Abstract
Telomerase elongates telomeres and is crucial for maintaining genomic stability. While stem cells and cancer cells display high telomerase activity, normal somatic cells lack telomerase activity primarily due to transcriptional repression of telomerase reverse transcriptase (TERT), the catalytic component of telomerase. Transcription factor binding, chromatin status as well as epigenetic modifications at the TERT promoter regulates TERT transcription. Myc is an important transcriptional regulator of TERT that directly controls its expression by promoter binding and associating with other transcription factors. In this review, we discuss the current understanding of the molecular mechanisms behind regulation of TERT transcription by Myc. We also discuss future perspectives in investigating the regulation of Myc at TERT promoter during cancer development.
Keywords: TERT, Myc, telomerase, transcription, cancer
Introduction
Telomerase is a reverse transcriptase that elongates telomeres (Blackburn and Collins, 2011). It is a ribonucleoprotein composed of a catalytic subunit telomerase reverse transcriptase (TERT), an RNA template Terc and accessory proteins (Cohen et al., 2007; Venteicher et al., 2009). Telomerase activity is detected in highly proliferating cells like stem cells, immune cells and germ cells (Feng et al., 1995; Avilion et al., 1996; Nakamura et al., 1997). However, most somatic human tissues lack telomerase activity (Aisner et al., 2002; Cong et al., 2002). Absence of telomerase activity results in telomere shortening with successive replication cycles. When telomeres become critically short, the DNA damage response is initiated followed by senescence or cell death (Blackburn et al., 2015). While Terc and other accessory proteins are ubiquitously expressed, TERT is transcriptionally downregulated, thereby limiting telomerase activity in somatic cells (Avilion et al., 1996; Heiss et al., 1998; Holzmann et al., 1998; Parfait et al., 2000; Wang and Zhu, 2004; Wang et al., 2009). For cancer cells arising from somatic cells, telomere maintenance becomes imperative in order to acquire immortality. Telomere length is maintained by telomerase in 90% of human cancers, while 10% of cancers utilize an alternative mechanism of telomere lengthening termed ALT (Kim et al., 1994; Bryan et al., 1995, 1997; Shay and Bacchetti, 1997). Besides TERT gene amplification and alternate splicing, TERT promoter regulation represents the key mechanism to regulate telomerase activity (Daniel et al., 2012). Thus, investigating transcriptional regulation of TERT is important for understanding cancer development.
Transcriptional regulation of TERT is extremely complex and not completely understood. Several transcription factors and signaling pathways are known to regulate TERT expression. The human TERT promoter contains binding sites for several transcription factors like Myc, SP1, ER, AP1, ETS, HIFs, and upstream stimulatory factors (USFs) (Kyo et al., 1999, 2000; Takakura et al., 1999, 2005; Wu et al., 1999; Goueli and Janknecht, 2003; Anderson et al., 2006; Xu et al., 2008). Amongst these transcription factors, Myc represents an important regulator of TERT transcription (Wu et al., 1999; Takahashi et al., 2007; Marion et al., 2009). Important insights about the interplay between Myc and TERT have been derived using a mouse model of lymphomagenesis in which lymphoma is specifically driven by Myc (Koh et al., 2015).
In the first part of this review, we discuss the role of Myc in modulating TERT transcription. We then describe how multiple cellular signals impinge on Myc to regulate TERT transcription. Lastly, we present correlation analyses between Myc and TERT in several cancers. In summary, we discuss the way forward to better understand the interplay between Myc and TERT in normal physiology and cancer.
Direct activation of TERT transcription by Myc
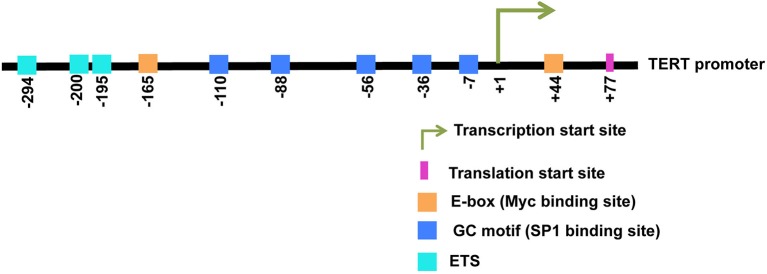
The TERT core promoter contains two canonical E-box consensus sites (5′-CACGTG-3′) at positions −165 and +44 nucleotide position relative to the transcription start site (TSS) (Horikawa et al., 1999; Takakura et al., 1999; Wick et al., 1999). A schematic representation of the TERT promoter with E-boxes and binding sites for several regulatory transcription factors is presented in Figure 1. E-boxes represent the known binding sites for E-box binding proteins like the Myc/Max/Mad1 superfamily and USFs. Amongst E-box binding proteins, Myc represents an important regulator of TERT transcription. Myc was the first transcription factor to be reported to directly activate TERT transcription in primary fibroblasts as well as in normal epithelial cells (Wu et al., 1999). Myc induces TERT expression independent of cellular proliferation and de novo protein synthesis, implying its direct role in regulating TERT transcription. Myc generally activates transcription by recruiting chromatin-modifying complexes like transformation/transcription domain associated protein (TRRAP) and associated histone acetyl transferases (HATs). It can also recruit P-TEFb, which is a kinase that phosphorylates the C-Terminus of RNA polymerase II, thereby helping in promoter clearance. However, the mechanism employed by Myc to activate the TERT promoter is not known (Zhao et al., 2014). The importance of E-box-driven TERT expression has also been investigated in the context of chromatin environment. Mutation of endogenous E-boxes in the TERT promoter can counteract TERT activation by Myc. It is suggested that E-boxes can also function to de-repress the TERT promoter (Zhao et al., 2014).
