Abstract
Background
Armillaria is a globally distributed mushroom-forming genus composed primarily of plant pathogens. Species in this genus are prolific producers of rhizomorphs, or vegetative structures, which, when found, are often associated with infection. Because of their importance as plant pathogens, understanding the evolutionary origins of this genus and how it gained a worldwide distribution is of interest. The first gasteroid fungus with close affinities to Armillaria—Guyanagaster necrorhizus—was described from the Neotropical rainforests of Guyana. In this study, we conducted phylogenetic analyses to fully resolve the relationship of G. necrorhizus with Armillaria. Data sets containing Guyanagaster from two collecting localities, along with a global sampling of 21 Armillaria species—including newly collected specimens from Guyana and Africa—at six loci (28S, EF1α, RPB2, TUB, actin-1 and gpd) were used. Three loci—28S, EF1α and RPB2—were analyzed in a partitioned nucleotide data set to infer divergence dates and ancestral range estimations for well-supported, monophyletic lineages.
Results
The six-locus phylogenetic analysis resolves Guyanagaster as the earliest diverging lineage in the armillarioid clade. The next lineage to diverge is that composed of species in Armillaria subgenus Desarmillaria. This subgenus is elevated to genus level to accommodate the exannulate mushroom-forming armillarioid species. The final lineage to diverge is that composed of annulate mushroom-forming armillarioid species, in what is now Armillaria sensu stricto. The molecular clock analysis and ancestral range estimation suggest the most recent common ancestor to the armillarioid lineage arose 51 million years ago in Eurasia. A new species, Guyanagaster lucianii sp. nov. from Guyana, is described.
Conclusions
The armillarioid lineage evolved in Eurasia during the height of tropical rainforest expansion about 51 million years ago, a time marked by a warm and wet global climate. Species of Guyanagaster and Desarmillaria represent extant taxa of these early diverging lineages. Desarmillaria represents an armillarioid lineage that was likely much more widespread in the past. Guyanagaster likely evolved from a gilled mushroom ancestor and could represent a highly specialized endemic in the Guiana Shield. Armillaria species represent those that evolved after the shift in climate from warm and tropical to cool and arid during the late Eocene. No species in either Desarmillaria or Guyanagaster are known to produce melanized rhizomorphs in nature, whereas almost all Armillaria species are known to produce them. The production of rhizomorphs is an adaptation to harsh environments, and could be a driver of diversification in Armillaria by conferring a competitive advantage to the species that produce them.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-017-0877-3) contains supplementary material, which is available to authorized users.
Keywords: Armillaria root rot, Cameroon, Eocene, Fungal taxonomy, Gasteromycetation, Guiana Shield, Melanin, Mushroom evolution, Physalacriaceae, Systematics
Background
The globally distributed mushroom-forming genus Armillaria contains species that are frequently encountered as tree pathogens in natural forests [1, 2] as well as silvicultural and agronomic systems [3, 4], and are the causal agent of the root disease Armillaria root rot [5]. Besides being pathogens, Armillaria species also play a critical role as decomposers. Extensive infection observed in stumps and roots and long possession of the substrate suggest that Armillaria species contribute significantly to decomposition and mineral cycling within many forests [6].
Armillaria species form basidiomes—in this case, mushrooms—which serve as reproductive structures that fruit seasonally when conditions are optimal. Each of these basidiomes produces their reproductive propagules, or basidiospores, on an exposed spore-bearing surface, or hymenium. At maturity, the basidiospores are propelled into the air column via a mechanism of forcible spore discharge known as ballistospory. Basidiospores of other fungi capable of ballistospory have been shown to disperse between continents [7–9]. Based on molecular clock analyses, the origin of Armillaria post-dates the Gondwana break-up, leading to the hypothesis that several such long distance basidiospore dispersal events led to the global distribution of Armillaria [10].
One change in basidiome form that has occurred repeatedly within mushroom-forming lineages is the shift from producing basidiospores on an exposed hymenium to producing them within an enclosed hymenium. This loss of an exposed hymenium has resulted in the loss of ballistospory [11]. Fungi that undergo this change will hereinafter be referred to as gasteroid fungi as they go through a process called gasteromycetation. A suite of changes in basidiome morphology, including the development of a fully enclosed basidiospore-bearing mass (termed a gleba) that is encased in a specialized covering (termed a peridium), typically take place during gasteromycetation. Gasteromycetation is believed to be a unidirectional process, as gasteroid fungi have never been observed to regain the mechanism of ballistospory [11]. In order to persist, gasteroid fungi have evolved a diverse array of dispersal mechanisms that engage exogenous forces. Examples include animal mycophagy for volatile-producing underground truffle-like basidiomycetes [12] and the “bellows” mechanism of puffballs [13].
Gasteroid fungi have evolved independently many times from ballistosporic ancestors within the Agaricomycetidae (e.g. [14–20]). These ancestors of gasteroid fungi include agaricoid lineages (i.e. those with lamellate hymenophores in the Agaricales and Russulales) and boletoid lineages (i.e. those with porose-tubulose hymenophores in the Boletales). To date, gasteroid fungi have only been documented as ectomycorrhizal (ECM) or saprotrophic in their ecology (e.g. [11, 21–26]).
The first known gasteroid fungus closely related to Armillaria was described in 2010 [27]. The monotypic genus, Guyanagaster T. W. Henkel, Aime & M. E. Sm. and the species G. necrorhizus T. W. Henkel, Aime & M. E. Sm. (Basidiomycota; Agaricales; Physalacriaceae) are only known from the Pakaraima Mountains of Guyana in the Guiana Shield region of South America [27]. This fungus produces subhypogeous basidiomes that are often attached to decaying tree roots, has a thick black peridium with pyramidal warts, a reduced stipe, and a tough, gelatinous gleba that changes from white to pink to brick red with maturation [27]. This species is remarkable in the sense that it does not possess the morphological hallmarks of gasteroid fungi adapted for any of the aforementioned dispersal mechanisms (e.g. mammal or mechanical dispersal).
To our knowledge, until the discovery of Guyanagaster, no known gasteroid fungus has been demonstrated to have evolved from a pathogenic lineage. Guyanagaster necrorhizus basidiomes are almost always found fruiting from dead and decaying tree roots, in this case primarily of the forest-dominating trees in the genus Dicymbe Spruce ex Benth. (Fabaceae subfam. Caesalpinioideae). Roots to which G. necrorhizus are attached display signs of white rot, indicating a wood decay capability for this fungus. Given the association of G. necrorhizus to dead and decaying roots and its close relationship to Armillaria species, it is conjectured that it is also has pathogenic capabilities, but no experimental evidence exists to confirm this.
Prior multi-gene phylogenetic analyses by Henkel et al. were equivocal in that some loci suggested G. necrorhizus was sister to Armillaria, while other loci showed G. necrorhizus derived from within Armillaria [27]. Resolving the phylogenetic placement of G. necrorhizus in relation to Armillaria will create a more complete picture of armillariod (a term to denote both Armillaria and Guyanagaster species) evolution through time and allow us to understand traits that led to the success of this group. A resolved phylogeny will also give us a robust framework in which to understand the evolution of Guyanagaster.
Here we use molecular data from G. necrorhizus and Armillaria species from every major infrageneric lineage, including Armillaria puiggarii Speg.—the only Armillaria species to be collected in the same region as G. necrorhizus—to provide a fully resolved phylogenetic hypothesis for armillarioid fungi. We also erect a new genus, Desarmillaria, composed of the exannulate mushroom-forming armillarioid species, to accommodate those species formerly placed in Armillaria subgenus Desarmillaria. A second species of Guyanagaster, Guyanagaster lucianii, is described as new to science. Using these data we produce a time-calibrated phylogeny, which we analyze in combination with ancestral range estimations, in order to examine the morphological and biogeographic evolution of armillarioid fungi.
Methods
Collecting and morphological analyses
Collecting expeditions to the Upper Potaro River Basin (hereafter referred to as the Potaro) in the west-central Pakaraima Mountains of Guyana were conducted during the rainy seasons of May–July of 2002–2013. Fungi were collected within a 15-km radius of a previously established base camp (5°18'04.80"N, 59°54'40.40"W) in forests dominated by Dicymbe corymbosa Spruce ex Benth. and Dicymbe altsonii Sandw. (Fabaceae subfam. Caesalpinioideae) [28, 29]. In December 2013–January 2014, a collecting expedition to the Mabura Ecological Reserve (hereafter referred to as Mabura) in Mabura Hill, Guyana—approximately 125 km from the Potaro—was conducted. Fungi were collected within a 3-km radius of the field station (5°10'09.26"N, 58°42'14.40"W) in forests with high densities of D. altsonii [30]. New collections of African Armillaria species were made in the Dja Biosphere Reserve in the East Province of Cameroon during the rainy season of August–September 2014 within a 2-km radius of the Dja base camp (3°21'29.80"N, 12°43'46.90"W).
Fresh morphological characteristics and substratum relationships were described in the field. Color was described subjectively and coded according to Kornerup and Wanscher [31], with color plates noted in parentheses. The descriptions of the collected Armillaria specimens were compared to the descriptions of validly published species. Additionally, attempts to obtain specimen cultures were made in the field by placing small pieces of unexposed basidiome tissue on Potato Dextrose Agar (PDA) (BD Difco™, Franklin Lakes, New Jersey, USA) and grown for one month at room temperature. Specimens were then field-dried with silica gel.
Micromorphological features on dried specimens were examined in the laboratory using an Olympus BH2-RFCA compound microscope. Two dried Armillaria specimens from Africa, TH 9926 and THDJA 91, and four dried Guyanagaster specimens, MCA 4424, RAK 84, RAK 88 and RAK 89, were rehydrated in 70% ethanol and then sectioned by hand and mounted in water, Melzer’s reagent and 1% aqueous Congo red. Twenty randomly selected basidiospores were measured per collection under a 100× objective. Length/width Q values for basidiospores are reported as Qr (range of Q values over 20 basidiospores measured) and Qm (mean of Q values ± SD). Specimens were deposited in the following herbaria: PUL (Kriebel Herbarium), BRG (Guyana National Herbarium), HSC (Humboldt State University) and YA (National Herbarium of Cameroon).
