Summary
Highly active antiretroviral therapy (HAART) has proved efficient in increasing CD4 counts in many randomized clinical trials. Because randomized trials have some limitations (e.g., short duration, highly selected subjects), it is interesting to assess the effect of treatments using observational studies. This is challenging because treatment is started preferentially in subjects with severe conditions. This general problem had been treated using Marginal Structural Models (MSM) relying on the counterfactual formulation. Another approach to causality is based on dynamical models. We present three discrete-time dynamic models based on linear increments models (LIM): the first one based on one difference equation for CD4 counts, the second with an equilibrium point, and the third based on a system of two difference equations, which allows jointly modeling CD4 counts and viral load. We also consider continuous-time models based on ordinary differential equations with non-linear mixed effects (ODE-NLME). These mechanistic models allow incorporating biological knowledge when available, which leads to increased statistical evidence for detecting treatment effect. Because inference in ODE-NLME is numerically challenging and requires specific methods and softwares, LIM are a valuable intermediary option in terms of consistency, precision and complexity. We compare the different approaches in simulation and in illustration on the ANRS CO3 Aquitaine Cohort and the Swiss HIV Cohort Study.
Keywords: Dynamic mechanistic models, Linear Increment Models (LIM), Marginal Structural Models (MSM), Non-Linear Mixed Effect Models (NLME), Observational study, Ordinary Differential Equation (ODE)
1. Introduction
Randomized clinical trials often have short durations and include highly selected subjects, thus assessing the effect of a treatment using observational studies is useful. However, it is challenging because the treatment may change, and covariate history of a subject up to time t may influence treatment given after t, and may also influence the outcome of interest, which induces a time-dependent confounding. For instance, one may wish to assess the effect of antiretroviral therapy in HIV infected subjects. As CD4+ T-lymphocytes (CD4, in short) are the main target cells of the HIV virus, it is possible to assess the effect of a treatment on the blood concentration of these cells: CD4 counts are measurements of this concentration. In observational studies, however, the decision to start an antiretroviral therapy may depend on CD4 counts as well as on other covariates. In this setting, it has been demonstrated that a conventional regression analysis leads to biased estimates of the treatment effect, typically underestimating it, and possibly (wrongly) indicating a negative effect. This is called “confounding by indication” (Walker, 1996).
Marginal structural models (MSM) (Robins et al., 2000) have been proposed for dealing with this issue; this is based on choosing a causal model in terms of potential responses, which are often counterfactual, to the different treatment histories. The parameters of a MSM can be estimated through a weighted approach but other methods exist (Petersen et al., 2006). The weights are the inverse probability of treatment assignment and are obtained through a “treatment model” which includes the covariates linked to the outcome. Because data are correlated, we use an inverse probability weighted generalized estimating equation (GEE). This approach has been applied by Hernán et al. (2002) and Cole et al. (2005) for estimating the effect of zidovudine and of highly active antiretroviral therapy (HAART) on CD4 count.Cole et al. (2007) and Sterne et al. (2005) used it for estimating the effect of HAART on viral load and on AIDS or death.
An alternative view to causality, that does not use the potential responses representation is to use dynamic models. This has been pioneered by Granger (1969); Dawid (2000), and further developed by Didelez (2008), Commenges and Gégout-Petit (2009), Gégout-Petit and Commenges (2010) and Eichler and Didelez (2010). Assumptions needed for a causal interpretation of dynamic models have been presented in Arjas and Parner (2004) and Commenges and Gégout-Petit (2015). Dynamical models in discrete time, and in particular linear increment models (LIM), have been proposed by Diggle et al. (2007) and Ho et al. (2014). Aalen et al. (2012) have suggested that such models can be useful for studying the HAART effect on CD4 counts or viral load. Discrete-time models, however, may not be completely satisfactory because the processes of interest most often exist in continuous time. Systems of differential equations in continuous time can also be used to model the interaction between HIV and CD4 cells populations. Models based on differential equations, called “mechanistic”, considerably helped in understanding some important features of the infection: see Perelson (2002) for a review. In our setting, it is possible to model the treatment effect from a biological perspective. Introducing random effects is an efficient way to model differences between subjects with a minimum of additional parameters (Wu, 2005; Guedj et al., 2007; Lavielle et al., 2011). Mechanistic models have mostly been used to analyze data from clinical trials. Using mechanistic models to estimate the effect of HAART based on data of large observational cohorts is possible, but to our knowledge, has never been attempted.
