Abstract
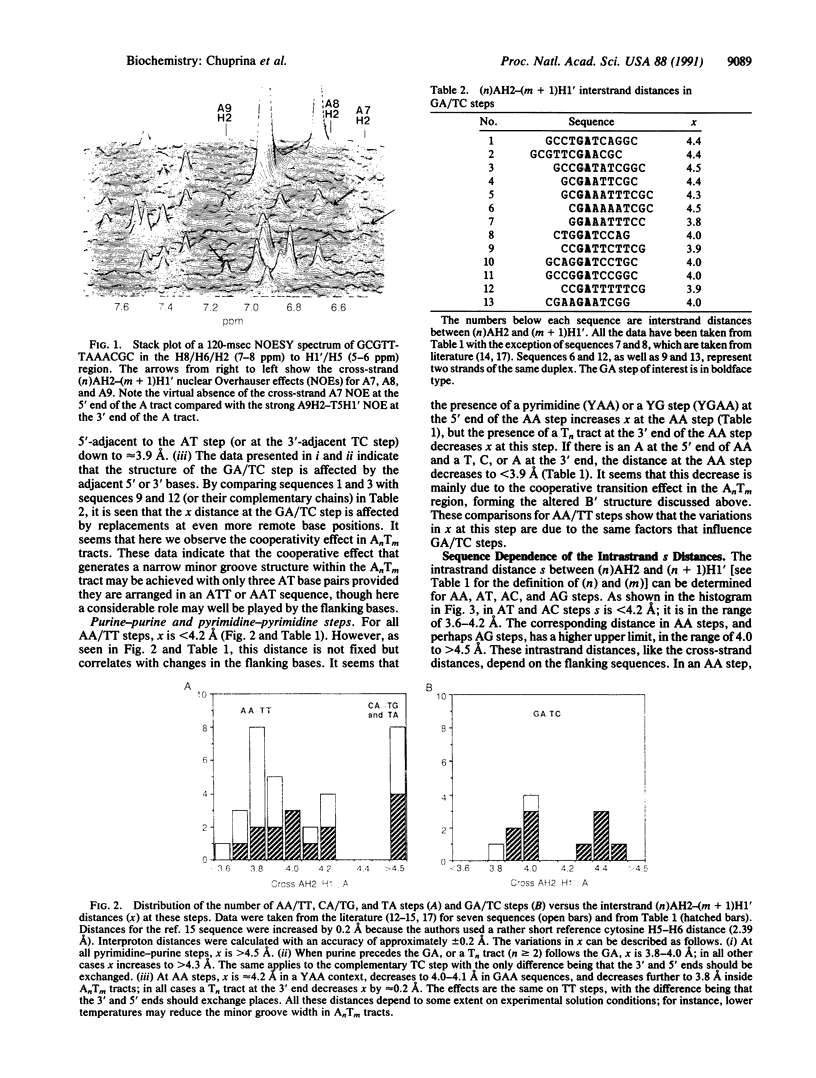
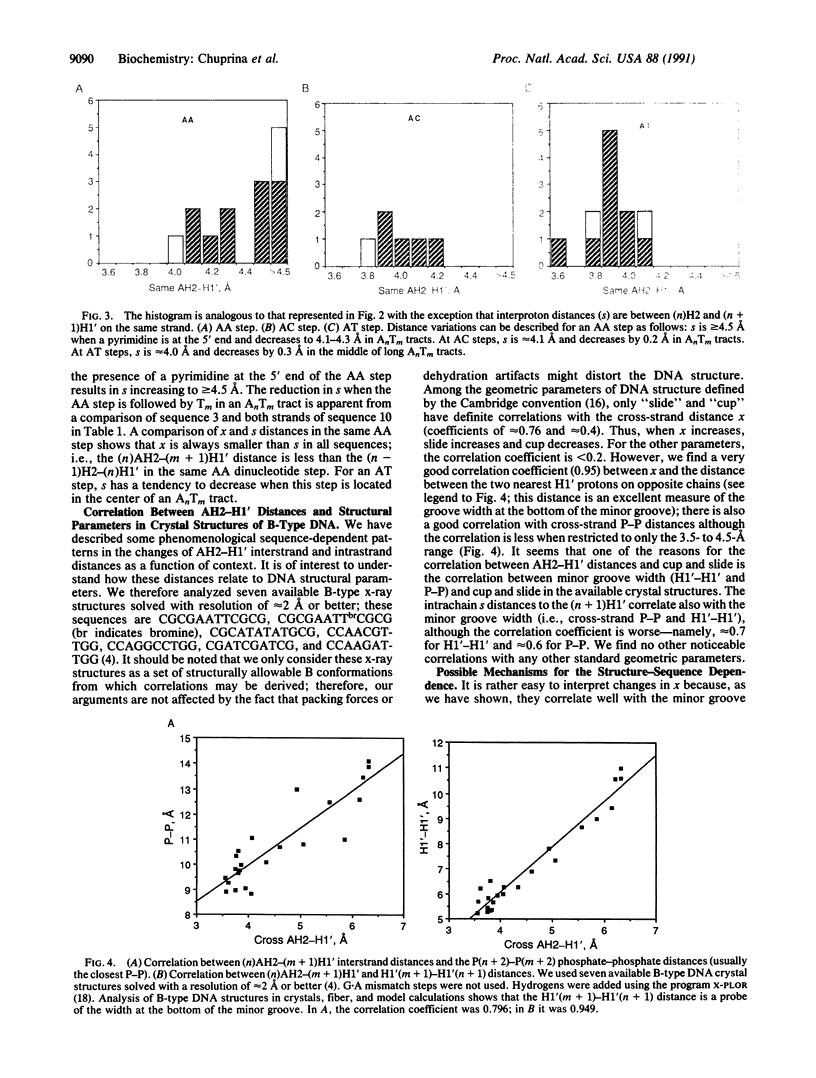
Nuclear Overhauser effect-derived distances between adenine H2 protons and anomeric H1' protons on the same strand or on the complementary strand are presented for several different DNA duplexes. The cross-strand (n)AH2 to (m + 1)H1' distances [designated as x, where (n) and (m) are complementary residues] vary by up to 1 A depending on the sequence. In all possible A-containing pyrimidine-purine steps (CA, TG, and TA), x is greater than 4.5 A. In GA steps, x varies within rather wide limits in the range 3.8-4.5 A, whereas in AA steps the lower limit is 3.7 A and the upper limit is approximately 4.2 A. In purine-purine steps, x is affected by at least three factors: (i) adjacent pyrimidine-purine steps at the 5' end [e.g., YRA sequences (where Y = T or C and R = G or A)], or a pyrimidine-purine step at the 3' end of the pyrimidine-pyrimidine step on the complementary strand, cause x to increase, (ii) an AT step at the 3' end of a purine-purine step (e.g., RAT) causes x to decrease, and (iii) substitution of bases at the next-nearest neighbor position leads to changes in x at GA and AA steps. The latter factor seems to be due to a cooperative effect arising from formation of the "anomalous" B' structure when the substitution produces an AnTm tract (which always produces a decrease in x). The data indicate that (n)AH2-(n + 1)H1' distances on the same strand (designated as s) are also sequence dependent. Thus on AA steps, neighboring substitutions produce the same effect on s as on the cross-strand x distances. The results lead to the ability to predict changes in AH2-H1' distances depending on the DNA sequence. By using high-resolution x-ray B-type structures as a set of allowable B conformations, a very good correlation was found between x and the minor groove width parameters P-P or H1'-H1'. Thus, the x distances are a direct probe of the minor groove width in B-type DNA, and changes in this distance therefore reflect changes in the minor groove width. Since many of the sequences studied are sites of protein recognition, the observed sequence-structure dependence in DNA probably plays an important role in the process of recognition by proteins and minor groove ligands such as drugs.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P., Abagyan R. A. Structural basis of stable bending in DNA containing An tracts. Different types of bending. J Biomol Struct Dyn. 1988 Aug;6(1):121–138. doi: 10.1080/07391102.1988.10506486. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P. Anomalous structure and properties of poly (dA).poly(dT). Computer simulation of the polynucleotide structure with the spine of hydration in the minor groove. Nucleic Acids Res. 1987 Jan 12;15(1):293–311. doi: 10.1093/nar/15.