Abstract.
Intraoperative tissue classification is one of the prerequisites for providing context-aware visualization in computer-assisted minimally invasive surgeries. As many anatomical structures are difficult to differentiate in conventional RGB medical images, we propose a classification method based on multispectral image patches. In a comprehensive ex vivo study through statistical analysis, we show that (1) multispectral imaging data are superior to RGB data for organ tissue classification when used in conjunction with widely applied feature descriptors and (2) combining the tissue texture with the reflectance spectrum improves the classification performance. The classifier reaches an accuracy of 98.4% on our dataset. Multispectral tissue analysis could thus evolve as a key enabling technique in computer-assisted laparoscopy.
Keywords: tissue classification, multispectral laparoscopy, multispectral texture analysis
1. Introduction
In contrast to traditional open surgery, laparoscopy is less invasive, causing smaller incisions and providing shorter recovery periods. One recent direction of research focuses on offering context-aware guidance (e.g., Ref. 1), where computer assistance is provided, depending on the current phase within the medical procedure. Situation recognition from sensor data, however, is extremely challenging and requires a good understanding of the scene. In this context, the classification of tissue may provide important cues with respect to what is currently happening.
In recent years, several methods for tissue or organ classification, which are based on gray-value or RGB images, have already been proposed (e.g., Refs. 2 and 3). More recently, multispectral (or hyperspectral) imaging techniques have achieved success in cancer detection and tissue classification.4 Multispectral images generally have tens or hundreds of channels, each of which corresponds to the reflection of light within a certain wavelength band. Therefore, they can provide high spectral resolution and reveal optical tissue characteristics. Multispectral tissue classification methods, as mentioned in the literature so far, mainly use the image pixel, which corresponds to a reflectance spectrum at a specific position, as their feature descriptor.5,6 Given the recent success of multispectral texture analysis outside of the field of laparoscopy,7,8 the hypothesis proposed by this paper is that texture-based methods can improve multispectral organ in minimally-invasive surgery.
A preliminary version of this paper was presented at the SPIE Medical Imaging Conference 2016.9 To our knowledge, we are the first to address the problem of tissue classification based on multispectral texture analysis for intraoperative laparoscopy. The contribution is twofold: (1) we investigate organ tissue classification in a laparoscopic setup and perform a comprehensive ex vivo study, showing that multispectral images are superior to RGB images for tissue classification in laparoscopy, and (2) we propose a feature descriptor combining texture and spectral information based on only a small number of specified bands.
2. Materials and Methods
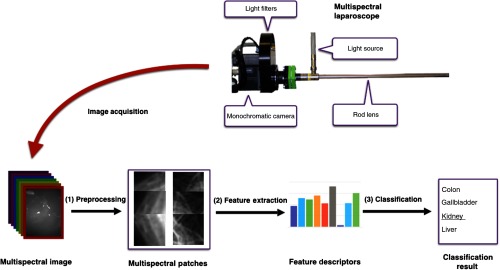
This section encompasses three central parts of our work: the multispectral image acquisition using a custom-built laparoscope (Sec. 2.1), feature extraction and classification (Sec. 2.2), and description of the experiments in Sec. 2.3. An overview of the proposed approach is shown in Fig. 1.
Fig. 1.
Concept for multispectral tissue classification. After multispectral image acquisition, noise is removed and the resulting image is cropped into patches (1). From each of these patches, the LBP texture feature and the AS are calculated (2) and fed into an SVM model to classify the organ characterized by the patch under investigation (3).
2.1. Hardware
Multispectral images are captured using a custom-built multispectral laparoscope, which is shown in Fig. 1. It combines a Richard Wolf (Knittlingen, Germany) laparoscope and light source with the 5 Mpixel Pixelteq Spectrocam (Largo, Florida). Following the recommendation by Wirkert et al.,10 we use light filters with the central wavelengths of 470, 480, 511, 560, 580, 600, 660, and 700 nm. The full width at half maximum (FWHM) of the bands is 20 nm, except for the 480-nm band, where it is 25 nm. The camera runs at 20 fps and thus records multispectral image stacks at 2.5 Hz. The pixel size is . The opening angle of the laparoscopic optics is 85 deg.
As the camera does not provide RGB images directly, we select the channels of 470, 560, and 700 nm and regard them as blue, green, and red, respectively. Note that our synthetic RGB image contains more specific information than a true RGB image as it is composed of bands with a FWHM of 20 nm, thus being much narrower than true RGB bands.
