Abstract
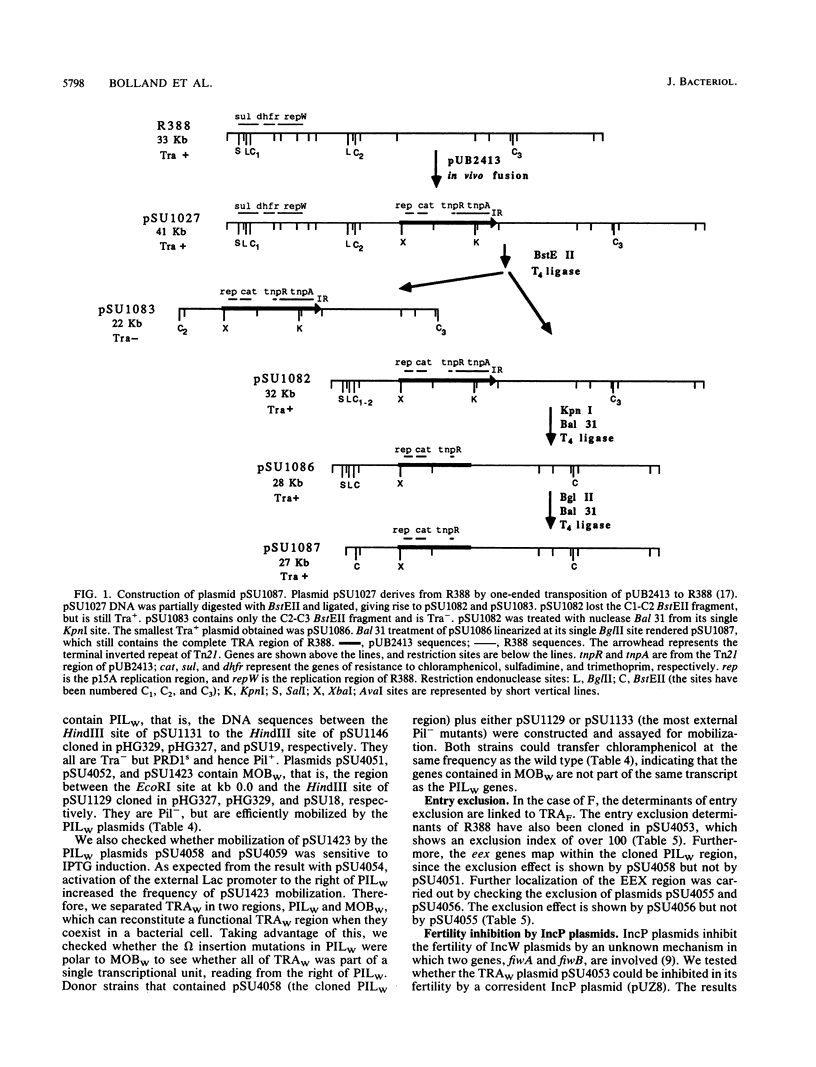
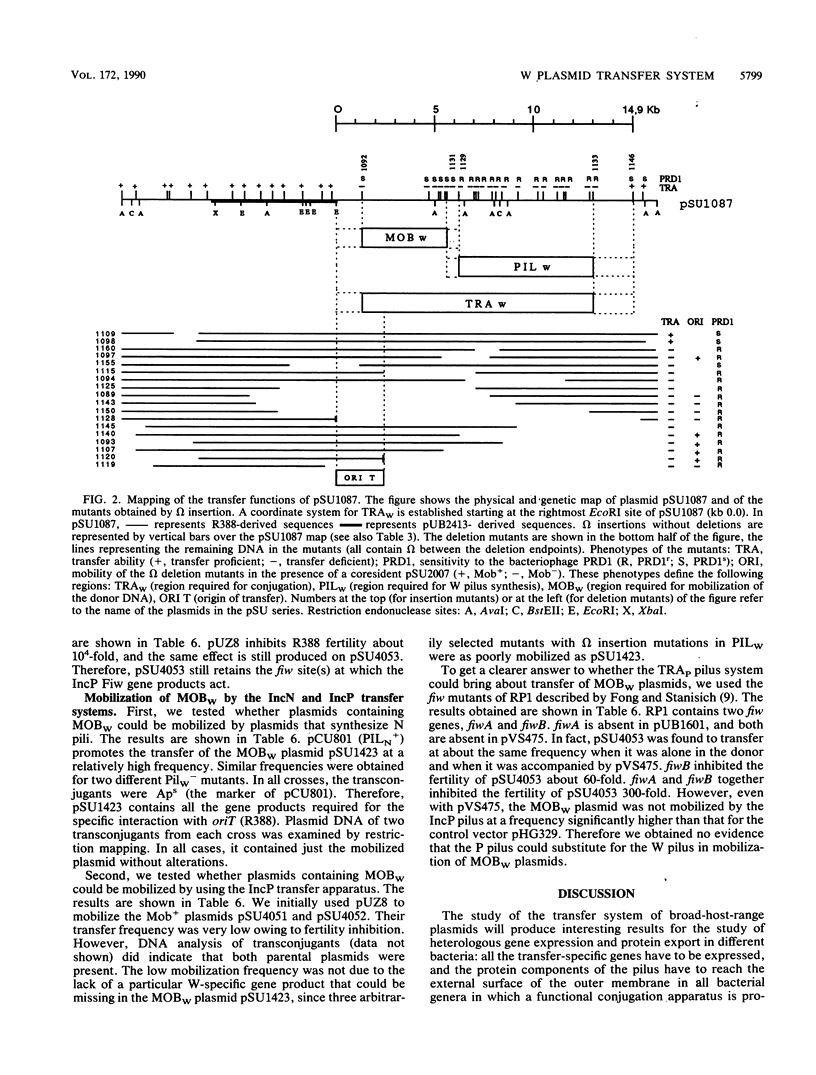
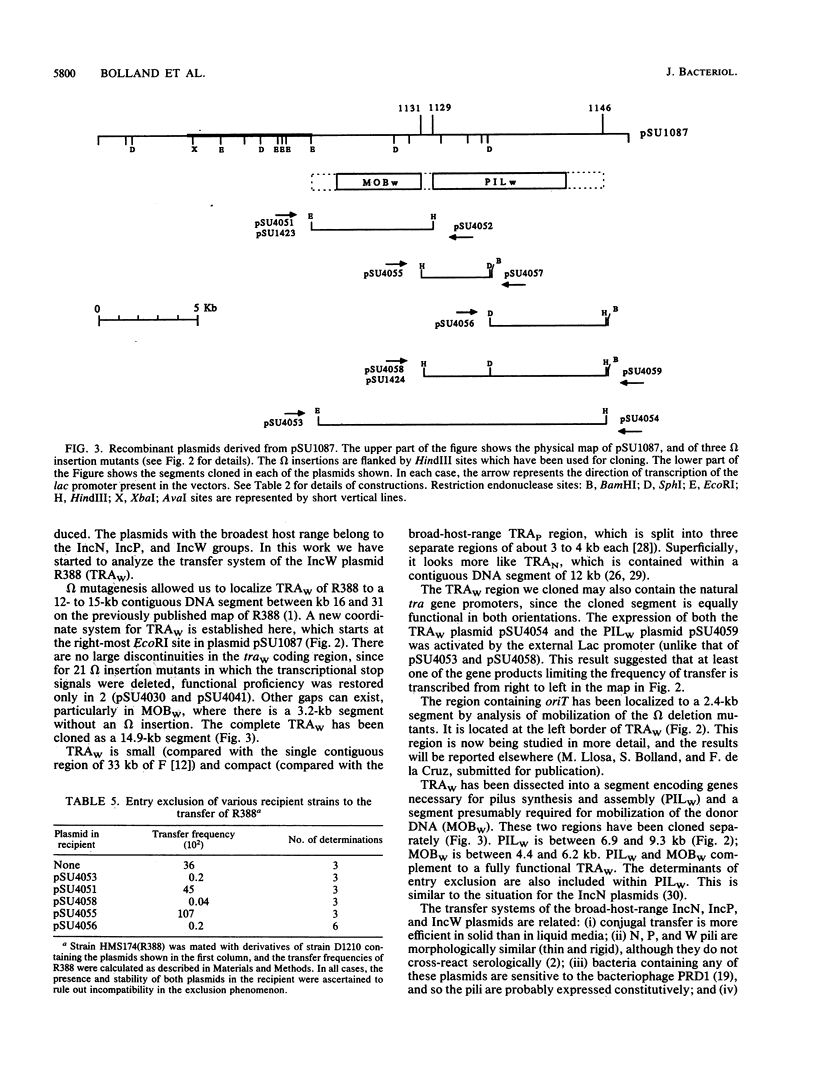
The complete conjugal transfer gene region of the IncW plasmid R388 has been cloned in multicopy vector plasmids and mapped to a contiguous 14.9-kilobase segment by insertion mutagenesis. The fertility of the cloned region could still be inhibited by a coresident IncP plasmid. The transfer region has been dissected into two regions, one involved in pilus synthesis and assembly (PILW), and the other involved in conjugal DNA metabolism (MOBW). They have been separately cloned. PILW also contains the genes involved in entry exclusion. MOBW contains oriT and the gene products required for efficient mobilization by PILW. MOBW plasmids could also be mobilized efficiently by PILN, the specific pilus of the IncN plasmid pCU1, but not by PILP, the specific pilus of the IncP plasmid RP1.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avila P., de la Cruz F. Physical and genetic map of the IncW plasmid R388. Plasmid. 1988 Sep;20(2):155–157. doi: 10.1016/0147-619x(88)90019-4. [DOI] [PubMed] [Google Scholar]
- Bradley D. E. Morphological and serological relationships of conjugative pili. Plasmid. 1980 Sep;4(2):155–169. doi: 10.1016/0147-619x(80)90005-0. [DOI] [PubMed] [Google Scholar]
- Bradley D. E., Taylor D. E., Cohen D. R. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J Bacteriol. 1980 Sep;143(3):1466–1470. doi: 10.1128/jb.143.3.1466-1470.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. L., Richardson C. C., Studier F. W. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc Natl Acad Sci U S A. 1978 May;75(5):2276–2280. doi: 10.1073/pnas.75.5.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. T., Miller R. H. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 1988 Apr 25;16(8):3580–3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier M., Bex F., Bergquist P. L., Maas W. K. Identification and classification of bacterial plasmids. Microbiol Rev. 1988 Sep;52(3):375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J Gen Microbiol. 1972 Sep;72(2):349–355. doi: 10.1099/00221287-72-2-349. [DOI] [PubMed] [Google Scholar]