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuprina V. P., Fedoroff OYu, Reid B. R. New insights into the structure of An tracts and B'-B' bends in DNA. Biochemistry. 1991 Jan 15;30(2):561–568. doi: 10.1021/bi00216a034. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P. Regularities in formation of the spine of hydration in the DNA minor groove and its influence on the DNA structure. FEBS Lett. 1985 Jul 1;186(1):98–102. doi: 10.1016/0014-5793(85)81347-8. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M. Determination of three-dimensional structures of proteins and nucleic acids in solution by nuclear magnetic resonance spectroscopy. Crit Rev Biochem Mol Biol. 1989;24(5):479–564. doi: 10.3109/10409238909086962. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Haran T. E., Nadeau J. G. Intrinsically bent DNA. J Biol Chem. 1990 May 5;265(13):7093–7096. [PubMed] [Google Scholar]
- DiGabriele A. D., Sanderson M. R., Steitz T. A. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E. Definitions and nomenclature of nucleic acid structure parameters. J Biomol Struct Dyn. 1989 Feb;6(4):627–634. doi: 10.1080/07391102.1989.10507726. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Goodsell D. S., Kopka M. L., Pjura P. E. The effect of crystal packing on oligonucleotide double helix structure. J Biomol Struct Dyn. 1987 Dec;5(3):557–579. doi: 10.1080/07391102.1987.10506413. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Hare D. R., Reid B. R. Three-dimensional structure of a DNA hairpin in solution: two-dimensional NMR studies and distance geometry calculations on d(CGCGTTTTCGCG). Biochemistry. 1986 Sep 9;25(18):5341–5350. doi: 10.1021/bi00366a053. [DOI] [PubMed] [Google Scholar]
- Herrera J. E., Chaires J. B. A premelting conformational transition in poly(dA)-Poly(dT) coupled to daunomycin binding. Biochemistry. 1989 Mar 7;28(5):1993–2000. doi: 10.1021/bi00431a006. [DOI] [PubMed] [Google Scholar]
- Katahira M., Sugeta H., Kyogoku Y., Fujii S., Fujisawa R., Tomita K. One- and two-dimensional NMR studies on the conformation of DNA containing the oligo(dA)oligo(dT) tract. Nucleic Acids Res. 1988 Sep 12;16(17):8619–8632. doi: 10.1093/nar/16.17.8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennard O., Hunter W. N. Oligonucleotide structure: a decade of results from single crystal X-ray diffraction studies. Q Rev Biophys. 1989 Aug;22(3):327–379. doi: 10.1017/s0033583500002997. [DOI] [PubMed] [Google Scholar]