2.2. Tissue Classification
Given the captured images, the workflow consists of three steps: (1) preprocessing, (2) feature extraction, and (3) classification. Each step is described in the following paragraphs.
2.2.1. Preprocessing
Image preprocessing converts multispectral raw images into multispectral patches, involving noise removal and patch extraction. Since Gaussian noise usually exists in the raw image, we apply total variation11 in this paper, which is able to remove Gaussian noise while preserving sharp edges. Afterward, we select several rectangular patches from the multispectral images. These patches all have the same size.
2.2.2. Feature extraction
In the feature extraction step, a feature vector is extracted from the spatial neighborhood and spectral profile of each pixel, serving as a (multispectral) fingerprint to enable tissue classification. The appearance of the tissue at a pixel’s location can be represented by texture because the tissue surface usually has characteristic patterns. The spectral profile of the tissue at the pixel location is influenced by optical characteristics, such as absorption and scattering properties.12 As laparoscopic images are captured from various viewpoints under various illumination conditions, the extracted features should be robust to the pose of the endoscope as well as to the lighting conditions. Furthermore, they should be computationally cheap to enable real-time image processing in future applications. Numerous texture descriptors have been proposed in the literature,13 but only a few of them are suitable for our purposes. In this paper, we use the local binary pattern (LBP)14 to extract texture information; we also use the average spectrum (AS) to extract spectral information.
Local binary pattern
LBP14 is a robust texture representation method, which encodes local primitive microstructures in the image. It is already being successfully used for other purposes such as face detection.15,16 In a 2-D gray-scale image , a circle with radius is centered at every pixel, and points on the circle are compared with the center pixel. One can then extract a binary string at every location within the image and obtain an LBP map, , which can be formulated as follows:
| (1) |
with
| (2) |
where is the point on the circle, the gray value of which is computed via interpolation. The occurrence histogram of the LBP map is regarded as the feature descriptor.
LBP is gray-scale invariant and provides low computational complexity, which is beneficial to the implementation of real-time uses. The study of Ojala et al.14 proposes an advanced version named , which provides rotational invariance and only contains uniform patterns for representative microstructures. One can refer to the study of Lahdenoja et al.17 for more insights. In this paper, we use the uniform and rotationally invariant version with multiresolution analysis and name it briefly as LBP. To improve the robustness, we normalize the feature vector from each occurrence histogram.
Average spectrum
Spectral reflectances of one location within a multispectral image could be directly extracted and used as the feature descriptor. Such a type of spectral feature descriptor would offer a high spatial resolution. However, it would only be feasible if all bands were already precisely aligned and if no noise existed.
We average all the spectral reflectances around one location in each channel, sacrificing high spatial resolution but improving the robustness against noise. Given one location within a multispectral image , the spectral feature vector is given by
| (3) |
where is the neighbor set of , and is the band index. One can find a similar feature descriptor in the literature.6 Since multispectral image patches are used in this paper, we set the point at the center of the patch with the neighbor set containing all remaining locations. To compensate for scaling in cases where the illumination condition changes, we normalize the feature vector to the unit length.
AS + LBP: Since the texture information mainly represents spatial characteristics in each band and the spectral information mainly represents underlying optical properties, we hypothesize that AS and LBP represent complementary information. We therefore propose the combined feature descriptor AS + LBP, which is a concatenation of the two feature vectors.
2.2.3. Classification
In this paper, we apply a support vector machine (SVM) model with a Gaussian kernel to discriminate tissues. Due to its advantages mentioned in the literature,18 it is suitable to address the classification problem in our scenario. First, it is less prone to the “curse-of-dimensionality.” Since high-dimensional data such as ours are explicitly handled by the kernel function, parameter proliferation is prevented in the high-dimensional feature space, leading to trackable computation and limited over-fitting. Second, derived from the statistical learning theory, the SVM model can provide complex decision functions and therefore can fit the data well. Third, the SVM solution is only determined by the support vectors, so its performance is potentially repeatable when the training data have small disturbances.
Given a set of training data with , the SVM model can be given by its dual formula as19
| (4) |
and the decision function can be given by
| (5) |
where the threshold can be computed by an averaging procedure, the kernel function , and is the hyperparameter governing the regularization weight.