- Fong S. T., Stanisich V. A. Location and characterization of two functions on RP1 that inhibit the fertility of the IncW plasmid R388. J Gen Microbiol. 1989 Mar;135(3):499–502. doi: 10.1099/00221287-135-3-499. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Deiss C., Simnad V., Yee L., Pansegrau W., Lanka E. Mutagenesis of the Tra1 core region of RK2 by using Tn5: identification of plasmid-specific transfer genes. J Bacteriol. 1989 Jul;171(7):4100–4103. doi: 10.1128/jb.171.7.4100-4103.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Ippen-Ihler K. A., Minkley E. G., Jr The conjugation system of F, the fertility factor of Escherichia coli. Annu Rev Genet. 1986;20:593–624. doi: 10.1146/annurev.ge.20.120186.003113. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay R., McPherson J. Hybrid pUC vectors for addition of new restriction enzyme sites to the ends of DNA fragments. Nucleic Acids Res. 1987 Mar 25;15(6):2778–2778. doi: 10.1093/nar/15.6.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E., de la Cruz F. Transposon Tn21 encodes a RecA-independent site-specific integration system. Mol Gen Genet. 1988 Feb;211(2):320–325. doi: 10.1007/BF00330610. [DOI] [PubMed] [Google Scholar]
- Mötsch S., Schmitt R., Avila P., de la Cruz F., Ward E., Grinsted J. Junction sequences generated by 'one-ended transposition'. Nucleic Acids Res. 1985 May 10;13(9):3335–3342. doi: 10.1093/nar/13.9.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., Shipley P. L. RP1 properties and fertility inhibition among P, N, W, and X incompatibility group plasmids. J Bacteriol. 1975 Jul;123(1):28–35. doi: 10.1128/jb.123.1.28-35.