Figure 1.
A schematic representation of the human TERT promoter is shown. Binding sites for various transcription factors are displayed. Specific nucleotide positions for all binding sites are indicated. +1 indicates the transcription start site and +77 indicates the translation start site.
Myc functions as a heterodimer with Max to activate transcription of genes. The Myc/Max heterodimer is known to activate transcription by inducing topological changes in DNA (Wechsler and Dang, 1992). Structural studies using atomic force microscopy indicate that Myc/Max dimers bind equally and specifically to both E-boxes present in the TERT promoter (Lebel et al., 2007). Although, Myc/Max dimers can oligomerize to form tetramers, the physiological existence of these higher order forms in the context of TERT promoter regulation has not been experimentally verified (Nair and Burley, 2003; Lebel et al., 2007).
Myc acts as a central signaling factor downstream of major cellular pathways in the regulation of TERT transcription
Myc functions as a downstream effector of several cellular signaling pathways to regulate TERT expression. There are several GC motifs present in the TERT promoter that are recognized by SP1 transcription factors (Kyo et al., 2000). The location of these GC motifs is shown in Figure 1. Transactivation of the TERT promoter by Myc/Max heterodimers is attenuated upon mutation of SP1 sites, suggesting that SP1 and Myc cooperatively function to modulate TERT expression (Kyo et al., 2000). The NF-κB pathway is also reported to regulate TERT transcription. The mammalian NF-κB family comprises of five members, RelA (also termed p65), RelB, c-Rel, NF-κB1 (also termed p50), and NF-κB2 (also termed p52). They associate with each other to form diverse dimeric complexes that regulate numerous target genes via binding to the κB enhancer (Cildir et al., 2016). The canonical NF-κB pathway involves inducible degradation of the IκBs, particularly IκBα, leading to nuclear translocation of various NF-κB complexes, predominantly the p50/RelA dimer (Hayden and Ghosh, 2004). The non-canonical NF-κB pathway activates the RelB/p52 NF-κB complex using a mechanism that does not involve degradation of IκBα, but involves inducible processing of p100 to generate p52 (Sun, 2011). In HTLV-I transformed cells, the viral protein Tax upregulates the canonical NF-κB pathway (Sinha-Datta et al., 2004). This activation of the NF-κB signaling pathway has been indicated to upregulate Myc and SP1 and thus indirectly induce TERT activation. Similarly, an increase in TERT expression by NF-κB-induced activation of Myc/Max has been reported in T lymphocytes activated by PKC θ and in smooth muscle cells stimulated with fibroblast growth factors (Sheng et al., 2006; Bu et al., 2010). The epidermal growth factor (EGF) has also been reported to upregulate TERT transcription via SP1- and Myc/Max-dependent activation of Pyk2/ERK1/2 (Bermudez et al., 2008). In non-small cell lung cancer cell lines, EGF has been reported to activate TERT transcription by inducing direct binding of ETS2 and Myc/Max to the TERT promoter (Hsu et al., 2015). Estrogen has been shown to increase Myc expression and thus indirectly induce TERT transcription, although direct binding of the estrogen receptor (ER) to the TERT promoter has also been demonstrated in ER-positive cancers (Kyo et al., 1999; Cha et al., 2008; Grasselli et al., 2008).
Isocitrate dehydrogenase (IDH) mutations commonly occur in low-grade gliomas (Hartmann et al., 2009; Yan et al., 2009). Mutant IDH1 produces 2-hydroxyglutarate, resulting in altered chromatin modifications, thereby driving gliomagenesis. Recently, it has been shown that mutant IDH1 can reactivate the TERT promoter in human astrocytes by altering the chromatin status and enabling Myc/Max binding (Ohba et al., 2016). This represents a novel mechanism whereby a mutant protein can activate the TERT promoter by modulating its epigenetic status and enabling Myc binding to drive its expression.