The Gaussian kernel-based SVM only has two hyperparameters, namely the Gaussian kernel size and the regularization weight , to specify and usually achieves satisfactory performances in practice. To perform multiclass classification, we use a “one-against-one” scheme for SVM. Since SVM is sensitive to the data scale, we normalize the feature vector of each sample to the unit length and perform standardization within each feature dimension.
2.3. Experiments
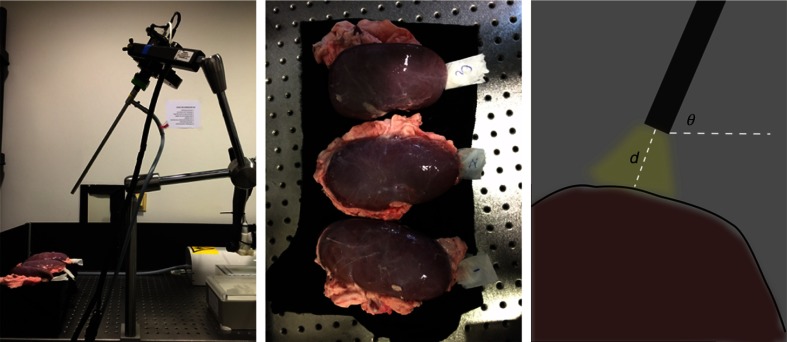
The goal of our experiments was to evaluate our method with respect to our hypothesis, which combined textural and spectral features extracted from multispectral images to benefit organ tissue classification. For these purposes, we discriminated in our experiment four types of porcine organ tissues typically encountered during hepatic laparoscopic surgeries: liver, gallbladder, colon, and kidney, which have been collected from three different pigs. All the porcine organs used in our experiments were collected from the butcher. The interval between organ dissection and imaging was less than 4 h. During this period, all these organs were covered with ice and stored in a cooling chamber. The laparoscopic setup is shown in Fig. 2, showing the process of capturing multispectral images of kidney tissue. When capturing images, the light was only provided by the laparoscopic light source. We targeted the rod lens to a smooth region of each organ whose mean surface normal was approximately perpendicular to the horizontal plane and captured images by varying the camera pose. The camera pose is defined by , where is the angle between the rod lens and the horizontal plane, and is the distance between the lens tip and the organ surface. We specify , as these distances and angles are typically encountered during laparoscopic surgeries.20 Therefore, we obtained an image set containing 27 subsets for each organ denoted by in which the images feature diverse anatomical structures and illumination conditions.
Fig. 2.
Setup for capturing multispectral images of kidney tissue. From left to right: the multispectral laparoscope. The three porcine kidneys originating from three different pigs. Camera pose, where the red region denotes the tissue and the black bar denotes the rod lens. Additionally, the yellow region denotes the light, and the dark background indicates that images are captured in a dark environment.
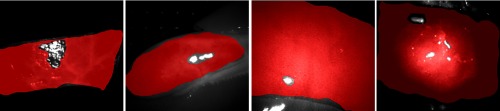
After capturing multispectral raw images, we performed the preprocessing procedures mentioned in Sec. 2.2.1. For training purposes, we annotated regions in each multispectral image as reference and marked invalid regions such as exposure caused by specular reflection. We then assigned a label to the image reference. Some image annotation results are shown in Fig. 3. Each multispectral image was cropped into several patches of a size . From these, 100 patches were randomly selected, following the criterion that at least 80% of each image patch needed to overlap with the annotated reference. These selected patches were then stored in a multispectral patch set named , which is shown by Fig. 4. Consequently, the patch set is well balanced.
Fig. 3.
Image annotation is completed by excluding areas not covered by tissue and high/low exposure regions. These exposure regions (e.g., specular reflections) do not contain tissue-specific information and can be excluded automatically21 in future work. The red overlay indicates regions to classify. From left to right: colon, gallbladder, liver, and kidney.
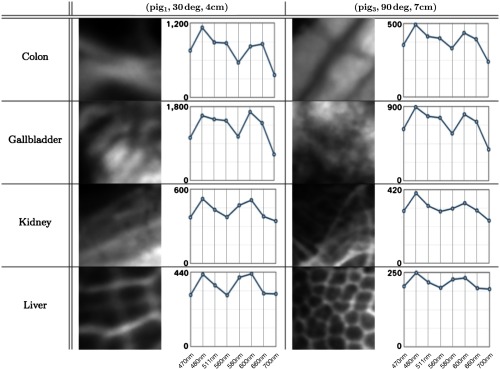
Fig. 4.
