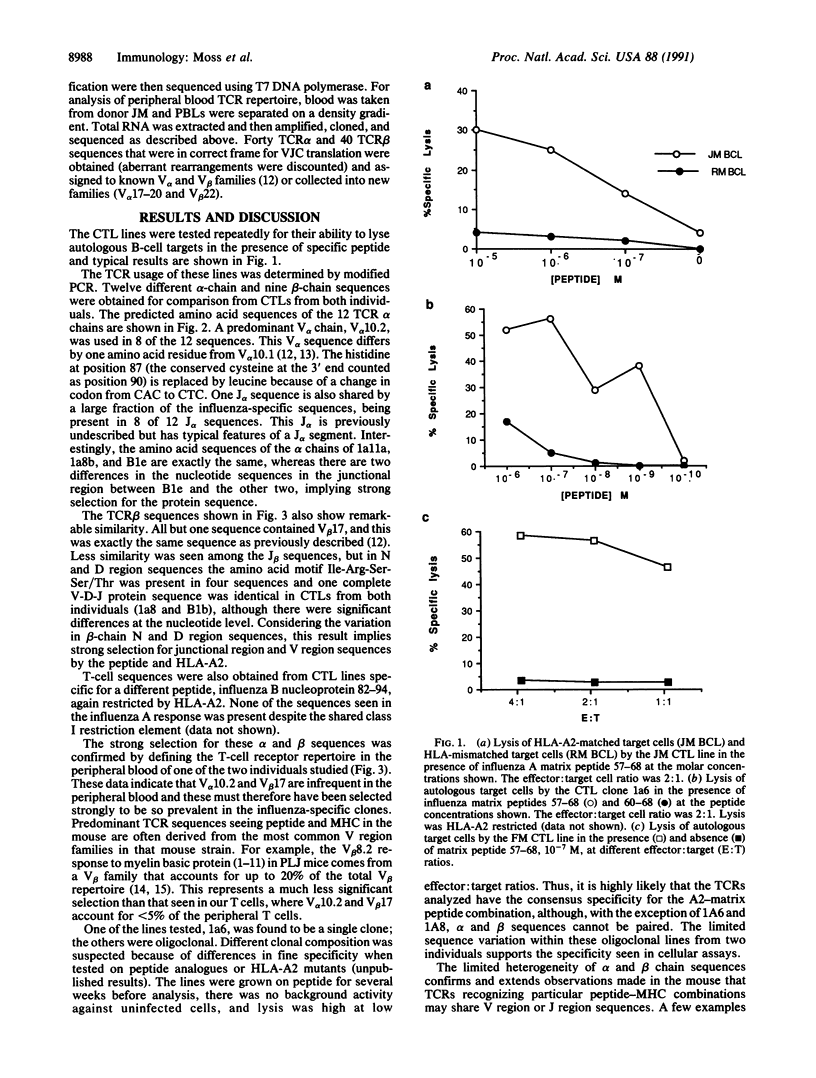
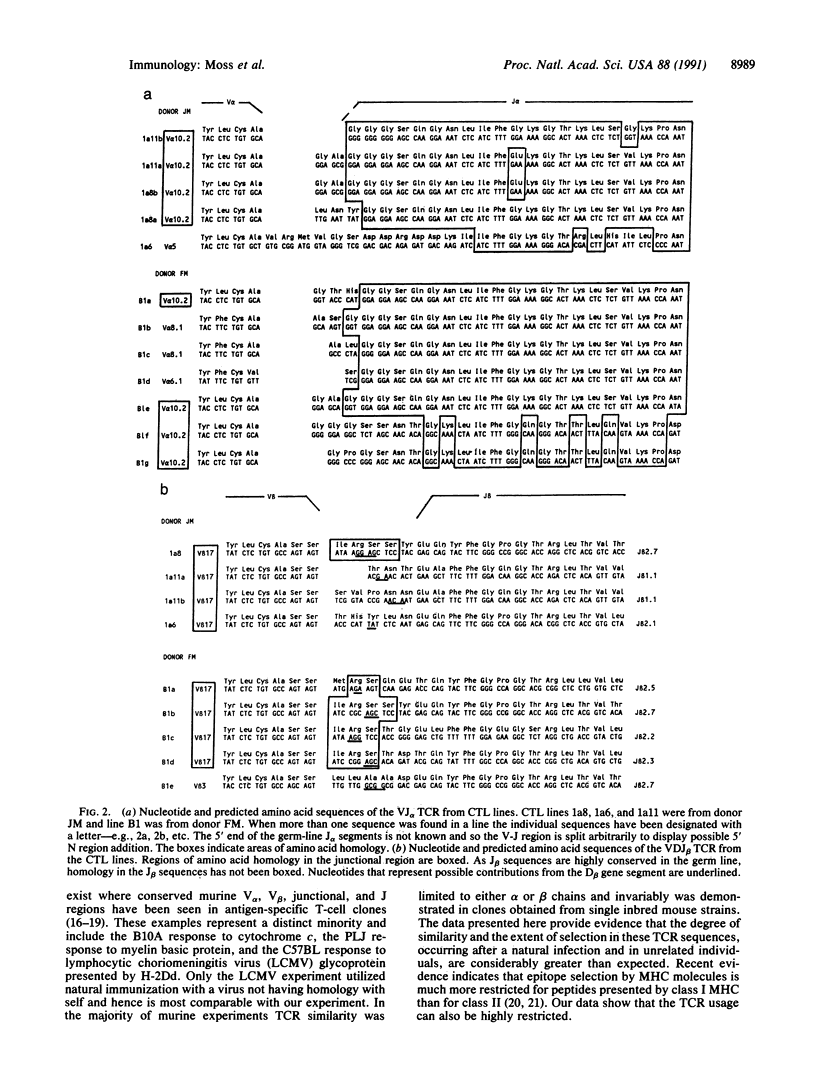
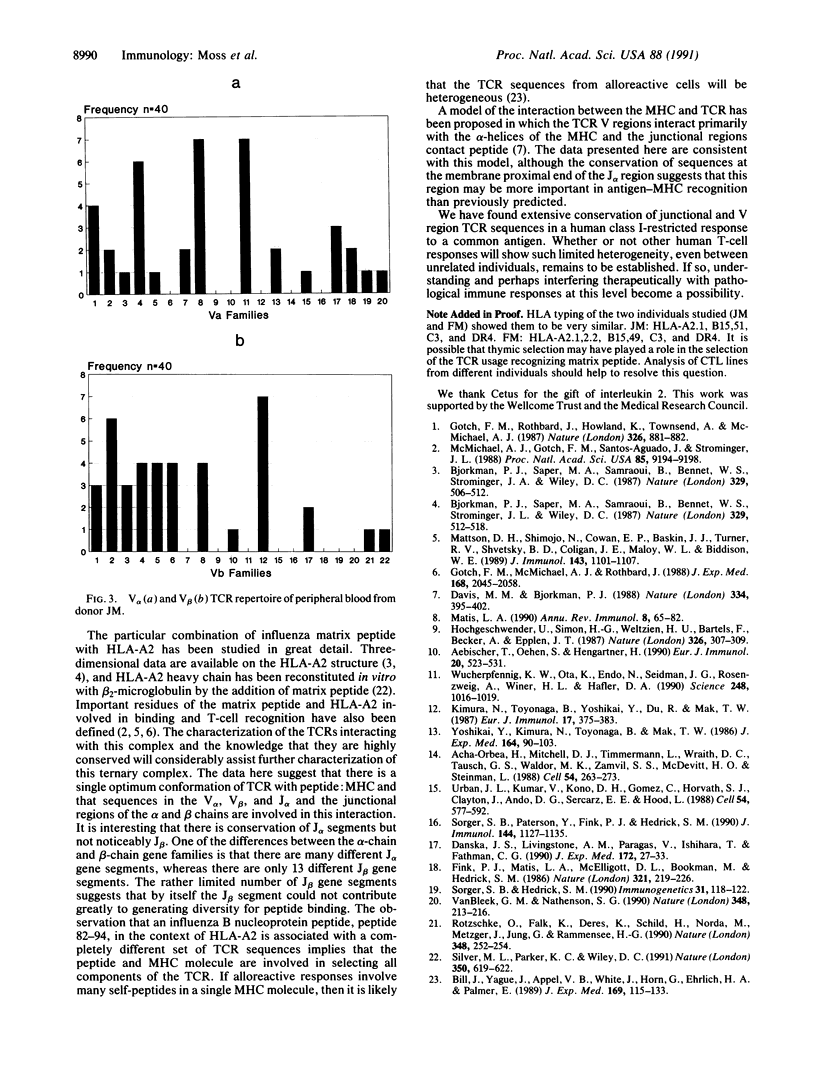
Abstract
The major histocompatibility complex class I molecule HLA-A2.1 presents the influenza A virus matrix peptide 57-68 to cytotoxic T lymphocytes in all individuals with this common HLA type and is among the most thoroughly studied immune responses in humans. We have studied the T-cell receptor (TCR) heterogeneity of T cells specific for HLA-A2 and influenza A matrix peptide using the polymerase chain reaction. The usage of V alpha and V beta sequences seen on these T cells is remarkably conserved as are certain junctional sequences associated with alpha and beta chains. Furthermore, two unrelated HLA-A2 individuals have a similar pattern of TCR usage, implying that this is a predominant response in HLA-A2 populations. Analysis in one individual showed that the conserved TCR V alpha and V beta genes are minor members of the peripheral blood TCR repertoire. The sequences provide important information on the TCR necessary for the final structural analysis of this ternary complex.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Acha-Orbea H., Mitchell D. J., Timmermann L., Wraith D. C., Tausch G. S., Waldor M. K., Zamvil S. S., McDevitt H. O., Steinman L. Limited heterogeneity of T cell receptors from lymphocytes mediating autoimmune encephalomyelitis allows specific immune intervention. Cell. 1988 Jul 15;54(2):263–273. doi: 10.1016/0092-8674(88)90558-2. [DOI] [PubMed] [Google Scholar]
- Aebischer T., Oehen S., Hengartner H. Preferential usage of V alpha 4 and V beta 10 T cell receptor genes by lymphocytic choriomeningitis virus glycoprotein-specific H-2Db-restricted cytotoxic T cells. Eur J Immunol. 1990 Mar;20(3):523–531. doi: 10.1002/eji.1830200310. [DOI] [PubMed] [Google Scholar]