The ETS family of transcription factors comprises more than 30 members that contain a characteristic DNA-binding domain known as the erythroblast transformation specific (ETS) domain (Dwyer and Liu, 2010). ETS factors have been shown to contribute to the regulation of telomerase (Maida et al., 2002; Xiao et al., 2003; Goueli and Janknecht, 2004). ETS2 is important for driving TERT gene expression and breast cancer cell proliferation (Dwyer et al., 2007; Xu et al., 2008). Silencing of ETS2 results in a reduction of TERT gene expression leading to increased human breast cancer cell death, while reconstitution with recombinant TERT reverses that effect (Xu et al., 2008). ETS2 has been shown to interact with Myc using co-immunoprecipitation and glutathione S-transferase pull-down assays (Xu et al., 2008). Immunological depletion of ETS2 or mutation of the ETS DNA-binding motif hampers c-Myc binding to the E-box. Further depletion of c-Myc or mutation of the E-box also attenuates ETS2 binding to the ETS DNA-binding motif (Xu et al., 2008). These results suggest that the interaction of ETS2 with c-Myc regulates TERT gene expression, which in turn affects breast cancer cell proliferation. However, this has not been reported in other cancer cell types. Recently, two somatic hotspot mutations were discovered in the TERT promoter that specifically occurred in a subset of cancers (Horn et al., 2013; Huang et al., 2013; Akincilar et al., 2016b). The mutations observed were cytosine to thymine transitions in the TERT promoter and were mutually exclusive (Horn et al., 2013; Huang et al., 2013). These mutations created de novo binding sites for ETS transcription factors on the TERT promoter (Bell et al., 2015; Li et al., 2015). GABP, an ETS factor, has been shown to specifically associate with the de novo ETS motif on the TERT promoter (Bell et al., 2015). Besides GABP, Li et al. demonstrated that ETS1/2 factors bind cooperatively with NF-kB2 at the mutant TERT promoter during non-canonical NF-kB signaling to drive telomerase reactivation in glioblastomas. These studies suggest that regulation of mutant TERT promoters may be context-dependent in different cancer types and can involve binding of other regulators other than a single ETS factor (Li et al., 2015).
Long-range chromatin interactions mediated by GABP have also been reported to specifically regulate activation of the mutant TERT promoter (Akincilar et al., 2016a). However, cooperation or crosstalk between Myc and GABP to regulate the mutant TERT promoter has not yet been reported.
TERT has also been shown to regulate Myc protein stability by directly interacting with Myc (Koh et al., 2015). This study suggests the existence of a feed-forward loop between Myc and TERT in Myc-driven cancers like lymphoma, wherein Myc upregulates TERT transcription and TERT, in turn, stabilizes Myc protein levels to promote lymphomagenesis. Apart from regulating Myc stability, TERT can also be recruited to Myc target promoters in Myc-driven lymphoma cells probably through association with Myc (Khattar et al., 2016). This represents another novel aspect of the crosstalk between Myc and TERT in cancers.
In contrast to the role of Myc as an activator of TERT transcription, it has also been shown to play a key role in repressing TERT transcription. Inhibition of Myc expression has been reported to increase TERT transcription in normal cells (Zhao et al., 2014). This effect is independent of E-boxes and results in an increase in active histone marks on the TERT promoter.
E2F1 is a direct transcriptional target of Myc and it inhibits TERT transcription by directly associating with the TERT promoter (Crowe et al., 2001; Elliott et al., 2008; Lacerte et al., 2008; Zhang et al., 2012). This exemplifies the dual nature of Myc in regulating TERT transcription. This may also represent another mechanism operating in normal cells to control Myc-dependent oncogenic signals.
A switch from Myc/Max to Max/Mad1 complexes has been shown to repress TERT transcription (Xu et al., 2001). TERT transcription is repressed when stem cells differentiate to specific lineages. The Myc/Max to Max/Mad1 switch is reported to arise during differentiation and has been proposed as one of the mechanisms of repressing TERT transcription.
Peroxisome proliferator-activated receptor γ (PPARγ) ligands inhibit TERT mRNA expression in colon cancer cells via modulation of the Myc/Max/Mad network (Toaldo et al., 2010). PPARγ ligands reduce the levels of Myc and TERT and simultaneously upregulate Mad1 levels, thereby inducing a switch from Myc/Max to Mad1/Max complexes on the TERT promoter. However, the exact molecular mechanism behind the transcriptional changes in Myc and Mad1 levels induced by PPARγ ligands has not been characterized.
Correlation between Myc and TERT expression in cancers
Alterations in Myc expression are commonly observed during cancer initiation and progression. They occur via chromosomal translocations, amplifications and gene mutations (Dang, 2012). Telomerase reconstitution by reactivating TERT expression is known to occur in cancers by various mechanisms that involve oncogenic transcription factors, gene amplification, promoter mutations, crosstalk with oncogenic pathways or epigenetic regulation (Akincilar et al., 2016b; Li and Tergaonkar, 2016; Li et al., 2016; Tergaonkar, 2016). The correlation between Myc and TERT mRNA expression has been extensively investigated in cancers for two reasons. The first being that simultaneous deregulation of Myc and TERT is a frequent genetic event occurring in several cancers. And secondly, TERT is a transcriptional target of Myc.
