresponding markers.
(A) Heat map showing that the unbiased HC well separates the single-cell gene expression profiles of 71 individual cells isolated by the C1 system and analyzed for expression of 46 gene transcripts by qRT-PCR. Top colored bars indicate clusters identified as clusters I (green), II (orange), and III (purple). (B) PCA of the full qRT-PCR results of 71 tendon-derived cells. Each dot represents a cell, colored according to its subpopulation as determined by HC. Cells are plotted against the first two PCs. (C) Genes projected onto the first two PC loadings. (D) Different genes with expression that was on or off in each group (cluster II/cluster III and cluster I/remainder of cluster II). The proportion differences of cells between clusters II and III, in descending order from top to bottom, are shown. Purple indicates genes that are overrepresented in cells of cluster III. Orange indicates genes that are overrepresented in cells of cluster II. Green represents genes that are overrepresented in cells of cluster I. (E) Correlation analysis of nestin (NES). (F) Violin plots represent expression of a number of genes (fold change above background) in individual cells. (G) SPADE from single-cell expression pattern of 46 genes. Overlaid expression patterns of different genes contribute in defining distinct cell populations.
Nestin expression is implicated in tendon differentiation
Analysis of nestin expression in GEO datasets obtained from forelimbs and hindlimbs during mouse embryogenesis showed that nestin expression steadily increased from E10.5 (embryonic day 10.5) to E13.5, which is correlated with up-regulated expression of teno-lineage–specific markers (Tnmd, Thbs4, Mkx, and Egr1) during limb development (fig. S1A). The datasets obtained from tendon cells that were isolated on the basis of Scx expression at different stages of development also showed that Nes expression steadily increased from E11.5 to E14.5 (Fig. 2A). The E11.5 stage was chosen for the selection of tendon progenitors, and the E14.5 stage was used to target tendon-differentiated cells (14). Expression of tendon lineage–specific genes Tnmd, Thbs4, Mkx, and Egr1 was significantly elevated from E11.5 to E14.5 stage. In addition, the RNA sequencing (RNA-seq) datasets generated from isolated cells from the developing mouse limbs at E11.5, E13.5, and E15.5 exhibited markedly increased expression of nestin (fig. S1B). Notably, the expression of tendon markers (Tnmd, Thbs4, Mkx, and Egr1) was gradually increased in Scx+ tendon cells from E11.5 to E15.5 (fig. S1B). Furthermore, comparison of the gene expression profiles of Scx-GFP+ and Scx-GFP− cells in the E13.5 limb revealed enhanced expression of nestin in Scx-GFP+ cells, which is also consistent with the up-regulation of other tendon marker genes (Tnmd, Thbs4, and Mkx) (Fig. 2B). Hence, the aforementioned results from three independent transcriptomic analyses indicated a positive correlation between nestin and teno-lineage–specific genes during tendon development. Subsequently, immunohistochemistry (IHC) analyses of postnatal tendons showed that nestin was highly expressed between postnatal days 10 and 14 in rat Achilles tendon (fig. S1C). Similarly, the number of Nes-GFP+ cells reached a peak on postnatal days 10 to 14 in Nes-GFP transgenic mouse Achilles tendon, which then decreased with age (Fig. 2C). Thus, these data suggest the involvement of nestin in specific stages of limb and tendon development. We also monitored the mRNA levels of nestin and transcription factors related to the tendon using qRT-PCR analyses. The expression level of nestin was gradually decreased with age. Consistent with previous microarray data, the expression profiles of the tendon-specific transcription factors Mkx and Scx, together with Tnmd and Eln, resembled the expression profile of nestin in the Achilles tendon (Fig. 2D). The expression of nestin peaks at specific stages of tendon postnatal development, indicating that nestin may play a role in tendon tissue differentiation and that nestin+ cells more likely serve as TSPCs. We compared nestin expression in three cell sources, and the qPCR results showed that TSPCs express a significantly higher level of nestin than do tenocyte and bone marrow MSC (BMSC) (fig. S2A). TSPCs also express significantly higher tendon-related markers, such as Mkx and Scx, than do BMSCs (fig. S2A). Immunofluorescence staining of nestin also showed that there were more nestin+ cells in TSPCs than in BMSCs and tenocytes (fig. S2B).
Fig. 2. Nestin is implicated in tendon development.
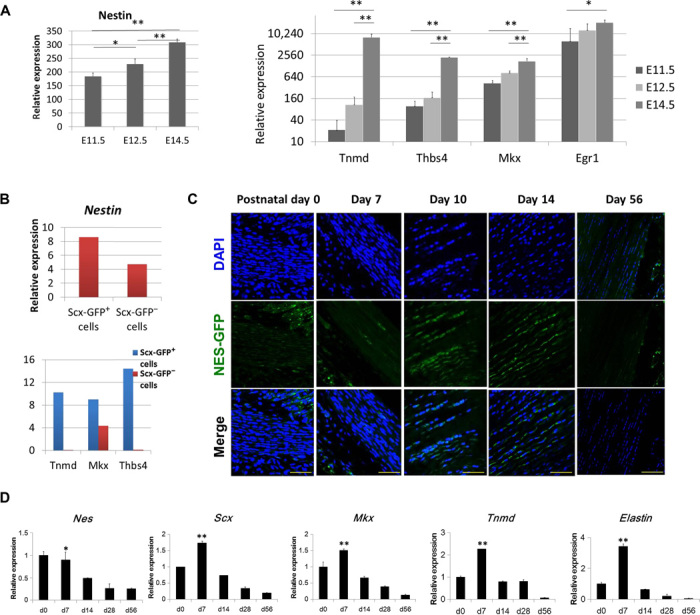
(A) Expression of nestin and teno-lineage marker genes obtained from Scx-GFP forelimb tendon during mouse embryogenesis. (B) Expression of nestin and teno-lineage marker genes in Scx-GFP+ cells and Scx-GFP− cells obtained from E13.5 forelimb samples. Data are presented as means ± SD. (C) Nes-GFP expression in the developing mouse Achilles tendon between postnatal days 0 and 56 (n = 5). DAPI, 4′,6-diamidino-2-phenylindole. (D) Nestin and teno-lineage marker gene expression in the developing mouse Achilles tendon between postnatal days 0 and 56. Data are presented as means ± SD. Scale bars, 50 μm.
Nestin+ cells participate in endogenous tendon injury repair
Although nestin+ cells in tendon tissue gradually decreased with age, there were nestin+ cells located mainly in the endotenon and peritenon regions of adult human tendon tissues, particularly in the perivascular region (Fig. 3A). It was shown that these nestin+ cells coexpressed the teno-lineage marker Scx by using Scx-GFP transgenic mice (Fig. 3B). Additionally, immunofluorescence staining showed that nestin+ cells in human tendon tissue also coexpressed the multipotent stem cell marker CD146 (Fig. 3C) (31). Aside from examining the physiological expression of nestin, we also analyzed the nestin expression pattern under pathological conditions in a transgenic Nes-GFP mouse model of Achilles tendon injury. Examination of GFP fluorescence revealed that Nes-GFP+ cells accumulated at the injury site 1 week after injury and that the number of Nes-GFP+ cells decreased gradually with time (Fig. 3D). Furthermore, the Nes-GFP+ cells also expressed the tendon stem cell markers CD146 and CD105 (Fig. 3E) (3, 32). Collectively, these data suggest that endogenous tendon stem cells are activated upon injury and that nestin could be a reliable marker for functional TSPCs.