- Bill J., Yagüe J., Appel V. B., White J., Horn G., Erlich H. A., Palmer E. Molecular genetic analysis of 178 I-Abm12-reactive T cells. J Exp Med. 1989 Jan 1;169(1):115–133. doi: 10.1084/jem.169.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987 Oct 8;329(6139):506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987 Oct 8;329(6139):512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- Danska J. S., Livingstone A. M., Paragas V., Ishihara T., Fathman C. G. The presumptive CDR3 regions of both T cell receptor alpha and beta chains determine T cell specificity for myoglobin peptides. J Exp Med. 1990 Jul 1;172(1):27–33. doi: 10.1084/jem.172.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. M., Bjorkman P. J. T-cell antigen receptor genes and T-cell recognition. Nature. 1988 Aug 4;334(6181):395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- Fink P. J., Matis L. A., McElligott D. L., Bookman M., Hedrick S. M. Correlations between T-cell specificity and the structure of the antigen receptor. Nature. 1986 May 15;321(6067):219–226. doi: 10.1038/321219a0. [DOI] [PubMed] [Google Scholar]
- Gotch F., McMichael A., Rothbard J. Recognition of influenza A matrix protein by HLA-A2-restricted cytotoxic T lymphocytes. Use of analogues to orientate the matrix peptide in the HLA-A2 binding site. J Exp Med. 1988 Dec 1;168(6):2045–2057. doi: 10.1084/jem.168.6.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotch F., Rothbard J., Howland K., Townsend A., McMichael A. Cytotoxic T lymphocytes recognize a fragment of influenza virus matrix protein in association with HLA-A2. 1987 Apr 30-May 6Nature. 326(6116):881–882. doi: 10.1038/326881a0. [DOI] [PubMed] [Google Scholar]