Myc expression is usually measured at the protein level using immunohistochemistry. For measurement of TERT expression, some studies use mRNA expression, while others measure telomerase enzyme activity since TERT mRNA levels positively correlate with telomerase activity (Armstrong et al., 2000; Kirkpatrick et al., 2003a). Table 1 lists all the studies that report an association between Myc and TERT expression. In most cancers, high expression of Myc and TERT (measured as mRNA expression or telomerase activity) occur together, suggesting positive correlation. The breast cancer and hepatocellular cancer studies listed in Table 1 show no correlation between Myc and TERT expression. This suggests that there may be additional, as yet undiscovered mechanisms operating for regulation of TERT expression through Myc- dependent or independent means in these cancer types. The reported correlation studies should be interpreted with caution because of the small sample sizes. Further, TERT expression and telomerase activity measurement is not standardized and can therefore vary with different protocols.
Table 1.
This table summarizes correlation analyses between: (a) Myc and TERT expression, (b) Myc expression and telomerase activity in various types of cancers.
| Cancer type (number of studies) | Correlation between TERT and Myc mRNA expressiona | Correlation between telomerase activity and Myc mRNA expressionb | References |
|---|---|---|---|
| Breast Cancer (n = 4) | No correlation | Not reported | Kirkpatrick et al., 2003b, 2004; Elkak et al., 2005; Bodvarsdottir et al., 2007 |
| Cervical cancer (n = 1) | Positive | Not reported | Sagawa et al., 2001 |
| Hepatocellular carcinoma (n = 2) | No correlation | Not reported | Chen et al., 2002; Liu et al., 2004 |
| Ovarian cancer (n = 1) | No correlation | Positive | Wisman et al., 2003 |
| Non-small cell lung cancer (n = 1) | Positive | Not reported | Geng et al., 2003 |
| Lymphoma (n = 1) | Positive | Positive | Klapper et al., 2003 |
| Malignant lipomatous tumors (n = 1) | Positive | Positive | Schneider-Stock et al., 2003 |
| Colon cancer (n = 1) | Positive | Not reported | Georgakopoulos et al., 2013 |
| Gastric cancer (n = 1) | Positive | Not reported | Silva et al., 2012 |
| Prostate cancer (n = 1) | Positive | Not reported | Latil et al., 2000 |
Positive correlation: high Myc expression co-occurs with high TERT mRNA expression or telomerase activity. No correlation: Myc levels and TERT mRNA expression or Myc levels and telomerase activity show no significant association. Not reported: the investigating group did not examine the correlation.
Perspectives
Myc and TERT play crucial roles during normal development as well as in carcinogenesis. Therefore, understanding their interplay in both these processes is important. Myc can activate TERT transcription directly or by cooperating with other transcription factors like SP1, ER and ETS. The factors that discriminate Myc-dependent TERT regulation during normal development from cancer are not known and need further investigation. Mutations in the TERT promoter represent the most common non-coding somatic mutations in cancer and it would be interesting to investigate the role of Myc in regulating the mutant TERT promoter. The existing correlation studies between Myc and TERT are not very conclusive and need further analysis.
Author contributions
EK conceived, designed and wrote the manuscript. EK and VT revised the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Agency for Science Technology and Research, Singapore (A*STAR) for funding and support to the V.T. laboratory.
References
- Aisner D. L., Wright W. E., Shay J. W. (2002). Telomerase regulation: not just flipping the switch. Curr. Opin. Genet. Dev. 12, 80–85. 10.1016/S0959-437X(01)00268-4 [DOI] [PubMed] [Google Scholar]
- Akincilar S. C., Unal B., Tergaonkar V. (2016b). Reactivation of telomerase in cancer. Cell. Mol. Life Sci. 73, 1659–1670. 10.1007/s00018-016-2146-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akincilar S., Khattar E., Boon P. L., Unal B., Fullwood M. J., Tergaonkar V. (2016a). Long-range chromatin interactions drive mutant Tert promoter activation. Cancer Discov. 6, 1276–1291. 10.1158/2159-8290.CD-16-0177 [DOI] [PubMed] [Google Scholar]
- Anderson C. J., Hoare S. F., Ashcroft M., Bilsland A. E., Keith W. N. (2006). Hypoxic regulation of telomerase gene expression by transcriptional and post-transcriptional mechanisms. Oncogene 25, 61–69. 10.1038/sj.onc.1209011 [DOI] [PubMed] [Google Scholar]
- Armstrong L., Lako M., Lincoln J., Cairns P. M., Hole N. (2000). mTert expression correlates with telomerase activity during the differentiation of murine embryonic stem cells. Mech. Dev. 97, 109–116. 10.1016/S0925-4773(00)00423-8 [DOI] [PubMed] [Google Scholar]
