Abstract
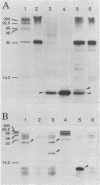
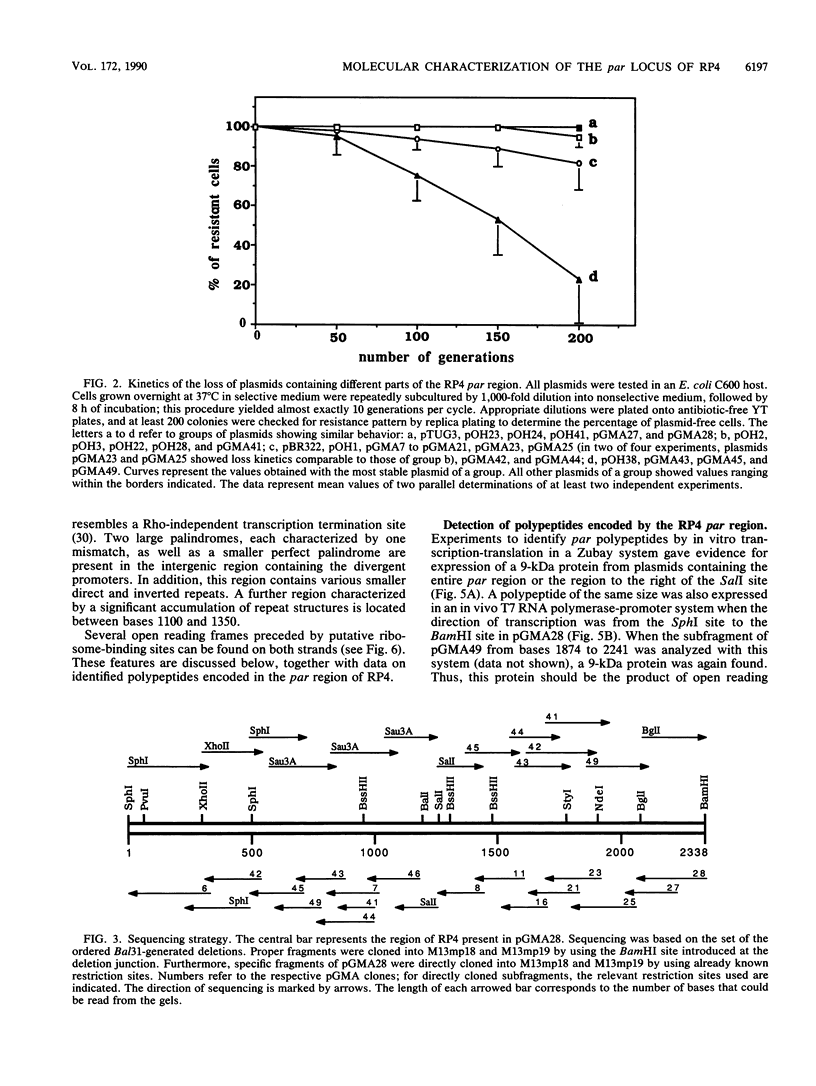
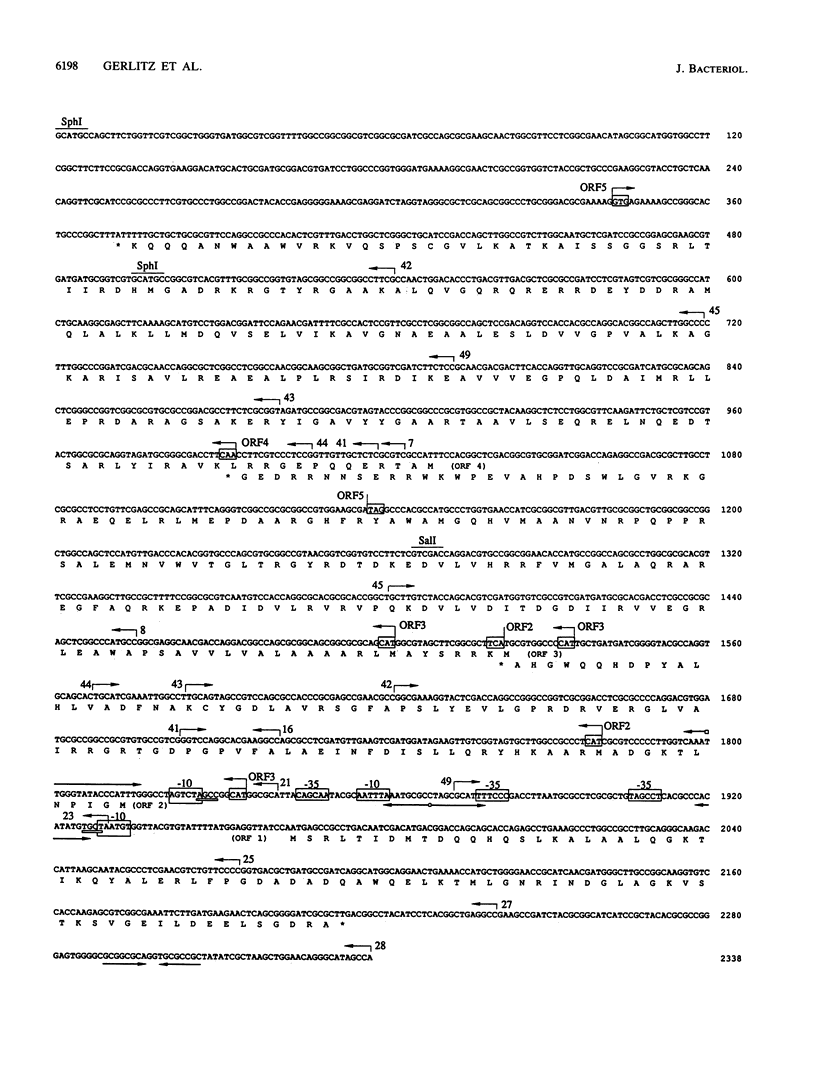
The broad-host-range plasmid RP4 encodes a highly efficient partitioning system (par) that was previously mapped within the 6.2-kb PstI C fragment. The essential functions were assigned to a region of 2.2 kb between fiwA and IS21 (IS8). On the basis of the nucleotide sequence data of the entire par locus and of in vitro and in vivo expression studies, three distinct loci encoding polypeptides of 9, 18, and 24 kDa were identified. Evidence for the expression of another polypeptide was found. A putative divergent promoter was localized in an intergenic region and is suggested to be responsible for transcription of these genes. It was found that the RP4 par region includes a function resolving plasmid dimers. The 24-kDa polypeptide is considered to function as a resolvase, since its predicted amino acid sequence shows homology to sequences of resolvases of the Tn3 family. Furthermore, palindromes present in the intergenic region containing the divergent promoter resemble repeat structures specific for res sites of Tn3-related transposons. However, it was found that dimer resolution itself was not sufficient for stabilization; additional functions, including the other two polypeptides, seemed to play an important role. These results suggested that RP4 contains a complex stabilization system involving resolution of plasmid dimers during cell division, thus ensuring the delivery of at least one copy to each daughter cell.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Friedman S. A., Austin S. J. Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J Mol Biol. 1985 Sep 20;185(2):261–272. doi: 10.1016/0022-2836(85)90402-4. [DOI] [PubMed] [Google Scholar]
- Altenbuchner J., Schmitt R. Transposon Tn1721: site-specific recombination generates deletions and inversions. Mol Gen Genet. 1983;190(2):300–308. doi: 10.1007/BF00330655. [DOI] [PubMed] [Google Scholar]
- Austin S. J. Plasmid partition. Plasmid. 1988 Jul;20(1):1–9. doi: 10.1016/0147-619x(88)90001-7. [DOI] [PubMed] [Google Scholar]
- Austin S., Ziese M., Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell. 1981 Sep;25(3):729–736. doi: 10.1016/0092-8674(81)90180-x. [DOI] [PubMed] [Google Scholar]
- Davis M. A., Austin S. J. Recognition of the P1 plasmid centromere analog involves binding of the ParB protein and is modified by a specific host factor. EMBO J. 1988 Jun;7(6):1881–1888. doi: 10.1002/j.1460-2075.1988.tb03021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depicker A., De Block M., Inzé D., Van Montagu M., Schell J. IS-like element IS8 in RP4 plasmid and its involvement in cointegration. Gene. 1980 Sep;10(4):329–338. doi: 10.1016/0378-1119(80)90153-5. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R. A., James A. A., Kolodner R. recA-independent general genetic recombination of plasmids. Nature. 1981 Nov 12;294(5837):184–186. doi: 10.1038/294184a0. [DOI] [PubMed] [Google Scholar]