Eight patches from two subsets in and the associated AS of each patch. The patches on the left and right side correspond to tissue areas of and , respectively. They had been extracted from the image corresponding to the central wavelength of 470 nm. In each spectrum plot, the -axis denotes the wavelengths (470, 480, 511, 560, 580, 600, 660, and 700 nm) and the -axis denotes the image intensity.
For each multispectral image patch, we extracted its texture feature LBP, its spectral feature AS, and its combined textural/spectral feature AS + LBP, where we extracted patterns with three combinations, namely , , and , for multiple resolutions14 and repeated this computation at each image channel to capture texture information at every spectral band. For comparison purposes, we also used two other commonly used feature description methods: the Gabor filter bank (GFB) and the gray-level cooccurrence matrix (GLCM).13 Their hyperparameters were selected heuristically according to the natural image analysis literature, while considering the computational speed, memory consumption, and the final recognition accuracy in our scenario. The GFB used frequencies ranging from 0.1 to 0.6 and four orientations: 0, , , and . The parameters of GLCM were set to and , and we chose contrast, dissimilarity, homogeneity, angular second moment, energy, and correlation as the property set. The pixel window of GFB was automatically determined by “Python scikit-image” according to the frequencies. As with LBP, these methods were applied to each channel, and the feature vectors were concatenated. The number of features for each descriptor and each image type is given in Table 1.
Table 1.
The number of features for different image types and descriptors used in our experiments.
| AS | LBP | AS + LBP | CLGM | GFB | |
|---|---|---|---|---|---|
| Multispectral image | 8 | 432 | 440 | 48 | 320 |
| RGB image | 3 | 162 | 165 | 18 | 120 |
The classification test was performed on every subset of in turn, i.e., when the performance was tested on , the classifier is trained on . To test the influence of camera pose changes, we excluded the camera pose in the testing set from the training set. Specifically, when testing on , the classifier was trained on .
During the training phase, the two hyperparameters and in the SVM model were optimized via grid search and cross validation on all 27 subsets. The parameter had 10 candidates ranging from to 10 in the two-based logarithm coordinate, and the parameter had 10 candidates ranging from to 3 in the two-based logarithm coordinate (in our Python implementation, we specified and ). The determined values for the hyperparameters were subsequently used in the testing phase. The classification performance was evaluated by the accuracy rate which is the ratio of correctly classified samples to all samples in the testing set. This evaluation did not give biased results in our scenario since our dataset was balanced. Note that C and gamma would ideally have been optimized separately on each training data set used in this study. However, the focus of the experiments was on relative changes in accuracy when using multispectral vs. RGB data rather than on absolute accuracy rates. Further, varying the parameters had only a very moderate effect on the classification results. Finally, any other classification algorithm (e.g., random forests) could have been applied instead of SVMs for our purposes.
In our experiments, images were annotated manually using the Medical Imaging Interaction Toolkit (MITK) software.22 After the image patch set was established, feature extraction and classification algorithms were implemented using Python, based on the modules “scikit-image” and “scikit-learn.”
3. Results
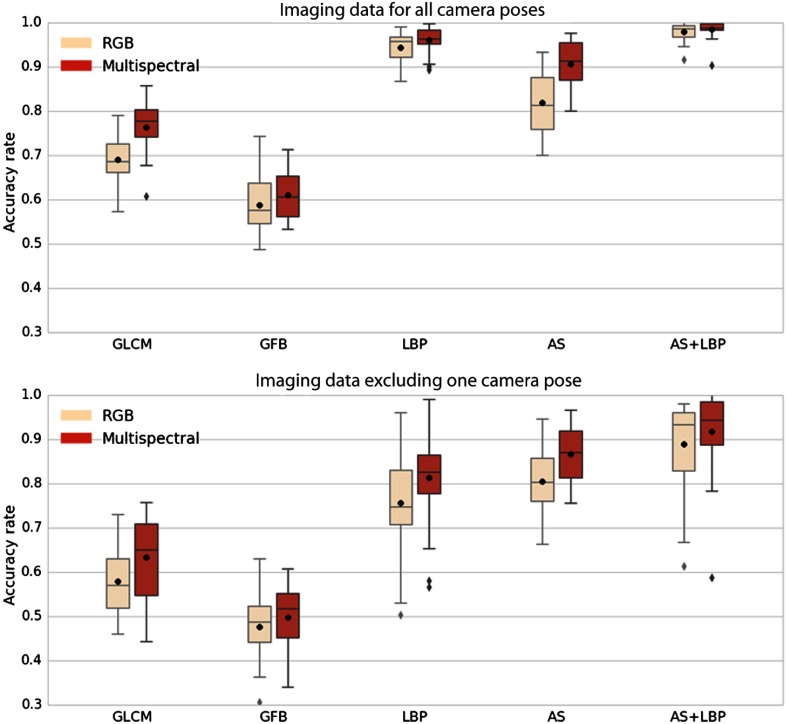
The accuracy rates are shown in terms of box plots in Fig. 5. Comparing the two box plots in each row, one can observe that: (1) the performance using the multispectral imaging data is better than the performance using the RGB imaging data for all feature descriptors; (2) when excluding the camera pose used in the testing data from the training data, all performances deteriorate; and (3) the proposed feature descriptor AS + LBP performs best in each case.