- Avilion A. A., Piatyszek M. A., Gupta J., Shay J. W., Bacchetti S., Greider C. W. (1996). Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res. 56, 645–650. [PubMed] [Google Scholar]
- Bell R. J., Rube H. T., Kreig A., Mancini A., Fouse S. D., Nagarajan R. P., et al. (2015). Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 348, 1036–1039. 10.1126/science.aab0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez Y., Yang H., Cheng J. Q., Kruk P. A. (2008). Pyk2/ERK 1/2 mediate Sp1- and c-Myc-dependent induction of telomerase activity by epidermal growth factor. Growth Factors 26, 1–11. 10.1080/08977190802001389 [DOI] [PubMed] [Google Scholar]
- Blackburn E. H., Collins K. (2011). Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb. Perspect. Biol. 3:a003558. 10.1101/cshperspect.a003558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H., Epel E. S., Lin J. (2015). Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science 350, 1193–1198. 10.1126/science.aab3389 [DOI] [PubMed] [Google Scholar]
- Bodvarsdottir S. K., Steinarsdottir M., Hilmarsdottir H., Jonasson J. G., Eyfjord J. E. (2007). MYC amplification and TERT expression in breast tumor progression. Cancer Genet. Cytogenet. 176, 93–99. 10.1016/j.cancergencyto.2007.04.002 [DOI] [PubMed] [Google Scholar]
- Bryan T. M., Englezou A., Dalla-Pozza L., Dunham M. A., Reddel R. R. (1997). Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3, 1271–1274. [DOI] [PubMed] [Google Scholar]
- Bryan T. M., Englezou A., Gupta J., Bacchetti S., Reddel R. R. (1995). Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14, 4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu D. X., Johansson M. E., Ren J., Xu D. W., Johnson F. B., Edfeldt K., et al. (2010). Nuclear factor {kappa}B-mediated transactivation of telomerase prevents intimal smooth muscle cell from replicative senescence during vascular repair. Arterioscler. Thromb. Vasc. Biol. 30, 2604–2610. 10.1161/ATVBAHA.110.213074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha Y., Kwon S. J., Seol W., Park K. S. (2008). Estrogen receptor-alpha mediates the effects of estradiol on telomerase activity in human mesenchymal stem cells. Mol. Cells 26, 454–458. [PubMed] [Google Scholar]
- Chen C. J., Tsai N. M., Liu Y. C., Ho L. I., Hsieh H. F., Yen C. Y., et al. (2002). Telomerase activity in human hepatocellular carcinoma: parallel correlation with human telomerase reverse transcriptase (hTERT) mRNA isoform expression but not with cell cycle modulators or c-Myc expression. Eur. J. Surg. Oncol. 28, 225–234. 10.1053/ejso.2001.1237 [DOI] [PubMed] [Google Scholar]
- Cildir G., Low K. C., Tergaonkar V. (2016). Noncanonical NF-kappaB Signaling in Health and Disease. Trends Mol. Med. 22, 414–429. 10.1016/j.molmed.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Cohen S. B., Graham M. E., Lovrecz G. O., Bache N., Robinson P. J., Reddel R. R. (2007). Protein composition of catalytically active human telomerase from immortal cells. Science 315, 1850–1853. 10.1126/science.1138596 [DOI] [PubMed] [Google Scholar]
- Cong Y. S., Wright W. E., Shay J. W. (2002). Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 66, 407–425. 10.1128/MMBR.66.3.407-425.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe D. L., Nguyen D. C., Tsang K. J., Kyo S. (2001). E2F-1 represses transcription of the human telomerase reverse transcriptase gene. Nucleic Acids Res. 29, 2789–2794. 10.1093/nar/29.13.2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V. (2012). MYC on the path to cancer. Cell 149, 22–35. 10.1016/j.cell.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel M., Peek G. W., Tollefsbol T. O. (2012). Regulation of the human catalytic subunit of telomerase (hTERT). Gene 498, 135–146. 10.1016/j.gene.2012.01.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer J., Li H., Xu D., Liu J. P. (2007). Transcriptional regulation of telomerase activity: roles of the the Ets transcription factor family. Ann. N.Y. Acad. Sci. 1114, 36–47. 10.1196/annals.1396.022 [DOI] [PubMed] [Google Scholar]
- Dwyer J. M., Liu J. P. (2010). Ets2 transcription factor, telomerase activity and breast cancer. Clin. Exp. Pharmacol. Physiol. 37, 83–87. 10.1111/j.1440-1681.2009.05236.x [DOI] [PubMed] [Google Scholar]