- Fong S. T., Stanisich V. A. Location and characterization of two functions on RP1 that inhibit the fertility of the IncW plasmid R388. J Gen Microbiol. 1989 Mar;135(3):499–502. doi: 10.1099/00221287-135-3-499. [DOI] [PubMed] [Google Scholar]
- Funnell B. E. Mini-P1 plasmid partitioning: excess ParB protein destabilizes plasmids containing the centromere parS. J Bacteriol. 1988 Feb;170(2):954–960. doi: 10.1128/jb.170.2.954-960.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie D. R., Kado C. I. Agrobacterium tumefaciens pTAR parA promoter region involved in autoregulation, incompatibility and plasmid partitioning. J Mol Biol. 1987 Feb 5;193(3):465–478. doi: 10.1016/0022-2836(87)90260-9. [DOI] [PubMed] [Google Scholar]
- Gerdes K., Bech F. W., Jørgensen S. T., Løbner-Olesen A., Rasmussen P. B., Atlung T., Boe L., Karlstrom O., Molin S., von Meyenburg K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986 Aug;5(8):2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Molin S. Partitioning of plasmid R1. Structural and functional analysis of the parA locus. J Mol Biol. 1986 Aug 5;190(3):269–279. doi: 10.1016/0022-2836(86)90001-x. [DOI] [PubMed] [Google Scholar]
- Hakkaart M. J., van den Elzen P. J., Veltkamp E., Nijkamp H. J. Maintenance of multicopy plasmid Clo DF13 in E. coli cells: evidence for site-specific recombination at parB. Cell. 1984 Jan;36(1):203–209. doi: 10.1016/0092-8674(84)90090-4. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S., Jaffé A., Ogura T., Mori H., Takahashi H. F plasmid ccd mechanism in Escherichia coli. J Bacteriol. 1986 Apr;166(1):100–104. doi: 10.1128/jb.166.1.100-104.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusukawa N., Mori H., Kondo A., Hiraga S. Partitioning of the F plasmid: overproduction of an essential protein for partition inhibits plasmid maintenance. Mol Gen Genet. 1987 Jul;208(3):365–372. doi: 10.1007/BF00328125. [DOI] [PubMed] [Google Scholar]
- Lane D., de Feyter R., Kennedy M., Phua S. H., Semon D. D protein of miniF plasmid acts as a repressor of transcription and as a site-specific resolvase. Nucleic Acids Res. 1986 Dec 22;14(24):9713–9728. [PMC free article] [PubMed] [Google Scholar]
- Lanka E., Lurz R., Fürste J. P. Molecular cloning and mapping of SphI restriction fragments of plasmid RP4. Plasmid. 1983 Nov;10(3):303–307. doi: 10.1016/0147-619x(83)90047-1. [DOI] [PubMed] [Google Scholar]