Molecular methods
DNA was extracted from basidiome tissue using the Wizard® Genomic DNA Purification kit (Promega Co., Madison, Wisconsin, USA). Specimens housed in the Kriebel Herbarium at Purdue University were used to obtain DNA if sequence data for equivalent taxa were not available on the NCBI Nucleotide database. PCR reactions included 12.5 μL of MeanGreen 2× Taq DNA Polymerase PCR Master Mix (Syzygy Biotech, Grand Rapids, Michigan, USA), 1.25 μL of each primer (at 10 μM) and approximately 100 ng of DNA. The final PCR reaction volume was 25 μL. The recommended cycling conditions for each primer pair we used were followed.
To determine the identity of the recently collected Armillaria specimens from Guyana and Africa, as well as to confirm the identity of the recently collected Guyanagaster specimens, PCR was performed to acquire sequence data from the internal transcribed spacer (ITS) region (inclusive of ITS1, 5.8S and ITS2 regions), using the primer pair ITS1F/ITS4B [32]. To analyze the phylogenetic relationship of the armillarioid fungi, PCR was performed on all specimens at the three following loci: nuclear ribosomal large subunit DNA (28S), Elongation Factor 1-α (EF1α) and RNA polymerase II (RPB2) genes using the following primer pairs, respectively: LROR/LR6 [33, 34], 987 F/2218R [35] and bRPB2-6 F/bRPB2-7.1R [36].
Uncleaned PCR products were sent to Beckman Coulter, Inc. (Danvers, Massachusetts, USA) for sequencing. Sequences were manually edited using Sequencher 5.2.3 (Gene Codes Corporation, Ann Arbor, Michigan, USA). The ITS sequences we generated were used as queries in the sequence similarity search tool, NCBI BLAST, to search the NCBI Nucleotide database.
Phylogenetic analyses
In order to resolve the phylogeny of Guyanagaster and Armillaria, we compiled a dataset composed of sequence data from both Armillaria and Guyanagaster specimens. Two closely related species in the Physalacriaceae, Oudemansiella mucida and Strobilurus esculentus, served as outgroup taxa. Six loci were used (28S, EF1α, RPB2, Actin-1 (actin-1), Glyceraldehyde-3-Phosphate Dehydrogenase (gpd) and Beta-Tubulin (TUB)) to assess Guyanagaster and Armillaria systematic relationships through phylogenetic analysis. Not all loci were available for all specimens. DNA sequences that were not generated in this study were obtained from GenBank or published whole genome sequences. Collection information for all specimens and GenBank accession numbers for the included sequences are compiled in Additional file 1.
Sequences were aligned in Mega 5.0 [37] using the MUSCLE algorithm [38] with refinements to the alignment done manually. The introns of EF1α were alignable across the Armillaria/Guyanagaster dataset and were therefore included. Phylogenies were reconstructed using maximum likelihood (ML) and Bayesian methods. The GTR + G model of molecular evolution was selected for all data sets as determined by PartitionFinder v1.1.0 [39]. Maximum likelihood bootstrap analysis for phylogeny and assessment of the branch support by bootstrap percentages (BS%) was performed using RAxML v2.2.3 [40]. One thousand bootstrap replicates were produced. Each locus was analyzed separately as well as all together in a supermatrix data set using ML. Bayesian analyses for the reporting of Bayesian posterior probability (BPP) support for branches was conducted on individual loci as well as the six-gene data set using the program Mr. Bayes v3.2.2 [41]. Four simultaneous, independent runs each with four Markov chain Monte Carlo (MCMC) chains, were initiated and run at a temperature of 0.15 for 20 million generations, sampling trees every 1000 generations until the standard deviation of the split frequencies reached a final stop value of 0.01. We discarded the initial 10% of trees as burn in and produced a maximum clade credibility tree from the remaining trees; 0.95 BPP represents a well-supported lineage.
Divergence time estimation
Divergence ages within the armillarioid clade were estimated using the fossil calibration approach described and implemented by [24, 42–44]. Molecular clock analysis was performed using BEAST v1.8.3 [45] with XML files containing BEAST commands and priors assembled in BEAUTi v1.8.3. These files included the following analytical settings: GTR + G model of evolution, uncorrelated relaxed clock with lognormal rate distribution; tree prior was set to speciation birth-death process, running 100 million generations, sampling every 1000th tree. The analysis was run three times. The first 10% of trees were removed as burn-in after ensuring a minimum effective sample size (ESS) of 200 was reached for all of the parameters. The remaining trees—representing the posterior distribution from all Bayesian analyses—were combined into a single file using LogCombiner v1.8.3. This file was used to produce a summary tree using Tree Annotator v1.8.3. The mean ages of supported nodes, as well as the corresponding 95% highest posterior densities (HPDs), were examined from BEAST logfiles using Tracer v1.6 [46].
Taxa used in this analysis are in bold in Additional file 1. It includes 21 Armillaria species (one representative specimen from each of the species in the first analysis), two Guyanagaster specimens (one representative from each collecting locality), nine Physalacriaceae species (one representative from nine different genera that encompass the major lineages within this family fide [47]), while six species from Agaricales families closely related to the Physalacriaceae fide [48] served as outgroup taxa. The alignment was analyzed using five partitions: 28S was analyzed in a single partition, while two codon partitions ((1 + 2), 3) were utilized for both EF1α and RPB2. The marasmioid fungi (Marasmius alliaceus, Marasmius rotula and Mycena amabilissima) were calibrated based on a 90-Ma fossil Archaeomarasmius leggetti from mid-Cretaceous amber [49], following the parameter settings of [24]. In BEAUTi, this prior was set as a lognormal distribution with a mean of 10, log standard deviation of 1 and offset of 90, with mean in real space, truncated on the lower end at 90 and at the upper end at 200.
Ancestral range estimation
To estimate the ancestral range of the armillarioid clade and lineages within, we used dispersal-extinction-cladogenesis (DEC) analysis in the package Lagrange [50] and implemented in the program RASP [51]. Six areas were defined: North America, Europe + Asia (hereinafter referred to as Eurasia), Africa, tropical South America (composed of the area covered by tropical rainforest in South America), Australasia (composed of Australia, New Zealand and southeast Asia) and temperate South America (composed of the area not covered by tropical rainforest in South America). Each taxon in our data set was assigned to areas based on its current known range. In the scenario we tested, movement between any area at any time was unconstrained. Under this model, all areas are treated as equally probable ancestral ranges. This model was tested under both two and three area constraints. The consensus tree produced during the divergence time analysis was used for all Lagrange calculations and the root node of the armillarioid clade was calibrated to 51 Ma.
Results
Sequences, collections and cultures
Eight ITS, eleven 28S, twelve EF1α and five RPB2 sequences were generated during this study (Additional file 1). The size of the sequences ranged from 514–713, 705–1031, 796–1233, and 553–704 bp, respectively. After the ends of the individual alignments were trimmed, the size of the aligned datasets were as follows: 28S was 906 bp; EF1α was 906 bp; RPB2 was 638 bp; actin-1 was 607 bp; gpd was 499 bp; TUB was 911 bp. The six-gene dataset was composed of a total of 52 Armillaria specimens, representing 21 species, as well as four Guyanagaster specimens, representing two species. The number of taxa in each of the single-locus phylogenies is as follows: 28S had 42 taxa; EF1α had 57 taxa; RPB2 had 32 taxa; actin-1 had19 taxa; gpd had 19 taxa; TUB had 12 taxa. Phylogenies for each locus, as well as a combination of loci, are available in Additional file 2.
Two Armillaria specimens were collected from Guyana—MCA 3111 and TH 9751. ITS sequences from both shared 99% identity with Armillaria puiggarii (GenBank: FJ664608). No significant morphological differences were found when comparing our field descriptions to the species description by [52]. Two Armillaria specimens were collected from Cameroon—TH 9926 and TH DJA 91. ITS sequences from both shared 99% identity with an unidentified Armillaria species collected from Zimbabwe (GenBank: AY882982). Additionally, there was variation between the two sequences. No significant morphological differences were found when comparing TH 9926 to the species description of Armillaria camerunensis [53, 54], but there was some morphological variation when comparing TH DJA 91. For now, we conservatively hypothesize that both of these specimens represent Armillaria camerunensis. Morphological descriptions of TH 9926 and TH DJA 91 are available in Additional file 3.
Pure vegetative cultures were obtained from Guyanagaster specimens MCA 3950 and RAK 88, as well as Armillaria puiggarii specimen TH 9751, with rhizomorphs produced in each (Fig. 1). After one month, differences in growth and branching pattern of the rhizomorphs can be observed between the cultures of the two Guyanagaster specimens, but the rhizomorphs of neither were observed to become melanized. In contrast, the rhizomorphs in the culture of A. puiggarii became melanized almost completely. Strain MCA 3950 is deposited in Centraalbureau voor Schimmelcultures with the strain accession number CBS 138623, while strains RAK 88 and TH 9751 are available from the authors upon request.
Fig. 1.

a Vegetative culture of Guyanagaster necrorhizus MCA 3950 showing unmelanized rhizomorphs. b Vegetative culture of Guyanagaster lucianii RAK 88 showing unmelanized rhizomorphs. c Vegetative culture of Armillaria puiggarii TH 9751 showing melanized rhizomorphs. Bar = one cm
Resolved Guyanagaster phylogenetic hypothesis
No single locus was sufficient to obtain a well-supported armillarioid phylogeny (see Additional file 2), but through the use of multiple loci, we were able to obtain a well-supported armillarioid phylogeny (Fig. 2). The six-gene analyses recovered a strongly supported armillarioid clade (100% BS and 1.00 BPP) (Fig. 2), which is composed of all known Armillaria and Guyanagaster species. The armillarioid clade is further composed of three well-supported lineages that were recovered from both analyses (Fig. 2): (1) the annulate lineage (100% BS and 1.00 BPP), which includes annulate mushroom-forming armillarioid species, (2) the exannulate lineage (91% BS and 1.00 BPP), composed of exannulate mushroom-forming armillarioid species formerly placed in Armillaria subgen. Desarmillaria and (3) the gasteroid lineage (100% BS and 1.00 BPP), composed of Guyanagaster species. The gasteroid lineage consists of two distinct species: G. necrorhizus from the Potaro region and an undescribed Guyanagaster species from the Mabura region of Guyana. These two species are geographically separated by mountains of elevations over 1000 m, as well as Kaieteur Falls and the Essequibo River, all within a distance of 125 km.