- Elkak A. E., Meligonis G., Salhab M., Mitchell B., Blake J. R., Newbold R. F., et al. (2005). hTERT protein expression is independent of clinicopathological parameters and c-Myc protein expression in human breast cancer. J. Carcinog. 4:17. 10.1186/1477-3163-4-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott K. A., Rickords L. F., Labrum J. M. (2008). Transduction of E2F-1 TAT fusion proteins represses expression of hTERT in primary ductal breast carcinoma cell lines. Mol. Cancer 7:28. 10.1186/1476-4598-7-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Funk W. D., Wang S. S., Weinrich S. L., Avilion A. A., Chiu C. P., et al. (1995). The RNA component of human telomerase. Science 269, 1236–1241. [DOI] [PubMed] [Google Scholar]
- Geng Z., Zhang D., Liu Y. (2003). Expression of telomerase hTERT in human non-small cell lung cancer and its correlation with c-myc gene. Chin. Med. J. 116, 1467–1470. [PubMed] [Google Scholar]
- Georgakopoulos G., Tsiambas E., Korkolopoulos P., Kavantzas N., Karameris A., Ragkos V., et al. (2013). c-MYC and h-TERT co-expression in colon adenocarcinoma: a tissue microarray digitized image analysis. J. BUON 18, 124–130. [PubMed] [Google Scholar]
- Goueli B. S., Janknecht R. (2003). Regulation of telomerase reverse transcriptase gene activity by upstream stimulatory factor. Oncogene 22, 8042–8047. 10.1038/sj.onc.1206847 [DOI] [PubMed] [Google Scholar]
- Goueli B. S., Janknecht R. (2004). Upregulation of the Catalytic Telomerase Subunit by the Transcription Factor ER81 and Oncogenic HER2/Neu, Ras, or Raf. Mol. Cell. Biol. 24, 25–35. 10.1128/MCB.24.1.25-35.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasselli A., Nanni S., Colussi C., Aiello A., Benvenuti V., Ragone G., et al. (2008). Estrogen receptor-alpha and endothelial nitric oxide synthase nuclear complex regulates transcription of human telomerase. Circ. Res. 103, 34–42. 10.1161/CIRCRESAHA.107.169037 [DOI] [PubMed] [Google Scholar]
- Hartmann C., Meyer J., Balss J., Capper D., Mueller W., Christians A., et al. (2009). Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 118, 469–474. 10.1007/s00401-009-0561-9 [DOI] [PubMed] [Google Scholar]
- Hayden M. S., Ghosh S. (2004). Signaling to NF-κB. Genes Dev. 18, 2195–2224. 10.1101/gad.1228704 [DOI] [PubMed] [Google Scholar]
- Heiss N. S., Knight S. W., Vulliamy T. J., Klauck S. M., Wiemann S., Mason P. J., et al. (1998). X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 19, 32–38. 10.1038/ng0598-32 [DOI] [PubMed] [Google Scholar]
- Holzmann K., Gerner C., Korosec T., Poltl A., Grimm R., Sauermann G. (1998). Identification and characterization of the ubiquitously occurring nuclear matrix protein NMP 238. Biochem. Biophys. Res. Commun. 252, 39–45. 10.1006/bbrc.1998.9604 [DOI] [PubMed] [Google Scholar]
- Horikawa I., Cable P. L., Afshari C., Barrett J. C. (1999). Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res. 59, 826–830. [PubMed] [Google Scholar]
- Horn S., Figl A., Rachakonda P. S., Fischer C., Sucker A., Gast A., et al. (2013). TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961. 10.1126/science.1230062 [DOI] [PubMed] [Google Scholar]
- Hsu C. P., Lee L. W., Tang S. C., Hsin I. L., Lin Y. W., Ko J. L. (2015). Epidermal growth factor activates telomerase activity by direct binding of Ets-2 to hTERT promoter in lung cancer cells. Tumour Biol. 36, 5389–5398. 10.1007/s13277-015-3204-x [DOI] [PubMed] [Google Scholar]
- Huang F. W., Hodis E., Xu M. J., Kryukov G. V., Chin L., Garraway L. A. (2013). Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959. 10.1126/science.1229259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattar E., Kumar P., Liu C. Y., Akincilar S. C., Raju A., Lakshmanan M., et al. (2016). Telomerase reverse transcriptase promotes cancer cell proliferation by augmenting tRNA expression. J. Clin. Invest. 126, 4045–4060. 10.1172/JCI86042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L., et al. (1994). Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick K. L., Clark G., Ghilchick M., Newbold R. F., Mokbel K. (2003a). hTERT mRNA expression correlates with telomerase activity in human breast cancer. Eur. J. Surg. Oncol. 29, 321–326. 10.1053/ejso.2002.1374 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick K. L., Newbold R. F., Mokbel K. (2004). There is no correlation between c-Myc mRNA expression and telomerase activity in human breast cancer. Int. Semin. Surg. Oncol. 1:2. 10.1186/1477-7800-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick K. L., Ogunkolade W., Elkak A. E., Bustin S., Jenkins P., Ghilchick M., et al. (2003b). hTERT expression in human breast cancer and non-cancerous breast tissue: correlation with tumour stage and c-Myc expression. Breast Cancer Res. Treat. 77, 277–284. 10.1023/A:1021849217054 [DOI] [PubMed] [Google Scholar]
