Abstract
NF-κB was discovered thirty years ago as a rapidly inducible transcription factor. Since that time it has been found to have a broad role in gene induction in diverse cellular responses, particularly throughout the immune system. Here we summarize elaborate regulatory pathways involving this transcription factor and use recent discoveries in human genetic diseases to place specific proteins within their relevant medical and biological contexts.
Introduction
Thirty years ago, Ranjan Sen and one of the authors did a series of experiments that identified a protein binding to a specific, conserved DNA sequence in nuclei of activated B lymphocytes (Sen and Baltimore, 1986). We named it for the cell type in which we identified it and the gene it affected: Nuclear Factor binding near the κ light chain gene in B cells, or NF-κB. Had we realized that NF-κB would have a wide role in inflammation and other natural and pathological processes, we might have found a simpler designation for ease of typing.
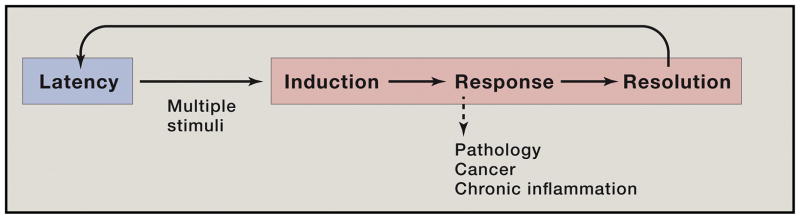
Because NF-κB was induced during B cell maturation, we suspected that it had a major role in B cell activation and development. This has proved true but it greatly underestimated the impact of NF-κB on biological systems (Baltimore, 2011; Hayden and Ghosh, 2008). First of all, it constitutes a paradigm of a rapid response factor, one held in latency in the cell (Figure 1). When an inflammatory or other insult impinges on a cell, it activates a whole pathway of orderly responses, starting immediately after stimulation. Once resolution of the stimulus is achieved, the pathway resets to latency. Second, not just B cells are activated; perhaps more importantly, cells of the innate immune system are the first to be activated through NF-κB at a wound or site of infection (Ben-Neriah, 2002). These cells provide the first line of defense, especially to bacterial invaders. Many minor incursions are cleared solely by innate immunity in mammals; for lower organisms, the innate system may be their only defense. More extensive inflammation will activate B and T cells of the adaptive immune system, again with the heavy involvement of NF-κB.
Figure 1.
The NF-κB paradigm of timely and flexible biochemical control of cell behavior.
As time has passed, what seemed a simple induction process mediated by such proinflammatory molecules as TNF or lipopolysaccharide has turned out to involve many intermediate factors and processes including protein-protein dimerization, phosphorylation of serine, threonine or tyrosine residues and polyubiquitin modifications (Hayden and Ghosh, 2008, 2012). Many inducers are known, each seemingly working through its individual pathway. Also, we have appreciated that NF-κB is not a single transcription protein but rather a family of related protein complexes working as hetero- and homo-dimers, drawing from a pool of 5 monomers, yielding up to 15 NF-κB complexes (Smale, 2012).
The remarkable capability of NF-κB to alter a cell’s biology results from hundreds of target genes that it activates or represses. Even with 15 potential forms, this is a daunting task because each component is needed at a characteristic concentration and time following inflammatory challenge. We know now that there is specificity information all around the target genes, from which other transcription factors may be bound along with NF-κB, to the specific binding site sequence and its effect on the detailed structure of NF-κB, to the particular form of NF-κB that best binds to a particular site, to the intrinsic half-life of the induced mRNA and to the rate of splicing of the induced pre-mRNA (Hayden and Ghosh, 2012). This is a rich field of biology, not yet fully plumbed.
In the following presentation, we will review the detailed molecular biology of NF-κB-mediated signaling. Against this background of understanding, we will then discuss mutational evidence for NF-κB’s function in particular situations, contrasting human genetic data with mouse studies and highlighting the differences between these two mammalian species. With such interesting differences, we can confidently predict that all species will, over time, have adapted the proteins we describe to their individual biology and ecological challenges, leaving much detail to be uncovered.
Biochemistry
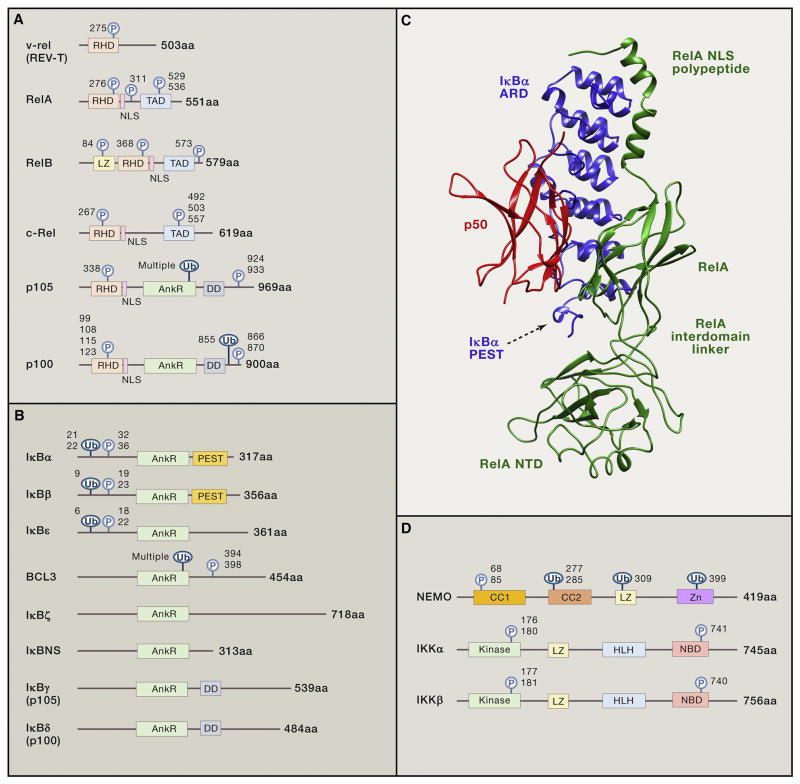
Hundreds of genes utilize NF-κB through variations of a nearly palindromic DNA sequence with a consensus of 5′ –GGGRNWYYCC – 3′ (N – any base; R - purine; W – adenine or thymine; and why – pyrimidine), termed κB, found in their enhancers or promoters (Gilmore, 2016 http://www.bu.edu/nf-kb/gene-resources/target-genes/) (Sen and Baltimore, 1986; Lenardo et al., 1987). The site binds dimers of 5 proteins comprising the Rel transcription factor family: p50, p52, Rel A (p65), Rel B, and c-Rel that share N-terminal homology with the v-Rel oncogene (Hayden and Ghosh, 2008)(Figure 2A). The Rel homology domain (RHD) is 300 amino acids and has 3 functions: sequence specific DNA-binding, dimerization, and inhibitory protein binding (Hayden and Ghosh, 2012; Smale, 2012). The RHD contains 2 subdomains with beta-barrel immunoglobulin (Ig)-like folds. The N-terminal subdomain resembles the p53 DNA-binding domain and specifies DNA recognition. The C-terminal subdomain has hydrophobic residues that form dimerization and inhibitory protein interfaces. The domains are joined at a hinge that clamps onto both sides of the cognate κB site across the major groove of the DNA. Following the RHD is a nuclear localization sequence (NLS) (Figure 2A). The 5 Rel family proteins fall into 2 classes. First, p50 and p52 arise from precursor proteins, p105 (NFKB1) and p100 (NFKB2), respectively, from which a C-terminal region containing ankyrin repeats (AnkR) is post-translationally cleaved. Second, Rel A, Rel B, c-Rel, are synthesized as mature proteins with transcription transactivation domains (TADs). The two paradigmatic dimers are p50:p65 and p52:RelB, although other combinations including p50:p50 and p52:p52 homodimers or p52:RELA or p50:RELB heterodimers exist with distinct functions (Smale, 2012). All-in-all, 13 of the 15 potential NF-κB family complexes have been demonstrated; the other 2 being undescribed and possibly unfavored (S. Ghosh, personal communication). TAD-containing heterodimers are transcriptional activators whereas p50 or p52 homodimers are repressors unless bound to secondary proteins. Since dimer combinations may recognize distinct variants of the κB site with different affinities, many gene regulatory patterns can be generated from this handful of proteins.
Figure 2.
A. Rel homology domain proteins in the NF-κB protein family and the v-Rel oncoprotein. The N-terminal regions of REL family proteins share the Rel homology domain (RHD), which has amino acid similarity to the product of the v-rel oncogene from the Reticuloendotheliosis virus REV-T and a nuclear localization sequence (NLS). RelB has a leucine zipper (LZ) and RelA, RelB, and c-Rel, harbor a transactivation domain (TAD). p105 and p100 have 5–7 tandem ankyrin repeats (AnkR) and a death domain (DD). Regulatory phosphorylation sites (P) and ubiquitination sites (Ub) are shown. B. Inhibitors of NF-κB (IκB) family of proteins. The eight described IκBs are characterized by their 5–7 tandem ankyrin repeats (AnkR) which mediate binding to NF-κB dimers. The N-terminal regions of the classical IκBs (IκBα, IκBβ, IκBε) contain two serine residues (P), which permit the accelerated ubiquitination (Ub) and degradation of the protein when phosphorylated. BCL3 has similar modification residues but IκBζ and IκBNS do not. The C-terminal regions of p105 and p100 function as IκBs for their linked RHD even before processing by forming large complexes with Rel proteins, including p50 and p52. PEST: Region rich in the amino acids proline, glutamic acid, serine, and threonine. DD: death domain. C. Structure of the IκB:NF-κB latent complex (PBD 1NFI). Ribbon diagram of the crystal structure of the isolated complex including IκBα (blue), showing its anchor and repeat domain (ARD) and PEST sequence rich in the amino acids proline, glutamic acid, serine, and threonine that acts as a signal for protein degradation, p50 (red), and the RelA (green), inter-domain linker, and N-terminal domain (NTD). D. IKK complex. Shown are the components of the IKK complex including two kinases: IKKα, IKKβ, and the non-enzymatic subunit NEMO. CC1: Coiled-coil domain 1; CC2: Coiled-coil domain 2; LZ: Leucine zipper; Zn: Zinc-finger; Kinase: Kinase domain; HLH: Helix-loop-helix region; NBD: NEMO-binding domain. Regulatory phosphorylation sites (P) and ubiquitination sites (Ub) are shown. The number of amino acids (aa) of each protein is shown at right. Adapted from (Hayden and Ghosh, 2008, 2012).
In unstimulated cells, such as “resting” lymphocytes before antigen encounter, NF-κBs are mainly cytoplasmic due to the binding of a dedicated set of inhibitory proteins comprising the “Inhibitor of κB” (IκB) family (Hayden and Ghosh, 2008). The intricate biochemical pathways controlling NF-κB/inhibitor complexes that have been reviewed authoritatively and we will only provide a brief summary (Hayden and Ghosh, 2012). The inhibitors, IκBα, IκBβ, IκBε, BCL-3, IκBz, IκBNS and the C-terminal portions of the precursor proteins p105 (IκBγ) and p100 (IκBδ), all contain 5–7 tandem ankyrin repeats (AnkRs) (Figure 2B). AnkRs are 33 amino acid ankyrin-like protein-protein association domains that extend as helices capable of binding to NF-κB covering the NLS. The C-terminal AnkR-containing, IκB-like regions of p100 and p105 undergo limited proteolysis to make p52 and p50 which transit to the nucleus. For p50, this occurs constitutively, but p52 is only generated after specific signaling events. The canonical p50/p65 heterodimer is regulated principally by IκBα (Figure 2C). Biochemical control of nuclear localization via IκB has two strategic consequences: 1) it becomes a principal control point for NF-κB-induced gene expression and, 2) it enables the preformed NF-κB transcription complexes to be launched into action instantaneously providing a rapid molecular switch for responses to pathogens or inflammatory stimuli. NF-κB also embodies an interesting developmental paradigm at least for B cells. One conceptual problem with tissue specific gene expression is an infinite regression paradox. If a tissue-specific transcription factor (TSTF) is required for a set of tissue-specific genes, then does TSTF itself require a TSTF and so on? The NF-κB paradigm shows that lineage and developmental transcriptional regulators can be induced biochemically, thereby resolving the paradox.
The first step of NF-κB activation involves post-translational modification of IκB inhibitors. This occurs by a canonical pathway (CP) and an alternative pathway (AP) (Hayden and Ghosh, 2008). For the CP, a kinase complex called IκB kinase (IKK) specifically phosphorylates IκB proteins leading to their degradation (Ben-Neriah, 2002). This causes NF-κB dimers, which actively shuttle between the nucleus and cytosol, to stay nuclear and induce gene expression. The cytosolic IKK holoenzyme, which we’ll call “classical” IKK, contains a regulatory subunit, “NF-κB essential modifier” (NEMO, also called IKKγ/Fip-3/IKKAP), and two kinase subunits, IKKα and IKKβ (Figure 2D). NEMO is a non-catalytic subunit that tethers IKKα and IKKβ into a regulatory holocomplex and is required for ubiquitination reactions that beget protein oligomerization and signaling (Hayden and Ghosh, 2008). Activation involves signaling assemblies connecting upstream signal generation apparatuses to IKK using ubiquitin modifications as the mortar. The TNF receptor (TNFR) induces a well-understood IKK induction process. TNF stimulation of TNFR recruits the RIP-1, TRADD, TRAF2, cIAP, TAB, and TAK1 proteins into a megacomplex with the linear ubiquitin assembly complex (LUBAC) stabilized by linear and K63-linked polyubiquitin chains (Dondelinger et al., 2016). Then IKK is incorporated, causing ubiquitination of NEMO and phosphorylation of IKK that induce its kinase activity (Hayden and Ghosh, 2008). The specific composition of the megacomplex depends on specific ubiquitin modifications. This, in turn, determines whether TNFR engagement causes NF-κB induction and survival or an alternative fate in which recruitment of RIP kinases 1 and 3 induce cell death instead of NF-κB (Dondelinger et al., 2016). The RIP protein family, RIPs 1–7, are adaptors and kinases that transmit signals from a wide variety of surface receptors and intracellular stress sensors to transcriptional mediators including Jnk, Erk, and p38 and NF-κB as well as mediators of necrotic death (Meylan and Tschopp, 2005). Generally, these transduction complexes can send pleiotropic parallel signals besides NF-κB to coordinate necessary responses.
B and T lymphocytes induce NF-κB in adaptive immune responses through the CARD11:Bcl10:MALT1 (CBM) complex (Hayden and Ghosh, 2008). Newly expressed genes promote lymphocyte proliferation and specific immune functions including antibody production by B cells and the generation of cytokines and other anti-pathogen responses by T cells. Abnormalities of immune activation can foster immunodeficiency, autoimmune diseases, or lymphoid malignancies. The CBM complex is the shared central conduit of T-cell receptor (TCR) and B cell receptor (BCR) signaling essential for NF-κB (Blonska and Lin, 2011; Hayden and Ghosh, 2012). After receptor engagement, CARD11 (CARMA1) is recruited and phosphorylated by PKC-θ in T cells and PKC-β in B cells. In T cells, CARD11 nucleates the tripartite CBM on the inner leaflet of the membrane by attracting BCL10 and the paracaspase MALT1 (Hayden and Ghosh, 2012). Caspase-8 is recruited as a full-length, unprocessed, but weakly active form and ropes in IKK to the CBM. MALT1-dependent tethering of TRAF6 stimulates polyubiquitination of NEMO and the phosphorylation of BCL10, IKKα, and IKKβ. Then, casein kinase 1α brings IKKβ into the signaling complex and phosphorylates CARD11 to temper further signaling (Bidere et al., 2009). CARD9, CARD10 (CARMA3), and CARD14 (CARMA2) can form analogous biochemical complexes to activate NF-κB during specialized immunity (Blonska and Lin, 2011). Which specific CARD protein depends on the receptor and downstream effect. The BCL10/MALT1 dimer is shared by all CARDs. In addition to NF-κB, different CBM complexes can stimulate c-Jun N-terminal kinase (JNK), mechanistic target of rapamycin (mTOR), and other signal pathways (Blonska and Lin, 2011). However, lymphocytes require NF-κB. For B lymphocytes, BAFF/BAFF receptor (BAFFR) causes maturation and survival through NF-κB (Gerondakis and Siebenlist, 2010; Mackay and Cancro, 2006). For T lymphocytes, many cytokines and their receptors — ones that control and are controlled by NF-κB — govern proliferation, specialization into functional subsets, and survival (Gerondakis and Siebenlist, 2010).