Fig. 3. Localization of nestin+ cells in tendon tissue and endogenous tendon injury repair.
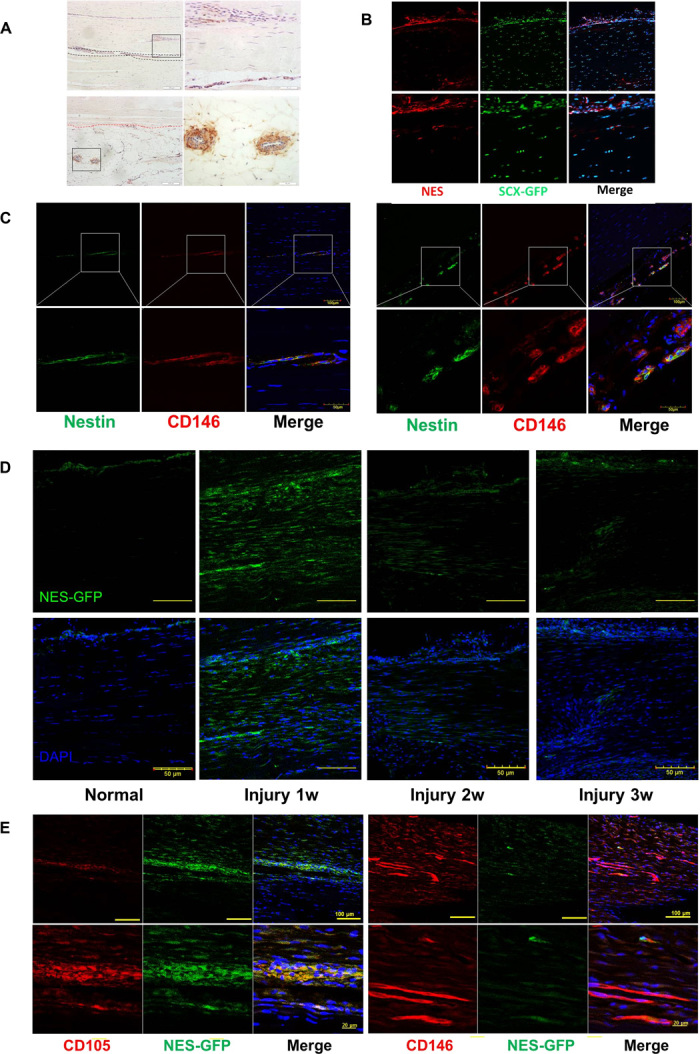
(A) Immunofluorescence staining of nestin expression in the human Achilles tendon endotenon (region between the black dashed lines) and surrounding blood vessels in the peritenon (area below the red dashed line). Scale bars, 200 μm (right); 50 μm (left). (B) nestin expression in Scx-GFP mouse Achilles tendon. (C) Nestin and CD146 expression in human Achilles tendon. Scale bars, 100 μm (top); 50 μm (bottom). (D) Nes-GFP expression in normal and injured Achilles tendon 1, 2, and 3 weeks after surgery (n = 5). Scale bars, 50 μm. (E) Immunofluorescence staining of CD105 and CD146 expression at the injured tendon of Nes-GFP mice 1 week after injury (n = 5). Scale bars, 100 μm (top); 20 μm (bottom).
Nestin+ TSPCs exhibit strong capacity for self-renewal and tenogenesis
To investigate the phenotypic differences between nestin+ and nestin− TSPCs, we first isolated TSPCs and demonstrated that they were CD34−CD18−CD44+CD90+CD105+ (fig. S3A), as previously reported (3), and then isolated nestin+ and nestin− colonies from TSPCs on the basis of nestin expression at the transcriptional level (Fig. 4A). The nestin+ TSPCs expressed significantly higher levels of CD146 (Fig. 4B), as well as other stem/progenitor cell–related surface marker genes, such as CD105, CD90, CD51, CD44, and CD29 (fig. S3B), as compared to nestin− TSPCs. Immunofluorescence staining for nestin and CD146 in TSPCs and individual colonies showed their correlated expression at the protein level (Fig. 4C). The colony formation assay revealed that nestin+ TSPCs exhibited significantly higher self-renewal capacity than did nestin− TSPCs (Fig. 4, D and E). However, the proliferation capacity of these two groups of cells was similar (Fig. 4F). Moreover, three-lineage differentiation potential toward the osteogenic, adipogenic, and chondrogenic lineages was evaluated, and there was no significant differences between the nestin+ and nestin− groups, as evidenced by staining with alkaline phosphatase (ALP), Alizarin Red S, Oil Red O, and Safranin O (Fig. 4, G to I). Ultrastructural analyses demonstrated that the average diameter of collagen fibrils formed in vitro by the nestin+ group was 1.7-fold of that in the nestin− group (21.9 ± 6.5 nm versus 12.7 ± 2.7 nm, P < 0.01) (Fig. 4M). We also quantified the distribution of collagen fibril diameters in the two groups and found sharp unimodal distributions of diameters centered around 12 nm in the nestin− group, whereas heterogeneous diameters were observed in the nestin+ group (Fig. 4, K and L). Consistent with the knockdown results, the expression of nestin led to strong induction of Scx and Mkx, as well as the tendon-related matrix genes Eln, Col I, and Col XIV (Fig. 4J), indicating that nestin is essential for tenogenesis of TSPCs. Together, the nestin+ TSPCs displayed a significantly higher capacity for colony formation and tenogenesis than did nestin− TSPCs.
Fig. 4. Nestin+ TSPCs displayed stronger capacity for tenogenesis than did nestin− TSPCs.
