Abstract
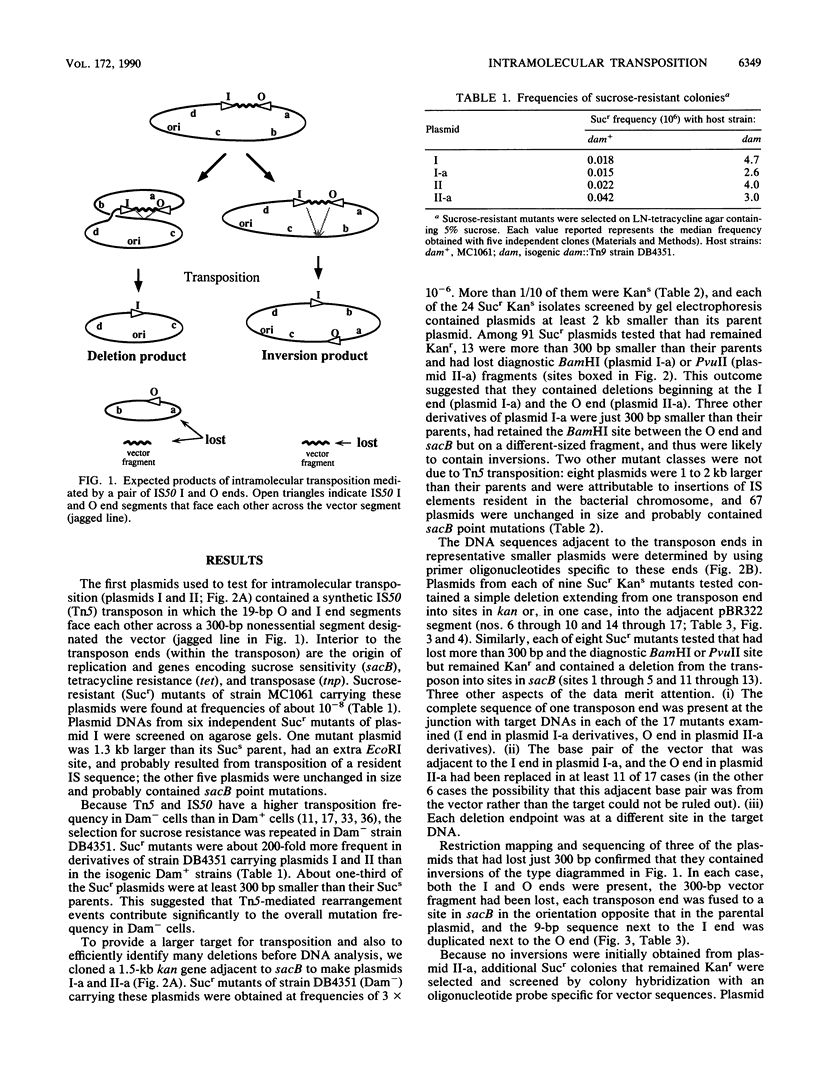
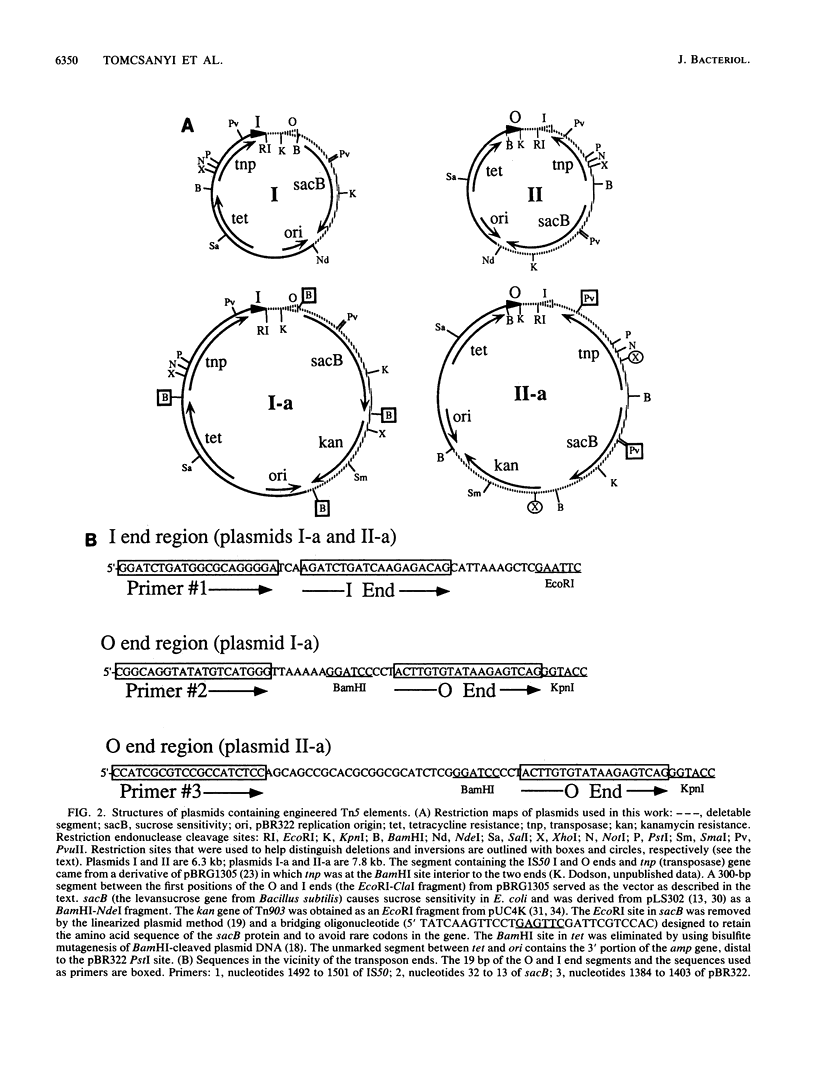
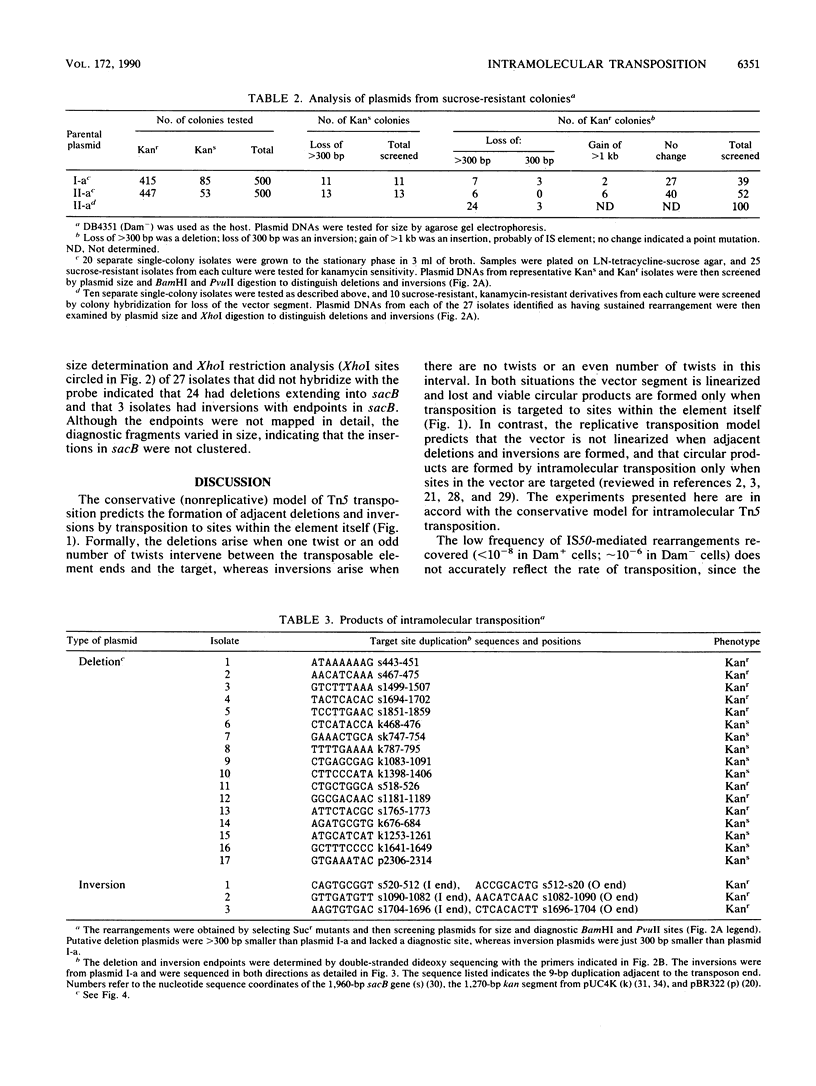
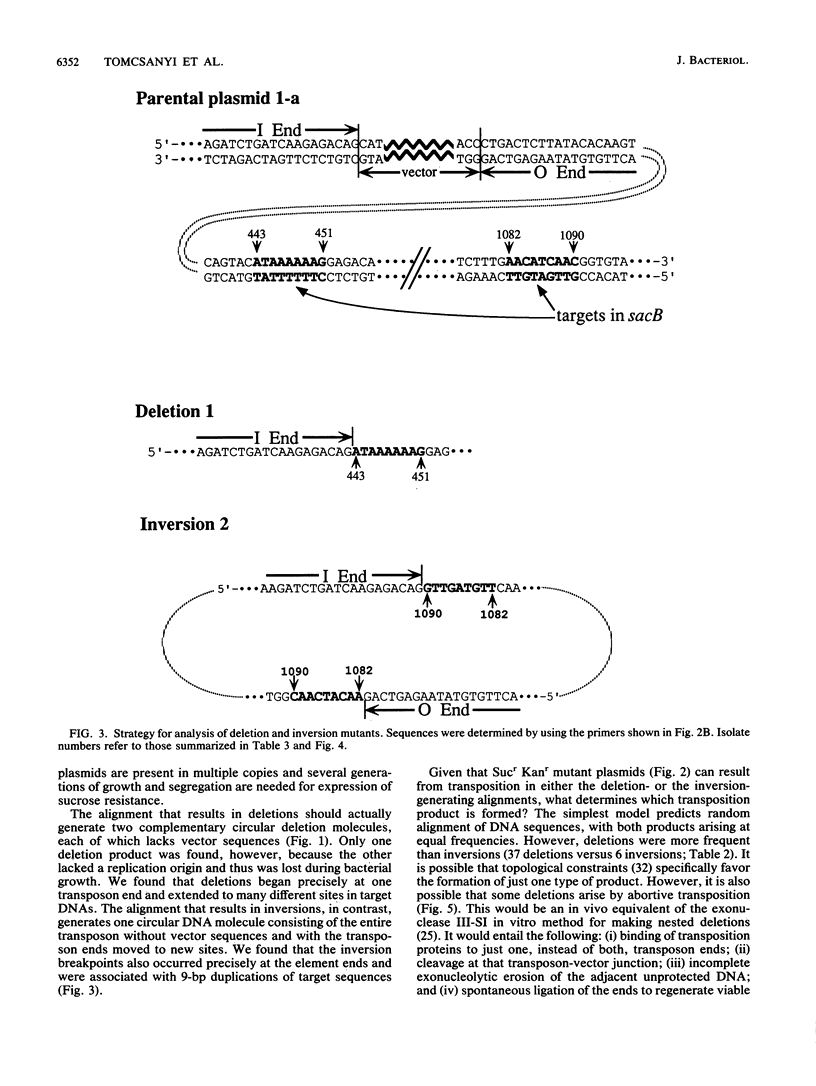
We report the formation of deletions and inversions by intramolecular transposition of Tn5-derived mobile elements. The synthetic transposons used contained the IS50 O and I end segments and the transposase gene, a contraselectable gene encoding sucrose sensitivity (sacB), antibiotic resistance genes, and a plasmid replication origin. Both deletions and inversions were associated with loss of a 300-bp segment that is designated the vector because it is outside of the transposon. Deletions were severalfold more frequent than inversions, perhaps reflecting constraints on DNA twisting or abortive transposition. Restriction and DNA sequence analyses showed that both types of rearrangements extended from one transposon end to many different sites in target DNA. In the case of inversions, transposition generated 9-bp direct repeats of target sequences.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed A. Use of transposon-promoted deletions in DNA sequence analysis. Methods Enzymol. 1987;155:177–204. doi: 10.1016/0076-6879(87)55016-9. [DOI] [PubMed] [Google Scholar]