The rapid and irreversible demolition of IκB proteins in the proteasome - the toggle for NF-κB – is triggered by phosphorylation and ubiquitination. In the CP, IKKβ phosphorylates IκBα on Ser 32 and Ser 36, or IκBβ on Ser 19 and Ser 23. Phosphorylated IκBs are then polyubiquitinated by SCFβ–TrCP E3 ubiquitin ligases, the signal for proteasome deposition (Ben-Neriah, 2002). Mutation of any of these serines will block IκB degradation and prevent signaling. By contrast, the AP utilizes a dimer of IKKα to phosphorylate p100 on serines 176 and 180 causing proteasomal processing to p52 (Hayden and Ghosh, 2008, 2012). p100 is mainly complexed with RelB with its AnkR domain as an IκB (sometimes called IκBδ) to retain nascent p52 and RelB in the cytoplasm. Phosphorylation of the p100 AnkR domain is independent of IKKβ, IKKγ, or the classical IKK complex. Instead, it requires the NF-κB-inducing kinase (NIK). NIK is unstable and a TRAF complex controls its ubquitination and proteasomal turnover. Receptors induce the AP by pulling away the TRAF complex, thereby stabilizing NIK so it can phosphorylate IKKα. This causes IKKα to specifically phosphorylate the cytoplasmic p100:RelB complex on serines 866 and 870 causing ubiquitination and proteolysis of the C-terminal AnkR domain and nuclear ingress of p52:RelB (Sun, 2011). Transcription by NF-κB is also enhanced by Rel protein phosphorylation (Hayden and Ghosh, 2008). Also, IκBα and IκBε can go to the nucleus and recapture DNA-bound NF-κB and halt transcription (Hayden and Ghosh, 2008). Moreover, NF-κB induces the IκBα gene establishing a cybernetic negative feedback (Hayden and Ghosh, 2012).
The physiological role of NF-κB is best delineated in the immune system. Knockout mice for the RHD proteins show predominantly immunological defects (Gerondakis et al., 2006). Also, a cornucopia of human genetic disorders affecting NF-κB show prominent immune pathology (Courtois et al., 2016). Correspondingly, CP inducers, mainly p50:RelA dimers, include all sorts of pro-inflammatory cytokines such as tumor necrosis factor (TNF) and interleukin-1 (IL-1), conventional humoral and cellular antigens, pathogen-associated molecular patterns (PAMPs) including many bacterial and viral products, cellbound and soluble immune mediators, and effector molecules. These potently trigger NF-κB in lymphocytes, macrophages, and dendritic cells and other minor immune cell types (Hayden and Ghosh, 2008, 2012). Inducing NF-κB turns on thousands of genes that re-sculpt the transcriptional program of immune cells from quiescence to a proliferating effector phenotype capable of fighting pathogens. The NF-κB-induced genes include nearly the entire armamentarium of immune guardians: chemokines, cytokines, adhesion molecules, inflammatory mediators, and apoptosis inhibitors, giving NF-κB a pivotal role in global immunity (Lenardo et al., 1989).
The AP of NF-κB activation is not regulated by classical IKK or IκBs and therefore doesn’t respond to CP inducers (Nishikori, 2005). Whereas the CP responds rapidly to immediate infectious threats, the AP has slow and sustained kinetics dependent on new protein synthesis consistent with a role in organogenesis (Baltimore, 2011; Hayden and Ghosh, 2008). Many inducers are a subgroup of the TNFR superfamily, perhaps any receptor that signals through TRAFs, including BAFFR, CD40, RANK, 4-1BB, HVEM, OX40, GITR, Fn14, TNFR2, and CD30, each of which likely relates to a different function of the AP (Gerondakis and Siebenlist, 2010; Mackay and Cancro, 2006). For example, B-cell-activating factor (BAFF) drives peripheral B cell differentiation and survival (Mackay and Cancro, 2006). CD40, is expressed broadly on immune cells and uses both the AP and CP to control B cell activation, maturation, germinal center formation, somatic mutation and class switching. Lymphotoxin (LT) and LIGHT trigger the LTβ receptor (LTβR) to mediate development of peripheral lymph nodes and Peyer’s patches (Sun, 2011). Finally, RANK regulates bone formation and dendritic cell functions through AP signals (Hayden and Ghosh, 2008). How each of these distinct responses is conveyed by the p52:RelB dimer is unknown. Specific microRNAs, miR-223, miR-15a, and miR-16, are negative regulators of IKKα, showing that there exist even more sophisticated regulatory mechanisms for gene control (Ma et al., 2011).
As these 30 years have gone on, the field has witnessed discovery of an increasing diversity of activators, signaling components, and responding genes for the NF-κB system that are updated continuously on the web (Gilmore, 2016 http://www.bu.edu/nf-kb/gene-resources/target-genes/). The central question persists: how this fairly simple regulatory system can respond to an enormous group of inducers and faithfully transpose those into appropriate patterns of gene expression in different tissues. A relatively new approach is to examine genetic defects affecting NF-κB in humans. Clinicians provide an extraordinarily detailed phenotype that can now be combined with whole genome analysis together with exhaustive systems analysis using transcriptomics and proteomics. We now review the human genetic data and compare it with our current understanding from biochemistry and mouse data.
Human genetics
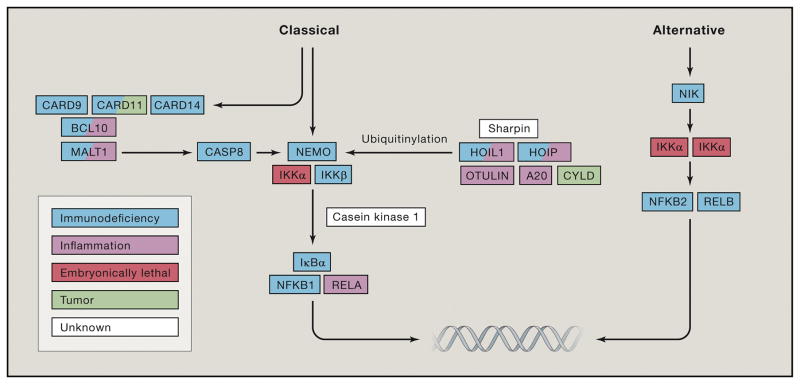
There are now human genetic diseases affecting a large number of NF-κB components and these are summarized in Figure 3 and Table 1.
Figure 3. The major monogenic diseases in the core pathways of both the classical (CP) and alternative (AP) pathways of NF-κB signaling.
Color coded boxes indicate the main feature of diseases.
Table 1.
Features of monogenic diseases in the core NF-κB pathways (in order of discovery)
| Year of discovery | Protein name | Gene name | Method of discovery | Type of disorder | Inheritance model | Defects in patients | Defects in mouse models | Ref |
|---|---|---|---|---|---|---|---|---|
| 2000 | CYLD | CYLD | Linkage study | familial cylindromatosis | AD | Cell hyperproliferation | T cell developmental defect, susceptibility to induced colonic inflammation and increased incidence of tumors | (Bignell et al., 2000; Massoumi et al., 2006; Zhang et al., 2006) |
| 2000 | NEMO | IKBKG | Candidate gene sequencing | Familial incontinentia pigmenti (IP) | XL female | Failures of NF-κB induction in integument and ectoderm-derived appendages | Dermatopathy similar to the human X-linked disorder incontinentia pigmenti (female mouse) | (Schmidt-Supprian et al., 2000; Smahi et al., 2000) |
| 2001 | NEMO | IKBKG | Candidate gene sequencing | Anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID) | XL male | Impaired CP responses to many inducers including IL-1β, IL-18, TNFα, or LPS. Normally developed T, B, and NK cells. | Reduced number of CD8+ but not CD4+ thymocytes. Complete absence of peripheral T cells (T linage conditional ko). | (Doffinger et al., 2001; Gerondakis et al., 2006) |
| 2002 | Caspase-8 | CASP8 | Candidate gene sequencing | Autoimmune lymphoproliferative syndrome with immunodeficiency | AR | Defective CD95/FASinduced apoptosis and TCR signaling. | Embryonic lethal. Lymphopenia and impaired responses to antigen (T cell specific casp8 deficiency). | (Chun et al., 2002; Salmena et al., 2003) |
| 2003 | IκBα | NFKBIA | Biochemistry | Anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID) | AD | Poor T cell responses to antigen stimuli, poor responses to TNFα and CD40L. | Similar EDA phenotype plus lacking lymph nodes, Peyer’s patches, splenic marginal zones, and germinal centers and exhibit significantly decreased responses to TLRs, TNF-α, LTβR, and BAFF. | (Courtois et al., 2003; Mooster et al., 2015) |
| 2009 | CARD9 | CARD9 | Linkage study | Chronic Mucocutaneous Candidiasis; Deep dermatophytosis | AR | Fungal infections, especially rare deep dermatophytoses, blunted GM-CSF responses, decreased Th17 cells. | Impaired NF-κB signals from Dectin-1 and Dectin-2 | (Blonska and Lin, 2011; Glocker et al., 2009; Lanternier et al., 2013) |
| 2010 | IKKα | IKBKA | Microarray | Severe Fetal Encasement Malformation | AR | Embryonic lethal. | Die at birth, skeletal and epidermal defects. | (Gerondakis et al., 2006; Lahtela et al., 2010) |
| 2012 | HOIL-1 | HOIL1 | NGS | Immunodeficiency, autoinflammation and amylopectinosis | AR | NEMO ubiquitination in response to TNF or IL-1. | Spontaneous amylopectin-like deposits in the myocardium, susceptibility to Listeria monocytogenes, Toxoplasma gondii, and Citrobacter rodentium infections, but not spontaneous autoinflammation | (Boisson et al., 2012; MacDuff et al., 2015) |
| 2012 | CARD14 | CARD14 | Linkage study | Psoriasis | AD | Increased expression and hyperactivation of NF-κB in endothelial cells. | N/A | (Fuchs-Telem et al., 2012; Jordan et al., 2012) |
| 2012 | CARD11 | CARD11 | RNA-Seq | Congenital B cell lymphocytosis | AD (GOF) | B cell lymphocytosis, hyper-responsiveness of B cells to BCR stimulation, CD40L, and BAFF, spontaneously aggregated CARD11 and NF-κB activation in T cells. | N/A | (Snow et al., 2012) |
| 2013 | CARD11 | CARD11 | NGS | Severe combined immunodeficiency (SCID) | AR (LOF) | Absent degradation of IκBα or phosphorylation of p65 in response to TCR or BCR stimulation. | Defective TCR, BCR, CD40 signaling, blockage of Treg development. | (Egawa et al., 2003; Greil et al., 2013; Hara et al., 2003; Stepensky et al., 2013) |
| 2013 | IKKβ | IKBKB | Linkage study | Severe combined immunodeficiency (SCID) | AR | Defective T and B cell activation/proliferation, reduced B cell response to CD40L. | Embryonic lethal, TNFainduced hepatocyte apoptosis, liver degeneration, lung deformation, skin inflammation. | (Pannicke et al., 2013; Pasparakis et al., 2002; Tanaka et al., 1999) |
| 2013 | P100/52 | NFKB2 | NGS | Common variable immune deficiency (CVID) with adrenal insufficiency | AD | Poor antibody responses, predominantly unswitched naive B cells in the periphery. Abrogated processing of p100 to p52 in patient cells before or after CD40-CD40L signaling. | Defective secondary lymphoid organ development, impaired B-cell development; enhanced DC function. | (Chen et al., 2013; Gerondakis et al., 2006) |
| 2013 | MALT1 | MALT1 | Linkage study + NGS | Combined immunodeficiency (CID) + inflammatory cell infiltration | AR | Impaired NF-κB responses to PMA + ionomycin, PHA, and anti-CD3 stimulation, severely decreased Treg counts. | Similar to human without Treg disfunction. | (Jabara et al., 2013; Ruefli-Brasse et al., 2003) |
| 2014 | BCL10 | BCL10 | NGS | Combined immunodeficiency (CID)+ autoimmunity | AR | Recurrent respiratory viral infections, and oral candidiasis. Normal lymphocyte counts with overwhelmingly naive phenotypes and undetectable Tregs. Impaired TLR signaling. | Defective B cell maturation, decreased marginal zone B cells, susceptibility to bloodborne bacteria. | (Torres et al., 2014; Xue et al., 2003) |
| 2014 | NIK | MAP3K14 | Linkage study + NGS | Combined immunodeficiency (CID) | AR | Defective maturation and functions of T, B, and NK cells. | Severe structural defects in lymph nodes, Peyer’s patches, splenic and thymic structures found in the latter | (Willmann et al., 2014; Yin et al., 2001) |
| 2015 | P105/50 | NFKB1 | NGS | Common variable immune deficiency (CVID) | Haploinsufficiency? | B cell dysfunctions. | No phenotype in heterozygous mouse. Homozygous mouse however showed B cell dysfunctions. | (Fliegauf et al., 2015; Sha et al., 1995) |
| 2015 | RelB | RELB | Linkage study + NGS | Combined immunodeficiency (CID) | AR | Increased p65 nuclear translocation and hyperactivation in response to TCR stimulation, decreased c-REL nuclear translocation after CD40 stimulation in B cells. | Inflammatory phenotype and hematopoietic abnormalities | (Gerondakis et al., 2006; Mericoa et al., 2015) |
| 2015 | HOIP | HOIP | Linkage study + NGS | Immunodeficiency, autoinflammation and amylopectinosis | AR | Absent response to CD40L, impaired responses to IL-1β and TNFα. | Embryonic lethal. | (Boisson et al., 2015; Peltzer et al., 2014) |
| 2016 | A20 | TNFAIP3 | NGS | Behcet like autoimmunity | Haploinsufficiency | Increased K63-ubiquitinated NEMO, increased phosphorylation of IκBα and IKKs, and prolonged nuclear residence of NF-κB. Increased pro IL-1β, NLRP3 and activated caspase-1 and increased production of mature IL-1β after LPS stimulation | Spontaneous NLRP3 inflammasome activity to LPS. | (Vande Walle et al., 2014; Zhou et al., 2016) |
| 2016 | OTULIN | OTULIN | NGS | Inflammatory syndromes | AR | Persisting IKK activity and IκBα phosphorylation in response to TNF stimulation stemming from a lack of NEMO deubiquitination | Embryonic lethal. | (Damgaard et al., 2016) |
AR: autosomal recessive; AD: autosomal dominant; XL: X-linked; GOF: gain-of-function; LOF: loss-of-function; NGS: next generation sequencing.
IκB regulatory complex
1. NEMO
NEMO/IKBKG mutations were the first genetic defects discovered in the NF-κB pathway and showed unique phenotypes caused by different mutations. Because IKBKG is on the X chromosome, disease was more severe in males. Complete loss-of-function (LOF), i.e. null or amorphic, mutations cause incontinentia pigmenti (IP) in females and prenatal lethality in males except when the latter show mosaicism or an XXY karyotype. Thus, it is an essential gene. Hypomorphic mutations cause a different skin disease, anhidrotic ectodermal dysplasia with Immunodeficiency (EDA-ID) in males, and mild IP in females. The severity of phenotypes dictates that most mutations are de novo.