The aim of this paper is to propose dynamic models in discrete and continuous time for assessing the causal effect of a treatment on a marker in observational studies. Specifically, we aim to estimate the HAART effect in HIV infected patients. We present several possible dynamic models, as well as MSMs, and compare them using simulations and real data. Although we worked out analytics in simple cases (see Appendix), the comparison is mainly empirical and aims to describe the assumptions and variability of results provided by the various methods. In Section 2, we present the statistical models: the naive model, the MSMs, the discrete-time dynamic models and the mechanistic approach. In Section 3, we compare the results of these models in simulation, where the data are generated from a complex mechanistic model. Section 4 is the application on the data of two cohorts of HIV infected patients: the Swiss HIV Cohort Study (SHCS) and the ANRS CO3 Aquitaine cohort. Section 5 concludes.
2. Modeling the treatment effect in observational studies
2.1 Notations and the naive model
We denote the value of a physiological marker for subject i at time t by , say the CD4 count. The value of a treatment given at time t to subject i is denoted by , say HAART. For sake of simplicity, we only model two treatment states: when treatment is not given, and when treatment is given, and we assume that once initiated the treatment is not interrupted; however, one could expand the model to accommodate different levels of treatment. If treatment is started at time t then and . We use overbars to represent the histories of these processes: for instance . We denote by the cumulative time under treatment until time t. In the absence of confounding by indication, the simplest model would be to regress on . Cole et al. (2005) notes the advantages of a piecewise linear regression model that allows a change in the effect of treatment after one year. Thus, our Model 1 is a naive model with different treatment effects before and after one year:
| (2.1) |
where is the cumulative time under treatment up to time t minus one year: (with the convention for t < 0). The β’s can be estimated by conventional GEE (Liang and Zeger, 1986) because we are interested in the population average. We use the independence working correlation structure in our analyses; otherwise results could be biased because of the presence of time-varying covariates (Pepe and Anderson, 1994).
2.2 Marginal structural models (MSM)
Because treatment is given to subjects with low CD4 counts, treated subjects tend to have low CD4 counts. Thus, the true value of parameter β1 in Model 1 cannot be interpreted as the causal effect. MSMs are designed to estimate the causal effect of a treatment given time dependent confounding. It is assumed that to each particular value of treatment history of , a potential outcome is associated (possibly contrary to the treatment actually received). A model is postulated to describe how the potential outcomes vary as a function of the different treatment trajectories. Hernán et al. (2002) and Cole et al. (2005) proposed benchmark models that also adjust for confounders such as time (t) and baseline value of biomarkers (Y0) in the regression. To duplicate this, our Model 2 is::
| (2.2) |
Robins et al. (2000) showed that the causal parameters of this model can be estimated with a suitably weighted GEE. The weights represent the inverse probability of treatment. The probability of treatment at time t depends on the history up to time t of a vector of variables L denoted typically includes include . The probability of treatment is estimated at each point in time using a treatment model (generally a logistic model) and the weights are the product over time of these probabilities; one often use stabilized weights as in Equation 2.3. An extension allows for censoring (Cole et al., 2005; Cole and Hernán,2008), however, the most important correction is generally for the probability of treatment (Ko et al., 2003). Stabilized inverse probability of treatment weights are defined as:
| (2.3) |
Results from Model 2 are consistent if the treatment model corresponds to the true treatment assignment mechanism; i.e. if all the confounders (factors influencing both the outcome of interest and treatment assignment) have been taken into account. We defined L for treatment model in Model 2 as baseline and time-varying CD4 count in categories (< 200, [200; 400], > 400), viral load in categories (< 401, 401 – 10000 and > 10000), and an indicator of undetectable viral load. In Models 1 and 2 the effect of treatment on CD4 counts during the first year is given by β1 and the effect after one year of treatment is given by β1 + β2.