- Klapper W., Krams M., Qian W., Janssen D., Parwaresch R. (2003). Telomerase activity in B-cell non-Hodgkin lymphomas is regulated by hTERT transcription and correlated with telomere-binding protein expression but uncoupled from proliferation. Br. J. Cancer 89, 713–719. 10.1038/sj.bjc.6601112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh C. M., Khattar E., Leow S. C., Liu C. Y., Muller J., Ang W. X., et al. (2015). Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J. Clin. Invest. 125, 2109–2122. 10.1172/JCI79134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyo S., Takakura M., Kanaya T., Zhuo W., Fujimoto K., Nishio Y., et al. (1999). Estrogen activates telomerase. Cancer Res. 59, 5917–5921. [PubMed] [Google Scholar]
- Kyo S., Takakura M., Taira T., Kanaya T., Itoh H., Yutsudo M., et al. (2000). Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT). Nucleic Acids Res. 28, 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerte A., Korah J., Roy M., Yang X. J., Lemay S., Lebrun J. J. (2008). Transforming growth factor-beta inhibits telomerase through SMAD3 and E2F transcription factors. Cell. Signal. 20, 50–59. 10.1016/j.cellsig.2007.08.012 [DOI] [PubMed] [Google Scholar]
- Latil A., Vidaud D., Valeri A., Fournier G., Vidaud M., Lidereau R., et al. (2000). Htert expression correlates with MYC over-expression in human prostate cancer. Int. J. Cancer 89, 172–176. [DOI] [PubMed] [Google Scholar]
- Lebel R., McDuff F. O., Lavigne P., Grandbois M. (2007). Direct visualization of the binding of c-Myc/Max heterodimeric b-HLH-LZ to E-box sequences on the hTERT promoter. Biochemistry 46, 10279–10286. 10.1021/bi700076m [DOI] [PubMed] [Google Scholar]
- Li Y., Cheng H. S., Chng W. J., Tergaonkar V. (2016). Activation of mutant TERT promoter by RAS-ERK signaling is a key step in malignant progression of BRAF-mutant human melanomas. Proc. Natl. Acad. Sci. U.S.A. 113, 14402–14407. 10.1073/pnas.1611106113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Tergaonkar V. (2016). Telomerase reactivation in cancers: mechanisms that govern transcriptional activation of the wild-type vs. mutant TERT promoters. Transcription 7, 44–49. 10.1080/21541264.2016.1160173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhou Q. L., Sun W., Chandrasekharan P., Cheng H. S., Ying Z., et al. (2015). Non-canonical NF-κB signalling and ETS1/2 cooperatively drive C250T mutant TERT promoter activation. Nat. Cell Biol. 17, 1327–1338. 10.1038/ncb3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. C., Chen C. J., Wu H. S., Chan D. C., Yu J. C., Yang A. H., et al. (2004). Telomerase and c-myc expression in hepatocellular carcinomas. Eur. J. Surg. Oncol. 30, 384–390. 10.1016/j.ejso.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Maida Y., Kyo S., Kanaya T., Wang Z., Yatabe N., Tanaka M., et al. (2002). Direct activation of telomerase by EGF through Ets-mediated transactivation of TERT via MAP kinase signaling pathway. Oncogene 21, 4071–4079. 10.1038/sj.onc.1205509 [DOI] [PubMed] [Google Scholar]
- Marion R. M., Strati K., Li H., Tejera A., Schoeftner S., Ortega S., et al. (2009). Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell 4, 141–154. 10.1016/j.stem.2008.12.010 [DOI] [PubMed] [Google Scholar]
- Nair S. K., Burley S. K. (2003). X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112, 193–205. 10.1016/S0092-8674(02)01284-9 [DOI] [PubMed] [Google Scholar]
- Nakamura T. M., Morin G. B., Chapman K. B., Weinrich S. L., Andrews W. H., Lingner J., et al. (1997). Telomerase catalytic subunit homologs from fission yeast and human. Science 277, 955–959. [DOI] [PubMed] [Google Scholar]
- Ohba S., Mukherjee J., Johannessen T. C., Mancini A., Chow T. T., Wood M., et al. (2016). Mutant IDH1 expression drives tert promoter reactivation as part of the cellular transformation process. Cancer Res. 76, 6680–6689. 10.1158/0008-5472.CAN-16-0696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfait B., Giovangrandi Y., Asheuer M., Laurendeau I., Olivi M., Vodovar N., et al. (2000). Human TIP49b/RUVBL2 gene: genomic structure, expression pattern, physical link to the human CGB/LHB gene cluster on chromosome 19q13.3. Ann. Genet. 43, 69–74. 10.1016/S0003-3995(00)01016-9 [DOI] [PubMed] [Google Scholar]
- Sagawa Y., Nishi H., Isaka K., Fujito A., Takayama M. (2001). The correlation of TERT expression with c-myc expression in cervical cancer. Cancer Lett. 168, 45–50. 10.1016/S0304-3835(01)00501-8 [DOI] [PubMed] [Google Scholar]
- Schneider-Stock R., Boltze C., Jager V., Epplen J., Landt O., Peters B., et al. (2003). Elevated telomerase activity, c-MYC-, and hTERT mRNA expression: association with tumour progression in malignant lipomatous tumours. J. Pathol. 199, 517–525. 10.1002/path.1315 [DOI] [PubMed] [Google Scholar]
- Shay J. W., Bacchetti S. (1997). A survey of telomerase activity in human cancer. Eur. J. Cancer 33, 787–791. 10.1016/S0959-8049(97)00062-2 [DOI] [PubMed] [Google Scholar]