Fig. 2.

Phylogram generated from the analysis of six gene regions (28S, EF1α, RPB2, TUB, gpd and actin-1) from 58 taxa. Guyanagaster is the earliest diverging lineage and is sister to the mushroom-forming armillarioid species. The exannulate armillarioid species are the next to diverge, and compose Desarmillaria. The annulate armillarioid species form a monophyletic lineage and compose Armillaria sensu stricto. The major lineages within Armillaria are indicated in bold text at the corresponding node. The exannulate armillarioid species are sister to the annulate armillarioid species. Strobilurus esculentus and Oudemansiella mucida were selected as outgroup taxa. Black circles represent support of 90% (maximum likelihood bootstrap values, shown as percentages) and 0.95 BPP (Bayesian posterior probabilities) or greater, grey circles represent support of 0.95 BPP or greater and white circles represent support of 75% or greater. Images: (a) Armillaria sinapina; (b) Armillaria hinnulea; (c) Armillaria puiggarii; (d) Armillaria mellea; (e) Armillaria camerunensis; (f) Desarmillaria tabescens; (g) Desarmillaria ectypa, (h–j) Guyanagaster necrorhizus. (Photo credits: (a) Christian Schwarz; (b) J. J. Harrison; (c) Todd F. Elliott; (d, h–j) Rachel A. Koch; (e) Terry W. Henkel; (f) Stephen D. Russell; (g) Tatyana Svetasheva)
The annulate lineage is further composed of four well-supported lineages (Fig. 2), with species compositions as follows: (1) African lineage composed of Armillaria fuscipes and A. camerunensis; (2) melleioid lineage composed of A. mellea; (3) northern hemisphere lineage composed of A. altimontana, A. borealis, A. calvescens, A. cepistipes, A. gallica, A. gemina, A. sinapina and A. solidipes; and (4) Australasian + temperate South American lineage composed of A. affinis, A. fumosa, A. hinnulea, A. limonea, A. luteobubalina, A. novae-zelandiae, A. pallidula and A. puiggarii.
The most glaring conflict between single gene and multi-gene analyses is the placement of the African lineage: in gpd, actin-1 and EF1α this lineage is nested within or sister to the Australasian + temperate South American lineage; in TUB and RPB2, it is sister to A. mellea; in 28S it is sister to Guyanagaster. In the multi-gene analyses, it is consistently recovered as diverging earlier than the A. mellea lineage. In all of the single gene analyses except for 28S, a close relationship between the exannulate and the gasteroid lineages was recovered.
Age and ancestral range of Armillaria and Guyanagaster
Table 1 has a complete list of the divergence dates for the well-supported nodes recovered in the phylogenetic analysis. Figure 3 is the time-calibrated phylogeny for the group. The time to most recent common ancestor (tMRCA) of the armillarioid clade estimated in the BEAST analysis was 51 Ma (node 1; 95% HPD 30–73 Ma). The tMRCA of the exannulate lineage was estimated at 41 Ma (node 4; 95% HPD 24–59 Ma). The tMRCA of the annulate armillarioid lineage was estimated at 33 Ma (node 5; 95% HPD 19–47 Ma). The date of the divergence between the two Guyanagaster species was estimated at 8 Ma (node 2; 95% HPD 3–14 Ma).
Table 1.
Ages for nodes, confidence intervals and posterior probability on the phylogenetic tree presented in Fig. 3
| Node | Lineage | Mean Age | 95% HPD [min, max] | Posterior Probability |
|---|---|---|---|---|
| 1 | Armillarioid | 50.81 | 30.03, 72.32 | 1.00 |
| 2 | Guyanagaster | 7.88 | 2.70, 13.91 | 1.00 |
| 3 | Desarmillaria + Armillaria | 41.32 | 24.47, 59.07 | 0.90 |
| 4 | Desarmillaria | 31.74 | 15.83, 47.71 | 1.00 |
| 5 | Armillaria | 33.00 | 18.90, 46.98 | 1.00 |
| 6 | African lineage | 15.49 | 6.84, 24.76 | 1.00 |
| 7 | Melleioid lineage | 30.12 | 17.73, 43.43 | 0.90 |
| 8 | Northern hemisphere + Australasian + temperate South American lineage | 27.52 | 17.13, 37.60 | 0.94 |
| 9 | Northern hemisphere | 12.07 | 6.12, 18.87 | 1.00 |
| 10 | A. borealis + A. solidipes + A. gemina | 6.66 | 2.41, 11.51 | 1.00 |
| 11 | A. altimontana + A. calvescens + A. gallica | 8.58 | 3.80, 14.02 | 1.00 |
| 12 | A. cepistipes + A. sinapina | 6.44 | 2.22, 11.24 | 0.52 |
| 13 | Australasian + temperate South American | 19.27 | 11.18, 27.99 | 0.98 |
| 14 | A. novae-zelandiae + A. affinis + A. puiggarii | 8.96 | 4.04, 14.47 | 1.00 |
| 15 | A. hinnulea + A. pallidula + A. fumosa + A. limonea + A. luteobubalina | 18.13 | 10.30, 26.29 | -- |
Fig. 3.
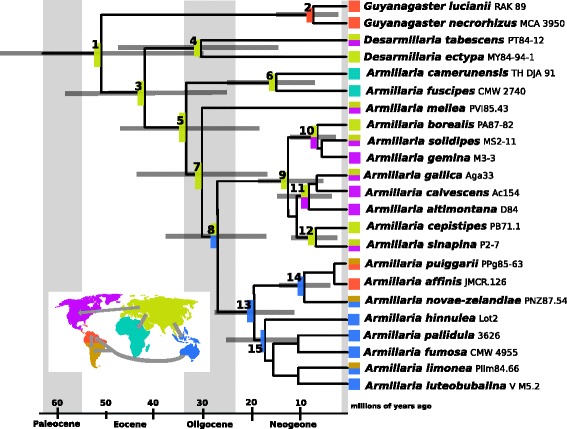
Time-calibrated phylogeny generated from Bayesian analysis of three gene regions (28S, EF1α, RPB2) from 19 Armillaria species, two Desarmillaria species and two Guyanagaster species. Geographic origin of each specimen is indicated by the box to the left of the name: red = Neotropics, blue = Australasia, brown = temperate South America, lime green = Eurasia, purple = North America, and turquoise = Africa. Boxes at ancestral nodes correspond to the most probable ancestral range at that node as presented in Additional file 4. Geologic epochs are noted above the time scale. Numbers at nodes correspond to lineages in Table 1 and Additional file 4. Dark grey bars correspond to the 95% HPD and correspond to the values in Table 1. The map in the lower left hand corner represents the proposed dispersal pattern
We estimated the ancestral ranges for 15 well-supported clades. The most probable area was the same under range constraints of ≤ 2 and ≤ 3 for all nodes. The most probable ancestral range for the most recent common ancestor (MRCA) of the armillarioid clade is Eurasia. Eurasia is also the most probable ancestral range for all of the annulate mushroom-forming lineages containing northern hemisphere taxa. Migration to both Africa and Australasia from Eurasia is the most probable scenario as estimated from this analysis. Additional file 4 provides the probabilities for each area being the ancestral range under both area constraints; numbers in bold represent the most probable region at that node.
Discussion
Our phylogenetic analyses, coupled with morphological data, support a three-genus composition of the armillarioid clade: Armillaria contains the annulate mushroom-forming species, Desarmillaria contains the exannulate mushroom-forming species, and Guyanagaster contains the known gasteroid species. Our estimates of the tMRCA of the armillarioid clade suggest it arose 51 Ma, which is in accord with a previous estimate [10], even though a different set of taxa, loci and calibration methods were used. During this time, Earth was experiencing one of the warmest intervals of the past 65 million years known as the Paleocene-Eocene Thermal Maximum (PETM) [55]. This warm and humid climate led to the widespread expansion of area covered in tropical rainforests. The ancestral range estimation suggests the most probable area for the origin of the MRCA is the Eurasian subcontinent (Additional file 4), much of which contained tropical rainforests at that time [56].
Biogeography and evolution of Guyanagaster and Desarmillaria
Our phylogenetic reconstruction suggests that Guyanagaster species evolved within the earliest diverging lineage in the armillarioid clade. The process of gasteromycetation is believed to be unidirectional; gasteroid fungi evolve from a ballistosporic ancestor, and there is no evidence that this transformation has ever been reversed [11]. With this in mind, the ancestor to the armillarioid lineage was likely a gilled mushroom. According to our estimations, between 51 and 8 Ma the mushroom progenitor to Guyanagaster underwent the gasteromycetation process, resulting in the evolution of Guyanagaster. At one point in time, this ancient lineage was composed of mushroom-forming species—all of which likely went extinct—as the only known extant members of this lineage are Guyanagaster species.
The next lineage to diverge is composed of the two described Desarmillaria species: D. ectypa and D. tabescens. Neither of the species in this genus have an annulus at maturity, in contrast to all other known mushroom-forming armillarioid species. Desarmillaria ectypa is different from all other armillarioid species by virtue of its ecology: whereas Armillaria species are pathogens and wood decomposers, D. ectypa is restricted to peat bogs and associated with Sphagnum [57]. The ecology of D. tabescens is much more typical of Armillaria species. It is a primary pathogen towards introduced Eucalyptus species in France, as well as a secondary pathogen to Quercus species, but in many cases it is a saprotroph, colonizing stumps of Quercus species [58].
Both Guyanagaster and Desarmillaria evolved within the oldest lineages in the armillarioid clade and both genera contain two known species. Armillaria represents the most recently diverged genus-level lineage within the armillarioid clade, yet has over 35 described species (see [59] for species counts). The older ages of the lineages from which Guyanagaster and Desarmillaria evolved, coupled with the fact that they contain far fewer extant species, suggest these two genera are on a different evolutionary trajectory compared to Armillaria. As stated above, our ancestral range estimation suggests the most probable ancestral range for the MRCA to the armillarioid clade is Eurasia, whereas species of Guyanagaster have only so far been found in Guyana. One question is how the mushroom progenitor to Guyanagaster dispersed from Eurasia to Guyana. One possibility is that the mushroom progenitor to Guyanagaster was widely dispersed within the humid, tropical rainforest habitat that predominated at the time it arose. It is possible that Guyanagaster evolved within the Guiana Shield and represents a highly specialized endemic to this region.