- Mori H., Kondo A., Ohshima A., Ogura T., Hiraga S. Structure and function of the F plasmid genes essential for partitioning. J Mol Biol. 1986 Nov 5;192(1):1–15. doi: 10.1016/0022-2836(86)90459-6. [DOI] [PubMed] [Google Scholar]
- Nordström K., Austin S. J. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pansegrau W., Miele L., Lurz R., Lanka E. Nucleotide sequence of the kanamycin resistance determinant of plasmid RP4: homology to other aminoglycoside 3'-phosphotransferases. Plasmid. 1987 Nov;18(3):193–204. doi: 10.1016/0147-619x(87)90062-x. [DOI] [PubMed] [Google Scholar]
- Pinkney M., Thomas C. M. Replication and maintenance of promiscuous plasmids of gram-negative bacteria. Microbiol Sci. 1987 Jun;4(6):186–191. [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Reimmann C., Moore R., Little S., Savioz A., Willetts N. S., Haas D. Genetic structure, function and regulation of the transposable element IS21. Mol Gen Genet. 1989 Feb;215(3):416–424. doi: 10.1007/BF00427038. [DOI] [PubMed] [Google Scholar]
- Roberts R. C., Burioni R., Helinski D. R. Genetic characterization of the stabilizing functions of a region of broad-host-range plasmid RK2. J Bacteriol. 1990 Nov;172(11):6204–6216. doi: 10.1128/jb.172.11.6204-6216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowsky P., Schmitt R. Tn1721-encoded resolvase: structure of the tnpR gene and its in vitro functions. Mol Gen Genet. 1985;200(1):176–181. doi: 10.1007/BF00383332. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab H., Saurugger P. N., Lafferty R. M. Occurrence of deletion plasmids at high rates after conjugative transfer of the plasmids RP4 and RK2 from Escherichia coli to Alcaligenes eutrophus H16. Arch Microbiol. 1983 Nov;136(2):140–146. doi: 10.1007/BF00404789. [DOI] [PubMed] [Google Scholar]
- Stirling C. J., Stewart G., Sherratt D. J. Multicopy plasmid stability in Escherichia coli requires host-encoded functions that lead to plasmid site-specific recombination. Mol Gen Genet. 1988 Sep;214(1):80–84. doi: 10.1007/BF00340183. [DOI] [PubMed] [Google Scholar]
- Summers D. K., Sherratt D. J. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984 Apr;36(4):1097–1103. doi: 10.1016/0092-8674(84)90060-6. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi A., Min Y. N., Kim C. K., Fan Y. L., Womble D. D., Rownd R. H. Genetic organization and nucleotide sequence of the stability locus of IncFII plasmid NR1. J Mol Biol. 1988 Aug 5;202(3):511–525. doi: 10.1016/0022-2836(88)90282-3. [DOI] [PubMed] [Google Scholar]
- Tsuchimoto S., Ohtsubo H., Ohtsubo E. Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J Bacteriol. 1988 Apr;170(4):1461–1466. doi: 10.1128/jb.170.4.1461-1466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]