Fig. 5.
Accuracy obtained for different descriptors using RGB data (rose) and eight channels of multispectral data (red). Each box extends from the first quartile to the third quartile. The whiskers denote the range from the fifth percentile to the 95th percentile. Outliers are visualized as well. In each figure, the horizontal axis shows the feature description methods. The vertical axis indicates the accuracy rate. The black point and the bar within each box denote the mean and the median of accuracy rates, respectively. The first row: all camera poses are incorporated in the training set. The second row: the camera pose used in testing set is excluded from the training set.
The median accuracy rates given in Table 2 correspond to the bands inside the boxes in Fig. 5. For each feature, the percentage change of recognition accuracy from the RGB image to the multispectral image is equal to the accuracy rate difference divided by the accuracy rate of RGB data. The median improvement in accuracy obtained by using multispectral data as opposed to RGB data is 7% on average.
Table 2.
Median accuracy rates, which correspond to the bands inside the boxes in Fig. 5.
| All camera poses are considered |
One camera poses is excluded |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GLCM (%) | GFB (%) | LBP (%) | AS (%) | AS + LBP (%) | GLCM (%) | GFB (%) | LBP (%) | AS (%) | AS + LBP (%) | |
| RGB | 68.5 | 57.5 | 95.8 | 81.3 | 97.8 | 57.0 | 48.7 | 74.8 | 80.2 | 93.3 |
| Multispectral | 77.7 | 60.5 | 96.3 | 91.2 | 98.4 | 65.0 | 51.7 | 82.5 | 87.0 | 94.3 |
| Improvement | 13.4 | 5.2 | 0.52 | 12.2 | 0.61 | 14.0 | 6.2 | 10.3 | 8.5 | 1.1 |
One can also see the benefit of combining texture information and spectral information. For multispectral imaging data, the feature AS + LBP outperforms AS by 8% and LBP by 2% when all camera poses are considered. When one camera pose is excluded from training, the feature AS + LBP outperforms AS by 8% and LBP by 14%.
We investigated differences in classification performance among various feature sets. For this purpose, we computed the mean differences along with bootstrapped 95% confidence intervals (bootstrapping was applied due to the skewness of the underlying data). Moreover, the Wilcoxon signed-rank test (for paired samples) was used to test the null hypothesis that the difference is zero. To assure a nominal significance level of 5%, Bonferroni adjustment was applied and values obtained from the Wilcoxon signed-rank test were multiplied by the number of independent tests to yield corrected values . We observed that the proposed method outperformed the other investigated methods by at least two percentage points. In all experiments, the null hypothesis of no difference could be rejected. The quantitative results of our comparisons can be found in Table 3.
Table 3.
Comparison of our proposed classifier to variants. The 95% confidence interval is shown in parenthesis. All and values are below our 5% significance threshold.
| All camera poses considered |
One camera pose is excluded |
|||||
|---|---|---|---|---|---|---|
| AS + LBP multispectral (MS) compared to: | AS MS | LBP MS | AS + LBP RGB | AS MS | LBP MS | AS + LBP RGB |
| Mean improvement (percentage points) | 7 (6, 9) | 3 (2, 4) | 2 (1, 2) | 8 (5, 10) | 11 (8, 14) | 2 (1, 4) |
| -value | ||||||
| -value | 0.0002 | 0.0003 | 0.0002 | 0.006 | ||
4. Discussion
To our knowledge, we are the first to investigate organ tissue classification in a laparoscopic setup based on multispectral texture analysis. Our method classifies image patches based on both textural (LBP) and spectral (AS) features. The experimental results show that when using equal feature descriptors and classification methods, the multispectral image leads to statistically significant higher accuracy rates than the RGB image in all of the cases. We therefore conclude that the multispectral image is superior to the RGB image in our scenario. This fact further indicates that the multispectral image contains more information than the RGB image.