The two species of Desarmillaria are thought to be more thermophilic than their sympatric Armillaria species [57, 60]. This temperature adaptation could be a residual characteristic of their origin during the PETM and could also explain the paucity of species in this genus. As the climate changed, species adapted for a cool and arid climate could have competitively displaced the species adapted for a warm and humid climate, in this case, species of Desarmillaria and the mushroom progenitor to Guyanagaster. The novel morphology of Guyanagaster species suggests that they are highly specialized for their current habitat, and as stated above, the restriction of D. ectypa to peat bogs is a specialized habitat for armillarioid fungi. Habitat specialization has been shown to act as buffer from extinction [61], and could be what has allowed species in these early diverging lineages to avoid competitive displacement by better-adapted armillarioid fungi. Whether D. tabescens occupies a specialized niche that has aided in its persistence to the present remains unknown.
Biogeography and spread of Armillaria
According to our analyses, Armillaria sensu stricto started diversifying approximately 33 Ma in Eurasia, at which time the Earth’s climate shifted from the relatively ice-free world to one with glacial conditions; the tropical climate during which the MRCA of the armillarioid clade arose was replaced by a period of severe cooling and drying as well as an increase in seasonality [55]. This led to a contraction in the area covered by tropical rainforests [62] and the geographic expansion of species-poor deciduous vegetation [63]. Therefore, we hypothesize that Armillaria species represent annulate mushrooms that survived, diversified and radiated with their hosts as they adapted to the drier, cooler and seasonal climate (i.e. temperate) of the late Eocene and beyond.
Armillaria species in Africa form a monophyletic lineage (100% and 1.00 BPP). Single gene analyses and multi-gene analyses were ambiguous as to where this lineage diverged within the armillarioid clade (Additional file 2). In all multi-gene combinations, the African Armillaria lineage is resolved as the earliest diverging within Armillaria sensu stricto, which is similar to previous work that has also suggested that this lineage is divergent compared to the rest of the armillarioid lineage [10, 64]. Our ancestral area estimation suggests that the ancestral region of the African lineage is Eurasia. During the Eocene and Oligocene, a shallow seaway separated Africa from Eurasia, acting as a barrier to dispersal. In the early Miocene (19–12 Ma), the seaway drained, effectively linking the biotas in these two regions [65]. Our molecular clock analysis suggests that extant species in Africa started diversifying approximately 15 Ma, which fits a migration from Eurasia during this time.
Many northern hemisphere Armillaria species are found in both North America and Eurasia (see [58, 59]). In a study of A. mellea, which is a species distributed throughout the northern hemisphere, four distinct lineages were identified, corresponding to their locality: 1) western North America, 2) eastern North America, 3) western Eurasia and 4) eastern Eurasia [66]. They suggest that this species was once widespread and is now in the process of speciation [66]. Within this backdrop, it is possible that Armillaria species facilitated their dispersal as pathogens on the diverse flora that existed during this time. As available pathways disappeared, ultimately ceasing migration, the species started to diverge. From our ancestral range estimation, we can see that there were multiple armillarioid introductions into North America (Fig. 3), suggesting a dynamic process of dispersal across the northern hemisphere. A more comprehensive study on northern hemisphere Armillaria species is necessary to determine when and how they migrated between the major landmasses.
Armillaria species started diversifying in the temperate southern hemisphere (outside of Africa) about 19 Ma (Fig. 3, node 13). Our results suggest that the most probable dispersal pathway was from Eurasia to Australasia. In a plant biogeographic meta-analysis of the Australasian region, it was found that after 25 Ma, there was an increase of flora inputs to Australia from the IndoMalayan region, representing the time when the continental shelf containing Southeast Asia collided with that containing New Guinea and Australia [67]. The authors of this study hypothesized that habitat instability in the region at that time generated vacant niches and increased the probability of successful establishment of dispersed lineages compared with the relatively stable, saturated states existing prior to approximately 25 Ma [67]. In this scenario both the greater dispersal distance and relative difficulty in establishing viable populations together contributed to the infrequency of successful exchanges prior to 25 Ma. This too follows our hypothesis that Armillaria species dispersed as pathogens on flora that existed during this time.
Many of the same Armillaria species are found in both Australasia and temperate South America [68]. This pattern fits with an ancient Gondwanan distribution, but the lineage is far too young for this to be the case. Alternatively, the discovery of fossilized Nothofagus wood in Antarctic deposits from the Pliocene [69] suggests this could have been an overland dispersal route until 2 Ma. Many of the fungi in this Australasian + temperate South American lineage are thought to be close associates with Nothofagus [68], suggesting again that they could have dispersed with their plant associates through Antarctica. Alternatively, long distance basidiospore dispersal events could account for the geographic disjunction in closely related Armillaria species. Because appropriate hosts—species of Nothofagus—occur in both Australasia and temperate South America [70], establishment after a long-distance basidiospore dispersal event could be more probable.
The rise of rhizomorphs: morphological developments in the armillarioid clade
Our phylogenetic hypothesis sheds light on the evolution of rhizomorphs in the armillarioid clade. Rhizomorphs are discrete, filamentous aggregations that extend from a resource base into substrates that may not support their growth, foraging for new resource bases [71]. Individuals that produce rhizomorphs can occupy huge swaths of habitat and prevent other organisms from establishing, as is the case with the “humungous fungus” [72]. All armillarioid species have the capacity to form rhizomorphs in culture, but only the later diverging lineages have been observed to form them in nature (see Table 2, Fig. 1).
Table 2.
Trophic strategy, rhizomorph production in nature and known geographic range of species used in this study
| Species | Facultative Necrotroph? | Characterization | Rhizomorphs in nature? | Hosts | Known range | References |
|---|---|---|---|---|---|---|
| Armillaria affinis | Unknown | Unknown | Central America | [59] | ||
| Armillaria altimontana | Unknown | Yes | Hardwoods and conifers | Higher elevation forests of Western interior of North America | [83] | |
| Armillaria borealis | Yes | Weakly pathogenic, opportunistic | Yes | Birch, wild cherry | Europe | [58, 84] |
| Armillaria calvescens | Yes | Opportunistic pathogen on stressed trees | Yes | Hardwoods, particularly sugar maple | Eastern North America | [85, 86] |
| Armillaria camerunensis | Unknown | Observed in a disease center, but unknown if it is the causal agent | None observed | Unknown | Africa | This study |
| Armillaria cepistipes | Yes | Weakly pathogenic | Yes | Conifers | Europe, North America | [58, 87] |
| Armillaria fumosa | Unknown | Unknown | Australia | [59] | ||
| Armillaria fuscipes | Yes | Particularly pathogenic to exotic species | None observed | Pine forest plantations, Acacia and Cordia species | Africa, India | [74, 75, 88] |
| Armillaria gallica | Yes | Weakly or secondarily pathogenic | Yes | Hardwoods | Europe, North America, Asia | [58] |
| Armillaria gemina | Yes | Primary pathogen | Yes | Maples, beech Birch | Eastern North America | [86] |
| Armillaria hinnulea | Yes | Secondary pathogen | Yes | In wet sclerophyll forests | Australia, New Zealand | [89, 90] |
| Armillaria limonea | Yes | Pathogenic to pine seedlings (introduced tree) | Yes | Pine | Argentina, Chile, New Zealand | [68, 91, 92] |
| Armillaria luteobubalina | Yes | Primary pathogen in native forests | Yes | Eucalyptus | Australia, Tasmania | [68, 93–95] |
| Armillaria mellea | Yes | Highly pathogenic | Yes | Over 600 ornamentals, hardwood and orchard trees | Europe, North America, Asia | [58, 96] |
| Armillaria novae-zelandiae | Yes | Pathogenic to pine seedlings (introduced tree) | Yes | Pine | Argentina, Australia, Chile, New Zealand |
[91, 92, 95] |
| Armillaria pallidula | Unknown | Unknown | Australia | [59] | ||
| Armillaria puiggarii | Unknown | Observed in a disease center, but unknown if it is the causal agent | Melanized rhizomorphs observed in the field | Dicymbe spp. | Argentina, Bolivia Caribbean, Guyana |
[95] and this study |
| Armillaria sinapina | Yes | Weakly pathogenic | Yes | Conifers | North America, Japan | [97] |
| Armillaria solidipes | Yes | Highly pathogenic | Yes | Conifers | Cooler regions of North America, Europe, China | [58, 98] |
| Desarmillaria ectypa | Unknown | Saprotrophic on decaying peat moss | No | Sphagnum moss | Europe, Russia, Japan, China | [57] |
| Desarmillaria tabescens | Yes | Highly pathogenic | No | Eucalyptus, Quercus | Asia, Europe, North America | [58, 73] |
| Guyanagaster lucianii | Unknown | Only saprotrophic stage observed | No | Eperua spp. | Guyana | This study |
| Guyanagaster necrorhizus | Unknown | Only saprotrophic stage observed | No, but short non-melanized hyphal cords may be produced | Dicymbe spp. | Guyana | [27] |
Species in the earliest diverging genera in the armillarioid clade, Desarmillaria and Guyanagaster, have not been observed to form rhizomorphs in nature, but they do produce unmelanized rhizomorphs in culture [27, 57, 58, 73] (Fig. 1). Rhizomorph production of Armillaria species from Africa, the next lineage to diverge, has been scantily observed in nature [74], but the production of melanized rhizomorphs does occur in culture [75]. The next lineage to diverge is that composed of A. mellea, which produces short-lived melanized rhizomorphs with limited growth in the soil [58]. The most species rich armillarioid lineage, the northern hemisphere and Australasian/temperate South America lineage, produce melanized rhizomorphs in nature (see Table 1).
The capacity of Armillaria species to produce melanized rhizomorphs in nature is thought to be an adaptation to harsh environments [71]. Melanized rhizomorphs can confer advantages like protection against other microbial competitors, translocation of resources, growth from a suitable resource base into an environment that may not support growth, as well as enhancement of inoculum potential [71]. Additionally, melanin is thought to promote longevity and survival of rhizomorphs within the soil [76], and has also been found to help other fungi survive in extreme environments [77]. Although all species in the armillarioid clade appear to retain the trait of rhizomorph production, only the most recently diverged appear to be adapted for melanized rhizomorph production in the current environment. In a controlled environment, it was found that only under high oxygen availability and near saturated moisture would D. tabescens produce melanized rhizomorphs [78]. It was concluded that these environmental conditions are sufficiently stringent in the climate of today, and could explain why D. tabescens has not been observed to form rhizomorphs in nature. It is possible that the production of melanized rhizomorphs could be a driver of diversification in Armillaria by conferring a competitive advantage to the species that produce them.