- Hochgeschwender U., Simon H. G., Weltzien H. U., Bartels F., Becker A., Epplen J. T. Dominance of one T-cell receptor in the H-2Kb/TNP response. Nature. 1987 Mar 19;326(6110):307–309. doi: 10.1038/326307a0. [DOI] [PubMed] [Google Scholar]
- Kimura N., Toyonaga B., Yoshikai Y., Du R. P., Mak T. W. Sequences and repertoire of the human T cell receptor alpha and beta chain variable region genes in thymocytes. Eur J Immunol. 1987 Mar;17(3):375–383. doi: 10.1002/eji.1830170312. [DOI] [PubMed] [Google Scholar]
- Matis L. A. The molecular basis of T-cell specificity. Annu Rev Immunol. 1990;8:65–82. doi: 10.1146/annurev.iy.08.040190.000433. [DOI] [PubMed] [Google Scholar]
- Mattson D. H., Shimojo N., Cowan E. P., Baskin J. J., Turner R. V., Shvetsky B. D., Coligan J. E., Maloy W. L., Biddison W. E. Differential effects of amino acid substitutions in the beta-sheet floor and alpha-2 helix of HLA-A2 on recognition by alloreactive viral peptide-specific cytotoxic T lymphocytes. J Immunol. 1989 Aug 15;143(4):1101–1107. [PubMed] [Google Scholar]
- McMichael A. J., Gotch F. M., Santos-Aguado J., Strominger J. L. Effect of mutations and variations of HLA-A2 on recognition of a virus peptide epitope by cytotoxic T lymphocytes. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9194–9198. doi: 10.1073/pnas.85.23.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rötzschke O., Falk K., Deres K., Schild H., Norda M., Metzger J., Jung G., Rammensee H. G. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature. 1990 Nov 15;348(6298):252–254. doi: 10.1038/348252a0. [DOI] [PubMed] [Google Scholar]