- Sheng W. Y., Chen Y. R., Wang T. C. (2006). A major role of PKC theta and NFkappaB in the regulation of hTERT in human T lymphocytes. FEBS Lett. 580, 6819–6824. 10.1016/j.febslet.2006.11.044 [DOI] [PubMed] [Google Scholar]
- Silva T. C., Leal M. F., Calcagno D. Q., de Souza C. R., Khayat A. S., dos Santos N. P., et al. (2012). hTERT, MYC and TP53 deregulation in gastric preneoplastic lesions. BMC Gastroenterol. 12:85. 10.1186/1471-230X-12-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha-Datta U., Horikawa I., Michishita E., Datta A., Sigler-Nicot J. C., Brown M., et al. (2004). Transcriptional activation of hTERT through the NF-kappaB pathway in HTLV-I-transformed cells. Blood 104, 2523–2531. 10.1182/blood-2003-12-4251 [DOI] [PubMed] [Google Scholar]
- Sun S. C. (2011). Non-canonical NF-kappaB signaling pathway. Cell Res. 21, 71–85. 10.1038/cr.2010.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Takakura M., Kyo S., Inoue M., Wright W. E., Shay J. W. (2005). Function of AP-1 in transcription of the telomerase reverse transcriptase gene (TERT) in human and mouse cells. Mol. Cell. Biol. 25, 8037–8043. 10.1128/MCB.25.18.8037-8043.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakura M., Kyo S., Kanaya T., Hirano H., Takeda J., Yutsudo M., et al. (1999). Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 59, 551–557. [PubMed] [Google Scholar]
- Tergaonkar V. (2016). NFκB drives TERT promoter reactivation in cancer. Cell Cycle 15, 156–157. 10.1080/15384101.2015.1118908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toaldo C., Pizzimenti S., Cerbone A., Pettazzoni P., Menegatti E., Daniela B., et al. (2010). PPARgamma ligands inhibit telomerase activity and hTERT expression through modulation of the Myc/Mad/Max network in colon cancer cells. J. Cell. Mol. Med. 14, 1347–1357. 10.1111/j.1582-4934.2009.00966.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher A. S., Abreu E. B., Meng Z., McCann K. E., Terns R. M., Veenstra T. D., et al. (2009). A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323, 644–648. 10.1126/science.1165357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zhao Y., Hu C., Zhu J. (2009). Differential repression of human and mouse TERT genes during cell differentiation. Nucleic Acids Res. 37, 2618–2629. 10.1093/nar/gkp125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zhu J. (2004). The hTERT gene is embedded in a nuclease-resistant chromatin domain. J. Biol. Chem. 279, 55401–55410. 10.1074/jbc.M411352200 [DOI] [PubMed] [Google Scholar]
- Wechsler D. S., Dang C. V. (1992). Opposite orientations of DNA bending by c-Myc and Max. Proc. Natl. Acad. Sci. U.S.A. 89, 7635–7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick M., Zubov D., Hagen G. (1999). Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT). Gene 232, 97–106. [DOI] [PubMed] [Google Scholar]
- Wisman G. B., Hollema H., Helder M. N., Knol A. J., Van der Meer G. T., Krans M., et al. (2003). Telomerase in relation to expression of p53, c-Myc and estrogen receptor in ovarian tumours. Int. J. Oncol. 23, 1451–1459. 10.3892/ijo.23.5.1451 [DOI] [PubMed] [Google Scholar]
- Wu K. J., Grandori C., Amacker M., Simon-Vermot N., Polack A., Lingner J., et al. (1999). Direct activation of TERT transcription by c-MYC. Nat. Genet. 21, 220–224. 10.1038/6010 [DOI] [PubMed] [Google Scholar]
- Xiao X., Athanasiou M., Sidorov I. A., Horikawa I., Cremona G., Blair D., et al. (2003). Role of Ets/Id proteins for telomerase regulation in human cancer cells. Exp. Mol. Pathol. 75, 238–247. 10.1016/S0014-4800(03)00092-3 [DOI] [PubMed] [Google Scholar]
- Xu D., Dwyer J., Li H., Duan W., Liu J. P. (2008). Ets2 maintains hTERT gene expression and breast cancer cell proliferation by interacting with c-Myc. J. Biol. Chem. 283, 23567–23580. 10.1074/jbc.M800790200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Popov N., Hou M., Wang Q., Bjorkholm M., Gruber A., et al. (2001). Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc. Natl. Acad. Sci. U.S.A. 98, 3826–3831. 10.1073/pnas.071043198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Parsons D. W., Jin G., McLendon R., Rasheed B. A., Yuan W., et al. (2009). IDH1 and IDH2 mutations in gliomas. N.Engl. J. Med. 360, 765–773. 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen L., Yang S., Fang D. (2012). E2F1: a potential negative regulator of hTERT transcription in normal cells upon activation of oncogenic c-Myc. Med. Sci. Monit. 18, RA12–RA15. 10.12659/MSM.882192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Cheng D., Wang S., Zhu J. (2014). Dual roles of c-Myc in the regulation of hTERT gene. Nucleic Acids Res. 42, 10385–10398. 10.1093/nar/gku721 [DOI] [PMC free article] [PubMed] [Google Scholar]