Amorphic NEMO mutations
IP patients have skin inflammation combined with varying abnormalities of hair, nails, teeth, retinal vascularization, and the central nervous system. The disease was clinically recognized in the 1920s and genetically linked to Xq28 in the late 1980s (Wieacker et al., 1985). In 2000, only two years after it was cloned and mapped to the same region, NEMO was identified as the causal gene of IP (Smahi et al., 2000). All IP mutations are amorphic. A deletion of exons 4 through 10 of NEMO occurs in approximately 65% of IP patients. Only heterozygous female survive. The prominent skin lesions suggest a vital role of NEMO and NF-κB in skin cells. Due to X-inactivation/lyonization, female heterozygotes have half of their skin cells with normal NEMO expression and the other half deficient. The defective half die quickly after birth and leave behind damaged skin and skewed X-inactivation in the residual keratinocytes. Skewed X-inactivation in leukocytes also implies that NEMO is essential for their survival.
Hypomorphic NEMO mutations
Milder disease is caused by hypomorphic NEMO mutations manifesting as anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID) (Doffinger et al., 2001). EDA-ID studies also revealed a requirement for NF-κB in keratinocyte proliferation and differentiation. A paradox of NEMO deficiency is that the B, T, and NK subsets develop normally in most EDA-ID patients, but Ig production is often low and severe gram-positive cocci, gram-negative bacilli, or mycobacterial infections lead to high mortality at an early age. Studies in patient’s cells showed that defective NEMO impairs the CP response to many immune mediators including IL-1β, IL-18, TNF, or LPS (Doffinger et al., 2001; Hubeau et al., 2011). TNF signaling stimulates both apoptosis and activation pathways. Achieving the appropriate immune response requires the NF-κB induction of apoptosis inhibitory genes (Karin and Lin, 2002). Deficiencies of NEMO and other pathway components that disrupt NF-κB cause TNF-induced apoptosis to predominate which impairs immunity. Interestingly, NEMO-deficient patient cells also responded poorly to CD40 ligand (CD40L/CD154) that activates both CP and AP through multiple TRAFs possibly predisposing to mycobacterial infections. The NEMO-dependent CP rapidly activates NF-κB, as an immediate response to inflammatory stimuli. Conversely, the NEMO-independent AP acts slowly and continuously to promote cell maturation and differentiation. Thus, dendritic cells (DCs) from NEMO-deficient patients exhibited poor production of IL-12 in response to CD40L, while peripheral B cell differentiation dependent on CD40L was relatively normal (Filipe-Santos et al., 2006). Moreover, CD40L activates the MAPK pathway in a NEMO-and ubiquitination-dependent fashion, revealing NF-κB-independent functions for NEMO (Elgueta et al., 2009).
NEMO mutations found in EDA-ID are heterozygous, hypomorphic, and have varying phenotypes due to effects on different protein domains as well as non-coding regions involved in splicing and mRNA stability. There are two broad classes of disease: EDA-ID or immunodeficiency without EDA. Mutations causing decreased protein expression, folding, or stability, such as mutations in the 5′ untranslated region or exon 1B, usually cause immunodeficiency without EDA; the A288G mutation, which affects protein stability, leads to EDA-ID. By contrast, mutations that do not affect protein levels, but abolish one of the ubiquitin binding sites, lead to EDA-ID (e.g. D306N, D311G, D311N, C417R) (Hubeau et al., 2011). Thus, abnormal ubiquitination patterns may cause aberrant signaling complexes that affect keratinocyte differentiation in ways that go beyond a simple lack of NEMO protein.
2. IKKα/IKK1/IKBKA
The two enzymatic components of the IKK complex, IKKα and IKKβ, share 50% overall sequence identity and 65% kinase domain identity but cause drastically different human diseases, neither of which resembles NEMO deficiency (Hayden and Ghosh, 2008). IKKα deficiency leads to lethal fetal malformations, whereas IKKβ deficiency causes severe combined immunodeficiency (SCID) (Lahtela et al., 2010; Pannicke et al., 2013).
Autosomal recessive LOF mutations of IKKα lead to fetal encasement malformation (also called Cocoon Syndrome because of the appearance of the fetuses), the most severe and fatal phenotype of any NF-κB gene deficiency. IKKα deficiency was confirmed by gene expression profiling in aborted fetuses. The severe phenotype of IKKα deficiency is surprising because in vitro binding assays show IKKβ has higher affinity for NEMO than IKKα and IKKβ can form homodimers and bind to NEMO without IKKα. So, in theory, IKKα deficiency should impair the IKK complex less than IKKβ deficiency. It is surprising that IKKβ cannot compensate for the lack of IKKα. However, consistent with the in vitro assays, ikkα knockout mouse died soon after birth, a slightly milder phenotype than the embryonic lethal phenotype of the ikkβ knockout mouse (Gerondakis et al., 2006). Even more confounding, mice with an unactivatable mutant but not complete loss of ikkα are viable.
Interestingly, LOF mutations of receptor-interacting protein kinase 4 (RIP4) cause the same but less severe spectrum of craniofacial, skin, and limb deformities as IKKα LOF mutations, in a disease called Bartsocas-Papas syndrome or Popliteal Pterygium Syndrome (Kalay et al., 2012). RIP4 is an adaptor for upstream signaling cassettes to the IKK complex with the participation of TRAF proteins (Hayden and Ghosh, 2008). Among the seven human RIPs, each of which signals to distinct gene induction programs, RIP4 regulates keratinocyte differentiation and wound healing through NF-κB. RIP4 mutations produce proteins that are catalytically inactive and fail to elicit NF-κB (Kalay et al., 2012). Thus, IKKα mutations and RIP4 mutations likely affect NF-κB but the prominent skin manifestations no doubt also reflect effects on non-NF-κB pathways. For example, IKKα deficiency also potentially reveals NF-κB-independent functions. IKKα can translocate to the nucleus and phosphorylate histone H3. It also acts on chromatin independently of its kinase activity altering the expression of specific genes, for example 14-3-3δ, which influences the G2/M checkpoint. Also, both IKKα and IKKβ regulate β-catenin stability in the Wnt pathway. Moreover, nuclear IKKα induces cell cycle arrest and terminal differentiation by regulating Smad2/3 target genes (Liu et al., 2012).
3. IKKβ/IKK2/IKBKB
IKKβ-deficient patients appear normal at birth but develop SCID. This is strikingly different than knockout mice, which die as embryos due to liver degeneration, lung deformation, skin inflammation, and many other dysmorphic features. IKKβ deficient patients lack signs of developmental or gene regulatory defects in other organs (Gerondakis et al., 2006). This mouse-human discordance indicates a true difference in physiological roles because human fibroblasts and peripheral blood cells from patients had no detectable IKKβ protein equivalent to a mouse “knockout.” Curiously, patient cells also had reduced NEMO and IKKα protein levels despite normal mRNA levels (Mousallem et al., 2014; Pannicke et al., 2013), suggesting that IKKβ confers stability on all three proteins in the classical IKK complex in humans. This was not seen in ikkβ deficient mice (Tanaka et al., 1999).
IKKβ-deficient patients reveal the biological differences between IKKα and IKKβ. Normally, purified classical IKK contains NEMO:IKKα:IKKβ in a 2:1:1 stoichiometry, although an alternate IKK composed of NEMO and IKKβ homodimers occurs in T cells (Hayden and Ghosh, 2008). IKKα homodimers complexed with NEMO can phosphorylate IκB in vitro, although it is uncertain whether they exist in vivo. In IKKβ deficient cells, however, IKK incorporates IKKα homodimers which alters responses to certain stimuli. Phosphorylation and degradation of IκBα and IL-6 induction were absent to TLR5 stimulation by flagellin, drastically reduced in response to TNF stimulation, and only marginally affected in response to IL-1β (Pannicke et al., 2013). Thus, IKKα homodimers can mediate CP signaling for certain NF-κB-activating receptors, perhaps because IKKα associates more weakly with NEMO than IKKβ. Receptor selectivity has also been confirmed by IKKβ knockdowns in human fibroblasts. Hence, IKKα and IKKβ homodimers might serve different receptors. Moreover, the full response to TNF was reconstituted by restoring IKKβ in patient cells, confirming that IKKβ has unique functions not compensated by IKKα (Pannicke et al., 2013).
Although NF-κB was first discovered for κ light-chain expression, it has proved indispensable for many lymphocyte responses (Hayden and Ghosh, 2012). Patients with IKKβ deficiency had normal T and B cell development but the activation and proliferation of mature T and B cells was severely impaired, fostering a SCID phenotype (Mousallem et al., 2014). Normal activation and differentiation into effector or memory cells is blocked and mostly naive lymphocytes remain. The activation block can be partly overcome by stronger stimulation with phorbol myristate acetate (PMA), or phytohemagglutinin (PHA). Patient B cells also showed reduced responses to CD40L stimulation with IL-21.
4. IκB family (IκBα)/NFKBIA
IκBα (NFKBIA) is the prototype of the IκB family and remains the only one identified in a human monogenic disease (Hayden and Ghosh, 2008). All human IκBα mutations are heterozygous gain-of-function (GOF) mutations that augment NF-κB inhibition causing autosomal dominant disease that blends the clinical features of NEMO and IKKβ deficiency (Courtois et al., 2003; Schimke et al., 2013). As in hypomorphic NEMO mutations, EDA is observed. Like IKKβ deficiency, peripheral T cells respond poorly to antigen(Courtois et al., 2003). TNF or CD40L evoke little nuclear translocation of NF-κB. Thus, the GOF mutations in IκBα have a dominant negative effect. A mouse model was generated recently to mimic the heterozygous human S32I IκBα mutation. Heterozygous mice develop EDA comparable to human patients, but also lack lymph nodes, Peyer’s patches, splenic marginal zones, and germinal centers and exhibit significantly decreased responses to TLRs, TNF, LTβR, and BAFF. Thus, mice, unlike humans, show impairment of both the CP and AP (Mooster et al., 2015). S
Most dominant interfering NFKBIA mutations are either substitutions affecting the phosphorylation of residues Ser32 and Ser36 or N-terminal protein truncations removing these serines. The mutant IκBα proteins are expressed but cannot undergo phosphorylation-driven degradation. Disregarding induction signals, they remain constitutively bound to NF-κB and freeze the pathway. This has been elegantly verified by overexpressing mutant IκBα proteins encoded by patient alleles and showing that they override the wildtype protein and block NF-κB-induced gene expression (Courtois et al., 2003). Certain truncation mutations have variable blocking effects perhaps due to differential protein stability or the efficiency of utilizing alternative ATG start sites downstream of premature mutant stop codons. For example, a very early truncating mutation, E14X, showed strong dominant inhibition due to a very efficient internal ATG that facilitated high mutant protein expression (Lopez-Granados et al., 2008). However, another heterozygous early truncating mutation, W11X, showed no dominant negative effect. The IκBα protein from the wild-type allele was normally phosphorylated and degraded so that there was only haploinsufficiency of NF-κB resulting in a milder disease phenotype (McDonald et al., 2007). Further studies might reveal how the kinetics of IκBα degradation, resynthesis, and accumulation of protein from the WT allele is affected by the presence of a mutant protein. Recently, a homozygous IκBNS/NFKBID mutation has been found in a patient with combined immunodeficiency, and marked B and CD8+ T cell lymphopenia (Vanessa Bryant, unpublished data). There are no disease associations yet for IκBβ or IκBε.
5. Rel homology domain protein family
Mutations in three of the five RHD protein genes: p50 (NFKB1), p65 (RELA), and c-Rel (REL), have been chiefly associated with humoral immunodeficiency. The p52 and RelB components of the AP will be discussed further below. Heterozygous NFKB1 (p105/p50) gene mutations cause common variable immunodeficiency (CVID) (Fliegauf et al., 2015). CVID is milder than SCID, prevails in older patients, and manifests as defective antibody responses. Patients with NFKB1 mutations experience recurrent respiratory infections, hypogammaglobulinemia with low IgG, IgM, and IgA (reflecting deficient Ig isotype class-switching), Epstein-Barr virus (EBV)-associated lymphoproliferation, low peripheral B cell counts and, occasionally, other symptoms such as granulomatous lung disease, neutropenia, and Idiopathic thrombocytopenic purpura (ITP). The disease has variable expressivity, a feature commonly observed in haploinsufficiency conditions. For example, the father of a young woman with severe respiratory infections and EBV-lymphoproliferative disease had the same mutation but only mild respiratory symptoms not requiring medical attention (Boztug et al., 2016). In large kindreds, heterozygous individuals exhibited widely varying antibody responses, infection, lymphoma, or inflammatory bowel diseases (Fliegauf et al., 2015). In haploinsufficient immunological diseases, whether this variation in clinical phenotype is due to independent genetic variants or environmental influences remains a fascinating question.
NFKB1 mutations cause partial reduction, i.e. haploinsufficiency, of both p105 and p50 in the resting state and severely reduced p105 phosphorylation after stimulation (Boztug et al., 2016). Several multiplex kindreds with splicing or frameshift mutations in NFKB1 caused haploinsufficiency in a dominant inheritance pattern due to reduced levels of p105 and p50 protein indicating that the WT allele cannot ramp up expression to compensate for the mutant allele. In other patients, whose heterozygous mutation generated a truncated unstable protein, nuclear translocation of NF-κB was impaired by an unknown dominant negative mechanism (Fliegauf et al., 2015). Considering that varied RHD dimers exist in lymphocytes, changes in p105 or p50 levels might alter the balance of other dimers causing surprising outcomes. For example, in the family mentioned above, the father with less severe disease had substantially greater levels of p50 and p105 protein than the daughter for undiscovered reasons (Boztug et al., 2016). A new study of Finnish families with heterozygous LOF mutations in NFKB1 revealed variants affecting protein stability, subunit phosphorylation, or nuclear translocation and causing a wide range of immune disorders (Seppanen Mikko, unpublished data).
Completely different clinical phenotypes are found with genetic alterations of the different RHD proteins. A heterozygous de-novo nonsense mutation in RELA/p65 is associated with lymphadenopathy and idiopathic thrombocytopenic purpura. The mutation results in roughly 1/2 of the normal protein expression, but how haploinsufficiency leads to abnormal lymphocyte homeostasis and autoimmunity is still under investigation (Comrie and Lenardo, unpublished observations).
6. CARD-BCL10-MALT1 signalosome (MALT1, BCL10, CARD9, CARD11, and CARD14)
The CBM complex is affected by many genetic defects causing immune dysregulation and malignancy. Interestingly, BCL10 and MALT1 deficiencies are similar since they generally work together, whereas the clinical disorders caused by deficiencies with CARD9, 11, and 14 are different because each plays a different role in immunity.
MALT1
MALT1 (MALT lymphoma translocation protein 1) was first discovered in mucosa-associated lymphoid tissue (MALT) lymphoma in 1999 (Dierlamm et al., 1999), in which a chromosome translocation spawns a cIAP2-MALT1 fusion gene that constitutively activates NF-κB. Members of 3 families with LOF MALT1 mutations show CID, enteropathy, and severe eczema together with dysmorphic features. Most patients have hypogammaglobulinemia but paradoxically increased levels of IgE, termed hyper-IgE. T regulatory cell (Treg) counts were decreased causing autoimmune features. Patient cells showed impaired NF-κB responses to PMA + ionomycin, PHA, and anti-CD3 stimulation marked by decreased BCL10 cleavage and IκBα degradation leading to immunodeficiency (Charbit-Henrion et al., 2016; Jabara et al., 2013). How specific gene expression changes cause the particular confluence of immunodeficiency and autoimmunity has not been explained.