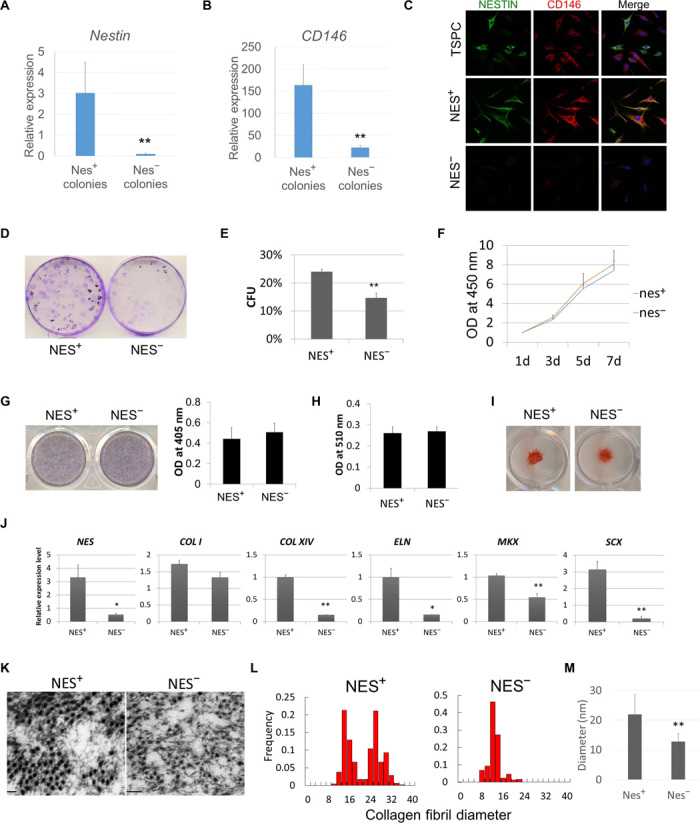
qPCR analyses of nestin (A) and CD146 (B) expression in selected nestin+ and nestin− colony-forming TSPCs (n = 6, representative data from three independent experiments). (C) Immunofluorescence staining of nestin and CD146 expression in TSPCs, selected nestin+ TSPCs, and nestin− TSPCs. (D) CFU assay for assessing the self-renewal of nestin+ and nestin− TSPCs. (E) Quantification data of the CFU assay (n = 6). (F) Proliferation rate of nestin+ and nestin− TSPCs (n = 6). OD, optical density. (G) ALP staining and quantification of Alizarin Red S staining of nestin+ and nestin− TSPCs after osteogenic induction (n = 6). (H) The quantification of accumulated lipid vacuoles by Oil Red O staining showed no difference in adipogenic potential between nestin+ and nestin− TSPCs (n = 6). (I) Safranin O staining of nestin+ and nestin− TSPCs after chondrogenic induction. (J) Tendon-associated gene expression in cell sheet formed by nestin+ and nestin− TSPCs (n = 6). (K) Transmission electron micrographs of cross sections of collagen fibrils within cell sheets formed by nestin+ and nestin− TSPCs. (L) Histogram showing the distribution range of collagen fibril diameters in nestin+ and nestin− TSPCs. (M) Average diameter of collagen fibrils in nestin+ and nestin− TSPCs (n = 6). Results are presented as means ± SD.
Effects of nestin knockdown on colony-forming unit capacity and morphology of TSPCs
To determine how nestin expression influences the phenotypic characteristics of TSPCs, we silenced its expression with two lentiviral vectors encoding short hairpin RNA (shRNA) sequences against nestin, and TSPCs displayed a marked reduction in nestin expression, as shown by qPCR and Western blot (Fig. 5A). Colony-forming unit (CFU) assays revealed that nestin knockdown significantly decreased the self-renewal capacity of TSPCs, which was 2.5-fold lower than that of the scrambled shRNA control treated in parallel (Fig. 5B). The unique spindle shape was determined to be a crucial marker in evaluating successful teno-lineage differentiation (33). Another marked effect of nestin knockdown is the loss of the typical spindle-like shape of TSPCs, as evidenced by a smaller radius ratio compared to the control (Fig. 5, C and D). The average diameter of floating TSPCs was significantly increased for both shRNA sequences against nestin (fig. S4A). Under confocal fluorescence microscopy, the change in adherent cell morphology was also apparent with tetramethyl rhodamine isothiocyanate–phalloidin–stained actin (Fig. 5C), with obvious enlargement of TSPCs subjected to nestin knockdown. The immunofluorescence staining of vinculin, which is a component of the focal adhesion complex, showed the presence of focal adhesions and their distribution (fig. S4B). The quantitative data showed that nestin knockdown enhanced focal adhesion formation in TSPCs (fig. S4C). The type IV IF protein nestin can copolymerize with other types of IF proteins because of its very short N-terminal head domain. The abundant expression of vimentin was comparable between nestin knockdown and scrambled shRNA control–treated cells (fig. S4B), thus suggesting that vimentin assembly and disassembly are not affected by nestin deficiency in TSPCs. There were no significant differences in the proliferation rates among these groups, upon carrying out the CCK8 and Edu assays (fig. S5). The cell viability assay showed that nestin expression was related to the sensitivity of TSPC to serum starvation–induced cell death, oxidant-induced cell death, and staurosporine-induced apoptosis (fig. S6). This indicated that nestin was a survival determinant in TSPCs. The multidifferentiation potential of TSPCs toward the adipogenic, osteogenic, and chondrogenic lineages was not affected by nestin knockdown.
Fig. 5. Nestin is important for self-renewal and phenotypic maintenance of TSPCs.
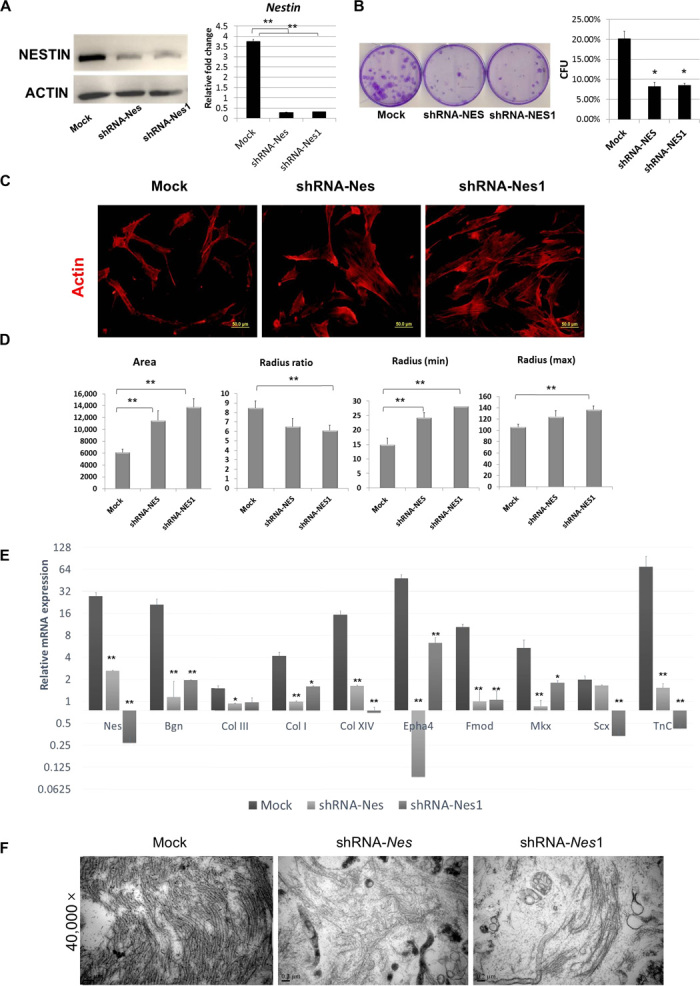
(A) Western blot and qPCR demonstrated successful knockdown of nestin expression in TSPCs. (B) CFU assay showed that self-renewal of TSPCs was decreased by the knockdown of nestin (n = 3). (C) Immunofluorescence staining of F-actin, expression in TSPCs infected with RNA interference (RNAi) against nestin (shRNA-Nes/shRNA-Nes1), or a control scrambled sequence (mock). (D) Quantitative data on cell morphology change of TSPCs under nestin knockdown. Results are presented as means ± SD (n = 40 to 60). Scale bars, 50 μm. (E) qPCR analyses showing that the expression of tendon-associated genes in TSPC cell sheets from the control group was significantly higher than that from the nestin knockdown group (n = 5). (F) Transmission electron micrographs of collagen fibrils in cell sheets from the control and nestin knockdown groups. *P < 0.05, **P < 0.01 (one-way ANOVA). Results are presented as means ± SD (n = 5). Scale bars, 50 μm (B); 200 nm (D).