Desarmillaria species lack an annulus at maturity, whereas Armillaria species have a robust annulus at maturity. An annulus is the remnant of a partial veil that remains after the pileus expands. Partial veils protect the immature hymenium [79]. The development of a more protected hymenium could be an adaptation of Armillaria to drier, more unpredictable habitats that were common when this lineage began to diversify.
Conclusions
Our analyses suggest that the armillarioid clade arose in Eurasia during the PETM, a time marked by a warm, tropical climate. Two lineages arose during this time: the earliest diverging lineage—which eventually led to the gasteroid genus Guyanagaster—and the exannulate mushroom-forming genus Desarmillaria. Besides being the oldest lineages in the armillarioid clade, they are also depauperate compared to Armillaria. Armillaria diverged after the shift to a cooler and more arid climate at the Eocene-Oligocene boundary. The production of melanized rhizomorphs in nature and the development of a protective partial veil could be adaptations that led to the subsequent dispersal and diversification of Armillaria in the much harsher, temperate climate. The success of Armillaria could have displaced now-extinct species in the exannulate and gasteroid lineages. Guyanagaster and Desarmillaria species have likely persisted to the present because they are highly specialized for their habitat.
Formal taxonomic descriptions
Desarmillaria (Herink) R. A. Koch & Aime gen. et stat. nov.
Basionym—Armillaria ss. Fries subgenus Desarmillaria Herink, Sympozium o václavce obecné (J. Hasek). 1972 September. Lesnicka fakulta VSZ Brno: 44. 1973.
Type—Armillaria socialis (DC. ex Fr.) Herink, Sympozium o václavce obecné (J. Hasek). 1972 September. Lesnicka fakulta VSZ Brno: 44. 1973.
MycoBank number—MB 819124 (genus).
Description—Basidiomata stipitate, usually in caespitose clusters on wood. Pileus convex to applanate, usually some shade of brown. Lamellae pallid, adnexed to adnate to subdecurrent with lamellulae. Stipe central, various colorations but often concolorous with cap, longitudinally striate, usually with fibrils on upper third. Annulus absent. Basidiospores ellipsoid to spherical, smooth, hyaline, inamyloid. Basidia clavate, four-sterigmate, hyaline. Cheilocystidia clavate, usually resembling basidioles. Pileipellis suprapellis composed of round, ellipsoid, cylindric, utriform, brown to hyaline, verrucose cells; subpellis composed of a layer of compact, shortened hyphae. Clamp connections absent. Rhizomorph production not observed in nature, while unmelanized production in culture has been observed. Saprotrophic to parasitic. Known only from the northern hemisphere.
Commentary—Desarmillaria includes mushroom-forming armillarioid species that lack an annulus. This difference in morphology led Singer [80, 81] to divide Armillaria (as Armillariella) into two sections based on the presence or absence of an annulus at maturity. Herink [82], then, recognized Armillaria as an annulate subgenus and Desarmillaria as an exannulate subgenus. Additionally, members of this genus have not been observed producing rhizomorphs in the field [57, 58], which is in contrast to most Armillaria species, most of which do produce rhizomorphs in the field (see Table 2). Rhizomorph production in nature appears to be a lost trait in this genus, as species are still able to form them in culture.
We have opted to elevate Desarmillaria to genus-level for two reasons. First, the absence of an annulus in Desarmillaria species and the presence of one in Armillaria species is a reliable characteristic to differentiate the two genera. Second, our phylogenetic and molecular clock analyses show Desarmillaria is on a separate evolutionary trajectory compared to Armillaria, meriting a separate genus.
Desarmillaria tabescens (Scop.) R. A. Koch & Aime comb. nov.
Basionym—Agaricus tabescens Scop., Scopoli 1772, Flora Carniolica Plantas Corniolae Indigenas no. 1537: 446.
MycoBank number—MB 819125 (species).
Desarmillaria ectypa (Fr.) R. A. Koch & Aime comb. nov.
Basionym—Agaricus ectypus Fr., Fries 1821, Syst. Mycol. 1: 108.
MycoBank number—MB 819126 (species).
Guyanagaster lucianii R. A. Koch & Aime sp. nov . —Holotype—BRG 41292, isotype—PUL F2891 (Figs. 1 and 3b–h). Mabura Ecological Reserve, (5°10'09.26"N, 58°42'14.40"W), 25 December 2013. R. A. Koch 89.
MycoBank number—MB 815807 (species).
Representative DNA barcode—RAK 89 (holotype), ITS—GenBank KU170950, 28S—GenBank KU170940, EF1α—GenBank KU289110.
Etymology—Lucianii = in honor of “Lucian” Edmund, a Patamona fungal parataxonomist, who discovered the type locality of G. lucianii and has since been invaluable in collecting Guyanagaster specimens in the field.
Description—Figures 2 and 4a–g. Basidiomata gasteroid, subhypogeous to hypogeous, scattered, or in linear troops, attached directly to woody roots of Eperua falcata and Dicymbe altsonii trees; 12–34 mm broad, 8–25 mm tall, globose to subglobose to ovoid and irregularly broadly lobate, dense, base with smooth, sterile concolorous stipe, 5–44 × 2–5 mm, attached directly to the substratum. Peridium dark brown (8 F4-8 F3) to black (8 F1) during all stages of development, moist, tough, covered in 4–6 sided polygonal pyramidal warts that come to a shallow point, these 0.8–3.1 mm broad, 0.5 mm tall, sharply differentiated from endoperidium; endoperidium white (8A1) to light pink (8A2), tough, 0.5–1 mm thick, composed of matted hyphae transitioning evenly to sterile, white hyphal veins between the glebal locules; gleba composed of well-defined locules and intervening veins of hyphae; locules globose to ovate to subangular, 0.2–2 mm broad, initially white (8A1), maturing in stages to gold (3A2-4A5), light orange-pink (7B6), to deep orange (8B8), often evenly, but rarely the locules that border the peridium and columella take on the darker shade first; hyphae separating locules initially white (8A1), hyaline and waterlogged in mature specimens; columella well-defined to not, base continuous with stipe, 6–13 × 3–6 mm, initially white (8A1), turning to brown (6D7), taking on the color of the gleba with age, cottony to gelatinous. Exoperidium individual hyphae of outer layer dark brown, individual hyphae curved to tortuously curved, thick-walled, non-gelatinous, 18–99 × 4–11 μm. Endoperidium 700–800 μm thick, well-differentiated from exoperidium and glebal locules, hyphae hyaline, 3–11 μm. Sterile hyphae separating locules similar to endoperidial hyphae, hyaline, thin-walled, branching, 3–5 um wide, organized in a parallel manner. Basidia not observed. Basidiospores globose, dextrinoid; 21.4–30.4 × 21.4–29.3 μm (mean = 25.1 ± 1.5 × 24.8 ± 1.5 μm, Qr = 0.95–1.10, Qm = 1.01 ± 0.04; n = 20); immature basidiospores hyaline when immature, then darkening to light pink and then rusty brown with maturity. Columella hyphae hyaline, thin-walled, not as tightly packed, some randomly inflated at septa, 4–13 μm wide, random apical cells inflated, 13–26 × 37–57 μm. Clamp connections absent in all tissues. Taste not tested. Odor not tested.
Fig. 4.

a–g Guyanagaster lucianii (BRG 41292 HOLOTYPE). a Exterior showing dark brown peridium and irregularly lobed shape. b Subhypogeous habit. c Longitudinal section of basidioma showing immature gleba, developed columella and elongated stipe. d Maturing gleba. e Basidiospores of immature basidioma. f Basidiospores of mature basidioma. g Mature gleba. Bar = one cm in a, b, c, d and g. Bar = 20 μm in e and f
Habit, habitat and distribution—Attached to decaying woody roots of Dicymbe altsonii and Eperua falcata in tropical rainforests. Known only from the type locality in the Mabura Ecological Reserve of Guyana.
Specimens examined—Guyana. Region 10 Upper Demerara-Berbice—Mabura Hill, elevation 161 m; vicinity of basecamp, Eduba stand 4, four basidiomata, 11 June 2011, MCA 4424 (BRG 41294, PUL F3439); vicinity of basecamp, patch 32, one basidioma found at the base of a dead Eperua falcata, 24 December 2013, RAK 84 (BRG 41230, PUL F3440); vicinity of basecamp, patch 35, four basidiomata found at the base of a dead Eperua falcata, 24 December 2013, RAK 88 (BRG 41293, PUL F2892); 2.5 km north of basecamp, patch 31, nine basidiomata found along the decaying roots of Eperua falcata, 25 December 2013, RAK 89 (BRG 41292, PUL F2891).
Commentary— Phylogenetic evidence suggests that the two lineages of Guyanagaster represent distinct species that diverged approximately 8 Ma. The sister species share 87% (777/893 bp) nucleotide identity at the ITS region and 96% (705/731 bp) nucleotide identity at LSU. The drastic morphological changes that occur during the maturation of Guyanagaster basidiomata (i.e. gleba coloration) as well as the morphological plasticity between individual basidiomata (e.g. basidioma size and shape, columella dimensions) make delimiting species using macromorphological characters unreliable. However, G. lucianii can be distinguished from G. necrorhizus by its consistently larger basidiospores. During all stages of maturation, the basidiospores of G. lucianii measure between 21–30 × 21–29 μm, compared to those of G. necrorhizus, which measure 15–19 × 15–18 μm. Additionally, the basidiospores of G. lucianii lack the pedicel that is readily apparent on the basidiospores of G. necrorhizus (Fig. 4e–f).
Ecologically, G. lucianii appears to occupy the same niche as G. necrorhizus. Basidiomata are hypogeous to subhypogeous and growing from the roots of dead and decaying trees. Both Guyanagaster species are white rotters as the decaying roots to which they are attached are all light-colored, spongy and sometimes gelatinous. The known habitat for G. necrorhizus and G. lucianii are separated by only 125 km, so elucidation of their dispersal strategy is needed to understand the evolutionary forces that led to their speciation.