2.3 Modeling the increment with a dynamical MSM
It is more relevant to model the change in the marker of interest, rather than its current value. This fits well with a causal thinking which considers that the change of a process depends on its present, and possibly past, states. Thus, we could model ; in order to account for non-equally spaced measurements, we might assume that the change in biomarker values is proportional to the time elapsed between two measurements; that is, , where is the tth calendar time of observation since the baseline measure of subject i. We define the biomarkers increments as . We take this approach when developing linear increment models in Section 2.4. However it is possible to express a MSM for the ’s as:
| (2.4) |
Our Model 3 is then the combination of this MSM for together with the treatment model (2.3). Note that with equally spaced measurements, Model 2 and 3 will be structurally identical if the same unit of time (e.g. year) is used for and the cumulative exposures (see Appendix). Inference approaches will be different, thus, Model 3 should be more suitable than Model 2 because we use an independence working correlation matrix for the ’s, which is much more acceptable than assuming independence for the ’s.
2.4 Full discrete-time dynamical models - linear increment models (LIM)
We can also fit a linear mixed-effects model for the ’s. The inter-subject variability is accounted for by a random effect b assumed normally distributed with zero expectation. Thus, LIM specify the distribution of Zi conditional on the bi. We propose three LIM; the simplest is Model 4, where the ε’s are i.i.d. normal variables with zero expectation:
| (2.5) |
In both Models 3 and 4, the effect of treatment on the CD4 counts is during the first year of treatment, and after one year, where is the mean of all the .
Many deterministic dynamical models have equilibrium points; similarly many stochastic dynamical models tend toward a stationary process. This property fits very well with the behavior of biological systems since concentrations of many molecules or cells have a tendency to return to the same value, a property called “homeostasis”. Difference equations of the type correspond to an autoregressive model of order one, denoted AR(1): with γ′ = (γ1 + 1). It is well known that if |γ′| < 1 this process converges toward a stationary process (in discrete time) with expectation ; this is always defined unless γ1 = 0, as is the case in Model 4 which does not have a finite stationary expectation. When using a model which has this convergence property, it may not be necessary to have a two-slope model. For example, we define Model 5, which tends to a stationary process with expectation for treated patients if −2 < β2 < 0 and β0 + β1 > 0:
| (2.6) |
A more realistic modeling of CD4 counts takes viral load into account. Here, we make a step toward mechanistic models because we know that the virus concentration and the CD4 concentration are inter-related processes. Thus, Model 6 is based on a system of two difference equations:
| (2.7) |
where , with the viral load at time t for patient i. The random effects di and bi, and the errors and are all i.i.d. and normally distributed with zero expectation. In Model 5 and 6, one year and subsequent years increase in CD4, as well as the long term change, are easily computed by solving the difference equations numerically. Moreover, for testing whether the treatment has an effect, it is convenient to test the hypotheses β1 = 0 and α1 = 0, by Wald tests for instance. As a reviewer notes, an interesting question is the correspondence between LIM and MSM. The Appendix shows that under some (rather strong) assumptions, Model 4 estimates the same causal parameters as Models 2 and 3 without using a treatment model.
2.5 Continuous Dynamical models, Mechanistic Models (ODE-NLME)
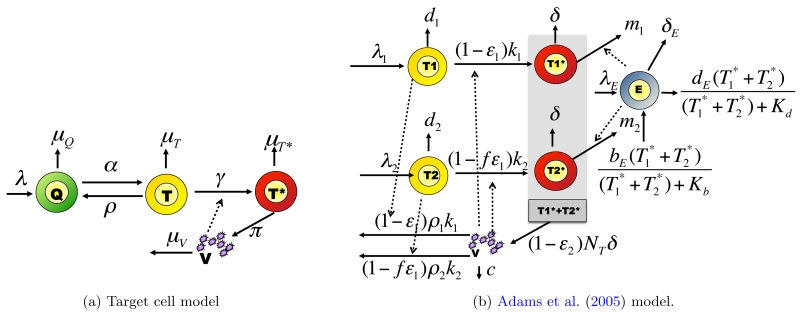
In reality, biomarkers processes exist in continuous time. A natural extension of a dynamic model in discrete time, as , is a model based on differential equations. (Perelson, 2002) give a review of some models for HIV dynamics based on ODE systems. In this article, we consider the “target cells model”, called Model 7 and described bellow, which proved to provide a good fit and prediction abilities (Prague et al., 2013).
Biological system
We know that only infected cells (T*) can produce viruses (V). The target cells model distinguishes between uninfected quiescent cells (Q) and target cells (T). The instantaneous change of concentrations of these populations at time t, for all real value of t > 0, is given by the ODE system:
| (2.8) |
The system is graphically represented in Figure 1a. Here, the parameters have biological meanings: λ is the production rate of new CD4 cells, the μ’s are death rates of different populations of cells, α and ρ are transition rates between quiescent and target cells, π is the rate of production of virions by infected cells, and γ is the infectivity parameter. The model assumes that the rate of infection of target cells is γVt.