MALT1 harbors a death domain (DD) and is distantly homologous to caspases in its C-terminal domain which confers caspase-like proteolytic function. Its substrates include BCL10, NIK, RELB, CYLD and A20, all of which are key NF-κB regulators. MALT1 enzymatic activation is induced by dimerization and substrate engagement, rather than processing as a zymogen like conventional caspases. Caspase function both promotes and inhibits specific NF-κB functions (Hachmann and Salvesen, 2016). Human mutations are found throughout the CARD, paracaspase, and Ig-like domains and they are all LOF (Charbit-Henrion et al., 2016; Jabara et al., 2013). These defects resemble the phenotypes found in the malt1 knockout mouse (Ruefli-Brasse et al., 2003). However, 5 out of 6 MALT1-deficient patients also had inflammatory disorders especially enteropathy (Charbit-Henrion et al., 2016). Inflammation is absent from malt1 knockout mice but emerged in genetically engineered paracaspase-dead mice indicating enzyme deficiency is involved (Yu et al., 2015).
BCL10
BCL10 deficiency, identified in one patient, resembles MALT1 deficiency (Torres et al., 2014). The consanguineous patient exhibited CID and autoimmunity with gastroenteritis, recurrent respiratory viral infections, and oral candidiasis. Lymphocyte counts were normal but exhibited overwhelmingly naive phenotypes and undetectable Tregs, implying disturbed CP activation. Patient fibroblasts showed impaired NF-κB responses to LPS (TLR4), zymosan (TLR2/6), and poly(I:C) (TLR3), marked by decreased nuclear translocation and deficient IL-6 and IL-8 production, hence innate immunity also failed. Interestingly, patient mononuclear cells showed normal innate responses which was a surprising difference in wiring between humans and mice because bcl10 knockout mice display impaired NF-κB in macrophages and DCs following innate stimuli.
CARD9
CARD9 deficiency illustrates how NF-κB connects different receptors and adaptors to specific cellular responses (Glocker et al., 2009). Homozygous nonsense (LOF) mutations in CARD9 were first found in a large consanguineous family and caused recurrent fungal infections especially in the brain. More recently, a large variety of patients with CARD9 deficiencies have been reported and they consistently show severe fungal infections especially rare deep dermatophytoses and central nerve system candidiasis. The patients carried LOF mutations such as stop-gain nonsense codons as well as missense mutations, although not all missense mutations have been tested rigorously for functional effects. Typically disease onset was in childhood, however, some patients presented in adulthood illustrating that there might be modifying genes or environmental influences involved (Lanternier et al., 2015).
A molecular understanding of the human phenotypes emerges from mouse knockout studies showing that CARD9 transduces NF-κB signals from Dectin-1 and Dectin-2, two receptors that recognize Candida albicans in unicellular and hyphal forms, respectively. If either receptor is activated, signaling through the SYK kinase leads to sequential assembly of the CARD9 CBM complex and downstream NF-κB signaling (Blonska and Lin, 2011). Cells from a patient with a p.Y92H variant in the CARD domain showed impaired phosphorylation of IκBα after stimulation with zymosan, a Dectin-1 inducer. However, when mutant and wildtype CARD9 proteins were co-expressed with BCL10 and MALT1, they both assembled CBM complexes. Therefore, it is still controversial whether the NF-κB signaling defect in the CARD9 deficient patient cells is CBM-dependent or not. Moreover, CARD9-deficient cells showed blunted GMCSF responses mediated by the ERK signaling pathway, reminiscent of defects in neutrophil killing and recruitment defects reported in CARD9 deficient patients (Gavino et al., 2016). CARD9 deficient patients also have decreased Th17 T cells, which are crucial for controlling mucosal, but not systemic candidiasis (Glocker et al., 2009).
CARD11/CARMA1
In 2006, CARD11, together with MALT1, were found to be targeted by somatic mutations that induce constitutive activation of NF-κB and survival signals for a particular subtype of human Diffuse Large B Cell Lymphoma (DLBCL) (Ngo et al., 2006) (see below). CARD11 missense mutations are found in 9.6% of DLBCL tumor biopsies. Introducing the mutant CARD11 into lymphoma cell lines constitutively induces NF-κB (Lenz et al., 2008) (see below). Germline CARD11 mutations can cause very different disease phenotypes. GOF mutations cause BENTA disease (B-cell expansion with NF-κB and T-cell anergy), whereas LOF mutations cause SCID. Families with hypomorphic, dominant negative CARD11 mutations also have significant atopic disease, including atopic dermatitis, allergy, high IgE and eosinophils, asthma, and immunodeficiency (Milner, J, Snow A, and Gelfand E. personal communication). Each disease provides a window into CBM signalosome function and potential therapeutic strategies (Greil et al., 2013; Snow et al., 2012).
The discovery of BENTA disease fits with the idea that CARD11 was a bona fide oncogene in DLBCL. Four different autosomal dominant GOF germline mutations of CARD11, G116S, E127G, G123D, and C49Y, have been discovered in 4 different families (Buchbinder et al., 2015; Snow et al., 2012). Three affect the CC domain and one variant, C49Y, is located in the N-terminal CARD. The mutant proteins aggregate, colocalize with MALT1, activate IKK, increase nuclear localization of NF-κB and transcription (Buchbinder et al., 2015; Snow et al., 2012). The germline mutations in the CC and CARD domains recapitulated or were identical to the somatic DLBCL mutations (Lenz et al., 2008). The germline GOF mutations resulted in polyclonal, sometimes even monoclonal, B cell lymphocytosis, expansion of transitional B cells, and hyper-responsiveness of B cells to BCR stimulation, CD40L, and BAFF, all through NF-κB. Similar to B cells, T cells from BENTA patients also showed spontaneously aggregated CARD11 and NF-κB activation. However, peripheral T cells were not expanded and poorly responsive to TCR stimulation, likely due to anergy mechanisms (Snow et al., 2012). Although BENTA patients did not develop T cell lymphocytosis and T cell lymphoma, GOF mutations of CARD11 have been detected in adult T-cell leukemia/lymphoma (ATLL) and in a form of cutaneous T-cell lymphoma (CTCL) called Sezary syndrome (Juilland and Thome, 2016). Also, one BENTA patient developed B cell chronic lymphocytic leukemia (Snow et al., 2012). The somatic and germline GOF mutations appear to be driver mutations indicating that CARD11 may be a promising therapeutic target for BENTA plus other lymphoid malignancies.
However, the discovery of CARD11 LOF mutations tells a different story. Patients with homozygous recessive LOF CARD11 mutations display severe immunodeficiency with predominantly naive T and B lymphocytes and a block in B cell maturation. Antibody production by B cells and cytokine production and proliferation by T cells are severely impaired (Greil et al., 2013; Stepensky et al., 2013). Patient lymphocytes showed essentially no degradation of IκBα or p65 phosphorylation in response to TCR or BCR stimulation (Stepensky et al., 2013).
Overall, the clinical and biochemical features of MALT1, BCL10, and CARD11 LOF mutations are similar, testimony to synergistic functions in the CBM. One characteristic feature of SCID caused by CBM deficiency is that the patients can have normal peripheral T and B cell counts, yet a predominantly naive lymphocyte phenotype indicative of failed TCR and BCR responses. Thus, there appear to be different requirements for NF-κB signaling in lymphocyte ontogeny and mature lymphocytes. The recent report of somatic reversion in a CARD11 LOF mutation patient, who presented as Omenn syndrome, underscored this point. In this patient, a second-site somatic reversion changed the original germline mutation (p.C150*) to a missense mutation (p.C150L), which partly restored CARD11 function. The somatic mutation appeared only in a subclone of T cells indicating a survival and expansion advantage conferred in a founder cell by reinstating NF-κB signaling. However, the expansion of a single T-cell clone perturbed immune homeostasis and led to massive T cell infiltration into the skin causing progressive eczema and erythroderma, accompanied by lymphadenopathy and hepatosplenomegaly. Furthermore, the oligoclonal T cells did not provide sufficiently broad immune protection against uncontrolled CMV infections and multiple bouts of sepsis caused by Staphylococcus aureus, Enterococcus, and Pseudomonas that eventually proved fatal to the child at 16 months of age (Fuchs et al., 2015).
CARD14
CARD14, a less well-studied member of the CARD-family, nucleates an alternative CBM complex and is the apparent causative gene in two related diseases: psoriasis 2 (PSORS2) and Familial Pityriasis Rubra Pilaris (PRP), a psoriasis variant that chiefly affects hair follicles (Fuchs-Telem et al., 2012; Jordan et al., 2012). Dominant GOF mutations lead to psoriasis with or without arthritis. GOF mutations in CARD14 can also lead to generalized pustular psoriasis (Sugiura, 2014). A coding SNP in CARD14 (R820W) has been associated with sporadic (non-congenital) psoriasis in a meta-genome-wide association study (GWAS) (Tsoi et al., 2012).
The discovery of heterozygous CARD14 mutations selectively in psoriatic diseases revealed an unexpected function of CARD14 in keratinocytes and innate immunity. CARD14 was thought to mediate NF-κB signaling like other CARD proteins. However, the specific skin manifestations are due to keratinocyte and not immune cell expression. More intriguingly, the CARD14 mutations increase expression in skin causing NF-κB hyperactivation in skin cells. Some patient mutations displayed autonomous activation in luciferase assays, whereas others showed increased TNF responses. However, CARD14 increases and NF-κB hyperactivation have also been observed in psoriasis without CARD14 mutations (Harden et al., 2015). Thus, these features may be a cause or consequence of psoriatic skin lesions or both.
7. LUBAC complex and its regulators (HOIL1, HOIP, A20, OTULIN, and CYLD)
Ubiquitination is a crucial post-translational protein modification governing NF-κB pathways [8, 113]. Many pathway components, including NEMO, IκBs, p100, and adaptors such as MALT1 and BCL10 rely on specific types of regulatory and degradative ubiquitination (Iwai, 2014). Specifically, non-degradative or “regulatory” ubiquitination is the attachment of polyubiquitin (joined at K63 residues) to signaling proteins. As mentioned above, this provides the reversible “glue” that facilitates the multiprotein complexes obligatory for NF-κB activation. Certain ubiquitin reactions are facilitated by TRAF proteins. The CP may also involve linear ubiquitin modifications catalyzed by the “linear ubiquitin assembly complex” (LUBAC) (Sasaki and Iwai, 2015). The enzyme holocomplex is composed of three key proteins. HOIL-1 interacting protein (HOIP) is the E3 ligase. Haem-oxidized IRP2 ubiquitin ligase-1 (HOIL1) and SHARPIN (SHANK-associated RH domain interacting protein) complete the active LUBAC complex that generates Methionine 1-linked linear polyubiquitin chains in the presence of several E2s to specifically during CP activation (Tokunaga, 2013). NEMO is a LUBAC target but there may be multiple, perhaps overlapping, ubiquitination processes in NF-κB activation (Sasaki and Iwai, 2015). Negative feedback for NF-κB activation is arbitrated by several deubiquitinases, including CYLD (CYLD lysine 63 deubiquitinase), OTULIN (OTU deubiquitinase with linear linkage specificity), and A20/TNFAIP3 (TNF alpha induced protein 3) (Tokunaga, 2013). Except for SHARPIN, all the other proteins in the LUBAC complex and its regulators are associated with monogenic diseases.
HOIL1
HOIL1/RBCK1 deficiency was found in three patients from two unrelated families with immunodeficiency, autoinflammation and amylopectinosis, a glycogen storage disease that causes liver failure. CP impairment accounted for the invasive pyogenic bacterial infections, but did not explain the autoinflammation and amylopectinosis. The mutations were autosomal recessive LOF leading to no detectable HOIL1 protein and decreased LUBAC complex stability. NEMO ubiquitination in response to TNF or IL-1 stimulation was defective. Consequently, less NEMO was recruited to the RIP1 or IRAK-1 adaptor proteins in cytokine receptor signaling complexes, IKK was underphosphorylated, and IκB degradation was delayed. HOIL1 overexpression in patient cells rescued the protein levels of HOIP and SHARPIN and generated a durable LUBAC complex which improved signaling (Boisson et al., 2012).
The mechanisms of autoinflammation and amylopectinosis in HOIL1 deficient patients were further investigated using Hoil1 knockout mice. These showed spontaneous amylopectin-like deposits in the myocardium, susceptibility to Listeria monocytogenes, Toxoplasma gondii, and Citrobacter rodentium infections, but not spontaneous autoinflammation (MacDuff et al., 2015). Immunodeficiency associated with this molecule affects innate immunity, because Hoil1 deficiency in Rag1-deficient mice with no lymphocytes still created a susceptibility to Listeria infection. Surprisingly, hoil1−/− mice displayed enhanced control of MHV68 and Mycobacterium tuberculosis infections compared to wildtype mice. Both serum IL-6 and TNF levels were increased in infected mice, which was also evident in HOIL1-deficient patients thereby providing insight into the autoinflammatory disorder. How defects in the LUBAC complex lead to dramatically variable susceptibilities to different pathogens is unknown. The animal models showed that chronic viral infections in the hoil1−/− mouse protected them from Listeria infection, possibly due to the increased production of the cytokines IL-6, TNF, and IL-12, which enhanced the ability of macrophages to kill Listeria (MacDuff et al., 2015). Therefore, HOIL1 deficiency in mice and humans revealed a homeostatic balance between inflammation and immune protection.
HOIP
A single case of autosomal recessive deficiency of HOIP has been reported, which provides unique information regarding the biological functions of HOIP since the hoip−/− mouse is embryonic lethal (Boisson et al., 2015; Peltzer et al., 2014). The HOIP patient has a homozygous missense mutation, which severely impaired LUBAC stability. Consequently, this patient phenocopies the HOIL-1 deficient patients with susceptibility to infections, amylopectinosis, and autoinflammation.
The biochemical defects in the HOIP deficient patient were different from the HOIL-1 deficient patients, although it might be difficult to draw conclusions based on a single patient with a missense mutation. The HOIP deficient patient showed severely decreased linear ubiquitination, decreased phosphorylation of IKK, and slightly delayed IκB degradation in response to IL-1β and TNF. However, when stimulated with CD40L, the patient B cells showed no NF-κB signaling, indicated by a failure of IKK phosphorylation and IκBα degradation. As a result, the patient’s B cells failed to induce CD27 or CD38 when activated with CD40L/IL-21. Interestingly, the defects in the patient B cells resembled CD40-deficient patients, suggesting that the CD40-CD40L signaling, at least in the CP, requires LUBAC. Although both IL-1β and TNF and induced NF-κB signaling were impaired in HOIP deficient cells, the defect in IL-1β was more severe than TNF, a phenomenon also observed in HOIL-1 deficient cells (Boisson et al., 2015).
A20/TNFAIP3
A20/TNFAIP3 is a ubiquitin removal enzyme regulating NF-κB [123–125]. It contains a C-terminal OTU (Ovarian Tumor) domain, which mediates its de-ubiquitylating (DUB) activity and seven zinc fingers (ZFs) that harbor its E3 ubiquitin binding and cleavage activities. A20 expression is induced by NF-κB and then decreases NF-κB signaling as a negative feedback loop. A20 reverses multiple NF-κB signaling pathways by cleaving the K63-linked polyubiquitin chains on activated RIP1, RIP2, TRAF6, TBK1, and NEMO. A20 also inhibits ubiquitin chain synthesis by blocking the binding of E2 and E3 proteins (Ma and Malynn, 2012). A20 also negatively regulates Nlrp3 inflammasome signaling by suppressing Nlrp3, proIL-1b, and proIL-18 production (Duong et al., 2015; Vande Walle et al., 2014). Thus, A20 connects NF-κB regulation to a broader spectrum of inflammation functions.
