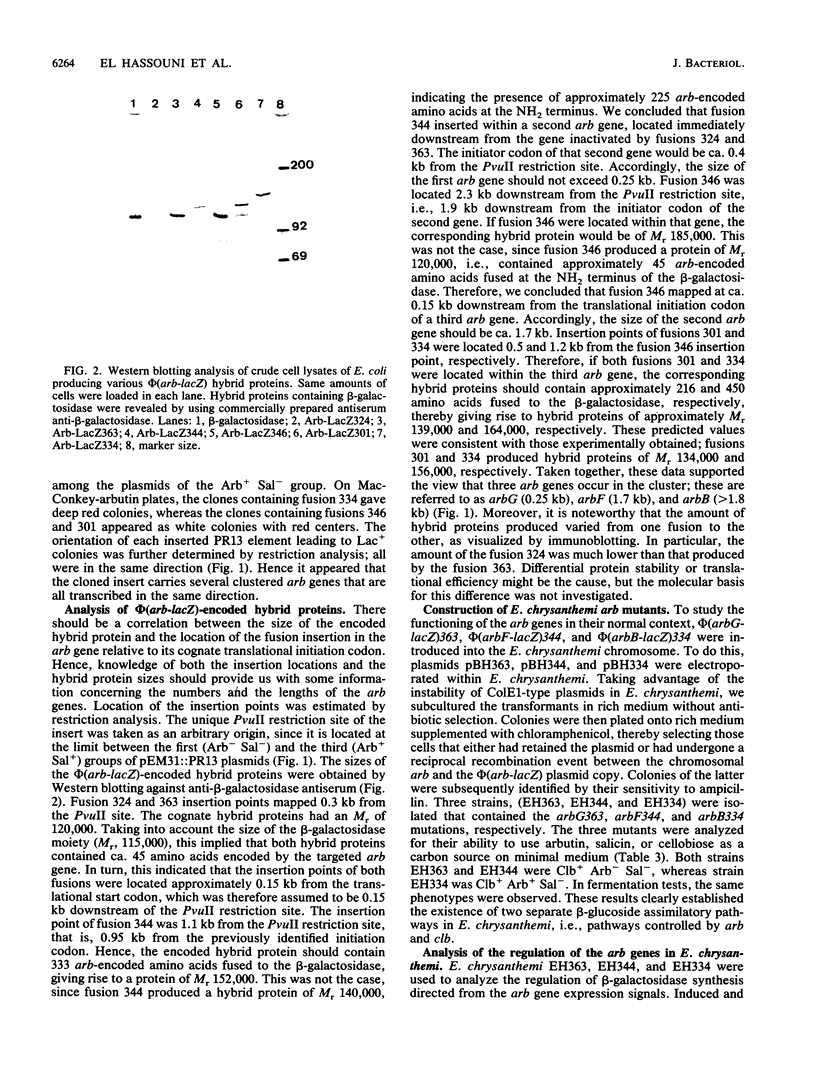
Abstract
Erwinia chrysanthemi is one of the few members of the family Enterobacteriaceae that is capable of metabolizing most of the naturally occurring beta-glucosides. We previously isolated the clb genes, which allow the use of the disaccharide cellobiose as well as the aromatic beta-glucosides arbutin and salicin. We report here the isolation of the arb genes, which permit fermentation of the aromatic beta-glucosides only. Establishment of a functional Arb system in Escherichia coli depended on the presence of the phosphotransferase system and on the activation by the cyclic AMP-cyclic AMP receptor protein complex. Strains carrying mini-Mu-induced LacZ fusions to the arb genes were used to analyze arb genes organization and function. Three arb genes (arbG, arbF, and arbB) were identified and organized in this order. Genetic and structural evidence allowed us to assign a phospho-beta-glucosidase and a permease activity to the ArbB and ArbF proteins, respectively. Several Lac+ arb-lacZ insertions were introduced into the E. chrysanthemi chromosome. Both ArbG- and ArbF- strains were unable to ferment the aromatic beta-glucosides, whereas ArbB- strains were impaired only in salicin fermentation. None of the mutations in the arb genes affected cellobiose metabolism. The expression of the arb genes was substrate inducible and required the ArbF permease and, possibly, the ArbG protein. Collectively, our results underline the resemblance between the naturally expressed E. chrysanthemi arbGFB and the cryptic E. coli bglGFB operons, yet the arbG gene product seemed unable to activate E. coli bgl operon expression.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amster-Choder O., Houman F., Wright A. Protein phosphorylation regulates transcription of the beta-glucoside utilization operon in E. coli. Cell. 1989 Sep 8;58(5):847–855. doi: 10.1016/0092-8674(89)90937-9. [DOI] [PubMed] [Google Scholar]
- Barras F., Chambost J. P., Chippaux M. Cellobiose metabolism in Erwinia: genetic study. Mol Gen Genet. 1984;197(3):486–490. doi: 10.1007/BF00329947. [DOI] [PubMed] [Google Scholar]
- Barras F., Lepelletier M., Chippaux M. Influence of gyrA mutation on expression of Erwinia chrysanthemi clb genes cloned in Escherichia coli. J Bacteriol. 1986 Apr;166(1):346–348. doi: 10.1128/jb.166.1.346-348.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barras F., el Hassouni M., Chambost J. P., Chippaux M. The beta-glucosides metabolism in Erwinia chrysanthemi: preliminary analysis and comparison to Escherichia coli systems. FEMS Microbiol Rev. 1989 Jun;5(1-2):143–147. doi: 10.1016/0168-6445(89)90018-1. [DOI] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. T., Miller R. H. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 1988 Apr 25;16(8):3580–3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Reuse H., Danchin A. The ptsH, ptsI, and crr genes of the Escherichia coli phosphoenolpyruvate-dependent phosphotransferase system: a complex operon with several modes of transcription. J Bacteriol. 1988 Sep;170(9):3827–3837. doi: 10.1128/jb.170.9.3827-3837.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Reuse H., Lévy S., Zeng G., Danchin A. Genetics of the PTS components in Escherichia coli K-12. FEMS Microbiol Rev. 1989 Jun;5(1-2):61–67. doi: 10.1016/0168-6445(89)90009-0. [DOI] [PubMed] [Google Scholar]