Figure 1.
Mechanistic models for HIV dynamics. Type of cells of interest are viruses (V), effector cells E and CD4 cells which may be quiescent (Q), target cells (T, T1, T2) or infected (T*,, ). Parameters are defined in Table 1.
Inter-individual variability
A model that allows parameters to vary between individual is a mixed effects model. We model the inter-individual variability on the log-transformed parameters denoted with a tilde to ensure positiveness during estimation. In this application, two random effects and are introduced (Prague et al., 2012): and . Biologically, the causal effect of treatment can be modeled as an effect on the infectivity parameter γ. The parameter γ depends on t through , where we expect β < 0, so that the treatment decreases the infectivity of the virus:
| (2.9) |
Observation model
One important consequence of using continuous time models is that we must distinguish between the biological system which exists in continuous time and observations which are made at discrete times. To make an additive model for measurement error acceptable, we use 4th-root transformation for CD4 and a log10 transformation for the viral load. With errors and both i.i.d. and normally distributed, the observation model is:
| (2.10) |
Inference
Inference is much more complex and computationally demanding than in discrete-time models. We base inference on a penalized maximum likelihood approach and to achieve identifiability, we include prior knowledge of mechanistic parameters. Our priors (Table 1) are based on estimates in the literature of these parameters (Prague et al., 2012). This approach has been implemented in the NIMROD program (Prague et al., 2013). Assessing the long-term treatment effect in Model 7 is possible by analytically computing the equilibrium point. One year and subsequent year increase in CD4 after treatment initiation can be computed by solving the ODE system for given values of the random effects. The marginal effect can be computed as the mean of the individual effects in the population. The β parameter in the infectivity definition gives the effect of treatment, and a Wald test can be used to test the no-effect hypothesis (β = 0).
Table 1.
Meaning of parameters in the dynamical models presented in Figure 1. The upper part gives prior means and standard deviations for normal a priori distributions used for estimation of mechanistic parameters in Model 7 for the ‘Target cell model’. The lower part gives parameter values used for data simulation from the Adams et al. (2005) model.
| Normal priors used for analysis* on the log value of the parameter |
||||
|---|---|---|---|---|
| Name | Description | mean | sd. | |
| λ | Natural production rate | 2.55 | 1.90 | |
| μ T* | Natural death rate of T* cells | −0.05 | 0.68 | |
| μ Q | Natural death rate of Q cells | −9.00 | 1.00 | |
| α | Transition rates between Q and T cells | −4.00 | 2.00 | |
| ρ | Transition rates between T and Q cells | −4.34 | 1.38 | |
| μ T | Virions natural death rate | −2.59 | 0.34 | |
| γ | Infectivity parameter | −5.76 | 4.02 | |
| π | Rate of production of virions by infected cells | 4.04 | 2.66 | |
| μ V | Natural death rate of T* cells | 2.83 | 0.68 | |
| Parameter Value used for simulations for each population (X)† |
||||||
|---|---|---|---|---|---|---|
| Name | Description | Units | Type 1 | Type 2 | Effectors | Virus |
| λ X | Natural production rate | 5000 | 31.98 | 1.0 | - | |
| (1-ϵX) | Treatment efficacy | no unit | 50% | 83% | - | - |
| dX | Natural death rate | 0.01 | 0.01 | 0.25 | - | |
| δ X | Infection-induced death rate | 0.7 | 0.7 | 0.1 | - | |
| ρ X | Number of virions infecting a cell | 1 | 1 | - | - | |
| mX | Immune-induced clearance rate | 1 × 10−5 | 1 × 10−5 | - | - | |
| kX | Infection rate | 8 × 10−7 | 1 × 10−4 | - | - | |
| c | Virions natural death rate | - | - | - | 13 | |
| NT | Virions production per infected cells | - | - | - | 100 | |
| Kb | Saturation constant cells birth | - | - | 100 | - | |
| Kd | Saturation constant cells death | - | - | 500 | - | |
| bE | Infection-induced birth rate for E cells | - | - | 0.3 | - | |
Reference and explanation for these choices can be found in Prague et al. (2012).