- Silver M. L., Parker K. C., Wiley D. C. Reconstitution by MHC-restricted peptides of HLA-A2 heavy chain with beta 2-microglobulin, in vitro. Nature. 1991 Apr 18;350(6319):619–622. doi: 10.1038/350619a0. [DOI] [PubMed] [Google Scholar]
- Sorger S. B., Hedrick S. M. Highly conserved T-cell receptor junctional regions. Evidence for selection at the protein and the DNA level. Immunogenetics. 1990;31(2):118–122. doi: 10.1007/BF00661222. [DOI] [PubMed] [Google Scholar]
- Sorger S. B., Paterson Y., Fink P. J., Hedrick S. M. T cell receptor junctional regions and the MHC molecule affect the recognition of antigenic peptides by T cell clones. J Immunol. 1990 Feb 1;144(3):1127–1135. [PubMed] [Google Scholar]
- Urban J. L., Kumar V., Kono D. H., Gomez C., Horvath S. J., Clayton J., Ando D. G., Sercarz E. E., Hood L. Restricted use of T cell receptor V genes in murine autoimmune encephalomyelitis raises possibilities for antibody therapy. Cell. 1988 Aug 12;54(4):577–592. doi: 10.1016/0092-8674(88)90079-7. [DOI] [PubMed] [Google Scholar]
- Van Bleek G. M., Nathenson S. G. Isolation of an endogenously processed immunodominant viral peptide from the class I H-2Kb molecule. Nature. 1990 Nov 15;348(6298):213–216. doi: 10.1038/348213a0. [DOI] [PubMed] [Google Scholar]
- Wucherpfennig K. W., Ota K., Endo N., Seidman J. G., Rosenzweig A., Weiner H. L., Hafler D. A. Shared human T cell receptor V beta usage to immunodominant regions of myelin basic protein. Science. 1990 May 25;248(4958):1016–1019. doi: 10.1126/science.1693015. [DOI] [PubMed] [Google Scholar]
- Yoshikai Y., Kimura N., Toyonaga B., Mak T. W. Sequences and repertoire of human T cell receptor alpha chain variable region genes in mature T lymphocytes. J Exp Med. 1986 Jul 1;164(1):90–103. doi: 10.1084/jem.164.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]