Nucleotide variants in A20 are associated in GWAS with multiple autoimmune diseases, including systemic lupus erythematosus (SLE), rheumatoid arthritis, psoriasis, type 1 diabetes, coeliac disease, Crohn’s disease, coronary artery disease in type 2 diabetes, and systemic sclerosis (Ma and Malynn, 2012). A20 mutations were identified in a monogenic syndrome that resembled Behcet’s disease, an autoinflammatory disease involving mucosal ulcers and eye inflammation (Zhou et al., 2016). All of the mutations were heterozygous and either frameshift or nonsense, which led to haploinsufficient A20 expression. After TNF stimulation, patient lymphocytes and fibroblasts showed increased K63-ubiquitinated NEMO, increased phosphorylation of IκBα and IKKs, and prolonged nuclear NF-κB. The patients had elevated serum TNF, IP-10, IL-17, IL-9, IFN, and IL-6. Similar to A20-deficient mice, the patients also had greater levels of pro IL-1β, NLRP3 and activated caspase-1 and increased production of mature IL-1β after LPS stimulation. Treatment with an IL-1 inhibitor, Anakinra, reversed systemic inflammation (Zhou et al., 2016). Additional patients with heterozygous missense mutations were discovered shortly after the first report, which raised the possibility that the mutant protein was dominant negative but this is not yet verified (Ohnishi et al., 2016; Shigemura et al., 2016).
OTULIN
OTULIN is another deubiquitinase that cleaves the N-terminal methionine-linked polyubiquitination chains generated by LUBAC (Keusekotten et al., 2013). Four families with homozygous mutations of OTULIN had autosomal recessive inflammatory disease with recurrent nodular panniculitis with fever and neutrophil infiltration, lipodystrophy, diarrhea, joint swelling, and failure to thrive. The mutations severely decreased protein levels and deubiquitinase activity. Patient cells showed persisting IKK activity and IκBα phosphorylation in response to TNF stemming from a lack of NEMO deubiquitination. Patients had increased cytokine profiles resembling the A20 deficient patients. Interestingly, conditional deletion of otulin in myeloid cells, but not T or B cells, causes systemic inflammation, neutrophil infiltration, and multi-organ inflammation in mouse models. Treatment with anti-TNF neutralizing antibodies ameliorated inflammation in patients and rescues mouse phenotypes, indicating that TNF is a key driver of the systemic disease (Damgaard et al., 2016). Considering the similar biochemical defects and cytokine profiles in A20 deficient and OTULIN deficient patients, TNF or IL-1 blockers may be an effective treatment.
CYLD
CYLD (cylindromatosis protein) was first mapped to chromosome 16q12-13 in 1995 by the familial skin cylidromatosis phenotype (Biggs et al., 1995). Later, a large cohort of cylidromatosis patients helped to identify CYLD as a tumor suppressor gene and NF-κB regulator (Bignell et al., 2000). CYLD contains three cytoskeletal-associated protein-glycine-conserved (CAP-GLY) domains that function as a deubiquitinating enzyme that targets key molecules such as NEMO. Moreover, CYLD contains a USP catalytic domain that acts on ubiquitin at specific lysine residues, and a molecular triumvirate of Cys601-His871-Asp889 in the α1 helical subdomain that mounts a nucleophilic attack on K63 residues in polyubiquitin.
The major biological function revealed by CYLD-deficient patients is the control of cell growth as indicated by profound cylindromatosis (Brooke-Spiegler syndrome), a benign tumor of the sweat glands around the head, face, and neck (Mathis et al., 2015). CYLD mutations are also associated with another skin disease, multiple familial trichoepithelioma, type 1 (MFT1), characterized by numerous firm skin papules that are trichoepitheliomas (follicular tumors) (Farkas et al., 2016). Why so many different CYLD mutations produce tumors of skin origin is not known. CYLD was first found to deubiquitinate NEMO, which inhibits the CP (Kovalenko et al., 2003). CYLD has been proposed to shape both adaptive and innate immunity by regulating TCR, BCR, and TLR signaling (Mathis et al., 2015). However, CYLD deficiency yields very different phenotypes compared to insufficiency of A20 and OTULIN even though both CYLD and OTULIN can operate together to regulate the LUBAC complex.
The difference may lie in the ability of CYLD to govern cell proliferation, which may not relate to its NF-κB regulatory function. For example, CYLD deficiency may cause overexpression of the c-myb oncogene as a feature of cylindromas (Corda and Sala, 2016). However, in addition to NEMO, activated CYLD can remove the lysine 63-linked polyubiquitin chains of BCL-3 (an atypical IκB), thus preventing BCL-3 translocation into the nucleus together with p52 and p50, which may also promote cell proliferation. Keratinocytes defective in CYLD showed increased nuclear translocation of BCL-3, p52, and p50 after chemical or UV treatment (Massoumi et al., 2006). Therefore, CYLD not only regulates the activation of canonical NF-κB in conditions of infection and inflammation, it also suppresses cell proliferation under stress conditions.
8. Caspase-8 (CASP8)
CASP8 deficiency revealed a surprising role for this enzyme in the NF-κB pathway. This was only verified when conditional knockouts in lymphocytes were made since the germline deficiency in mice is embryonic lethal (Lemmers et al., 2007). CASP8 belongs to the cysteine-aspartic acid protease (caspase) family and has a well-established role in the apoptosis signaling cascade from death receptors such as Fas (Price et al., 2014). However, rare patients with autosomal recessive disease were found to have homozygous LOF CASP8 mutations (Chun et al., 2002). The disease involved lymphadenopathy, splenomegaly, and defective CD95/FAS-induced apoptosis of peripheral blood lymphocytes. In addition, unlike related patients with the autoimmune lymphoproliferative syndrome, the disease also included immunodeficiency with recurrent sinopulmonary and herpes simplex virus (HSV) infections. T, B, and NK cells from patients responded poorly to stimuli through TCR, BCR, and Fc receptors (Niemela et al., 2015).
The immunodeficiency observed in the patients led to the discovery of CASP8 as a facilitatory molecule in the NF-κB pathway. During antigen receptor signaling, CASP8 is indispensable for full NF-κB induction and gene transcription by linking the IKK complex to IκBα in preparation for the phosphotransfer reaction. Consistently, CASP8-deficient patient cells showed severely impaired NF-κB nuclear translocation in responses to immune stimuli, however normal responses to CD40L stimulation (Su et al., 2005). Two more patients found recently confirmed the immunodeficiency but also broadened the phenotype with late-onset multi-organ lymphocytic infiltration with granulomas (Niemela et al., 2015). Why does CASP8 LOF gives a live human phenotype, whereas in mice it is embryonically lethal key? This is likely due to the highly homologous Caspase-10 gene that can function during embryogenesis in humans which has been lost in evolution in the mouse genome (Su and Lenardo, 2008).
Alternative pathway
9. NIK
Two NIK/MAP3K14 deficient patients were identified in one consanguineous family using next generation sequencing (NGS). The patients suffered recurrent and severe bacterial, viral and Cryptosporidium infections. Immune phenotyping showed multiple lymphocyte defects, including B cell lymphopenia, reduced class-switched memory B cells, impaired B cell survival, impaired ICOSL expression, defects in follicular helper T cells (Tfh) and memory T cells, and malfunctioning NK cells. This indicates that the triggering kinase for the AP greatly influences key lymphocyte functions.
The LOF mutation found in these patients abolished kinase activity against IKKα, (Willmann et al., 2014). The phenotype of patients was different from the NIK knockout mouse because it lacked the severe structural defects in lymph nodes, Peyer’s patches, splenic and thymic structures found in the latter (Yin et al., 2001). Nonetheless, bone marrow chimeras and conditional lymphocyte knockout mouse models recapitulated the patient phenotype, including decreased memory T cells, disrupted B cell maturation and activation, and defective DCs and NK cells (Brightbill et al., 2015; Katakam et al., 2015; Noma et al., 2015; Rowe et al., 2013). These results imply that NIK action in non-hematopoietic tissues is crucially different in humans and mice.
10. NFKB2/p100/p52
Heterozygous NFKB2/p100/p52 mutations have been identified in an autosomal-dominant type of CVID, complicated with autoimmunity and endocrine deficiency. The patients exhibited recurrent respiratory infections, hypogammaglobulinemia, alopecia areata, an autoimmune infiltration of the scalp that causes hair loss, and, frequently, adrenal insufficiency. They manifest poor antibody responses, predominantly unswitched naive B cells in the periphery, and some reports of T and NK cell activation defects (Chen et al., 2013; Lee et al., 2014; Lougaris et al., 2015). In resting lymphocytes, the NFKB2 protein predominantly exists as the p100 precursor form. Upstream signaling, especially TRAF3, activates NIK and IKKα dimers, which in turn phosphorylate the two regulatory serines in the NRD domain (S866 and S870). Consequently, the canonical acceptor site (K856) is ubiquitinated, which leads to the limited processing of p100 to p52 in the proteasome (Sun, 2012). So it is no surprise that all the mutations found so far (p.R853*, p.R853Afs*29, p.K855Sfs*7, p.D865G, p.D865Vfs*17, p.A867V, p.A867Cfs*19) affect the NRD domain and either substitute amino acids adjacent to S866, S870, K856, or remove the C-terminus that harbors these residues. These mutations abrogate the processing of p100 to p52 before or after CD40-CD40L signaling (Chen et al., 2013; Lee et al., 2014). The D865G mutation has a dominant effect over the wildtype protein. It interferes with the CP by blocking TNFα-induced NF-κB nuclear translocation (Lee et al., 2014). Unprocessed p100 can function as a CP inhibitor by excessive dimerization with p65 (Basak et al., 2007), which might explain how NFKB2 mutations cause effects similar to NFKB1 deficiency in terms of immune abnormalities. However, individuals carrying NFKB2 N-terminus mutations acting in a GOF manner present with a more severe CID phenotype (Hyesun Kuehn and Sergio Rosenzweig, unpublished data). Further investigation of these mutations may elucidate the dominant inheritance of the disease as well as the pathogenesis of the adrenal disorder.
11. RELB
Autosomal recessive RELB deficiency has been discovered in three patients from the same consanguineous family. The patients presented as CID from infancy, with recurrent infections, severe autoimmune skin diseases, and failure to thrive (Mericoa et al., 2015; Sharfe et al., 2015). The homozygous, LOF mutations led to absence of RELB expression but normal levels of other NF-κB/Rel proteins. Interestingly, the patient’s T cells exhibited increased p65 nuclear translocation and hyperactivation in response to TCR stimulation, whereas B cells showed decreased c-Rel nuclear translocation after CD40 stimulation (Mericoa et al., 2015). This implies a cell-specific homeostatic balance between the CP and AP. Moreover, the T and B cell defects in RELB deficient patients were not observed in relb knockout mice which had inflammatory and hematopoietic abnormalities (Gerondakis et al., 2006; Sun, 2012).
12. TRAFs and specific NF-κB pathways
The TRAF (Tumor necrosis factor receptor associated factor) family proteins connect receptors to both the CP and AP, as well as multiple other pathways (Hayden and Ghosh, 2008). There are seven TRAFs, which link various receptors to downstream pathways to induce specific signals. The six typical TRAF members (TRAF1-6) share similar functional domains, including a C-terminal TRAF domain that mediates homodimerization and interactions with other proteins, a zinc finger domain, and a RING finger domain with E3 ubiquitinase ligase activities (except for TRAF1). TRAFs 2,3,5, and 6 have E3 ubiquitinase ligase activity.
TRAFs are well-studied signaling adapters for the TNF receptors (TNFRs). They either directly bind the TRAF-interacting motif in the cytoplasmic domain of the TNFRs, or bind indirectly via other adaptor proteins like TRADD (TNFR1-associated death domain protein), which tethers them to specific TNFRs with cytoplasmic death domains. Upon binding to the receptors, TRAFs recruit and activate downstream signaling molecules such as TAK1 and IKK. TRAFs also have ubiquitinase activity. This function is particularly important in the AP to control the level of NIK. In cells prior to activation signals, TRAF induces ubiquitination of NIK by forming a complex with the E3 ligase cIAP. Activation leads to the degradation of TRAFs, which fosters NIK accumulation and AP activation.
The two TRAFs associated with monogenic diseases illustrate the biological functions of the entire pathway. Mutations affecting components in the same pathway produce similar phenotypes in patients. Mutations in the TLR3-TRIF-TRAF3-TBK1 pathway cause HSV encephalitis whereas those in the EDA-EDAR-EDARADD-TRAF6 pathway lead to EDA (also known as Hypohidrotic ectodermal dysplasia, HED) (Trzeciak and Koczorowski, 2016), which resembles hypomorphic NEMO mutations.
TRAF3 and the TLR3 signaling
TRAF3 (Tumor necrosis factor receptor associated factor 3)
TRIF/TICAM1 (toll like receptor adaptor molecule 1)
TLR3 (Toll like receptor 3)
TBK1 (TANK binding kinase 1)
Mutations in TLR3, TRIF, TRAF3, and TBK1 cause patients to be highly susceptible to HSV (Herpes simplex virus) encephalitis, despite robust health otherwise. This is an astonishingly specific presentation considering the broad function of the pathway (Guo et al., 2011; Herman et al., 2012; Perez de Diego et al., 2010; Sancho-Shimizu et al., 2011). TLR3 recognizes dsDNA in a broad range of tissues and cell types. When activated by dsDNA, TLR3 is phosphorylated and forms dimers that bind to TRIF to trigger the downstream signals. The TLR3-TRIF complex recruits TRAF3, TBK1, and IKKε, which activates IRF3. The TLR3-TRIF complex also recruits RIP1, TAB2, and TAK1 and then phosphorylates IKK to activate the CP. In fact, the TLR3-TRIF complex supplies a common node for several signaling pathways, which together mediate essential antiviral responses in the brain (Lafaille et al., 2012). Patient cells defective of the TLR3-TRIF-TRAF3-TBK1 pathway show impaired activation of both IGF3 and NF-κB and decreased production of both IFN and IL-6. Both IRF3 and NF-κB seem to contribute greatly to the antiviral responses. On one hand, patients with IRF3 mutations share the susceptibility to HSV encephalitis (Andersen et al., 2015). On the other hand, NEMO deficiency also creates susceptibility to HSV encephalitis (Audry et al., 2011).
TRAF6 and the EDA signaling
EDA (ectodysplasin A)
EDAR (ectodysplasin A receptor)
EDARADD (EDAR associated death domain)
TRAF6 (Tumor necrosis factor receptor associated factor 6)
Mutations in EDA, EDAR, EDARADD, and TRAF6 all manifest EDA/HED with a clinical phenotype similar to patients with hypomorphic NEMO mutations, although they lack immunodeficiency (Headon et al., 2001; Kere et al., 1996; Wisniewski and Trzeciak, 2012a, b). EDA, which belongs to the tumor necrosis factor family, is expressed on the ectoderm interfollicular cells. EDA activates EDAR on the surface of follicular cells in mesenchyme, which in turn binds to EDARADD and TRAF6 to induce downstream signaling. The EDAR-EDARADD-TRAF6 complex recruits TAB1, TAB2, and TAK1, which activate the CP by IKK phosphorylation. The fact that EDA-EDAR-EDARADD-TRAF6-NEMO mutations all present as EDA/HED revealing a prominent function in ectoderm development (Trzeciak and Koczorowski, 2016). Although TRAF6 has been implicated in multiple NF-κB pathways, sometimes synergistic with TRAF3, the disease due to TRAF6 mutations shows no resemblance to syndromes due to other NF-κB components including TRAF3. The exclusive dependence of EDA signaling on TRAF6, together with the exclusive dependence of TLR3 signaling on TRAF3, demonstrates that different receptors ignite specific responses through the common NF-κB signaling complexes. This specificity is still poorly understood at the molecular level.