Nestin knockdown resulted in the loss of tenogenic capacity of TSPCs
To investigate whether the tenogenic capacity of TSPCs is affected by nestin knockdown, the cell sheet–forming capacity of TSPCs was assessed in a previous report (11). Knockdown of nestin led to a strong reduction of tendon-specific gene expression, including that of the tendon matrix genes Bgn, Col I, Col III, Col XIV, Fmod, and TnC and the tendon-specific transcription factors Scx and Mkx. Among these, Bgn, Col I, Col XIV, Fmod, Epha4, TnC, and Mkx were significantly down-regulated in TSPCs silenced by both shRNA-Nes sequences, as compared to the control group (Fig. 5E). Because collagen fiber is the main extracellular matrix component of the tendon, we evaluated collagen fibril formation in these groups using transmission electron microscopy (TEM). The collagen fibril formation capacity of TSPCs was markedly hampered by nestin knockdown, as evidenced by hardly any observable collagen fibril bundles in the two shRNA-Nes groups (Fig. 5F). Together, these data indicate that nestin plays a crucial role in the tenogenic differentiation capacity of TSPCs.
Knockdown of nestin impaired tendon repair and regeneration in a rat model of patellar tendon defect
Patellar window defect is possible with clinical anterior cruciate ligament reconstruction in the middle one-third of the patellar tendon graft. We further evaluated the effects of nestin knockdown on the tenogenic differentiation capacity of TSPCs in a tendon regeneration model in vivo by surgically creating a rat model of patellar tendon window defect. After 2 weeks in vivo, a great number of cells exhibiting spindle-shaped morphology with organized collagen deposition were observed in the control group under hematoxylin and eosin (H&E) staining (Fig. 6A). In the nestin knockdown TSPC group, the cells displayed round shapes and random arrangement (Fig. 6A). Additionally, Masson’s trichrome staining showed that the arrangement of collagen fibril was less aligned in the nestin knockdown groups (Fig. 6A). The maturation of the repaired tendon was also assessed by histological scoring (Fig. 6B), and the nestin knockdown group displayed a significantly higher score than did the control group at both 2 and 4 weeks after implantation (Fig. 6B; P < 0.05). Histological examination revealed the formation of the regenerated tendon with the typical structure of a mature tendon at the repair site in the TSPC group 4 weeks after implantation (Fig. 6C). H&E staining showed that, in contrast to the polycellular collagen fibrils of the nestin knockdown TSPC group, the repaired tendons of the control TSPC group consisted of bands of collagen fibrils with small crimped patterns and tenocytes with pronounced spindle-shaped morphology 4 weeks after implantation (Fig. 6C). Moreover, polarized light microscopy showed that the collagen fibrils were more mature in the TSPC group, as compared to the nestin knockdown TSPC group at both 2 and 4 weeks after implantation (Fig. 6, A and C). These data thus indicate that the quality of the repaired tendon in the nestin knockdown groups was worse than the control group. Additionally, less collagen was deposited in the nestin knockdown group, as demonstrated by Masson’s trichrome staining and collagen content assay (Fig. 6, C and D). Furthermore, cross sections of the repaired tendon revealed a heterogeneous pattern of collagen fibrils in the control TSPC group and homogeneous patterns in the nestin knockdown groups 4 weeks after implantation, as shown by the measurements of collagen fibril diameters and their distribution (Fig. 6E). The average diameter of the collagen fibrils in the control group was 1.2-fold of that of the nestin knockdown group (47.79 ± 5.56 nm versus 39.75 ± 7.04 nm, P < 0.05) 4 weeks after implantation. Additionally, it was observed that the control TSPC group had better biomechanical properties than the nestin knockdown TSPC group. The failure force, modulus, and stress at failure were 1.48-fold (P < 0.05), 1.77-fold (P < 0.05), and 1.89-fold (P < 0.01) higher, respectively, in the control TSPC group than in the nestin knockdown group (Fig. 6F). Last, the expression of the tendon-specific transcription factors Six2 and Egr1 was significantly higher in the control group than in the nestin knockdown group 2 weeks after surgery (Fig. 6G). The other teno-lineage marker gene Tnmd and the tendon matrix genes Col I, Dcn, and TnC were also significantly more highly expressed in the control group than in the nestin knockdown group during the early stage of tendon tissue repair and regeneration (Fig. 6G). Collectively, the data thus indicated that nestin expression is crucial for tendon regeneration in vivo.
Fig. 6. Knockdown of nestin impaired tendon repair and regeneration in a rat model of patellar tendon defect.
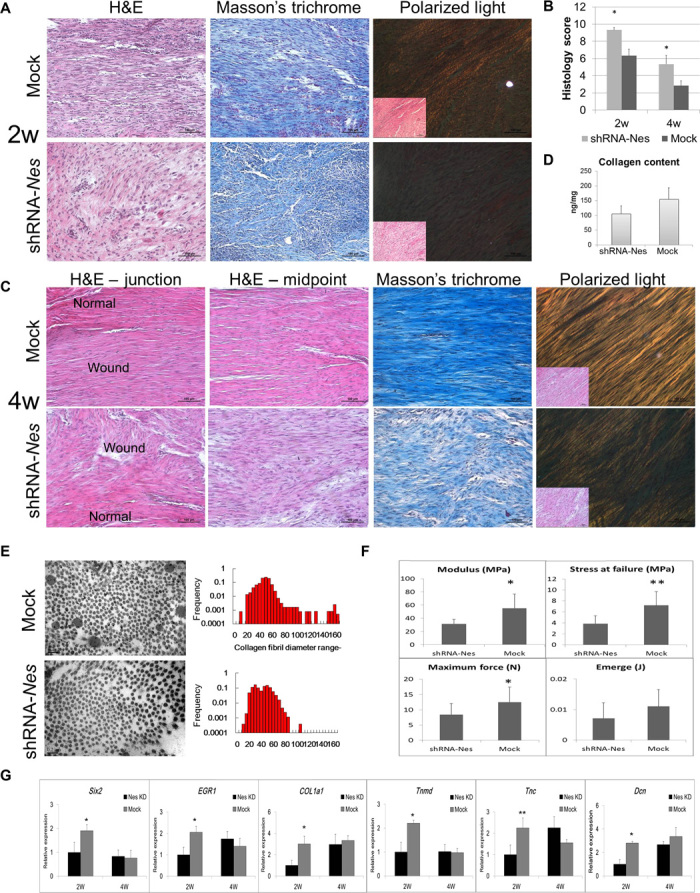
(A) Morphology of the repaired tissue site along the axis of the tendon in the control and nestin knockdown groups. H&E staining and polarized light microscopy images showing the collagen fibrils 2 weeks after implantation (n = 6). (B) Maturation of repaired tendon was assessed by histology scoring (n = 6). (C) H&E staining showing the histology of the junction between normal tendon and neo-tendon, as well as the midpoint of neo-tendon formation 4 weeks after implantation. Masson’s trichrome staining showing the deposited collagen at the repaired tissue site 4 weeks after implantation. Polarized light microscopy image showing the maturation of the newly formed collagen fibrils (n = 6). (D) Quantitative analyses of the collagen content. (E) Transmission electron micrographs of cross sections of collagen fibrils in the control and nestin knockdown groups from repaired tendons 4 weeks after implantation. Distribution of collagen fibril diameters of repaired tendons in the control and nestin knockdown groups 4 weeks after implantation (n = 6). (F) Biomechanical properties of the repaired tendon (failure force, energy absorbed at failure, stress at failure, and modulus) in the control and nestin knockdown groups 4 weeks after implantation (n = 6). Results are presented as means ± SD. (G) qPCR analyses showing the expression of tendon-associated genes in the repaired tendon (n = 3). Results are presented as means ± SEM. *P < 0.05, **P < 0.01 (Student’s t test). Scale bars, 100 μm (A and D); 200 nm (E).