Acknowledgements
The authors wish to thank the following for assisting with this study: Dillon Husbands functioned as the Guyanese local counterpart and assisted with field collecting, descriptions and specimen processing. Todd F. Elliott, J. J. Harrison, Stephen D. Russell, Christian Schwarz and Tatyana Svetasheva provided photographs for Fig. 2. Additional field assistance in Guyana was provided by C. Andrew, V. Joseph, P. Joseph, F. Edmond, L. Edmond and L. Williams. In Cameroon, Dr. Jean Michel Onana, Head of The National Herbarium of Cameroon (Institute of Agricultural Research for Development, IRAD), provided much logistical assistance. The Conservator of the Dja Biosphere Reserve, Mr. Mengamenya Goue Achille, and his staff greatly assisted the fieldwork in the Dja. Field assistance in Cameroon was provided by Alamane Gabriel (a.k.a. Sikiro), Abate Jackson, Mamane Jean-Pierre, Mei Lin Chin, Todd Elliott, and Camille Truong. Finally, the authors wish to thank Brandon Matheny and four anonymous reviewers for their insightful comments on an earlier version of this manuscript. Research permits were granted by the Guyana Environmental Protection Agency and by the Cameroon Ministry of Research and Scientific Innovation. This paper is number 217 in the Smithsonian Institution’s Biological Diversity of the Guiana Shield Program publication series.
Funding
The authors wish to thank the following for financial support: National Science Foundation DEB-0918591, NSF DEB-0732968, NSF DEB-1556412 and Linnaean Society Systematics Research grant to MCA, the National Geographic Society’s Committee for Research and Exploration grant 9235–13 to TWH, Explorer’s Club Exploration Fund, Mycological Society of America Fungal Forest Ecology grant, American Philosophical Society Lewis and Clark Fund for Exploration and the T. Woods Thomas award to RAK.
Availability of data and materials
The alignment and tree files are available in TreeBase and can be accessed at http://purl.org/phylo/treebase/phylows/study/TB2:S20469. Sequences generated in this study were deposited in GenBank and accession numbers for those sequences are available in Additional file 1.
Authors’ contributions
RAK, AWW and MCA conceived and developed the study. RAK, OS, TWH and MCA were involved in specimen collection and field descriptions. RAK performed all of the molecular work and RAK and AWW performed the analyses. RAK, AWW, and MCA wrote the paper. All authors read, advised on revisions and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Additional files
Single-locus and multi-gene phylogenies for the six loci used. (PDF 3262 kb)
Morphological description of Armillaria camerunensis TH DJA 91 and TH 9926. (DOCX 108 kb)
Results from the ancestral range estimation analysis. (XLSX 38 kb)
Contributor Information
Rachel A. Koch, Email: koch17@purdue.edu
Andrew W. Wilson, Email: andrew.wilson@botanicgardens.org
Olivier Séné, Email: olivier_sene@yahoo.fr.
Terry W. Henkel, Email: terry.henkel@humboldt.edu
M. Catherine Aime, Email: maime@purdue.edu.
References
- 1.Wargo PM. Armillariella mellea and Agrilus bilineatus and mortality of defoliated oak trees. Forest Sci. 1977;23:485–492. [Google Scholar]
- 2.Kile GA. Armillaria root rot in eucalypt forests: aggravated endemic disease. Pac Sci. 1983;37:457–464. [Google Scholar]
- 3.Podger FD, Kile GA, Watling R, Fryer J. Spread and effects of Armillaria luteobubalina sp. nov. in an Australian Eucalyptus regnans plantation. T Brit Mycol Soc. 1978;71:77–87. doi: 10.1016/S0007-1536(78)80009-6. [DOI] [Google Scholar]
- 4.Baumgartner K, Rizzo DM. Spread of Armillaria root disease in a California vineyard. Am J Enol Viticult. 2002;53:197–203. [Google Scholar]
- 5.Redfern DB, Filip GM. Inoculum and infection. In: Shaw CG, Kila GA, editors. Armillaria Root Disease. Agriculture Handbook 691. Washington DC: USDA Forest Service; 1991. pp. 48–61. [Google Scholar]
- 6.Kile GA, McDonald GI, Byler JW. Ecology and Disease in Natural Forests. In: Shaw CG, Kila GA, editors. Armillaria Root Disease. Agriculture Handbook 691. Washington DC: USDA Forest Service; 1991. pp. 102–121. [Google Scholar]
- 7.Moncalvo JM, Buchanan PK. Molecular evidence for long distance dispersal across the Southern Hemisphere in the Ganoderma applanatum-australe species complex (Basidiomycota) Mycol Res. 2008;112(4):425–436. doi: 10.1016/j.mycres.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 8.James TY, Moncalvo JM, Li S, Vilgalys R. Polymorphism at the ribosomal DNA spacers and its relation to breeding structure of the widespread mushroom Schizophyllum commune. Genetics. 2001;157:149–161. doi: 10.1093/genetics/157.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geml J, Timling I, Robinson CH, Lennon N, Nusbaum HC, Brochmann C, et al. An arctic community of symbiotic fungi assembled by long-distance dispersers: phylogenetic diversity of ectomycorrhizal basidiomycetes in Svalbard based on soil and sporocarp DNA. J Biogeogr. 2012;39:74–88. doi: 10.1111/j.1365-2699.2011.02588.x. [DOI] [Google Scholar]
- 10.Coetzee MPA, Bloomer P, Wingfield MJ, Wingfield BD. Paleogene radiation of a plant pathogenic mushroom. PLoS One. 2011;6:e28545. doi: 10.1371/journal.pone.0028545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hibbett DS, Pine EM, Langer E, Langer G, Donoghue MJ. Evolution of gilled mushrooms and puffballs inferred from ribosomal DNA sequences. Proc Natl Acad Sci U S A. 1997;94:12002–12006. doi: 10.1073/pnas.94.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maser C, Maser Z, Molina R. Small-mammal mycophagy in rangelands of Central and Southeastern Oregon. J Range Manage. 1988;41:309–312. doi: 10.2307/3899385. [DOI] [Google Scholar]
- 13.Miller OK, Jr, Miller HH. Gasteromycetes—Morphological and developmental features with keys to the orders, families, and genera. Eureka: Mad River Press; 1988. [Google Scholar]
- 14.Bruns TD, Fogel R, White TJ, Palmer JD. Accelerated evolution of a false-truffle from a mushroom ancestor. Nature. 1989;339:140–142. doi: 10.1038/339140a0. [DOI] [PubMed] [Google Scholar]
- 15.Mueller GM, Pine EM. DNA data provide evidence on the evolutionary relationships between mushrooms and false truffles. McIlvainea. 1994;11:61–74. [Google Scholar]
- 16.Aime MC, Miller OK., Jr Proposal to conserve the name Chroogomphus against Brauniellula (Gomphidiaceae, Agaricales, Basidiomycota) Taxon. 2006;55:228–229. doi: 10.2307/25065549. [DOI] [Google Scholar]
- 17.Lebel T, Tonkin JE. Australasian species of Macowanites are sequestrate species of Russula (Russulaceae, Basidiomycota) Austral Syst Bot. 2007;20:355–381. doi: 10.1071/SB07007. [DOI] [Google Scholar]
- 18.Lebel T, Syme A. Sequestrate species of Agaricus and Macrolepiota from Australia: new combinations and species, and their position in a calibrated phylogeny. Mycologia. 2012;104:496–520. doi: 10.3852/11-092. [DOI] [PubMed] [Google Scholar]
- 19.Ge ZW, Smith ME. Phylogenetic analysis of rDNA sequences indicates that the sequestrate Amogaster viridiglebus is derived from within the agaricoid genus Lepiota (Agaricaceae) Mycol Prog. 2013;12:151–155. doi: 10.1007/s11557-012-0841-y. [DOI] [Google Scholar]
- 20.Pegler DN, Young TWH. The gasteroid Russulales. T Brit Mycol Soc. 1979;72:353–388. doi: 10.1016/S0007-1536(79)80143-6. [DOI] [Google Scholar]
- 21.Hopple JS, Jr, Vilgalys R. Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: divergent domains, outgroups, and monophyly. Mol Phylogenet Evol. 1999;13:1–19. doi: 10.1006/mpev.1999.0634. [DOI] [PubMed] [Google Scholar]
- 22.Peintner U, Bougher NL, Castellano MA, Moncalvo J-M, Moser MM, Trappe JM, Vilgalys R. Multiple origins of sequestrate fungi related to Cortinarius (Cortinariaceae) Am J Bot. 2001;88:2168–2179. doi: 10.2307/3558378. [DOI] [PubMed] [Google Scholar]
- 23.Justo A, Morgenstern I, Hallen-Adams HE, Hibbett DS. Convergent evolution of sequestrate forms in Amanita under Mediterranean climate conditions. Mycologia. 2010;102:675–688. doi: 10.3852/09-191. [DOI] [PubMed] [Google Scholar]
- 24.Wilson AW, Binder M, Hibbett DS. Diversity and evolution of ectomycorrhizal host associations in the Sclerodermatineae (Boletales, Basidiomycota) New Phytol. 2012;194:1079–1095. doi: 10.1111/j.1469-8137.2012.04109.x. [DOI] [PubMed] [Google Scholar]
- 25.Braaten CC, Matheny PB, Viess DL, Wood MG, Williams JH, Bougher NL. Two new species of Inocybe from Australia and North America that include novel secotioid forms. Botany. 2014;92:9–22. doi: 10.1139/cjb-2013-0195. [DOI] [Google Scholar]
- 26.Smith ME, Amese KR, Elliott TF, Obase K, Aime MC, Henkel TW. New sequestrate fungi from Guyana: Jimtrappea guyanensis gen. sp. nov., Castellanea pakaraimophila gen. sp. nov., and Costatisporus cyanescens gen. sp. nov. (Boletaceae, Boletales) IMA Fungus. 2015;6:297–317. doi: 10.5598/imafungus.2015.06.02.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henkel TW, Aime MC, Smith ME. Guyanagaster, a new wood-decaying sequestrate fungal genus related to Armillaria (Physalacriaceae, Agaricales, Basidiomycota) Am J Bot. 2010;97:1474–1484. doi: 10.3732/ajb.1000097. [DOI] [PubMed] [Google Scholar]
- 28.Isaacs R, Gillman MP, Johnston M, Marsh F, Wood BC. Size structure of a dominant Neotropical forest tree species, Dicymbe altsonii, in Guyana and some factors reducing seedling leaf area. J Trop Ecol. 1996;12:599–606. doi: 10.1017/S0266467400009810. [DOI] [Google Scholar]
- 29.Henkel TW. Mondominance in the ectomycorrhizal Dicymbe corymbosa (Caesalpiniaceae) from Guyana. J Trop Ecol. 2003;19:417–437. doi: 10.1017/S0266467403003468. [DOI] [Google Scholar]
- 30.Zagt RJ. Pre-dispersal and early post-dispersal demography, and reproductive litter production, in the tropical tree Dicymbe altsonii in Guyana. J Trop Ecol. 1997;13:511–526. doi: 10.1017/S0266467400010683. [DOI] [Google Scholar]
- 31.Kornerup A, Wanscher JH. Methuen handbook of colour. 3. London: Eyre Methuen; 1978. [Google Scholar]
- 32.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 33.Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moncalvo JM, Lutzoni FM, Rehner SA, Johnson J, Vilgalys R. Phylogenetic relationships of agaric fungi based on nuclear large subunit ribosomal DNA sequences. Syst Biol. 2000;49:278–305. doi: 10.1093/sysbio/49.2.278. [DOI] [PubMed] [Google Scholar]
- 35.Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- 36.Matheny PB. Improving phylogenetic inference of mushrooms using RPB1 and RPB2 sequences (Inocybe, Agaricales) Mol Phylogenet Evol. 2005;35:1–20. doi: 10.1016/j.ympev.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanfear R, Calcott B, Ho SYW, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29(6):1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 40.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 41.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skrede I, Engh IB, Binder M, Carlsen T, Kauserud H, Bendiksby M. Evolutionary history of Serpulaceae (Basidiomycota): molecular phylogeny, historical biogeography and evidence for a single transition of nutritional mode. BMC Evol Biol. 2011;11:230. doi: 10.1186/1471-2148-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryberg M, Matheny PB. Asynchronous origins of ectomycorrhizal clades of Agaricales. Proc Biol Sci. 2012;279:2003–2011. doi: 10.1098/rspb.2011.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson AW, Hosaka K, Mueller GM. Evolution of ectomycorrhizas as a driver of diversification and biogeographic patterns in the model mycorrhizal mushroom genus Laccaria. New Phytol. 2016;doi:10.1111/nph.14270. [DOI] [PMC free article] [PubMed]
- 45.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUTi and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v1.6. 2014. http://beast.bio.ed.ac.uk/Tracer. Accessed 6 June 2016.