14. Other monogenic diseases associated with NF-κB pathways
Immunodeficiency and dysregulation
Mutations in MYD88 (myeloid differentiation primary response 88) and IRAK4 (interleukin 1 receptor associated kinase 4) cause clinically indistinguishable phenotypes, characterized by life-threatening, recurrent pyogenic bacterial infections, especially by invasive pneumococci (Picard et al., 2003; von Bernuth et al., 2008). Patient cells exhibit failures in multiple TLRs (including TLR2, TLR3, TLR4, TLR5, TLR9) and two IL-1Rs (IL-1R and IL-18R). The activation of NF-κB and JNK pathways downstream of these signals were absent, demonstrated by the absence of IκB degradation and JNK phosphorylation, and this led to decreased IL-6 and IFN-β. This fits with cellular data showing that MYD88 and IRAKs mediate signaling from almost all TLRs (except for TLR3) and interleukin-1 receptor and related receptors (IL-1Rs). Patient cells showed a broad range of defects in response to stimulation of these receptors. This includes poor responsiveness to poly(I:C) which is thought to be recognized by TLR3. Mouse knockout models showed a broad vulnerability to essentially all pathogens tested (common bacteria, virus, parasites, and fungi). However, the restricted susceptibility to pyogenic bacteria indicates a more specialized function in humans.
Developmental disorders
TAK1/MAP3K7 (mitogen-activated protein kinase kinase kinase 7) is a member of the serine/threonine protein kinase family. TAK1 unites with TRAF6, TAB1, and TAB2 to induce NF-κB signaling. It also activates JNK in responses to TGF-β signaling. Mutations in TAK1 have been clinically associated with Frontometaphyseal Dysplasia (FMD), a progressive skeletal dysplasia affecting the long bones and skull (Basart et al., 2015; Wade et al., 2016).
TAB2 (TAK1-binding protein 2) is also a part of the TRAF6-TAB1/2-TAK1 complex, which activates NF-κB and JNK signaling in responses to TGF-β. This explains why TAB2 deficiency has been clinically associated with congenital heart defects including unusual valve dysplasia and Tetralogy of Fallot rather than a prominent immune phenotype (Weiss et al., 2015).
RIPK4/RIP4 (receptor interacting serine/threonine kinase 4) is a serine/threonine protein kinase that interacts with protein kinase C-delta. In the balance of inflammation vs. nonapoptotic cell death that characterizes the RIP family of kinases, RIP4 tips towards NF-κB signaling for inflammation especially in skin cells. Mutations in RIPK4 are associated with Popliteal pterygium syndrome (Bartsocas-Papas type). It is considered to be a close resembling of IKKα deficiency, which we discussed in section 2 (Kalay et al., 2012).
TRAPPC9/NIBP (trafficking protein particle complex, subunit 9/NIK and IKBKB-binding protein) was originally identified as a NIK and IKKβ interacting protein, which enhances the cytokine-induced NF-κB. The mutations in TRAPPC9 cause mental retardation (autosomal recessive type 13) (Mir et al., 2009; Mochida et al., 2009). However, the disease is more likely to be associated with the function of TRAPPC9 in the TRAPP complex (trafficking protein particle complex), which regulates the vesicle trafficking of ERgolgi, intra-golgi, and endosome-golgi trafficking (Hu et al., 2005; Zong et al., 2011).
Cancer
Somatic mutations, ones that arise in individual cells as a person ages, in genes affecting NF-κB have roles as “drivers” of the oncogenic process in specific malignancies. These mutations have two principle effects: stimulation of proliferative pathways and inhibition of apoptosis, affecting malignant cells together with supporting stroma and blood vessels. There are also corollary effects by NF-κB on inflammation and immunity that promote favorable niches for cancer cells to take root and expand. NF-κB involvement in the pathogenesis of a wide variety of tumors, anti-tumor immunity, and cancer therapies is reported in thousands of publications, so we will focus only on the principle malignancy of lymphoid cells, lymphoma, to illustrate a few important concepts. NF-κB is deranged in lymphomas by somatic mutations as well as by oncogenic viruses such as Epstein-Barr virus. One instructive example is the role of NF-kB in the “activated B cell” (ABC) subtype of diffuse large cell B cell lymphoma (DLBCL) (Young et al., 2015). DLBCL is the most prevalent lymphoid cancer and the ABC subtype is highly aggressive and resists chemotherapy. Constitutive NF-κB activation is “conditionally essential” meaning that the transformation phenotype requires it and without it, ABL lymphoma cells die (Bidere et al., 2009). Nontransformed B cells have no such requirement. This is one of the key concepts. When the conditions of transformation are wired into lymphoma cells of this subtype, certain pathways become turned on and the cell becomes dependent, even addicted, to this adrenaline rush of molecular stimulatory pathways for growth and survival. NF-κB, which is normally quiescent and dependent on very specific molecular signals, is turned on in an unrelenting drive towards a self-enforcing ABC lymphoma gene expression pattern. Key to uncontrolled lymphoma growth is an anti-apoptotic transcriptional program, with boosted expression of apoptosis inhibitors such as Bcl-2. This underpins the resistance of these malignancies to chemotherapeutic agents, which provoke apoptosis to have their toxic effect. Constitutive deregulation of NF-κB predictably alters other regulators such as Bcl-6, PRDM-1, IRF-4 and others, to give the specific B cell phenotype.
ABC cells lines may be paradigmatic of other forms of hematopoietic cancers in that experimental elimination of a whole range of NF-κB signaling wires including IKKβ, CARMA1, BCL10, MALT1 (or its caspase activity), or LUBAC components induce programmed death of the cancer cell. The same has been observed when removing components of the BCR signal generation apparatus including CD79A, CD79B, Btk, Syk, Blnk, PLCγ2, PKCβ. Hence, the life of an ABC lymphoma cell depends on inducing and maintaining active NF-κB. A principal culprit behind unrelenting NF-κB activation is BCR clustering reminiscent of the antigen-engaged receptor. The putative triggering antigen has not been identified but plausible candidates are endogenous autoantigens (Young et al., 2015). In addition, enhanced TLR signaling due to mutations in MyD88 and IRAK1 further aggravate uncontrolled NF-κB in ABC-DLBCL. Finally, mutations inactivating A20 and potentially other down-modulators add even more fuel to the NF-κB conflagration. Hence, entire pathways used in normal immune function become reconfigured in the service of malignancy. Also, the mutations that activate these pathways and induce NF-κB are selective in generating ABC-DLBCL; other variants, such as the “germinal center B cell” (GCB) form of DLBCL does not show these characteristic mutations and does not depend on NF-κB (Pasqualucci and Dalla-Favera, 2015). Germline mutations that promote NF-κB activation may also create an inherited or de novo predisposition to the development of ABC lymphoma. The key oncogenic role of NF-κB is evident in other types of lymphoma, especially Burkitt’s lymphoma, caused in part by EBV which induces NF-κB through its LMP-1 function (Neoptolemos et al., 2010). Finally, genes encoding RHD NF-κB proteins or their key signaling proteins are common at breakpoints of chromosomal translocations in lymphomas and leukemias (Nishikori, 2005).
NF-κB as a therapeutical target
Given its key role in immunity, cancer, and other aspects of human physiology, the concept prevails that NF-κB should be a valuable pharmaceutical target. In fact, drugs already marketed, such as raloxifene, apparently exert therapeutic effects by inhibiting NF-κB (Olivier et al., 2006). The widespread involvement in signaling means that many drug targets exist in the relevant pathway components and a single successful drug may have utility in multiple diseases. The centrality of NF-κB in innate and adaptive immune receptor signaling suggests that NF-κB inhibitory drugs could be widely efficacious against autoimmune and lymphoproliferative disorders. However, these crossover effects may also be a harbinger of unexpected adverse side effects. Inhibition of NF-κB with test therapeutics has been found to cause unexpected, even counterintuitive, effects such as neutrophilia, fever, and abnormal IL-1 release. We will illustrate this paradox with drug development in cancer. Since NF-κB inhibits apoptosis and this is obligatory for cell survival, for example, in ABC-DLBCL lymphoma, then blocking NF-κB should be an effective treatment (Young et al., 2015). Furthermore, upregulated NF-κB promotes cell proliferation, metastasis, metabolic changes and other abnormalities that favor the expansion and spread of malignancy (Kim et al., 2006; Perkins, 2012). Thus, strategies to suppress NF-κB have been clinically tested, most prominently proteasome blockers and IKK inhibitors. Bortezimib (Velcade) is approved for multiple myeloma, a plasma cell malignancy, whose aggressiveness depends in part on NF-κB (Kim et al., 2006). Also, thalidomide and other putative IKK inhibitors have been successful in myeloma. These agents, however, have been thwarted by side effects potentially due to NF-κB suppression including nephrotoxicity, neuropathy, and also the relapse of more aggressive forms of malignancy (Mina et al., 2016). Finally, as the pivotal role of the immune system in fighting malignancy has come into focus with new discoveries in immunotherapy, NF-κB inhibition is cast into a new light (Couzin-Frankel, 2013). Potent NF-κB inhibitors may emasculate T cells that antigenically recognize and kill tumor cells, thereby worsening disease. Thus, the broad role of NF-κB in cellular regulation makes its druggability complicated.
Conclusions and Future Perspectives
In the 30 years that we have known about the seminal roles NF-κBs, much has been learned about their biochemical properties, their responses to signaling events and their physiological roles. Given the enormous number of genes activated by NF-κBs and the diverse modes of signaling that impinge upon these transcription factors, there remains much to learn. Most of the basic work on NF-κB was done in mice and their cells. However, by taking advantage of the striking phenotypes of human mutants in NF-κBs and their related proteins, we can now generate data on the roles of NF-κB in human biology and compare that to mouse data. As this review shows, that has proven very useful.
The many investigators seeking human mutations have now found defects in almost all of the core molecules in the NF-κB pathways. This provides a rich set of tools to study signaling and transcription in humans by proteins related to infection and inflammation. Studies in humans that correlate biochemical alterations with physiological defects are difficult to perform but these “experiments of nature” offer the opportunity to make these connections.
It is striking how often the effects in humans differ from those in mice when similar or even identical mutations are compared. This is a testament to how evolution has used these proteins in different ways in the two species. In many cases, we do not know precisely what these differences are: they may be quantitative or qualitative. Because NF-κB is involved in infection control and the pathogens impinging on the lives of humans and mice are so different, evolution would appear to have adapted to the precise requirements of the two species. If similar work were possible with more species, an increasing richness of specificity would presumably emerge.
Acknowledgments
We apologize to the many hundreds of colleagues over the past 30 years whose work has created this rich tapestry of science but could not be cited due to space limitations. We thank Sankar Ghosh for critically reading the manuscript. This work was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen LL, Mork N, Reinert LS, Kofod-Olsen E, Narita R, Jorgensen SE, Skipper KA, Honing K, Gad HH, Ostergaard L, et al. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J Exp Med. 2015;212:1371–1379. doi: 10.1084/jem.20142274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audry M, Ciancanelli M, Yang K, Cobat A, Chang HH, Sancho-Shimizu V, Lorenzo L, Niehues T, Reichenbach J, Li XX, et al. NEMO is a key component of NF-kappaB- and IRF-3-dependent TLR3-mediated immunity to herpes simplex virus. J Allergy Clin Immunol. 2011;128:610–617. e611–614. doi: 10.1016/j.jaci.2011.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D. NF-kappaB is 25. Nat Immunol. 2011;12:683–685. doi: 10.1038/ni.2072. [DOI] [PubMed] [Google Scholar]
- Basak S, Kim H, Kearns JD, Tergaonkar V, O’Dea E, Werner SL, Benedict CA, Ware CF, Ghosh G, Verma IM, et al. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basart H, van de Kar A, Ades L, Cho TJ, Carter E, Maas SM, Wilson LC, van der Horst CM, Wade EM, Robertson SP, et al. Frontometaphyseal dysplasia and keloid formation without FLNA mutations. Am J Med Genet A. 2015;167:1215–1222. doi: 10.1002/ajmg.a.37044. [DOI] [PubMed] [Google Scholar]
- Ben-Neriah Y. Regulatory functions of ubiquitination in the immune system. Nat Immunol. 2002;3:20–26. doi: 10.1038/ni0102-20. [DOI] [PubMed] [Google Scholar]
- Bidere N, Ngo VN, Lee J, Collins C, Zheng L, Wan F, Davis RE, Lenz G, Anderson DE, Arnoult D, et al. Casein kinase 1alpha governs antigen-receptor-induced NF-kappaB activation and human lymphoma cell survival. Nature. 2009;458:92–96. doi: 10.1038/nature07613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs PJ, Wooster R, Ford D, Chapman P, Mangion J, Quirk Y, Easton DF, Burn J, Stratton MR. Familial cylindromatosis (turban tumour syndrome) gene localised to chromosome 16q12-q13: evidence for its role as a tumour suppressor gene. Nat Genet. 1995;11:441–443. doi: 10.1038/ng1295-441. [DOI] [PubMed] [Google Scholar]
- Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, et al. Identification of the familial cylindromatosis tumoursuppressor gene. Nat Genet. 2000;25:160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- Blonska M, Lin X. NF-kappaB signaling pathways regulated by CARMA family of scaffold proteins. Cell Res. 2011;21:55–70. doi: 10.1038/cr.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson B, Laplantine E, Dobbs K, Cobat A, Tarantino N, Hazen M, Lidov HG, Hopkins G, Du L, Belkadi A, et al. Human HOIP and LUBAC deficiency underlies autoinflammation, immunodeficiency, amylopectinosis, and lymphangiectasia. J Exp Med. 2015;212:939–951. doi: 10.1084/jem.20141130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson B, Laplantine E, Prando C, Giliani S, Israelsson E, Xu Z, Abhyankar A, Israel L, Trevejo-Nunez G, Bogunovic D, et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat Immunol. 2012;13:1178–1186. doi: 10.1038/ni.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boztug H, Hirschmugl T, Holter W, Lakatos K, Kager L, Trapin D, Pickl W, Forster-Waldl E, Boztug K. NF-kappaB1 Haploinsufficiency Causing Immunodeficiency and EBVDriven Lymphoproliferation. J Clin Immunol. 2016;36:533–540. doi: 10.1007/s10875-016-0306-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brightbill HD, Jackman JK, Suto E, Kennedy H, Jones C, 3rd, Chalasani S, Lin Z, Tam L, Roose-Girma M, Balazs M, et al. Conditional Deletion of NF-kappaB-Inducing Kinase (NIK) in Adult Mice Disrupts Mature B Cell Survival and Activation. J Immunol. 2015;195:953–964. doi: 10.4049/jimmunol.1401514. [DOI] [PubMed] [Google Scholar]