To represent texture information, we used LBP, which is gray-scale invariant, and accounts for multiple resolutions. We also investigated GLCM and GFB texture features as baseline methods. The GLCM lacks robustness to illumination and geometry variations in nature, which probably causes its weaker performance in comparison with LBP. The GFB also performs worse than LBP, which could be caused by the nonoptimal parameters of the filter bank. According to the literature,23 the filter bank parameters determining the feature quality are highly dependent on the texture type. In our tissue classification scenario, the texture is diverse from image to image, even from training images to test images. This issue causes parameter selection to be a highly challenging task. From the computational perspective, high computational load for GLCM24 and GFB23 are reported, while LBP can be calculated rapidly and has low memory consumption.14 Thus, we conclude that LBP is the best texture descriptor among the three commonly used texture descriptors, both from a performance and a computational standpoint.
The feature AS is proposed to capture spectral information. Due to the averaging operation, AS is potentially robust to noise and of low spatial resolution. Due to the applied l2-normalization, it is also robust to multiplicative changes in illumination. During computer-assisted surgeries, however, tissues usually deform in a nonrigid manner, and real-time image registration in this scenario is highly challenging.25 Future work should thus investigate whether simple averaging is sufficient for ensuring descriptor robustness in the presence of motion. Furthermore, we extract the feature AS from the square image patch at the current stage. We expect that using a circular patch would improve the robustness to rotation.
As shown in the experimental results, the proposed texture-spectral feature AS + LBP outperforms other features as well as its own individual components, verifying that the texture information complements the spectral information. In our experiments, we captured images from multiple viewpoints with different illumination conditions to simulate challenges encountered during laparoscopic interventions; however, the classifier cannot cover all geometrical variations encountered during surgeries. This is verified by the accuracy decreasing when one camera pose is excluded from the training data, as shown in Fig. 5 and Table 2. The results also suggest that complicated geometric variations can, and should, be learned.
We performed a comprehensive ex vivo study on organ tissue classification to provide a basis for tissue classification in the clinical intraoperative scenario. However, our study has some limitations. First, compared with the ex vivo experiment, in vivo laparoscopic interventions cause new challenges, such as organ deformation during image capturing, internal bleeding, and optical characteristic shifting due to tissue perfusion. To address these challenges, we aim to collect more data using an in vivo setting and to create a benchmark for multispectral organ classification in laparoscopy.
Second, although we have chosen efficient textural and spectral descriptors, an in-depth run-time analysis to investigate the real-time capabilities of the proposed approach has not yet been performed. These were not the focus of this study, which used Python implementation of the algorithms. The runtime analysis will be made after the approach has been ported to real clinical uses for which we expect the implementation to be optimized using an efficient programming language.
Third, it would be interesting to investigate different classifiers, such as random forests and deep neural networks, especially when the amount of data is large. Additionally, if in vivo and ex vivo data are combined, the covariate shift due to the translation to perfused tissue could be combated by applying domain adaptation methods.26
5. Conclusion
This paper provides a comprehensive study of organ tissue classification based on multispectral texture analysis. According to our experiments, we show that, compared to RGB feature descriptors multispectral descriptors is superior for tissue classification. Therefore, we suggest that using multispectral imaging data is beneficial for organ tissue classification in laparoscopy.
Acknowledgments
Funding for this work was provided by the European Research Council (ERC) starting grant COMBIOSCOPY (637960).
Biographies
Yan Zhang was a research assistant in Computer-Assisted Interventions, German Cancer Research Center in Heidelberg, Germany, and now moved to Institute of Neural Information Processing, University of Ulm. His research interests cover image analysis and computer vision, pattern recognition, and machine learning, as well as their applications on biomedical engineering and healthcare.
Sebastian J. Wirkert is currently a research assistant and PhD student in Computer-Assisted Interventions, German Cancer Research Center in Heidelberg, Germany. His research interests cover multispectral imaging technology, biophotonics, mathematical modeling, and machine learning and their applications on endoscopy.
Benjamin Mayer is a medical student in Medical Faculty at Heidelberg University and Department of Minimally Invasive surgery, Heidelberg University Hospital. His research interests cover implementation of 3-D video analysis technology in surgical practice and evaluation of computer-based assistance, navigation, and robotic systems.
Christian Stock holds a PhD in epidemiology (Heidelberg) and MSc degree in biostatistics (Heidelberg) and health services research (York, UK). He is currently head of the statistical modeling group at the Institute of Medical Biometry and Informatics, Heidelberg University. His main research focuses on the development of statistical models for complex observational data.
Neil T. Clancy is an Imperial College Junior Research Fellow at the Hamlyn Centre for Robotic Surgery and the Department of Surgery and Cancer, Imperial College London. His research interests focus on the implementation of biophotonic imaging techniques in endoscopic clinical application areas, including multi- and hyperspectral imaging, polarization, structured lighting, and illumination sources for minimally invasive surgery.
Daniel S. Elson is a professor in the Hamlyn Centre for Robotic Surgery, Department of Surgery and Cancer and the Institute of Global Health Innovation. His research interests are based around the development and application of photonics technology with endoscopy for surgical imaging applications, including multispectral imaging, polarization-resolved imaging, fluorescence imaging, and structured lighting. These devices are being applied in minimally invasive surgery and in the development of new flexible robotic-assisted surgery systems.
Lena Maier-Hein received her PhD from Karlsruhe Institute of Technology with distinction in 2009 and conducted her postdoctoral at the German Cancer Research Center (DKFZ) and at the Hamlyn Centre for Robotic Surgery, Imperial College London. As department head at the DKFZ, she is now working in the field of computer-assisted medical interventions with a focus on surgical data science and computational biophotonics.
Biographies for the other authors are not available.
Disclosures
All authors declare and confirm that there are no competing interests.
References
- 1.Katić D., et al. , “Bridging the gap between formal and experience-based knowledge for context-aware laparoscopy,” Int. J. Comput. Assisted Radiol. Surg. 11(6), 881–888 (2016). 10.1007/s11548-016-1379-2 [DOI] [PubMed] [Google Scholar]
- 2.Lee J., et al. , “Automatic classification of digestive organs in wireless capsule endoscopy videos,” in Proc. of the 2007 ACM Symp. on Applied Computing, pp. 1041–1045, ACM; (2007). [Google Scholar]
- 3.Azzopardi C., Hicks Y. A., Camilleri K. P., “Exploiting gastrointestinal anatomy for organ classification in capsule endoscopy using locality preserving projections,” in 35th Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society (EMBC 2013), pp. 3654–3657, IEEE; (2013). 10.1109/EMBC.2013.6610335 [DOI] [PubMed] [Google Scholar]
- 4.Lu G., Fei B., “Medical hyperspectral imaging: a review,” J. Biomed. Opt. 19(1), 010901 (2014). 10.1117/1.JBO.19.1.010901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Triana B., et al. , “Multispectral tissue analysis and classification towards enabling automated robotic surgery,” Proc. SPIE 8935, 893527 (2014). 10.1117/12.2040627 [DOI] [Google Scholar]
- 6.Delporte C., et al. , “Biological tissue identification using a multispectral imaging system,” Proc. SPIE 8659, 86590H (2013). 10.1117/12.2003033 [DOI] [Google Scholar]
- 7.Khelifi R., Adel M., Bourennane S., “Multispectral texture characterization: application to computer aided diagnosis on prostatic tissue images,” EURASIP J. Adv. Signal Process. 2012, 118 (2012). 10.1186/1687-6180-2012-118 [DOI] [Google Scholar]