- 47.Jenkinson TS, Perry BA, Schaefer RE, Desjardin DE. Cryptomarasmius gen. nov. established in the Physalacriaceae to accommodate members of Marasmius section Hygrometrici. Mycologia. 2014;106(1):86–94. doi: 10.3852/11-309. [DOI] [PubMed] [Google Scholar]
- 48.Dentinger BTM, Gaya E, O’Brien H, Suz LM, Lachlan R, Díaz-Valderrama JR, et al. Tales from the crypt: genome mining from fungarium specimens improves resolution of the mushroom tree of life. Biol J Linn Soc. 2016;117:11–32. doi: 10.1111/bij.12553. [DOI] [Google Scholar]
- 49.Hibbett DS, Grimaldi D, Donoghue MJ. Fossil mushrooms from Miocene and Cretaceous ambers and the evolution of Homobasidiomycetes. Am J Bot. 1997;84(8):981–991. doi: 10.2307/2446289. [DOI] [PubMed] [Google Scholar]
- 50.Ree RH, Smith SE. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- 51.Yu Y, Harris AJ, Blair C, He X. RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Mol Phylogenet Evol. 2015;87:46–49. doi: 10.1016/j.ympev.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 52.Singer R. Flora Neotropica monograph 3, Omphalinae. New York: Hafner Publishing; 1970. [Google Scholar]
- 53.Hennings P. Fungi camerunenses. I Bot Jb. 1895;22:70–111. [Google Scholar]
- 54.Watling R. Armillaria Staude in the Cameroon Republic. Persoonia. 1992;14:483–491. [Google Scholar]
- 55.Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 56.Wolfe JA. Distributions of major vegetation types during the Tertiary. In: Sundquist ET, Broekner WS, editors. The Carbon Cycle and Atmospheric CO2: Natural Variations, Archean to Present, American Geophysical Union Monograph 32. Washington, DC: American Geophysical Union; 1985. pp. 357–376. [Google Scholar]
- 57.Zolciak A, Bouteville R-J, Tourvieille J, Roeckel-Drevet P, Nicolas P, Guillaumin J-J. Occurrence of Armillaria ectypa (Fr.) Lamoure in peat bogs of the Auvergne—the reproduction system of the species. Cryptogamie Mycol. 1997;18(4):299–313. [Google Scholar]
- 58.Guillaumin J-J, Mohammed C, Anselmi N, Courtecuisse R, Gregory SC, Holdenrieder O, et al. Geographical distribution and ecology of the Armillaria species in western Europe. Eur J Forest Pathol. 1993;23:321–341. doi: 10.1111/j.1439-0329.1993.tb00814.x. [DOI] [Google Scholar]
- 59.Volk TJ, Burdsall HH., Jr . A nomenclatural study of Armillaria and Armillariella species. Synopsis Fungorum 8. Oslo: Fungiflora; 1995. [Google Scholar]
- 60.Antonín V, Jankovský L, Lochman J, Tomšovský M. Armillaria socialis—morphological-anatomical and ecological characteristics, pathology, distribution in the Czech Republic and Europe and remarks on its genetic variation. Czech Mycol. 2006;58(3–4):209–224. [Google Scholar]
- 61.Cassens I, Vicario S, Waddell VG, Balchowsky H, Van Belle D, Ding W, et al. Independent adaptation to riverine habitats allowed survival of ancient cetacean lineages. Proc Natl Acad Sci U S A. 2000;97(21):11343–11347. doi: 10.1073/pnas.97.21.11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morley RJ. Cretaceous and Tertiary climate change and the past distribution of megathermal rainforests. In: Bush MB, Flenley JR, Gosling WD, editors. Tropical rainforest responses to climatic change. Heidelberg: Springer Praxis; 2011. pp. 1–34. [Google Scholar]
- 63.Tiffney BH. Perspectives on the origin of the floristic similarity between eastern Asia and Eastern North America. J Arnold Arboretum. 1985;66:73–94. doi: 10.5962/bhl.part.13179. [DOI] [Google Scholar]
- 64.Maphosa L, Wingfield BD, Coetzee MPA, Mwenje E, Wingfield MJ. Phylogenetic relationships among Armillaria species inferred from partial elongation factor 1-alpha DNA sequence data. Australas Plant Pathol. 2006;35:513–520. doi: 10.1071/AP06056. [DOI] [Google Scholar]
- 65.Dercourt J, Zonenshain LP, Ricou L-E, Kazmin VG, Le Pichon X, Knipper AL, et al. Geological evolution of the Tethys belt from the Atlantic to the Pamirs since the Lias. Tectonophysics. 1986;123:241–315. doi: 10.1016/0040-1951(86)90199-X. [DOI] [Google Scholar]
- 66.Coetzee MPA, Wingfield BD, Harrington TC, Dalevi D, Coutinho TA, Wingfield MJ. Geographic diversity of Armillaria mellea s.s. based on phylogenetic analysis. Mycologia. 2000;92:105–113. doi: 10.2307/3761454. [DOI] [Google Scholar]
- 67.Crayn DM, Costion C, Harrington MG. The Sahul-Sunda floristic exchange: dated molecular phylogenies document Cenozoic intercontinental dispersal dynamics. J Biogeogr. 2015;42:11–24. doi: 10.1111/jbi.12405. [DOI] [Google Scholar]
- 68.Coetzee MPA, Wingfield BD, Bloomer P, Ridley GS, Wingfield MJ. Molecular identification and phylogeny of Armillaria isolates from South America and Indo-Malaysia. Mycologia. 2003;95(2):285–293. doi: 10.2307/3762039. [DOI] [PubMed] [Google Scholar]
- 69.Webb PN, Harwood DM. Pliocene fossil Nothofagus (Southern Beech) from Antarctica: phytogeography, dispersal strategies, and survival in high latitude glacial-deglacial environments. In: Alden JN, Mastrantonio JL, Ødum S, editors. Forest Development in Cold Climates, NATO ASI Series 244. New York: Springer US; 1993. pp. 135–166. [Google Scholar]
- 70.Heads M. Panbiogeography of Nothofagus (Nothofagaceae): analysis of the main species massings. J Biogeogr. 2006;33:1066–1075. doi: 10.1111/j.1365-2699.2006.01479.x. [DOI] [Google Scholar]
- 71.Garraway MO, Hütterman A, Wargo PM. Ontogeny and physiology. In: Shaw CG, Kile GA, editors. Armillaria Root Disease. Agriculture Handbook 691. Washington DC: USDA Forest Service; 1991. pp. 21–47. [Google Scholar]
- 72.Smith ML, Bruhn JN, Anderson JB. The fungus Armillaria bulbosa is among the largest and oldest living organisms. Nature. 1992;356(2):428–431. doi: 10.1038/356428a0. [DOI] [Google Scholar]
- 73.Rhoads AS. A comparative study of two closely related root-rot fungi, Clitocybe tabescens and Armillaria mellea. Mycologia. 1945;61:741–766. doi: 10.2307/3755134. [DOI] [Google Scholar]
- 74.Swift MJ. The ecology of Armillaria mellea Vahl (ex Fries) in the indigenous and exotic woodlands of Rhodesia. Forestry Oxford. 1972;45:67–86. doi: 10.1093/forestry/45.1.67. [DOI] [Google Scholar]
- 75.Coetzee MPA, Wingfield BD, Coutinho TA, Wingfield MJ. Identification of the causal agent of Armillaria root rot of Pinus species in South Africa. Mycologia. 2000;92:777–785. doi: 10.2307/3761435. [DOI] [Google Scholar]
- 76.Rizzo DM, Blanchette RA, Palmer MA. Biosorption of metal ions by Armillaria rhizomorphs. Can J Bot. 1992;70(8):1515–1520. doi: 10.1139/b92-190. [DOI] [Google Scholar]
- 77.Mironenko NV, Alekhina IA, Zhdanova NN, Bulat SA. Intraspecific variation in gamma-radiation resistance and genomic structure in the filamentous fungus Alternaria alternata: a case study of strains inhabiting Chernobyl reactor, 4. Ecotoxicol Environ Saf. 2000;45:177–187. doi: 10.1006/eesa.1999.1848. [DOI] [PubMed] [Google Scholar]
- 78.Mihail JD, Bruhn JN, Leininger TD. The effects of moisture and oxygen availability on rhizomorph generation by Armillaria tabescens in comparison with A. gallica and A. mellea. Mycol Res. 2002;106(6):697–704. doi: 10.1017/S0953756202005920. [DOI] [Google Scholar]
- 79.Watkinson SC, Boddy L, Money NP. The Fungi. 3. Cambridge: Academic; 2015. [Google Scholar]
- 80.Singer R. Agaricales in modern taxonomy. Lilloa. 1951;22:5–832. [Google Scholar]
- 81.Singer R. The Agaricales in modern taxonomy. 4. Königstein: Koeltz Scientific Books; 1986. [Google Scholar]
- 82.Herink J. Taxonomie václacky obecné—Armillaria mellea (Vahl ex Fr.) Kumm. In: Hasek J, editor. Sympozium o václavce obecné: Armillaria mellea (Vahl ex Fr.) Kumm. Brno, Czechoslovakia: Vysoká Skola Zemĕdĕlská v Brné; 1973. p. 21–48.