- Berg D. E., Schmandt M. A., Lowe J. B. Specificity of transposon Tn5 insertion. Genetics. 1983 Dec;105(4):813–828. doi: 10.1093/genetics/105.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E. Structural requirement for IS50-mediated gene transposition. Proc Natl Acad Sci U S A. 1983 Feb;80(3):792–796. doi: 10.1073/pnas.80.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparon M. G., Scott J. R. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell. 1989 Dec 22;59(6):1027–1034. doi: 10.1016/0092-8674(89)90759-9. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Dodson K. W., Berg D. E. Factors affecting transposition activity of IS50 and Tn5 ends. Gene. 1989;76(2):207–213. doi: 10.1016/0378-1119(89)90161-3. [DOI] [PubMed] [Google Scholar]
- Gay P., Le Coq D., Steinmetz M., Berkelman T., Kado C. I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985 Nov;164(2):918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg R. R., Syvanen M. Tn5 transposes independently of cointegrate resolution. Evidence for an alternative model for transposition. J Mol Biol. 1985 Mar 5;182(1):69–78. doi: 10.1016/0022-2836(85)90028-2. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Reznikoff W. S. DNA sequences at the ends of transposon Tn5 required for transposition. Nature. 1983 Jul 21;304(5923):280–282. doi: 10.1038/304280a0. [DOI] [PubMed] [Google Scholar]