Transcriptional analysis of nestin knockdown in TSPC
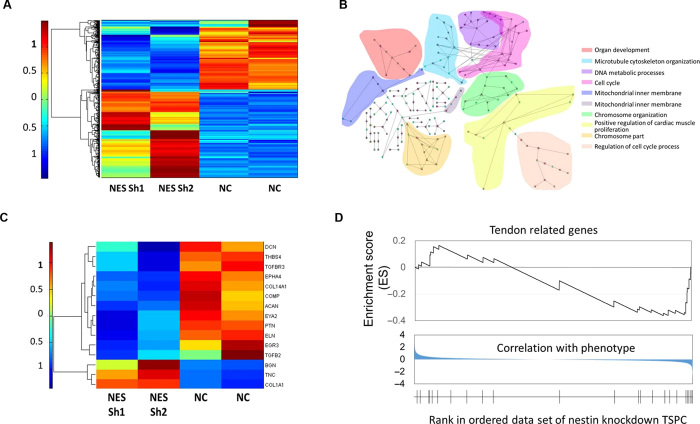
To investigate the function of nestin in TSPC, we performed global gene expression profiling of stable nestin knockdown human TSPC as compared to control cells (Fig. 7). Microarray analysis of the two different nestin shRNA–treated human TSPC revealed that the molecular signatures of nestin knockdown TSPCs were distinct and formed separate clusters, as compared to control cells (Fig. 7A). To find out the molecular and cellular functions of genes that changed after nestin knockdown, we performed gene set enrichment analysis (GSEA) of all genes in the Gene Ontology (GO) network. The top 80 GO network, which shows the relationship of down-regulated ontology entities, was analyzed according to the enrichment score (ES) of the GSEA of the whole genome. The GO was enriched in the parental GO involved in organ development, microtubule cytoskeleton organization, DNA metabolism, cell cycle, and the regulation of cell cycle process (Fig. 7B). Furthermore, several tendon-related genes, which are typically highly expressed in normal TSPCs, were substantially down-regulated after nestin knockdown (Fig. 7C). To pursue the potential significance of this observation, we performed the GSEA using a set of tendon-related genes. The tendon-related gene signature was negatively enriched with nestin-deficient TSPCs (Fig. 7D). This indicated that nestin is essential for TSPCs to maintain the tendon-lineage phenotype.
Fig. 7. Transcriptional analysis of nestin knockdown in TSPC.
(A) Heat map of the differentially expressed gene profiles of nestin knockdown in TSPCs. Two samples were used for each. (B) Top 80 GO network analysis according to the ES of the GSEA of the whole genome, which shows the relationship of down-regulated ontology. The GO was enriched in the parental GO involved in the organ development, DNA metabolism, and cell cycle, presented in different colors. (C) Heat map of tendon-related gene expression values showing that most of the tendon-related genes were down-regulated after nestin knockdown. (D) GSEA results of tendon-related genes, showing a depression.
DISCUSSION
The recent emergence of single-cell gene analysis has provided a means to elucidate previously hidden information among individual cells within a population (34–36). Here, we applied the single-cell gene analysis strategy in tendon research and revealed the heterogeneity of tendon-derived cells with three distinct cell subpopulations. One of these subpopulations is nestin+ and expressed the endothelial cell–specific marker CD31, suggesting an endothelial origin. We also observed nestin-expressing cells located in the vascular region of adult tendon tissue. Another nestin+ subpopulation in cluster II displayed relatively higher expression levels of the tendon-related genes Col I, TnC, Fmod, and Eln, as compared to nestin+ cells in cluster I. These are probably tendon stem cells, or tenoblasts, or tendon progenitor cells within tendon tissue. Because of the unclear definition and limited number of markers for different stages of tendon differentiation, the term TSPC may be appropriate. Similarly, a recent report demonstrated that nestin+ cells in postnatal bones were also heterogeneous populations, which comprised a range of cell types belonging to the osteoblast and endothelial lineages (37). The majority of the cell population of tendon tissue display high expression levels of teno-lineage genes and are almost negative for nestin and CD146 expression, suggesting that these cells are mature tenocytes. The high-content single-cell gene analysis is a powerful tool for identifying progenitor subpopulations and lineage hierarchies during tendon development, as well as for identifying lineage-specific regulatory factors (36, 38). Here, we isolated nestin+ and nestin− TSPC subpopulations in vitro, which have different tenogenic potential but similar propensity to differentiate into osteocytes, chondrocytes, and adipocytes. This demonstrates that the nestin-expressing TSPC subpopulation has multilineage mesodermal potential and would therefore fit the definition of teno-lineage stem cells.
Here, we showed that rat, mouse, and human tendon tissues contain a hitherto unrecognized distinct subpopulation of stem/progenitor cells that express the neural stem cell–specific marker nestin. During embryogenesis, nestin expression is not only found extensively within the central nervous system but is also present in the developing musculoskeletal tissue. Notably, most of the nestin+ cells in early development are stem/progenitor cell populations (17). Three datasets from independent studies by different research groups showed that nestin expression could be positively correlated with the expression of the tendon-specific genes Mkx, Egr1, Thbs4, and Tnmd during embryonic limb and tendon development. These findings thus suggest that nestin promotes tenogenesis at specific stages of limb and tendon development. We also demonstrated the existence of nestin+ cells in the adult tendon using IHC and Nes-GFP mice, which were mainly localized at the endotenon and peritenon, with some being found in the midsubstance of the tendon. In a previous study, iododeoxyuridine (IdU) was used to label stem cells within rat patellar tendons, and it was observed that there were more label-retaining stem cells in the peritenon compared with the midsubstance, particularly at the perivascular niche (39). This was consistent with the location of nestin+ TSPCs. In addition, the same study also showed label-retaining cells participating in tendon repair after injury via migration and proliferation, together with the activation of tenogenesis. It would be interesting to verify whether nestin is a marker for the label-retaining cells within tendon tissues in future studies. Nestin expression is frequently reactivated in adult tissues during pathological situations, such as central nervous system injury and regeneration of injured muscle tissue (40, 41). Here, the number of nestin+ cells was increased significantly upon tendon injury, and most of these cells also expressed characteristic tendon stem cell markers, such as CD146 and CD105. The healing process of an injured tendon can be divided into three main phases, which are the initial inflammatory stage, the reparative/proliferation stage, and the remolding stage. Our data showed that nestin is highly expressed during the proliferation stage and is essential for tenogenesis. These findings suggest that nestin-expressing cells may constitute a reservoir of stem/progenitor cells in adults, which would be activated to proliferate upon injury so as to participate in endogenous injury repair. Another study showed that tissue injury stimulated TGF-β release from the extracellular matrix and is essential for inducing the migration of nestin+ MSCs, which gave rise to both endothelial cells and myofibroblastic cells at the injury sites (42). Furthermore, TGF-β level was reported to be significantly elevated during tendon healing (1, 43). Hence, it is likely that TGF-β acts as an injury-activated signaling molecule for recruiting nestin+ TSPCs to participate in tendon repair, regeneration, and pathological remodeling.