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., Siak J. S., Gray R. H. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J Virol. 1974 Sep;14(3):689–699. doi: 10.1128/jvi.14.3.689-699.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombo E. A., Yusoff K., Stanisich V. A., Krishnapillai V., Willetts N. S. Cloning and genetic analysis of tra cistrons of the Tra 2/Tra 3 region of plasmid RP1. Plasmid. 1989 Jul;22(1):59–69. doi: 10.1016/0147-619x(89)90036-x. [DOI] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Sadler J. R., Tecklenburg M., Betz J. L. Plasmids containing many tandem copies of a synthetic lactose operator. Gene. 1980 Feb;8(3):279–300. doi: 10.1016/0378-1119(80)90005-0. [DOI] [PubMed] [Google Scholar]
- Stewart G. S., Lubinsky-Mink S., Jackson C. G., Cassel A., Kuhn J. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid. 1986 May;15(3):172–181. doi: 10.1016/0147-619x(86)90035-1. [DOI] [PubMed] [Google Scholar]
- Thatte V., Bradley D. E., Iyer V. N. N conjugative transfer system of plasmid pCU1. J Bacteriol. 1985 Sep;163(3):1229–1236. doi: 10.1128/jb.163.3.1229-1236.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Smith C. A. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- Winans S. C., Walker G. C. Conjugal transfer system of the IncN plasmid pKM101. J Bacteriol. 1985 Jan;161(1):402–410. doi: 10.1128/jb.161.1.402-410.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Walker G. C. Entry exclusion determinant(s) of IncN plasmid pKM101. J Bacteriol. 1985 Jan;161(1):411–416. doi: 10.1128/jb.161.1.411-416.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Lis J., Wu R. Elution of DNA from agarose gels after electrophoresis. Methods Enzymol. 1979;68:176–182. doi: 10.1016/0076-6879(79)68012-6. [DOI] [PubMed] [Google Scholar]
- de la Cruz F., Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982 Jul;151(1):222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]