- Kintanar A., Klevit R. E., Reid B. R. Two-dimensional NMR investigation of a bent DNA fragment: assignment of the proton resonances and preliminary structure analysis. Nucleic Acids Res. 1987 Jul 24;15(14):5845–5862. doi: 10.1093/nar/15.14.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahendrasingam A., Rhodes N. J., Goodwin D. C., Nave C., Pigram W. J., Fuller W., Brahms J., Vergne J. Conformational transitions in oriented fibres of the synthetic polynucleotide poly[d(AT)].poly[d(AT)] double helix. Nature. 1983 Feb 10;301(5900):535–537. doi: 10.1038/301535a0. [DOI] [PubMed] [Google Scholar]
- Metzler W. J., Wang C., Kitchen D. B., Levy R. M., Pardi A. Determining local conformational variations in DNA. Nuclear magnetic resonance structures of the DNA duplexes d(CGCCTAATCG) and d(CGTCACGCGC) generated using back-calculation of the nuclear Overhauser effect spectra, a distance geometry algorithm and constrained molecular dynamics. J Mol Biol. 1990 Aug 5;214(3):711–736. doi: 10.1016/0022-2836(90)90288-W. [DOI] [PubMed] [Google Scholar]
- Nadeau J. G., Crothers D. M. Structural basis for DNA bending. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2622–2626. doi: 10.1073/pnas.86.8.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilges M., Clore G. M., Gronenborn A. M., Brunger A. T., Karplus M., Nilsson L. Refinement of the solution structure of the DNA hexamer 5'd(GCATGC)2: combined use of nuclear magnetic resonance and restrained molecular dynamics. Biochemistry. 1987 Jun 16;26(12):3718–3733. doi: 10.1021/bi00386a068. [DOI] [PubMed] [Google Scholar]
- Nilges M., Clore G. M., Gronenborn A. M., Piel N., McLaughlin L. W. Refinement of the solution structure of the DNA decamer 5'd(CTGGATCCAG)2: combined use of nuclear magnetic resonance and restrained molecular dynamics. Biochemistry. 1987 Jun 16;26(12):3734–3744. doi: 10.1021/bi00386a069. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Reid B. R. Sequence-specific assignments and their use in NMR studies of DNA structure. Q Rev Biophys. 1987 Aug;20(1-2):1–34. doi: 10.1017/s0033583500004212. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Sequence-dependent helical periodicity of DNA. Nature. 1981 Jul 23;292(5821):378–380. doi: 10.1038/292378a0. [DOI] [PubMed] [Google Scholar]
- Suzuki E., Pattabiraman N., Zon G., James T. L. Solution structure of [d(A-T)5]2 via complete relaxation matrix analysis of two-dimensional nuclear Overhauser effect spectra and molecular mechanics calculations: evidence for a hydration tunnel. Biochemistry. 1986 Nov 4;25(22):6854–6865. doi: 10.1021/bi00370a019. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Curved DNA. CRC Crit Rev Biochem. 1985;19(2):89–106. doi: 10.3109/10409238509082540. [DOI] [PubMed] [Google Scholar]
- Zhurkin V. B. Sequence-dependent bending of DNA and phasing of nucleosomes. J Biomol Struct Dyn. 1985 Feb;2(4):785–804. doi: 10.1080/07391102.1985.10506324. [DOI] [PubMed] [Google Scholar]