- Kricker M., Hall B. G. Biochemical genetics of the cryptic gene system for cellobiose utilization in Escherichia coli K12. Genetics. 1987 Mar;115(3):419–429. doi: 10.1093/genetics/115.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopilato J., Bortner S., Beckwith J. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986 Nov;205(2):285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- Mahadevan S., Reynolds A. E., Wright A. Positive and negative regulation of the bgl operon in Escherichia coli. J Bacteriol. 1987 Jun;169(6):2570–2578. doi: 10.1128/jb.169.6.2570-2578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan S., Wright A. A bacterial gene involved in transcription antitermination: regulation at a rho-independent terminator in the bgl operon of E. coli. Cell. 1987 Jul 31;50(3):485–494. doi: 10.1016/0092-8674(87)90502-2. [DOI] [PubMed] [Google Scholar]
- Parker L. L., Hall B. G. A fourth Escherichia coli gene system with the potential to evolve beta-glucoside utilization. Genetics. 1988 Jul;119(3):485–490. doi: 10.1093/genetics/119.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma P. W., Lengeler J. W. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol Rev. 1985 Sep;49(3):232–269. doi: 10.1128/mr.49.3.232-269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. E., Felton J., Wright A. Insertion of DNA activates the cryptic bgl operon in E. coli K12. Nature. 1981 Oct 22;293(5834):625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- Reynolds A. E., Mahadevan S., LeGrice S. F., Wright A. Enhancement of bacterial gene expression by insertion elements or by mutation in a CAP-cAMP binding site. J Mol Biol. 1986 Sep 5;191(1):85–95. doi: 10.1016/0022-2836(86)90424-9. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr, Yamada M., Erni B., Suda K., Lengeler J., Ebner R., Argos P., Rak B., Schnetz K., Lee C. A. Sugar permeases of the bacterial phosphoenolpyruvate-dependent phosphotransferase system: sequence comparisons. FASEB J. 1988 Mar 1;2(3):199–208. doi: 10.1096/fasebj.2.3.2832233. [DOI] [PubMed] [Google Scholar]
- Schaefler S. Inducible system for the utilization of beta-glucosides in Escherichia coli. I. Active transport and utilization of beta-glucosides. J Bacteriol. 1967 Jan;93(1):254–263. doi: 10.1128/jb.93.1.254-263.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefler S., Malamy A. Taxonomic investigations on expressed and cryptic phospho-beta-glucosidases in Enterobacteriaceae. J Bacteriol. 1969 Aug;99(2):422–433. doi: 10.1128/jb.99.2.422-433.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz K., Rak B. Regulation of the bgl operon of Escherichia coli by transcriptional antitermination. EMBO J. 1988 Oct;7(10):3271–3277. doi: 10.1002/j.1460-2075.1988.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz K., Toloczyki C., Rak B. Beta-glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J Bacteriol. 1987 Jun;169(6):2579–2590. doi: 10.1128/jb.169.6.2579-2590.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]