For each simulated patient, every parameter got a random effect leading to 50% coefficient of variation
3. Simulation study
We simulated data with the Adams et al. (2005) model made up of two populations of target cells and a population of immune effectors such as cytotoxic T-lymphocytes (see Figure 1b and Table 1), which is much more complex than Model 7 (see Web-Supplementary Material A4). Individual variability was introduced by drawing parameters from normal distributions (with mean values listed in Table 1 and variances chosen to obtain a variation coefficient of 50%). By controlling the value of random effects, we ensured that the steady state baseline distributions of CD4 counts and viral load were consistent with the baseline values distributions found in Aquitaine cohort and SHCS dataset. See Web-supplementary Material A1 for details. We generated observations every 3 months; the standard deviations of the measurement errors were σVL = 0.6 and σCD4 = 0.1. Viral load was artificially made undetectable at the level of 50 copies/mL. Treatment assignment was done by simulating a CD4 count assessment at every observation and by fixing a probability of treatment assignment depending on the observed CD4 count. We took empirical probabilities from the Aquitaine cohort and SHCS dataset: treatment was attributed in 2%, 28% or 47% of patients where their CD4 count was > 400, [400, 200] or < 200. No other confounder was considered. Simulated patients are supposed fully observed for 5 years. We simulated n=200 and n=1500 patients. Table 3 gives a general description of both simulated and real datasets and there is no obvious difference between simulated and real cohort data in their descriptive statistics. We define the “average causal effect in treated patients” as the mean difference between the observed CD4 according to the observed treatment initiation and the counterfactual CD4 under no treatment initiation. The result of this computation was a 350 cells increase in CD4 after 1 year, a 362 cells increase after 2 years and an overall increase of 370 CD4 cells after an infinite (large) time. Technical details and code for analysis are described in Web-Supplementary Material C.
Table 3.
Data description for illustrations : Average viral load, CD4 counts and percentage of treatment assignment in the population are displayed for simulated data and real data from the Aquitaine cohort and the SHCS. Statistics displayed are mean [Q1;Q3].
| Simulated dataset |
Aquitaine Cohort |
SHCS | ||
|---|---|---|---|---|
|
| ||||
| n | 200 | 1500 | 1591 | 1726 |
| Missing data | ||||
| Administrative | - | - | 81.6% | 74.7% |
| Death | - | - | 12.7% | 6.4% |
| Lost of follow-up | - | - | 5.7% | 18.9% |
|
| ||||
| CD4 count | ||||
| Baseline | 428 [ 266 ; 545 ] | 420 [ 253 ; 530 ] | 471 [ 298 ; 612 ] | 536 [ 357 ; 670 ] |
| Follow-up untreated | 594 [ 485 ; 675 ] | 588 [ 478 ; 656 ] | 625 [ 440 ; 762 ] | 543 [ 363 ; 675 ] |
| Follow-up treated | 627 [ 417 ; 837] | 606 [ 405 ; 801 ] | 492 [ 315 ; 638 ] | 507 [ 300 ; 660 ] |
|
| ||||
| Viral Load | ||||
| Baseline | 3.9 [ 3.3 ; 4.6 ] | 4 [ 3.4 ; 4.7 ] | 4.2 [ 3.6 ; 4.8 ] | 4.0 [ 3.4 ; 4.6 ] |
| Follow-up untreated | 3.5 [ 2.9 ; 4.2 ] | 3.7 [ 3.1 ; 4.4 ] | 3.3 [ 2.3 ; 4.2 ] | 3.8 [ 3.1 ; 4.5 ] |
| Follow-up treated | 2.6 [ 1.7 ; 3.2 ] | 2.6 [ 1.7 ; 3.2 ] | 2.7 [ 1.7 ; 3.6 ] | 3.2 [ 2.4 ; 4.1 ] |
| % undetectable viral load (baseline,untreated, treated) |
(3%;4%;40%) | (2%;3%;38%) | (7%;22%;48%) | (10%;15%;57%) |
|
| ||||
| Treatment assignment | ||||
| Time (day) | 412 [ 1 ; 631 ] | 377 [ 91 ; 451 ] | 727 [ 1 ; 1281 ] | 548 [ 183 ; 752 ] |
| % treated | 69% | 65% | 64% | 34% |