Aside from neural stem cells, other nestin-expressing cells include a diverse variety of undifferentiated multipotent stem cells, such as hair follicle stem cells (44), stromal stem cells in the bone marrow (25), stem Leydig cells in the testis (24, 45), mesenchymal progenitor cells in the pancreatic islets (46), and quiescent satellite cells in the muscle (47). Hence, nestin can be used as a characteristic marker of multilineage progenitor cells, and its expression by any cell may indicate multipotentiality and regenerative potential. Here, we demonstrated that TSPCs express significantly higher levels of nestin than tenocytes, and the isolated nestin+ cells have the characteristics of TSPCs, such as the capacity for colony formation and multipotential differentiation. Together, these data suggest that nestin represents a characteristic marker of TSPCs with strong tenogenic and regenerative potential. Collectively, this study not only provides new insights into the subpopulation of tendon stem cells but also illustrates a novel role for nestin in tendon differentiation.
MATERIALS AND METHODS
Dataset analysis
The datasets of Nes, Mkx, Egr1, Thbs4, and Tnmd expression at different embryonic stages of limb development were downloaded from GSM1310181 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30138) (48). Expression data were available for two to five samples. The datasets of Nes, Mkx, Egr1, Tnmd, and Thbs4 expression in forelimbs from E11.5, E12.5, and E14.5 Scx-GFP transgenic mouse embryos were downloaded from GSM1310181 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM1310181) (14). Expression data were available for three samples. The datasets of Nes, Mkx, Tnmd, and Thbs4 expression in Scx-GFP+ cells and Scx-GFP− cells from E13.5 forelimb samples were downloaded from GSE65177 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65177) (49). For each sample, three pairs of six E13.5 forelimbs were pooled together. The datasets of Nes, Mkx, Egr1, Tnmd, and Thbs4 expression in Scx-GFP+ cells from E11.5, E13.5, and E15.5 hindlimb samples were downloaded from RNA-seq data GSE65179 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65179) (49). For E11.5, E13.5, and E15.5, 18, 12, and 11 hindlimbs were pooled together, respectively.
Transgenic mice
The homozygous transgenic mice of C57BL/6 strain that express GFP under the control of the nestin promoter (Nes-GFP) were provided by Cyagen Biosciences (26).
Cell isolation and monoclonal selection
TSPCs were isolated and cultured as previously reported (3), and details are given in the Supplementary Materials.
Multipotent differentiation
The differentiation potential of the nestin+ and nestin− TSPCs toward the osteogenic, adipogenic, and chondrogenic lineages was assessed, as previously described.
Cell sheets cultured from TSPCs
As described previously (11), upon reaching confluence, nestin+ and nestin− TSPCs were cultured in high-glucose Dulbecco’s modified Eagle’s medium supplemented with 10% (v/v) fetal bovine serum and ascorbic acid (50 μg/ml) (A8960, Sigma). A multilayered cell sheet was detached from the substratum by applying a small roll-up force after 2 weeks in culture. The cell sheets were harvested for TEM, RNA isolation, and implantation.
Inhibition of nestin expression by RNA interference
To stably knock down endogenous nestin expression, we used the lentiviral expression vector pGLVH1/GFP+Puro-encoding shRNA (purchased from GenePharma) to infect cells. The sequences of the shRNA target nestin and scrambled shRNA are provided in the Supplementary Materials.
Quantitative PCR
RNA isolation and RT-PCR were performed as previously described (50), and details are given in the Supplementary Materials.
Microarray analyses
TSPCs with stable nestin knockdown by two different nestin shRNA were harvested for microarray analyses using Human Genome U133 Plus 2.0 (Affymetrix). The data were further analyzed by the GSEA program MATLAB (MathWorks) (51, 52). GSEA was used to analyze both the whole GO enrichment status and the integrative alternation of genes relevant to tendon development, which has not been involved in GO. All the ESs of every GO were calculated by the GSEA program. Following this, the top 80 GO network analyses, according to the ES of the whole genome, were visualized. The gene set of teno-lineage differentiation includes the following genes: ACAN, BGN, BMP2, COL14A1, COL1A1, COL1A2, COL3A1, COMP, DCN, EGR1, EGR2, EGR3, ELN, EPHA4, EYA1, EYA2, FGF4, FGF8, FMOD, FN1, HOXA11, MKX, MSX2, NFATC4, PTN, SCX, SIX1, SIX2, SMAD9, TGFB1, TGFB2, TGFB3, TGFBR2, TGFBR3, THBS4, TNC, and TNMD. The ESs were calculated with the GSEA program.
Single-cell qRT-PCR
Passage 1 cells from human tendon were suspended, and single cells were captured on a microfluidic chip on the C1 system (Fluidigm). Amplified complementary DNA libraries were generated using the C1 Single-Cell Auto Prep Module 1 and Module 2 kits (Fluidigm) and a mixture of outer primers specific to 46 genes (table S1). Each gene was analyzed in duplicate. Genes were selected on the basis of established markers for multipotent stem cells and tendon lineage reported in the scientific literature, in addition to nestin and one reference gene (table S1). Single-cell qRT-PCR analyses were performed with inner (nested) primers on the BioMark platform (Fluidigm) using 96.96 Dynamic Array IFC chips (Fluidigm), according to the manufacturer’s instructions. HC, PCA, violin plots, ANOVA, and coexpressed gene identification were performed using the SINGuLAR Analysis Toolset 2.1. SPADE was performed on http://cytobank.org/. A background Ct of 28 was used.
Patellar tendon injury and repair animal model
To create the tendon defect, the central one-third of the patellar tendon (~1 mm in width) was removed from the distal apex of the patella to the insertion of the tibia tuberosity, according to a well-established protocol in our previous work (50). For further details, see the Supplementary Materials.
Histological examination
The harvested specimens were immediately fixed in 4% (w/v) paraformaldehyde in phosphate-buffered saline for 24 hours. H&E staining, Masson’s trichrome staining, and general histological scoring were performed, and detailed procedures are presented in the Supplementary Materials.
Mechanical testing
Mechanical testing was performed using an Instron tension/compression system with FastTrack software (model 5543, Instron), and details are given in the Supplementary Materials.
Transmission electron microscopy
Tissue specimens were fixed by standard procedures for TEM to assess collagen fibril diameters and alignment. For further details, see the Supplementary Materials. About 500 collagen fibrils were measured for each sample to obtain an accurate representation of the fibril diameter distribution.
Study approval
Human tendon tissue samples were obtained from the Second Affiliated Hospital, Zhejiang University. All procedures and protocols had informed consent and were approved by the Ethics Committee of the Second Affiliated Hospital, School of Medicine, Zhejiang University. All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee of Zhejiang University.
Statistical analysis
One-way ANOVA and Student’s t test were performed to assess whether there were statistically significant differences in the results between groups. Values of P < 0.05 were considered to be significantly different. The significance level is presented as either *P < 0.05 or **P < 0.01.
Supplementary Material
Acknowledgments
We are grateful to the Core Facilities of Zhejiang University School of Medicine for technical assistance. Funding: This work was supported by the National Key Research and Development Program of China (2016YFC1100204), the Zhejiang Provincial Natural Science Foundation of China (NSFC) (LR14H060001), NSFC grants (81330041, 81522029, 31570987, 31271041, 81401781, 81271970, 81572157, 81572115, and 81501937), the Key Scientific and Technological Innovation Team of Zhejiang Province (2013TD11), and Fundamental Research Funds for the Central Universities. Author contributions: Conceptualization: X.C. and H.-W.O.; formal analysis: C.-r.A., G.-j.G., and B.-b.W.; investigation: Z.Y., J.-j.H., L.Y., Z.-f.Z., C.Z., and W.-l.S.; resources: H.-H.L. and J.-l.C.; writing of the manuscript: Z.Y. and X.C.; review and editing of the manuscript: B.C.H. and H.-W.O.; supervision: X.C. and H.-W.O. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are available in the GEO (GSE70853) or present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/11/e1600874/DC1
Supplementary Materials and Methods
fig. S1. Nestin expression in the developing tendon.
fig. S2. Comparison of nestin expression in tenocytes, BMSCs, and TSPCs.
fig. S3. Cell surface marker expression in TSPCs.
fig. S4. Morphological changes of TSPC infected with RNAi against nestin.
fig. S5. Proliferation capability of TSPC infected with RNAi against nestin.
fig. S6. Cell viability tests of TSPC infected with RNAi against nestin.
table S1. Primers used in single-cell gene analysis.
REFERENCES AND NOTES
- 1.Docheva D., Müller S. A., Majewski M., Evans C. H., Biologics for tendon repair. Adv. Drug Deliv. Rev. 84, 222–239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaspar D., Spanoudes K., Holladay C., Pandit A., Zeugolis D., Progress in cell-based therapies for tendon repair. Adv. Drug Deliv. Rev. 84, 240–256 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Bi Y., Ehirchiou D., Kilts T. M., Inkson C. A., Embree M. C., Sonoyama W., Li L., Leet A. I., Seo B.-M., Zhang L., Shi S., Young M. F., Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat. Med. 13, 1219–1227 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Lovati A. B., Corradetti B., Lange C. A., Recordati C., Bonacina E., Bizzaro D., Cremonesi F., Characterization and differentiation of equine tendon-derived progenitor cells. J. Biol. Regul. Homeost. Agents 25, S75–S84 (2011). [PubMed] [Google Scholar]
- 5.Rui Y.-F., Lui P. P. Y., Li G., Fu S. C., Lee Y. W., Chan K. M., Isolation and characterization of multipotent rat tendon-derived stem cells. Tissue Eng. Part A 16, 1549–1558 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Zhang J., Wang J. H.-C., Characterization of differential properties of rabbit tendon stem cells and tenocytes. BMC Musculoskelet. Disord. 11, 10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J., Li J., Wang B., Zhang W. J., Zhou G., Cao Y., Liu W., The regulation of phenotype of cultured tenocytes by microgrooved surface structure. Biomaterials 31, 6952–6958 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Kovacevic D., Rodeo S. A., Biological augmentation of rotator cuff tendon repair. Clin. Orthop. Relat. Res. 466, 622–633 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole B. J., Gomoll A. H., Yanke A., Pylawka T., Lewis P., Macgillivray J. D., Williams J. M., Biocompatibility of a polymer patch for rotator cuff repair. Knee Surg. Sports Traumatol. Arthrosc. 15, 632–637 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Guerquin M.-J., Charvet B., Nourissat G., Havis E., Ronsin O., Bonnin M.-A., Ruggiu M., Olivera-Martinez I., Robert N., Lu Y., Kadler K. E., Baumberger T., Doursounian L., Berenbaum F., Duprez D., Transcription factor EGR1 directs tendon differentiation and promotes tendon repair. J. Clin. Invest. 123, 3564–3576 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H., Zhang C., Zhu S., Lu P., Zhu T., Gong X., Zhang Z., Hu J., Yin Z., Heng B. C., Chen X., Ouyang H. W., Mohawk promotes the tenogenesis of mesenchymal stem cells through activation of the TGFβ signaling pathway. Stem Cells 33, 443–455 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Rodeo S. A., Biologic augmentation of rotator cuff tendon repair. J. Shoulder Elbow Surg. 16, S191–S197 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Schlegel T. F., Hawkins R. J., Lewis C. W., Motta T., Turner A. S., The effects of augmentation with Swine small intestine submucosa on tendon healing under tension: Histologic and mechanical evaluations in sheep. Am. J. Sports Med. 34, 275–280 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Havis E., Bonnin M.-A., Olivera-Martinez I., Nazaret N., Ruggiu M., Weibel J., Durand C., Guerquin M.-J., Bonod-Bidaud C., Ruggiero F., Schweitzer R., Duprez D., Transcriptomic analysis of mouse limb tendon cells during development. Development 141, 3683–3696 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Guo G., Luc S., Marco E., Lin T.-W., Peng C., Kerenyi M. A., Beyaz S., Kim W., Xu J., Das P. P., Neff T., Zou K., Yuan G.-C., Orkin S. H., Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell 13, 492–505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang A. H., Lu H. H., Schweitzer R., Molecular regulation of tendon cell fate during development. J. Orthop. Res. 33, 800–812 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Wiese C., Rolletschek A., Kania G., Blyszczuk P., Tarasov K. V., Tarasova Y., Wersto R. P., Boheler K. R., Wobus A. M., Nestin expression—A property of multi-lineage progenitor cells?. Cell. Mol. Life Sci. 61, 2510–2522 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amoh Y., Katsuoka K., Hoffman R. M., The advantages of hair follicle pluripotent stem cells over embryonic stem cells and induced pluripotent stem cells for regenerative medicine. J. Dermatol. Sci. 60, 131–137 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Calderone A., Nestin+ cells and healing the infarcted heart. Am. J. Physiol. Heart Circ. Physiol. 302, H1–H9 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Castinetti F., Davis S. W., Brue T., Camper S. A., Pituitary stem cell update and potential implications for treating hypopituitarism. Endocr. Rev. 32, 453–471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Florio T., Adult pituitary stem cells: From pituitary plasticity to adenoma development. Neuroendocrinology 94, 265–277 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Ishiwata T., Matsuda Y., Naito Z., Nestin in gastrointestinal and other cancers: Effects on cells and tumor angiogenesis. World J. Gastroenterol. 17, 409–418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizzoti K., Adult pituitary progenitors/stem cells: From in vitro characterization to in vivo function. Eur. J. Neurosci. 32, 2053–2062 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Jiang M. H., Cai B., Tuo Y., Wang J., Zang Z. J., Tu X., Gao Y., Su Z., Li W., Li G., Zhang M., Jiao J., Wan Z., Deng C., Lahn B. T., Xiang A. P., Characterization of Nestin-positive stem Leydig cells as a potential source for the treatment of testicular Leydig cell dysfunction. Cell Res. 24, 1466–1485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Méndez-Ferrer S., Michurina T. V., Ferraro F., Mazloom A. R., MacArthur B. D., Lira S. A., Scadden D. T., Ma’ayan A., Enikolopov G. N., Frenette P. S., Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466, 829–834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park D., Xiang A. P., Mao F. F., Zhang L., Di C.-G., Liu X.-M., Shao Y., Ma B.-F., Lee J.-H., Ha K.-S., Walton N., Lahn B. T., Nestin is required for the proper self-renewal of neural stem cells. Stem Cells 28, 2162–2171 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Tempfer H., Wagner A., Gehwolf R., Lehner C., Tauber M., Resch H., Bauer H. C., Perivascular cells of the supraspinatus tendon express both tendon- and stem cell-related markers. Histochem. Cell Biol. 131, 733–741 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Carlsson L., Li Z., Paulin D., Thornell L.-E., Nestin is expressed during development and in myotendinous and neuromuscular junctions in wild type and desmin knock-out mice. Exp. Cell Res. 251, 213–223 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Vaittinen S., Lukka R., Sahlgren C., Rantanen J., Hurme T., Lendahl U., Eriksson J. E., Kalimo H., Specific and innervation-regulated expression of the intermediate filament protein nestin at neuromuscular and myotendinous junctions in skeletal muscle. Am. J. Pathol. 154, 591–600 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popov C., Burggraf M., Kreja L., Ignatius A., Schieker M., Docheva D., Mechanical stimulation of human tendon stem/progenitor cells results in upregulation of matrix proteins, integrins and MMPs, and activation of p38 and ERK1/2 kinases. BMC Mol. Biol. 16, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crisan M., Yap S., Casteilla L., Chen C.-W., Corselli M., Park T. S., Andriolo G., Sun B., Zheng B., Zhang L., Norotte C., Teng P. N., Traas J., Schugar R., Deasy B. M., Badylak S., Bühring H.-J., Giacobino J.-P., Lazzari L., Huard J., Péault B., A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3, 301–313 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Spanoudes K., Gaspar D., Pandit A., Zeugolis D. I., The biophysical, biochemical, and biological toolbox for tenogenic phenotype maintenance in vitro. Trends Biotechnol. 32, 474–482 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Hoffmann A., Pelled G., Turgeman G., Eberle P., Zilberman Y., Shinar H., Keinan-Adamsky K., Winkel A., Shahab S., Navon G., Gross G., Gazit D., Neotendon formation induced by manipulation of the Smad8 signalling pathway in mesenchymal stem cells. J. Clin. Invest. 116, 940–952 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grün D., Lyubimova A., Kester L., Wiebrands K., Basak O., Sasaki N., Clevers H., van Ounaarden A., Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Miyabara S., Yuda Y., Kasashima Y., Kuwano A., Arai K., Regulation of tenomodulin expression via Wnt/β-catenin signaling in equine bone marrow-derived mesenchymal stem cells. J. Equine Sci. 25, 7–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treutlein B., Brownfield D. G., Wu A. R., Neff N. F., Mantalas G. L., Espinoza F. H., Desai T. J., Krasnow M. A., Quake S. R., Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono N., Ono W., Mizoguchi T., Nagasawa T., Frenette P. S., Kronenberg H. M., Vasculature-associated cells expressing nestin in developing bones encompass early cells in the osteoblast and endothelial lineage. Dev. Cell 29, 330–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollen A. A., Nowakowski T. J., Shuga J., Wang X., Leyrat A. A., Lui J. H., Li N., Szpankowski L., Fowler B., Chen P., Ramalingam N., Sun G., Thu M., Norris M., Lebofsky R., Toppani D., Kemp D. W. II, Wong M., Clerkson B., Jones B. N., Wu S., Knutsson L., Alvarado B., Wang J., Weaver L. S., May A. P., Jones R. C., Unger M. A., Kriegstein A. R., West J. A. A., Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 32, 1053–1058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan Q., Lui P. P., Lee Y. W., In vivo identity of tendon stem cells and the roles of stem cells in tendon healing. Stem Cells Dev. 22, 3128–3140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Decimo I., Bifari F., Rodriguez F. J., Malpeli G., Dolci S., Lavarini V., Pretto S., Vasquez S., Sciancalepore M., Montalbano A., Berton V., Krampera M., Fumagalli G., Nestin- and doublecortin-positive cells reside in adult spinal cord meninges and participate in injury-induced parenchymal reaction. Stem Cells 29, 2062–2076 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michalczyk K., Ziman M., Nestin structure and predicted function in cellular cytoskeletal organisation. Histol. Histopathol. 20, 665–671 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Wan M., Li C., Zhen G., Jiao K., He W., Jia X., Wang W., Shi C., Xing Q., Chen Y.-F., Jan De Beur S., Yu B., Cao X., Injury-activated transforming growth factor β controls mobilization of mesenchymal stem cells for tissue remodeling. Stem Cells 30, 2498–2511 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Natsu-ume T., Nakamura N., Shino K., Toritsuka Y., Horibe S., Ochi T., Temporal and spatial expression of transforming growth factor-β in the healing patellar ligament of the rat. J. Orthop. Res. 15, 837–843 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Li L., Mignone J., Yang M., Matic M., Penman S., Enikolopov G., Hoffman R. M., Nestin expression in hair follicle sheath progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 100, 9958–9961 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinho S., Lacombe J., Hanoun M., Mizoguchi T., Bruns I., Kunisaki Y., Frenette P. S., PDGFRα and CD51 mark human Nestin+ sphere-forming mesenchymal stem cells capable of hematopoietic progenitor cell expansion. J. Exp. Med. 210, 1351–1367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallo R., Gambelli F., Gava B., Sasdelli F., Tellone V., Masini M., Marchetti P., Dotta F., Sorrentino V., Generation and expansion of multipotent mesenchymal progenitor cells from cultured human pancreatic islets. Cell Death Differ. 14, 1860–1871 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Day K., Shefer G., Richardson J. B., Enikolopov G., Yablonka-Reuveni Z., Nestin-GFP reporter expression defines the quiescent state of skeletal muscle satellite cells. Dev. Biol. 304, 246–259 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taher L., Collette N. M., Murugesh D., Maxwell E., Ovcharenko I., Loots G. G., Global gene expression analysis of murine limb development. PLOS ONE 6, e28358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu H., Xu J., Liu C.-F., Lan Y., Wylie C., Jiang R., Whole transcriptome expression profiling of mouse limb tendon development by using RNA-seq. J. Orthop. Res. 33, 840–848 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X., Song X.-H., Yin Z., Zou X.-H., Wang L.-L., Hu H., Cao T., Zheng M., Ouyang H. W., Stepwise differentiation of human embryonic stem cells promotes tendon regeneration by secreting fetal tendon matrix and differentiation factors. Stem Cells 27, 1276–1287 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Kerenyi M. A., Shao Z., Hsu Y.-J., Guo G., Luc S., O’Brien K., Fujiwara Y., Peng C., Nguyen M., Orkin S. H., Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. Elife 2, e00633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/11/e1600874/DC1
Supplementary Materials and Methods
fig. S1. Nestin expression in the developing tendon.
fig. S2. Comparison of nestin expression in tenocytes, BMSCs, and TSPCs.
fig. S3. Cell surface marker expression in TSPCs.
fig. S4. Morphological changes of TSPC infected with RNAi against nestin.
fig. S5. Proliferation capability of TSPC infected with RNAi against nestin.
fig. S6. Cell viability tests of TSPC infected with RNAi against nestin.
table S1. Primers used in single-cell gene analysis.