- 8.Lu G., et al. , “Spectral-spatial classification for noninvasive cancer detection using hyperspectral imaging,” J. Biomed. Opt. 19(10), 106004 (2014). 10.1117/1.JBO.19.10.106004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y., et al. , “Tissue classification for laparoscopic image understanding based on multispectral texture analysis,” Proc. SPIE 9786, 978619 (2016). 10.1117/12.2216090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wirkert S. J., et al. , “Endoscopic Sheffield index for unsupervised in vivo spectral band selection,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2014. Workshops Computer-Assisted and Robotic Endoscopy - CARE 2014, pp. 110–120, Springer; (2014). [Google Scholar]
- 11.Chambolle A., “An algorithm for total variation minimization and applications,” J. Math. Imaging Vision 20(1–2), 89–97 (2004). 10.1023/B:JMIV.0000011325.36760.1e [DOI] [Google Scholar]
- 12.Hidović-Rowe D., Claridge E., “Modelling and validation of spectral reflectance for the colon,” Phys. Med. Biol. 50(6), 1071–1093 (2005). 10.1088/0031-9155/50/6/003 [DOI] [PubMed] [Google Scholar]
- 13.Tou J. Y., Tay Y. H., Lau P. Y., “Recent trends in texture classification: a review,” in Symp. on Progress in Information & Communication Technology, Vol. 3, pp. 56–59 (2009). [Google Scholar]
- 14.Ojala T., Pietikäinen M., Mäenpää T., “Multiresolution gray-scale and rotation invariant texture classification with LBP,” IEEE Trans. Pattern Anal. Mach. Intell. 24(7), 971–987 (2002). 10.1109/TPAMI.2002.1017623 [DOI] [Google Scholar]
- 15.Ahonen T., Hadid A., Pietikainen M., “Face description with LBP: application to face recognition,” IEEE Trans. Pattern Anal. Mach. Intell. 28(12), 2037–2041 (2006). 10.1109/TPAMI.2006.244 [DOI] [PubMed] [Google Scholar]
- 16.Zhang W., et al. , “Local Gabor binary pattern histogram sequence (LGBPHS): a novel non-statistical model for face representation and recognition,” in 10th IEEE Int. Conf. on Computer Vision (ICCV 2005), Vol. 1, pp. 786–791, IEEE; (2005). 10.1109/ICCV.2005.147 [DOI] [Google Scholar]
- 17.Lahdenoja O., Poikonen J., Laiho M., “Towards understanding the formation of uniform LBP,” ISRN Mach. Vision 2013, 429347 (2013). 10.1155/2013/429347 [DOI] [Google Scholar]
- 18.Burges C. J., “A tutorial on support vector machines for pattern recognition,” Data Min. Knowl. Discovery 2(2), 121–167 (1998). 10.1023/A:1009715923555 [DOI] [Google Scholar]
- 19.Scholkopf B., Smola A. J., Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond, Adaptive Computation and Machine Learning, 644 p., MIT Press, Cambridge, Massachusetts: (2001). [Google Scholar]
- 20.Maier-Hein L., et al. , “Comparative validation of single-shot optical techniques for laparoscopic 3-d surface reconstruction,” IEEE Trans. Med. Imaging 33(10), 1913–1930 (2014). 10.1109/TMI.2014.2325607 [DOI] [PubMed] [Google Scholar]
- 21.Lu G., et al. , “Framework for hyperspectral image processing and quantification for cancer detection during animal tumor surgery,” J. Biomed. Opt. 20(12), 126012 (2015). 10.1117/1.JBO.20.12.126012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nolden M., et al. , “The medical imaging interaction toolkit: challenges and advances,” Int. J. Comput. Assisted Radiol. Surg. 8(4), 607–620 (2013). 10.1007/s11548-013-0840-8 [DOI] [PubMed] [Google Scholar]
- 23.Randen T., Husoy J. H., “Filtering for texture classification: a comparative study,” IEEE Trans. Pattern Anal. Mach. Intell. 21(4), 291–310 (1999). 10.1109/34.761261 [DOI] [Google Scholar]
- 24.Akoushideh A. R., Shahbahrami A., Maybodi B. M.-N., “High performance implementation of texture features extraction algorithms using FPGA architecture,” J. Real-Time Image Proc. 9(1), 141–157 (2014). 10.1007/s11554-012-0283-4 [DOI] [Google Scholar]
- 25.Crum W. R., Hartkens T., Hill D., “Non-rigid image registration: theory and practice,” Br. J. Radiol. 77(Suppl. 2), S140–S153(2004). 10.1259/bjr/25329214 [DOI] [PubMed] [Google Scholar]
- 26.Wirkert S. J., et al. , “Robust near real-time estimation of physiological parameters from megapixel multispectral images with inverse Monte Carlo and random forest regression,” Int. J. Comput. Assisted Radiol. Surg. 11(6), 909–917 (2016). 10.1007/s11548-016-1376-5 [DOI] [PMC free article] [PubMed] [Google Scholar]