Table 2 presents the estimates for Models 1-7 on the simulated datasets. The naive Model 1 largely underestimated the treatment effect. This was corrected by the MSM Models 2 and 3 (see details about weights in Web-Supplementary Material A2). Model 4 also yielded good estimates of the mean causal effect in treated patients. Moreover, it led to an increased significance of results compared to Model 3 for increase in CD4 count during the first year. Regarding increase in CD4 count in subsequent years, whereas the Model 3 underestimate its variability (for large samples, 12 ∉ [2; 6]), Model 4 is more reliable (for large samples, 12 ∈ [−1; 15]). This underestimation of the variance of Model 3 may be driven by an overfitting of inverse probability of treatment weights. This overfitting could have arisen because the data generation was based on CD4 cell count evolution and not on viral load, whereas the inverse probability of treatment weights were calculated from treatment models with both past CD4 cell count and viral load as predictors. We note that long-term increase in CD4 is infinite in Models 1 to 4, even though it happens at a really slow rate. On the contrary, Models 5-7 exhibit an equilibrium point which makes it possible to consider the long-term causal effect of treatment. All dynamic models gave an estimate of the long-term effect of treatment for which the true value was included in the 95% confidence interval. The initial increase in CD4 during the first year was not correctly reproduced by Model 5. Models 6 and 7 which both incorporate the dynamics of viral load gave a better estimate of the increase in CD4 count (see details about estimated values of biological parameters in the Model 7 in Web-Supplementary Material A3). All models found a significant effect of treatment on CD4 counts in the first year. Sandwich estimators were used for calculating the standard errors for GEE methods and Fisher information matrix was used for methods based on maximum likelihood. Altogether, the dynamic models (Model 4-7) have greater statistical evidence, with higher Z-statistics rejecting the no-effect hypotheses, than the GEE based models (Models 1-3). Finally, while fitting Models 1-6 took less than a minute on a typical laptop, fitting Model 7 took about 10 hours of parallel computing with 100 cores. All results and conclusions are similar in small and large samples.
Table 2.
Estimated treatment effect on CD4 counts from simulated data: Model 1: Naive regression; Model 2: MSM on ; Model 3: MSM on ; Model 4: Simple LIM; Model 5: autoregressive LIM; Model 6: LIM system; Model 7: mechanistic model.
| Simulated Dataset with Adams et al. (2005) model | |||||||
|---|---|---|---|---|---|---|---|
| n=200 |
n=1500 |
||||||
| Model | β treatment‡ | Effect | Sd. | Z-stat† | Effect | Sd. | Z-stat† |
| Model 1 | < 1 yr | 136 | 29 | 4.68 | 172 | 11 | 16.34 |
| > 1 yr | −11 | 6 | −1.99 | −12 | 2 | −5.07 | |
| ∞ | − ∞ | - | - | − ∞ | - | - | |
| Model 2 | < 1 yr | 327 | 31 | 10.64 | 325 | 11 | 28.81 |
| > 1 yr | −14 | 10 | −1.45 | −7 | 4 | −1.56 | |
| ∞ | − ∞ | - | - | − ∞ | - | - | |
|
| |||||||
| Model 3 | < 1 yr | 371 | 24 | 15.47 | 364 | 10 | 35.49 |
| > 1 yr | 8 | 3 | 2.45 | 4 | 1 | 3.2 | |
| ∞ | +∞ | - | - | +∞ | - | - | |
|
| |||||||
| Model 4 | < 1 yr | 362 | 17 | 21.60 | 378 | 6 | 61.35 |
| > 1 yr | 8 | 12 | 0.7 | 7 | 4 | 1.61 | |
| ∞ | +∞ | - | - | +∞ | - | - | |
| Model 5 | < 1 yr | 133 | * | * | 136 | * | * |
| > 1 yr | 84 | * | * | 86 | * | * | |
| ∞ | 359 | * | * | 370 | * | * | |
| β 1 | 149 | 5 | 31.24 | 154 | 2 | 89.36 | |
| Model 6 | < 1 yr | 325 | * | * | 334 | * | * |
| > 1 yr | 31 | * | * | 34 | * | * | |
| ∞ CD4 | 360 | * | * | 371 | * | * | |
| ∞ VL | −1.9 | * | * | −2 | * | * | |
| β 1 | 600 | 21 | 28.42 | 630 | 8 | 82.22 | |
| α 1 | −6.6 | 0.16 | −40.86 | −6.9 | 0.06 | −120.51 | |
|
| |||||||
| