Abstract
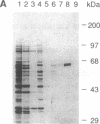
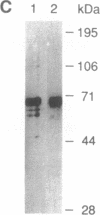
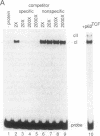
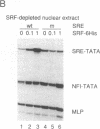
Vaccinia virus has been used as a vector to express foreign genes for the production of functional and posttranslationally modified proteins. A procedure is described here that allows the rapid native purification of vaccinia-expressed proteins fused to an amino-terminal tag of six histidines. Extracts from cells infected with recombinant vaccinia virus are loaded onto Ni2+.nitrilotriacetic acid (Ni2+.NTA)-agarose and histidine-tagged proteins are selectively eluted with imidazole-containing buffers. In the case of the human serum response factor (SRF), a transcription factor involved in the regulation of the c-fos protooncogene, the vaccinia-expressed histidine-tagged SRF (SRF-6His) could be purified solely by this step to greater than 95% purity. SRF-6His was shown to resemble authentic SRF by functional criteria: it was transported to the nucleus, bound specifically the c-fos serum response element, interacted with the p62TCF protein to form a ternary complex, and stimulated in vitro transcription from the serum response element. Thus, the combination of vaccinia virus expression and affinity purification by Ni2+.NTA chromatography promises to be useful for the production of proteins in a functional and posttranslationally modified form.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Esteban M. Rifampin and vaccinia DNA. J Virol. 1977 Feb;21(2):796–801. doi: 10.1128/jvi.21.2.796-801.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner F. G., Moss B. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J Virol. 1988 Jun;62(6):1849–1854. doi: 10.1128/jvi.62.6.1849-1854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentz R., Chen C. H., Rosen C. A. Bioassay for trans-activation using purified human immunodeficiency virus tat-encoded protein: trans-activation requires mRNA synthesis. Proc Natl Acad Sci U S A. 1989 Feb;86(3):821–824. doi: 10.1073/pnas.86.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounari F., De Francesco R., Schmitt J., van der Vliet P., Cortese R., Stunnenberg H. Amino-terminal domain of NF1 binds to DNA as a dimer and activates adenovirus DNA replication. EMBO J. 1990 Feb;9(2):559–566. doi: 10.1002/j.1460-2075.1990.tb08143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizani I., Kieny M. P., Lathe R., Clertant P. Characterization of polyoma virus early proteins expressed from vaccinia virus recombinants. Gene. 1988 Dec 15;73(1):163–173. doi: 10.1016/0378-1119(88)90322-8. [DOI] [PubMed] [Google Scholar]
- Hirschmann P., Vos J. C., Stunnenberg H. G. Mutational analysis of a vaccinia virus intermediate promoter in vivo and in vitro. J Virol. 1990 Dec;64(12):6063–6069. doi: 10.1128/jvi.64.12.6063-6069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänggi M., Bannwarth W., Stunnenberg H. G. Conserved TAAAT motif in vaccinia virus late promoters: overlapping TATA box and site of transcription initiation. EMBO J. 1986 May;5(5):1071–1076. doi: 10.1002/j.1460-2075.1986.tb04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S. Expression of foreign genes in insects using baculovirus vectors. Annu Rev Entomol. 1989;34:351–372. doi: 10.1146/annurev.en.34.010189.002031. [DOI] [PubMed] [Google Scholar]
- Moss B. Regulation of vaccinia virus transcription. Annu Rev Biochem. 1990;59:661–688. doi: 10.1146/annurev.bi.59.070190.003305. [DOI] [PubMed] [Google Scholar]
- Norman C., Runswick M., Pollock R., Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988 Dec 23;55(6):989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- Prywes R., Dutta A., Cromlish J. A., Roeder R. G. Phosphorylation of serum response factor, a factor that binds to the serum response element of the c-FOS enhancer. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7206–7210. doi: 10.1073/pnas.85.19.7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera V. M., Greenberg M. E. Growth factor-induced gene expression: the ups and downs of c-fos regulation. New Biol. 1990 Sep;2(9):751–758. [PubMed] [Google Scholar]
- Runkel L., Shaw P. E., Herrera R. E., Hipskind R. A., Nordheim A. Multiple basal promoter elements determine the level of human c-fos transcription. Mol Cell Biol. 1991 Mar;11(3):1270–1280. doi: 10.1128/mcb.11.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalasta G., Doppler C. Inhibition of c-fos transcription and phosphorylation of the serum response factor by an inhibitor of phospholipase C-type reactions. Mol Cell Biol. 1990 Oct;10(10):5558–5561. doi: 10.1128/mcb.10.10.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter H., Mueller C. G., Meese K., Nordheim A. Synergism in ternary complex formation between the dimeric glycoprotein p67SRF, polypeptide p62TCF and the c-fos serum response element. EMBO J. 1990 Apr;9(4):1123–1130. doi: 10.1002/j.1460-2075.1990.tb08218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P. E., Schröter H., Nordheim A. The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the human c-fos promoter. Cell. 1989 Feb 24;56(4):563–572. doi: 10.1016/0092-8674(89)90579-5. [DOI] [PubMed] [Google Scholar]