- Buchbinder D, Stinson JR, Nugent DJ, Heurtier L, Suarez F, Sukumar G, Dalgard CL, Masson C, Parisot M, Zhang Y, et al. Mild B-cell lymphocytosis in patients with a CARD11 C49Y mutation. J Allergy Clin Immunol. 2015;136:819–821. e811. doi: 10.1016/j.jaci.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbit-Henrion F, Jeverica AK, Begue B, Markelj G, Parlato M, Avcin SL, Callebaut I, Bras M, Parisot M, Jazbec J, et al. Deficiency in Mucosa Associated Lymphoid Tissue Lymphoma Translocation 1 (MALT1): A Novel Cause of Ipex-Like Syndrome. J Pediatr Gastroenterol Nutr. 2016 doi: 10.1097/MPG.0000000000001262. [DOI] [PubMed] [Google Scholar]
- Chen K, Coonrod EM, Kumanovics A, Franks ZF, Durtschi JD, Margraf RL, Wu W, Heikal NM, Augustine NH, Ridge PG, et al. Germline mutations in NFKB2 implicate the noncanonical NF-kappaB pathway in the pathogenesis of common variable immunodeficiency. Am J Hum Genet. 2013;93:812–824. doi: 10.1016/j.ajhg.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun HJ, Zheng L, Ahmad M, Wang J, Speirs CK, Siegel RM, Dale JK, Puck J, Davis J, Hall CG, et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature. 2002;419:395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- Corda G, Sala A. Cutaneous cylindroma: it’s all about MYB. J Pathol. 2016;239:391–393. doi: 10.1002/path.4746. [DOI] [PubMed] [Google Scholar]
- Courtois G, Pescatore A, Gautheron J, Fusco F, Ursini MV, Senegas A. NF-KB-related genetic diseases. New York, NY: Springer Berlin Heidelberg; 2016. [Google Scholar]
- Courtois G, Smahi A, Reichenbach J, Doffinger R, Cancrini C, Bonnet M, Puel A, Chable-Bessia C, Yamaoka S, Feinberg J, et al. A hypermorphic IkappaBalpha mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J Clin Invest. 2003;112:1108–1115. doi: 10.1172/JCI18714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzin-Frankel J. Breakthrough of the year 2013. Cancer immunotherapy. Science. 2013;342:1432–1433. doi: 10.1126/science.342.6165.1432. [DOI] [PubMed] [Google Scholar]
- Damgaard RB, Walker JA, Marco-Casanova P, Morgan NV, Titheradge HL, Elliott PR, McHale D, Maher ER, McKenzie AN, Komander D. The Deubiquitinase OTULIN Is an Essential Negative Regulator of Inflammation and Autoimmunity. Cell. 2016;166:1215–1230. e1220. doi: 10.1016/j.cell.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, De Wolf-Peeters C, Hagemeijer A, Van den Berghe H, Marynen P. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood. 1999;93:3601–3609. [PubMed] [Google Scholar]
- Doffinger R, Smahi A, Bessia C, Geissmann F, Feinberg J, Durandy A, Bodemer C, Kenwrick S, Dupuis-Girod S, Blanche S, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat Genet. 2001;27:277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- Dondelinger Y, Darding M, Bertrand MJ, Walczak H. Poly-ubiquitination in TNFR1-mediated necroptosis. Cell Mol Life Sci. 2016;73:2165–2176. doi: 10.1007/s00018-016-2191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong BH, Onizawa M, Oses-Prieto JA, Advincula R, Burlingame A, Malynn BA, Ma A. A20 restricts ubiquitination of pro-interleukin-1beta protein complexes and suppresses NLRP3 inflammasome activity. Immunity. 2015;42:55–67. doi: 10.1016/j.immuni.2014.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T, Albrecht B, Favier B, Sunshine MJ, Mirchandani K, O’Brien W, Thome M, Littman DR. Requirement for CARMA1 in antigen receptor-induced NF-kappa B activation and lymphocyte proliferation. Curr Biol. 2003;13:1252–1258. doi: 10.1016/s0960-9822(03)00491-3. [DOI] [PubMed] [Google Scholar]
- Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 2009;229:152–172. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K, Deak BK, Sanchez LC, Martinez AM, Corell JJ, Botella AM, Benito GM, Lopez RR, Vanecek T, Kazakov DV, et al. The CYLD p.R758X worldwide recurrent nonsense mutation detected in patients with multiple familial trichoepithelioma type 1, Brooke-Spiegler syndrome and familial cylindromatosis represents a mutational hotspot in the gene. BMC Genet. 2016;17:36. doi: 10.1186/s12863-016-0346-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe-Santos O, Bustamante J, Haverkamp MH, Vinolo E, Ku CL, Puel A, Frucht DM, Christel K, von Bernuth H, Jouanguy E, et al. X-linked susceptibility to mycobacteria is caused by mutations in NEMO impairing CD40-dependent IL-12 production. J Exp Med. 2006;203:1745–1759. doi: 10.1084/jem.20060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M, Bryant VL, Frede N, Slade C, Woon ST, Lehnert K, Winzer S, Bulashevska A, Scerri T, Leung E, et al. Haploinsufficiency of the NF-kappaB1 Subunit p50 in Common Variable Immunodeficiency. Am J Hum Genet. 2015;97:389–403. doi: 10.1016/j.ajhg.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S, Rensing-Ehl A, Pannicke U, Lorenz MR, Fisch P, Jeelall Y, Rohr J, Speckmann C, Vraetz T, Farmand S, et al. Omenn syndrome associated with a functional reversion due to a somatic second-site mutation in CARD11 deficiency. Blood. 2015;126:1658–1669. doi: 10.1182/blood-2015-03-631374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs-Telem D, Sarig O, van Steensel MA, Isakov O, Israeli S, Nousbeck J, Richard K, Winnepenninckx V, Vernooij M, Shomron N, et al. Familial pityriasis rubra pilaris is caused by mutations in CARD14. Am J Hum Genet. 2012;91:163–170. doi: 10.1016/j.ajhg.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavino C, Hamel N, Zeng JB, Legault C, Guiot MC, Chankowsky J, Lejtenyi D, Lemire M, Alarie I, Dufresne S, et al. Impaired RASGRF1/ERK-mediated GM-CSF response characterizes CARD9 deficiency in French-Canadians. J Allergy Clin Immunol. 2016;137:1178–1188. e1171–1177. doi: 10.1016/j.jaci.2015.09.016. [DOI] [PubMed] [Google Scholar]
- Gerondakis S, Grumont R, Gugasyan R, Wong L, Isomura I, Ho W, Banerjee A. Unravelling the complexities of the NF-kappaB signalling pathway using mouse knockout and transgenic models. Oncogene. 2006;25:6781–6799. doi: 10.1038/sj.onc.1209944. [DOI] [PubMed] [Google Scholar]
- Gerondakis S, Siebenlist U. Roles of the NF-kappaB pathway in lymphocyte development and function. Cold Spring Harb Perspect Biol. 2010;2:a000182. doi: 10.1101/cshperspect.a000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker EO, Hennigs A, Nabavi M, Schaffer AA, Woellner C, Salzer U, Pfeifer D, Veelken H, Warnatz K, Tahami F, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil J, Rausch T, Giese T, Bandapalli OR, Daniel V, Bekeredjian-Ding I, Stutz AM, Drees C, Roth S, Ruland J, et al. Whole-exome sequencing links caspase recruitment domain 11 (CARD11) inactivation to severe combined immunodeficiency. J Allergy Clin Immunol. 2013;131:1376–1383. e1373. doi: 10.1016/j.jaci.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Guo Y, Audry M, Ciancanelli M, Alsina L, Azevedo J, Herman M, Anguiano E, Sancho-Shimizu V, Lorenzo L, Pauwels E, et al. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J Exp Med. 2011;208:2083–2098. doi: 10.1084/jem.20101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachmann J, Salvesen GS. The Paracaspase MALT1. Biochimie. 2016;122:324–338. doi: 10.1016/j.biochi.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Wada T, Bakal C, Kozieradzki I, Suzuki S, Suzuki N, Nghiem M, Griffiths EK, Krawczyk C, Bauer B, et al. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity. 2003;18:763–775. doi: 10.1016/s1074-7613(03)00148-1. [DOI] [PubMed] [Google Scholar]
- Harden JL, Krueger JG, Bowcock AM. The immunogenetics of Psoriasis: A comprehensive review. J Autoimmun. 2015;64:66–73. doi: 10.1016/j.jaut.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Headon DJ, Emmal SA, Ferguson BM, Tucker AS, Justice MJ, Sharpe PT, Zonana J, Overbeek PA. Gene defect in ectodermal dysplasia implicates a death domain adapter in development. Nature. 2001;414:913–916. doi: 10.1038/414913a. [DOI] [PubMed] [Google Scholar]
- Herman M, Ciancanelli M, Ou YH, Lorenzo L, Klaudel-Dreszler M, Pauwels E, Sancho-Shimizu V, Perez de Diego R, Abhyankar A, Israelsson E, et al. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J Exp Med. 2012;209:1567–1582. doi: 10.1084/jem.20111316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WH, Pendergast JS, Mo XM, Brambilla R, Bracchi-Ricard V, Li F, Walters WM, Blits B, He L, Schaal SM, et al. NIBP, a novel NIK and IKK(beta)-binding protein that enhances NF-(kappa)B activation. J Biol Chem. 2005;280:29233–29241. doi: 10.1074/jbc.M501670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubeau M, Ngadjeua F, Puel A, Israel L, Feinberg J, Chrabieh M, Belani K, Bodemer C, Fabre I, Plebani A, et al. New mechanism of X-linked anhidrotic ectodermal dysplasia with immunodeficiency: impairment of ubiquitin binding despite normal folding of NEMO protein. Blood. 2011;118:926–935. doi: 10.1182/blood-2010-10-315234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K. Diverse roles of the ubiquitin system in NF-kappaB activation. Biochim Biophys Acta. 2014;1843:129–136. doi: 10.1016/j.bbamcr.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Jabara HH, Ohsumi T, Chou J, Massaad MJ, Benson H, Megarbane A, Chouery E, Mikhael R, Gorka O, Gewies A, et al. A homozygous mucosa-associated lymphoid tissue 1 (MALT1) mutation in a family with combined immunodeficiency. J Allergy Clin Immunol. 2013;132:151–158. doi: 10.1016/j.jaci.2013.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CT, Cao L, Roberson ED, Duan S, Helms CA, Nair RP, Duffin KC, Stuart PE, Goldgar D, Hayashi G, et al. Rare and common variants in CARD14, encoding an epidermal regulator of NF-kappaB, in psoriasis. Am J Hum Genet. 2012;90:796–808. doi: 10.1016/j.ajhg.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juilland M, Thome M. Role of the CARMA1/BCL10/MALT1 complex in lymphoid malignancies. Curr Opin Hematol. 2016;23:402–409. doi: 10.1097/MOH.0000000000000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalay E, Sezgin O, Chellappa V, Mutlu M, Morsy H, Kayserili H, Kreiger E, Cansu A, Toraman B, Abdalla EM, et al. Mutations in RIPK4 cause the autosomal-recessive form of popliteal pterygium syndrome. Am J Hum Genet. 2012;90:76–85. doi: 10.1016/j.ajhg.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- Katakam AK, Brightbill H, Franci C, Kung C, Nunez V, Jones C, 3rd, Peng I, Jeet S, Wu LC, Mellman I, et al. Dendritic cells require NIK for CD40-dependent cross-priming of CD8+ T cells. Proc Natl Acad Sci U S A. 2015;112:14664–14669. doi: 10.1073/pnas.1520627112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kere J, Srivastava AK, Montonen O, Zonana J, Thomas N, Ferguson B, Munoz F, Morgan D, Clarke A, Baybayan P, et al. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat Genet. 1996;13:409–416. doi: 10.1038/ng0895-409. [DOI] [PubMed] [Google Scholar]
- Keusekotten K, Elliott PR, Glockner L, Fiil BK, Damgaard RB, Kulathu Y, Wauer T, Hospenthal MK, Gyrd-Hansen M, Krappmann D, et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153:1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Hawke N, Baldwin AS. NF-kappaB and IKK as therapeutic targets in cancer. Cell Death Differ. 2006;13:738–747. doi: 10.1038/sj.cdd.4401877. [DOI] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- Lafaille FG, Pessach IM, Zhang SY, Ciancanelli MJ, Herman M, Abhyankar A, Ying SW, Keros S, Goldstein PA, Mostoslavsky G, et al. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature. 2012;491:769–773. doi: 10.1038/nature11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahtela J, Nousiainen HO, Stefanovic V, Tallila J, Viskari H, Karikoski R, Gentile M, Saloranta C, Varilo T, Salonen R, et al. Mutant CHUK and severe fetal encasement malformation. N Engl J Med. 2010;363:1631–1637. doi: 10.1056/NEJMoa0911698. [DOI] [PubMed] [Google Scholar]
- Lanternier F, Mahdaviani SA, Barbati E, Chaussade H, Koumar Y, Levy R, Denis B, Brunel AS, Martin S, Loop M, et al. Inherited CARD9 deficiency in otherwise healthy children and adults with Candida species-induced meningoencephalitis, colitis, or both. J Allergy Clin Immunol. 2015;135:1558–1568. e1552. doi: 10.1016/j.jaci.2014.12.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanternier F, Pathan S, Vincent QB, Liu L, Cypowyj S, Prando C, Migaud M, Taibi L, Ammar-Khodja A, Boudghene Stambouli O, et al. Deep dermatophytosis and inherited CARD9 deficiency. N Engl J Med. 2013;369:1704–1714. doi: 10.1056/NEJMoa1208487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CE, Fulcher DA, Whittle B, Chand R, Fewings N, Field M, Andrews D, Goodnow CC, Cook MC. Autosomal-dominant B-cell deficiency with alopecia due to a mutation in NFKB2 that results in nonprocessable p100. Blood. 2014;124:2964–2972. doi: 10.1182/blood-2014-06-578542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers B, Salmena L, Bidere N, Su H, Matysiak-Zablocki E, Murakami K, Ohashi PS, Jurisicova A, Lenardo M, Hakem R, et al. Essential role for caspase-8 in Toll-like receptors and NFkappaB signaling. J Biol Chem. 2007;282:7416–7423. doi: 10.1074/jbc.M606721200. [DOI] [PubMed] [Google Scholar]
- Lenardo M, Pierce JW, Baltimore D. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 1987;236:1573–1577. doi: 10.1126/science.3109035. [DOI] [PubMed] [Google Scholar]
- Lenardo MJ, Fan CM, Maniatis T, Baltimore D. The involvement of NF-kappa B in beta-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell. 1989;57:287–294. doi: 10.1016/0092-8674(89)90966-5. [DOI] [PubMed] [Google Scholar]
- Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- Liu F, Xia Y, Parker AS, Verma IM. IKK biology. Immunol Rev. 2012;246:239–253. doi: 10.1111/j.1600-065X.2012.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Granados E, Keenan JE, Kinney MC, Leo H, Jain N, Ma CA, Quinones R, Gelfand EW, Jain A. A novel mutation in NFKBIA/IKBA results in a degradation-resistant N-truncated protein and is associated with ectodermal dysplasia with immunodeficiency. Hum Mutat. 2008;29:861–868. doi: 10.1002/humu.20740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lougaris V, Tabellini G, Vitali M, Baronio M, Patrizi O, Tampella G, Biasini A, Moratto D, Parolini S, Plebani A. Defective natural killer-cell cytotoxic activity in NFKB2-mutated CVID-like disease. J Allergy Clin Immunol. 2015;135:1641–1643. doi: 10.1016/j.jaci.2014.11.038. [DOI] [PubMed] [Google Scholar]
- Ma A, Malynn BA. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat Rev Immunol. 2012;12:774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-kappaB signaling. J Mol Cell Biol. 2011;3:159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDuff DA, Reese TA, Kimmey JM, Weiss LA, Song C, Zhang X, Kambal A, Duan E, Carrero JA, Boisson B, et al. Phenotypic complementation of genetic immunodeficiency by chronic herpesvirus infection. Elife. 2015;4 doi: 10.7554/eLife.04494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay F, Cancro MP. Travelling with the BAFF/BLyS family: are we there yet? Semin Immunol. 2006;18:261–262. doi: 10.1016/j.smim.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- Mathis BJ, Lai Y, Qu C, Janicki JS, Cui T. CYLD-mediated signaling and diseases. Curr Drug Targets. 2015;16:284–294. doi: 10.2174/1389450115666141024152421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald DR, Mooster JL, Reddy M, Bawle E, Secord E, Geha RS. Heterozygous N-terminal deletion of IkappaBalpha results in functional nuclear factor kappaB haploinsufficiency, ectodermal dysplasia, and immune deficiency. J Allergy Clin Immunol. 2007;120:900–907. doi: 10.1016/j.jaci.2007.08.035. [DOI] [PubMed] [Google Scholar]