- 83.Brazee NJ, Ortiz-Santana B, Banik MT, Lindner DL. Armillaria altimontana, a new species from the western interior of North America. Mycologia. 2012;104(5):1200–1205. doi: 10.3852/11-409. [DOI] [PubMed] [Google Scholar]
- 84.Gregory SC, Watling R. Occurerence of Armillaria borealis in Britain. T Brit Mycol Soc. 1985;84(1):47–55. doi: 10.1016/S0007-1536(85)80219-9. [DOI] [Google Scholar]
- 85.Bauce E, Allen DC. Role of Armillaria calvescens and Glycobius speciosus in a sugar maple decline. Can J For Res. 1992;22:549–552. doi: 10.1139/x92-072. [DOI] [Google Scholar]
- 86.Dessureault M. Morphological studies of the Armillaria mellea complex: Two new species, A. gemina and A. calvescens. Mycologia. 1989;81(2):216–225. doi: 10.2307/3759703. [DOI] [Google Scholar]
- 87.Prospero S, Holdenrieder O, Rigling D. Comparison of the virulence of Armillaria cepistipes and Armillaria ostoyae on four Norway spruce provenances. Forest Pathol. 2004;24(1):1–14. doi: 10.1046/j.1437-4781.2003.00339.x. [DOI] [Google Scholar]
- 88.Mwenje E, Ride JP, Pearce RB. Distribution of Zimbabwean Armillaria groups and their pathogenicity on cassava. Plant Pathol. 1998;47:623–634. doi: 10.1046/j.1365-3059.1998.00275.x. [DOI] [Google Scholar]
- 89.Kile GA. Genotypes of Armillaria hinnulea in wet sclerophyll eucalypt forest in Tasmania. Trans Br Mycol Soc. 1986;87(2):312–314. doi: 10.1016/S0007-1536(86)80036-5. [DOI] [Google Scholar]
- 90.Ramsfield TD, Power MWP, Ridley GS. A comparison of populations of Armillaria hinnulea in New Zealand and Australia. N Z Plant Prot. 2008;61:41–47. [Google Scholar]
- 91.Shaw CG, III, MacKenzie M, Toes EHA, Hood IA. Cultural characteristics and pathogenicity to Pinus radiata of Armillaria novae-zelandiae and A. limonea. N Z J For Sci. 1981;11(1):65–70. [Google Scholar]
- 92.Shaw CG, III, Calderon S. Impact of Armillaria root rot in plantations of Pinus radiata established on sites converted from indigenous forest. N Z J For Sci. 1977;7:359–373. [Google Scholar]
- 93.Kile GA. Armillaria luteobubalina—a primary cause of decline and death of trees in mixed species eucalypt forests in central Victoria. Aust Forest Res. 1981;11(1):63–67. [Google Scholar]
- 94.Pareek M, Cole L, Ashford AE. Variations in structure of aerial and submerged rhizomorphs of Armillaria luteobubalina indicate that they may be organs of adsorption. Mycol Res. 2001;105:1377–1387. doi: 10.1017/S0953756201004622. [DOI] [Google Scholar]
- 95.Pildain MB, Coetzee MPA, Wingfield BD, Wingfield MJ, Rajchenberg M. Taxonomy of Armillaria in the Patagonian forests of Argentina. Mycologia. 2010;102(2):392–403. doi: 10.3852/09-105. [DOI] [PubMed] [Google Scholar]
- 96.Raabe RD. Host list of the root rot fungus Armillaria mellea. Hilgardia. 1962;32:25–88. doi: 10.3733/hilg.v33n02p025. [DOI] [Google Scholar]
- 97.Cleary MR, van der Kamp BJ, Morrison DJ. Pathogenicity and virulence of Armillaria sinapina and host response to infection in Douglas-fir, western hemlock and western red cedar in the southern Interior of British Columbia. Forest Pathol. 2012;42(6):481–491. doi: 10.1111/j.1439-0329.2012.00782.x. [DOI] [Google Scholar]
- 98.Burdsall HH, Jr, Volk TJ. Armillaria solidipes, an older name for the fungus called Armillaria ostoyae. N Am Fungi. 2008;3(7):261–267. doi: 10.2509/naf2008.003.00717. [DOI] [Google Scholar]
- 99.Moncalvo J-M, Vilgalys R, Redhead SA, Johnson JE, James TY, Aime MC, et al. One hundred and seventeen clades of eugarics. Mol Phylogenet Evol. 2002;23:357–400. doi: 10.1016/S1055-7903(02)00027-1. [DOI] [PubMed] [Google Scholar]
- 100.Brazee NJ, Hulvey JP, Wick RL. Evaluation of partial tef1, rpb2, and nLSU sequences for identification of isolates representing Armillaria calvescens and Armillaria gallica from northeastern North America. Fungal Biol. 2011;115(8):741–749. doi: 10.1016/j.funbio.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 101.Guo T, Wang HC, Xue WQ, Zhao J, Yang ZL. Phylogenetic analyses of Armillaria reveal at least 15 phylogenetic lineages in China, seven of which are associated with cultivated Gastrodia elata. PLoS One. 2016;11(5):e0154794. doi: 10.1371/journal.pone.0154794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wingfield BD, Ambler JM, Coetzee MPA, de Beer ZW, Duong TA, Joubert F, et al. Draft genome sequences of Armillaria fuscipes, Certocystiopsis minuta, Ceratocystis adiposa, Endoconidiophora laricicola, E. polonica and Penicillium freii DAOMC 242723. IMA Fungus. 2016;7(1):217–227. doi: 10.5598/imafungus.2016.07.01.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Binder M, Hibbett DS, Wang Z, Farnham WF. Evolutionary relationships of Mycaureola dilseae (Agaricales), a basidiomycete pathogen of subtidal rhodophyte. Am J Bot. 2006;93:547–556. doi: 10.3732/ajb.93.4.547. [DOI] [PubMed] [Google Scholar]
- 104.Matheny PB, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson RH, et al. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi) Mol Phylogenet Evol. 2007;43:430–451. doi: 10.1016/j.ympev.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 105.Assembling the Fungal Tree of Life. 1995. www.aftol.org. Accessed 6 June 2016.
- 106.Petersen RH, Hughes KW. The Xerula/Oudemansiella Complex (Agaricales) Nova Hedwigia Beih. 2010;137:625. [Google Scholar]
- 107.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucl Acids Res. 2013;42:D699–D704. [DOI] [PMC free article] [PubMed]
- 108.Hibbett DS, Gilbert L-B, Donoghue MJ. Evolutionary instability of ectomycorrhizal symbioses in basidiomycetes. Nature. 2000;407:506–508. doi: 10.1038/35035065. [DOI] [PubMed] [Google Scholar]
- 109.Wang Z, Binder M, Dai YC, Hibbett DS. Phylogenetic relationships of Sparassis inferred from nuclear and mitochondrial ribosomal DNA and RNA polymerase sequences. Mycologia. 2004;96(5):1015–1029. doi: 10.2307/3762086. [DOI] [PubMed] [Google Scholar]
- 110.Matheny PB, Curtis JM, Hofstetter V, Aime MC, Moncalvo JM, Ge ZW, et al. Major clades of Agaricales: a multilocus phylogenetic overview. Mycologia. 2006;98(6):982–995. doi: 10.3852/mycologia.98.6.982. [DOI] [PubMed] [Google Scholar]
- 111.Binder M, Larsson KH, Matheny PB, Hibbett DS. Amylocorticiales ord. nov. and Jaapiales ord. nov.: early diverging clades of Agaricomycetidae dominated by corticioid forms. Mycologia. 2010;102(4):865–880. doi: 10.3852/09-288. [DOI] [PubMed] [Google Scholar]
- 112.Ronikier M, Ronikier A. Rhizomarasmius epidryas (Physalacriaceae): phylogenetic placement of an arctic-alpine fungus with obligate saprobic affinity to Dryas spp. Mycologia. 2011;103(5):1124–1132. doi: 10.3852/11-018. [DOI] [PubMed] [Google Scholar]
- 113.Qin J, Hao Y-J, Yang ZL, Li Y-C. Paraxerula ellipsospora, a new Asian species of Physalacriaceae. Mycol Prog. 2014;13(3):639–647. doi: 10.1007/s11557-013-0946-y. [DOI] [Google Scholar]
- 114.Qin J, Yang ZL. Three new species of Physalacria from China, with a key to the Asian taxa. Mycologia. 2016;108(1):215–226. doi: 10.3852/15-166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The alignment and tree files are available in TreeBase and can be accessed at http://purl.org/phylo/treebase/phylows/study/TB2:S20469. Sequences generated in this study were deposited in GenBank and accession numbers for those sequences are available in Additional file 1.