- Krebs M. P., Reznikoff W. S. Transcriptional and translational initiation sites of IS50. Control of transposase and inhibitor expression. J Mol Biol. 1986 Dec 20;192(4):781–791. doi: 10.1016/0022-2836(86)90028-8. [DOI] [PubMed] [Google Scholar]
- Lodge J. K., Weston-Hafer K., Berg D. E. Transposon Tn5 target specificity: preference for insertion at G/C pairs. Genetics. 1988 Nov;120(3):645–650. doi: 10.1093/genetics/120.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandecki W. Oligonucleotide-directed double-strand break repair in plasmids of Escherichia coli: a method for site-specific mutagenesis. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7177–7181. doi: 10.1073/pnas.83.19.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis S. H., Berg D. E. Identification of base pairs in the outside end of insertion sequence IS50 that are needed for IS50 and Tn5 transposition. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9118–9122. doi: 10.1073/pnas.84.24.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis S. H., Berg D. E. recA-independent recombination between repeated IS50 elements is not caused by an IS50-encoded function. J Bacteriol. 1985 Mar;161(3):928–932. doi: 10.1128/jb.161.3.928-932.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried J. L., Collmer A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene. 1987;57(2-3):239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakawa C., Carle G. F., Berg D. E. Sequences essential for transposition at the termini of IS50. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7293–7297. doi: 10.1073/pnas.80.23.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Le Coq D., Aymerich S., Gonzy-Tréboul G., Gay P. The DNA sequence of the gene for the secreted Bacillus subtilis enzyme levansucrase and its genetic control sites. Mol Gen Genet. 1985;200(2):220–228. doi: 10.1007/BF00425427. [DOI] [PubMed] [Google Scholar]
- Taylor L. A., Rose R. E. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 1988 Jan 11;16(1):358–358. doi: 10.1093/nar/16.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomcsanyi T., Berg D. E. Transposition effect of adenine (Dam) methylation on activity of O end mutants of IS50. J Mol Biol. 1989 Sep 20;209(2):191–193. doi: 10.1016/0022-2836(89)90271-4. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weston-Hafer K., Berg D. E. Specificity of deletion events in pBR322. Plasmid. 1989 May;21(3):251–253. doi: 10.1016/0147-619x(89)90050-4. [DOI] [PubMed] [Google Scholar]
- Yin J. C., Krebs M. P., Reznikoff W. S. Effect of dam methylation on Tn5 transposition. J Mol Biol. 1988 Jan 5;199(1):35–45. doi: 10.1016/0022-2836(88)90377-4. [DOI] [PubMed] [Google Scholar]