- Mericoa D, NS, PHc, Jo-Anne Herbricka aCM, Roifman CM. RelB deficiency causes combined immunodeficiency. LymphoSign Journal. 2015;2 [Google Scholar]
- Meylan E, Tschopp J. The RIP kinases: crucial integrators of cellular stress. Trends Biochem Sci. 2005;30:151–159. doi: 10.1016/j.tibs.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Mina R, Cerrato C, Bernardini A, Aghemo E, Palumbo A. New pharmacotherapy options for multiple myeloma. Expert Opin Pharmacother. 2016;17:181–192. doi: 10.1517/14656566.2016.1115016. [DOI] [PubMed] [Google Scholar]
- Mir A, Kaufman L, Noor A, Motazacker MM, Jamil T, Azam M, Kahrizi K, Rafiq MA, Weksberg R, Nasr T, et al. Identification of mutations in TRAPPC9, which encodes the NIK- and IKK-beta-binding protein, in nonsyndromic autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:909–915. doi: 10.1016/j.ajhg.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida GH, Mahajnah M, Hill AD, Basel-Vanagaite L, Gleason D, Hill RS, Bodell A, Crosier M, Straussberg R, Walsh CA. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet. 2009;85:897–902. doi: 10.1016/j.ajhg.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooster JL, Le Bras S, Massaad MJ, Jabara H, Yoon J, Galand C, Heesters BA, Burton OT, Mattoo H, Manis J, et al. Defective lymphoid organogenesis underlies the immune deficiency caused by a heterozygous S32I mutation in IkappaBalpha. J Exp Med. 2015;212:185–202. doi: 10.1084/jem.20140979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousallem T, Yang J, Urban TJ, Wang H, Adeli M, Parrott RE, Roberts JL, Goldstein DB, Buckley RH, Zhong XP. A nonsense mutation in IKBKB causes combined immunodeficiency. Blood. 2014;124:2046–2050. doi: 10.1182/blood-2014-04-571265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neoptolemos JP, Stocken DD, Bassi C, Ghaneh P, Cunningham D, Goldstein D, Padbury R, Moore MJ, Gallinger S, Mariette C, et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA. 2010;304:1073–1081. doi: 10.1001/jama.2010.1275. [DOI] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- Niemela J, Kuehn HS, Kelly C, Zhang M, Davies J, Melendez J, Dreiling J, Kleiner D, Calvo K, Oliveira JB, et al. Caspase-8 Deficiency Presenting as Late-Onset Multi-Organ Lymphocytic Infiltration with Granulomas in two Adult Siblings. J Clin Immunol. 2015;35:348–355. doi: 10.1007/s10875-015-0150-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikori M. Classical and Alternative NF-kB Activation Pathways and Their Roles in Lymphoid Malignancies. Journal of clinical and experimental hematopathology. 2005;45:9. [Google Scholar]
- Noma H, Eshima K, Satoh M, Iwabuchi K. Differential dependence on nuclear factor-kappaB-inducing kinase among natural killer T-cell subsets in their development. Immunology. 2015;146:89–99. doi: 10.1111/imm.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi H, Kawamoto N, Seishima M, Ohara O, Fukao T. A Japanese family case with juvenile onset Behcet’s disease caused by TNFAIP3 mutation. Allergol Int. 2016 doi: 10.1016/j.alit.2016.06.006. [DOI] [PubMed] [Google Scholar]
- Olivier S, Close P, Castermans E, de Leval L, Tabruyn S, Chariot A, Malaise M, Merville MP, Bours V, Franchimont N. Raloxifene-induced myeloma cell apoptosis: a study of nuclear factor-kappaB inhibition and gene expression signature. Mol Pharmacol. 2006;69:1615–1623. doi: 10.1124/mol.105.020479. [DOI] [PubMed] [Google Scholar]
- Pannicke U, Baumann B, Fuchs S, Henneke P, Rensing-Ehl A, Rizzi M, Janda A, Hese K, Schlesier M, Holzmann K, et al. Deficiency of innate and acquired immunity caused by an IKBKB mutation. N Engl J Med. 2013;369:2504–2514. doi: 10.1056/NEJMoa1309199. [DOI] [PubMed] [Google Scholar]
- Pasparakis M, Courtois G, Hafner M, Schmidt-Supprian M, Nenci A, Toksoy A, Krampert M, Goebeler M, Gillitzer R, Israel A, et al. TNF-mediated inflammatory skin disease in mice with epidermis-specific deletion of IKK2. Nature. 2002;417:861–866. doi: 10.1038/nature00820. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Dalla-Favera R. The genetic landscape of diffuse large B-cell lymphoma. Semin Hematol. 2015;52:67–76. doi: 10.1053/j.seminhematol.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltzer N, Rieser E, Taraborrelli L, Draber P, Darding M, Pernaute B, Shimizu Y, Sarr A, Draberova H, Montinaro A, et al. HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Rep. 2014;9:153–165. doi: 10.1016/j.celrep.2014.08.066. [DOI] [PubMed] [Google Scholar]
- Perez de Diego R, Sancho-Shimizu V, Lorenzo L, Puel A, Plancoulaine S, Picard C, Herman M, Cardon A, Durandy A, Bustamante J, et al. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 2010;33:400–411. doi: 10.1016/j.immuni.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND. The diverse and complex roles of NF-kappaB subunits in cancer. Nat Rev Cancer. 2012;12:121–132. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
- Picard C, Puel A, Bonnet M, Ku CL, Bustamante J, Yang K, Soudais C, Dupuis S, Feinberg J, Fieschi C, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299:2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- Price S, Shaw PA, Seitz A, Joshi G, Davis J, Niemela JE, Perkins K, Hornung RL, Folio L, Rosenberg PS, et al. Natural history of autoimmune lymphoproliferative syndrome associated with FAS gene mutations. Blood. 2014;123:1989–1999. doi: 10.1182/blood-2013-10-535393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe AM, Murray SE, Raue HP, Koguchi Y, Slifka MK, Parker DC. A cell-intrinsic requirement for NF-kappaB-inducing kinase in CD4 and CD8 T cell memory. J Immunol. 2013;191:3663–3672. doi: 10.4049/jimmunol.1301328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruefli-Brasse AA, French DM, Dixit VM. Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science. 2003;302:1581–1584. doi: 10.1126/science.1090769. [DOI] [PubMed] [Google Scholar]
- Salmena L, Lemmers B, Hakem A, Matysiak-Zablocki E, Murakami K, Au PY, Berry DM, Tamblyn L, Shehabeldin A, Migon E, et al. Essential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunity. Genes Dev. 2003;17:883–895. doi: 10.1101/gad.1063703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho-Shimizu V, Perez de Diego R, Lorenzo L, Halwani R, Alangari A, Israelsson E, Fabrega S, Cardon A, Maluenda J, Tatematsu M, et al. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J Clin Invest. 2011;121:4889–4902. doi: 10.1172/JCI59259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Iwai K. Roles of linear ubiquitinylation, a crucial regulator of NF-kappaB and cell death, in the immune system. Immunol Rev. 2015;266:175–189. doi: 10.1111/imr.12308. [DOI] [PubMed] [Google Scholar]
- Schimke LF, Rieber N, Rylaarsdam S, Cabral-Marques O, Hubbard N, Puel A, Kallmann L, Sombke SA, Notheis G, Schwarz HP, et al. A novel gain-of-function IKBA mutation underlies ectodermal dysplasia with immunodeficiency and polyendocrinopathy. J Clin Immunol. 2013;33:1088–1099. doi: 10.1007/s10875-013-9906-1. [DOI] [PubMed] [Google Scholar]
- Schmidt-Supprian M, Bloch W, Courtois G, Addicks K, Israel A, Rajewsky K, Pasparakis M. NEMO/IKK gamma-deficient mice model incontinentia pigmenti. Mol Cell. 2000;5:981–992. doi: 10.1016/s1097-2765(00)80263-4. [DOI] [PubMed] [Google Scholar]
- Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- Sharfe N, Merico D, Karanxha A, Macdonald C, Dadi H, Ngan B, Herbrick JA, Roifman CM. The effects of RelB deficiency on lymphocyte development and function. J Autoimmun. 2015;65:90–100. doi: 10.1016/j.jaut.2015.09.001. [DOI] [PubMed] [Google Scholar]
- Shigemura T, Kaneko N, Kobayashi N, Kobayashi K, Takeuchi Y, Nakano N, Masumoto J, Agematsu K. Novel heterozygous C243Y A20/TNFAIP3 gene mutation is responsible for chronic inflammation in autosomal-dominant Behcet’s disease. RMD Open. 2016;2:e000223. doi: 10.1136/rmdopen-2015-000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smahi A, Courtois G, Vabres P, Yamaoka S, Heuertz S, Munnich A, Israel A, Heiss NS, Klauck SM, Kioschis P, et al. Genomic rearrangement in NEMO impairs NF-kappaB activation and is a cause of incontinentia pigmenti. The International Incontinentia Pigmenti (IP) Consortium. Nature. 2000;405:466–472. doi: 10.1038/35013114. [DOI] [PubMed] [Google Scholar]
- Smale ST. Dimer-specific regulatory mechanisms within the NF-kappaB family of transcription factors. Immunol Rev. 2012;246:193–204. doi: 10.1111/j.1600-065X.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- Snow AL, Xiao W, Stinson JR, Lu W, Chaigne-Delalande B, Zheng L, Pittaluga S, Matthews HF, Schmitz R, Jhavar S, et al. Congenital B cell lymphocytosis explained by novel germline CARD11 mutations. J Exp Med. 2012;209:2247–2261. doi: 10.1084/jem.20120831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepensky P, Keller B, Buchta M, Kienzler AK, Elpeleg O, Somech R, Cohen S, Shachar I, Miosge LA, Schlesier M, et al. Deficiency of caspase recruitment domain family, member 11 (CARD11), causes profound combined immunodeficiency in human subjects. J Allergy Clin Immunol. 2013;131:477–485. e471. doi: 10.1016/j.jaci.2012.11.050. [DOI] [PubMed] [Google Scholar]
- Su H, Bidere N, Zheng L, Cubre A, Sakai K, Dale J, Salmena L, Hakem R, Straus S, Lenardo M. Requirement for caspase-8 in NF-kappaB activation by antigen receptor. Science. 2005;307:1465–1468. doi: 10.1126/science.1104765. [DOI] [PubMed] [Google Scholar]
- Su HC, Lenardo MJ. Genetic defects of apoptosis and primary immunodeficiency. Immunol Allergy Clin North Am. 2008;28:329–351. ix. doi: 10.1016/j.iac.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura K. The genetic background of generalized pustular psoriasis: IL36RN mutations and CARD14 gain-of-function variants. J Dermatol Sci. 2014;74:187–192. doi: 10.1016/j.jdermsci.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Sun SC. Non-canonical NF-kappaB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC. The noncanonical NF-kappaB pathway. Immunol Rev. 2012;246:125–140. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Fuentes ME, Yamaguchi K, Durnin MH, Dalrymple SA, Hardy KL, Goeddel DV. Embryonic lethality, liver degeneration, and impaired NF-kappa B activation in IKK-beta-deficient mice. Immunity. 1999;10:421–429. doi: 10.1016/s1074-7613(00)80042-4. [DOI] [PubMed] [Google Scholar]
- Tokunaga F. Linear ubiquitination-mediated NF-kappaB regulation and its related disorders. J Biochem. 2013;154:313–323. doi: 10.1093/jb/mvt079. [DOI] [PubMed] [Google Scholar]
- Torres JM, Martinez-Barricarte R, Garcia-Gomez S, Mazariegos MS, Itan Y, Boisson B, Rholvarez R, Jimenez-Reinoso A, del Pino L, Rodriguez-Pena R, et al. Inherited BCL10 deficiency impairs hematopoietic and nonhematopoietic immunity. J Clin Invest. 2014;124:5239–5248. doi: 10.1172/JCI77493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzeciak WH, Koczorowski R. Molecular basis of hypohidrotic ectodermal dysplasia: an update. J Appl Genet. 2016;57:51–61. doi: 10.1007/s13353-015-0307-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, Ding J, Li Y, Tejasvi T, Gudjonsson JE, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Walle L, Van Opdenbosch N, Jacques P, Fossoul A, Verheugen E, Vogel P, Beyaert R, Elewaut D, Kanneganti TD, van Loo G, et al. Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature. 2014;512:69–73. doi: 10.1038/nature13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bernuth H, Picard C, Jin Z, Pankla R, Xiao H, Ku CL, Chrabieh M, Mustapha IB, Ghandil P, Camcioglu Y, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321:691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade EM, Daniel PB, Jenkins ZA, McInerney-Leo A, Leo P, Morgan T, Addor MC, Ades LC, Bertola D, Bohring A, et al. Mutations in MAP3K7 that Alter the Activity of the TAK1 Signaling Complex Cause Frontometaphyseal Dysplasia. Am J Hum Genet. 2016;99:392–406. doi: 10.1016/j.ajhg.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Applegate C, Wang T, Batista DA. Familial TAB2 microdeletion and congenital heart defects including unusual valve dysplasia and tetralogy of fallot. Am J Med Genet A. 2015;167A:2702–2706. doi: 10.1002/ajmg.a.37210. [DOI] [PubMed] [Google Scholar]
- Wieacker P, Zimmer J, Ropers HH. X inactivation patterns in two syndromes with probable X-linked dominant, male lethal inheritance. Clin Genet. 1985;28:238–242. doi: 10.1111/j.1399-0004.1985.tb00392.x. [DOI] [PubMed] [Google Scholar]
- Willmann KL, Klaver S, Dogu F, Santos-Valente E, Garncarz W, Bilic I, Mace E, Salzer E, Conde CD, Sic H, et al. Biallelic loss-of-function mutation in NIK causes a primary immunodeficiency with multifaceted aberrant lymphoid immunity. Nat Commun. 2014;5:5360. doi: 10.1038/ncomms6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski SA, Trzeciak WH. A new mutation resulting in the truncation of the TRAF6-interacting domain of XEDAR: a possible novel cause of hypohidrotic ectodermal dysplasia. J Med Genet. 2012a;49:499–501. doi: 10.1136/jmedgenet-2012-100877. [DOI] [PubMed] [Google Scholar]
- Wisniewski SA, Trzeciak WH. A rare heterozygous TRAF6 variant is associated with hypohidrotic ectodermal dysplasia. Br J Dermatol. 2012b;166:1353–1356. doi: 10.1111/j.1365-2133.2012.10871.x. [DOI] [PubMed] [Google Scholar]
- Xue L, Morris SW, Orihuela C, Tuomanen E, Cui X, Wen R, Wang D. Defective development and function of Bcl10-deficient follicular, marginal zone and B1 B cells. Nat Immunol. 2003;4:857–865. doi: 10.1038/ni963. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu L, Wesche H, Arthur CD, White JM, Goeddel DV, Schreiber RD. Defective lymphotoxin-beta receptor-induced NF-kappaB transcriptional activity in NIK-deficient mice. Science. 2001;291:2162–2165. doi: 10.1126/science.1058453. [DOI] [PubMed] [Google Scholar]
- Young RM, Shaffer AL, 3rd, Phelan JD, Staudt LM. B-cell receptor signaling in diffuse large B-cell lymphoma. Semin Hematol. 2015;52:77–85. doi: 10.1053/j.seminhematol.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Hoffman S, Beal AM, Dykon A, Ringenberg MA, Hughes AC, Dare L, Anderson AD, Finger J, Kasparcova V, et al. MALT1 Protease Activity Is Required for Innate and Adaptive Immune Responses. PLoS One. 2015;10:e0127083. doi: 10.1371/journal.pone.0127083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Stirling B, Temmerman ST, Ma CA, Fuss IJ, Derry JM, Jain A. Impaired regulation of NF-kappaB and increased susceptibility to colitis-associated tumorigenesis in CYLD-deficient mice. J Clin Invest. 2006;116:3042–3049. doi: 10.1172/JCI28746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Wang H, Schwartz DM, Stoffels M, Park YH, Zhang Y, Yang D, Demirkaya E, Takeuchi M, Tsai WL, et al. Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early-onset autoinflammatory disease. Nat Genet. 2016;48:67–73. doi: 10.1038/ng.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong M, Wu XG, Chan CW, Choi MY, Chan HC, Tanner JA, Yu S. The adaptor function of TRAPPC2 in mammalian TRAPPs explains TRAPPC2-associated SEDT and TRAPPC9-associated congenital intellectual disability. PLoS One. 2011;6:e23350. doi: 10.1371/journal.pone.0023350. [DOI] [PMC free article] [PubMed] [Google Scholar]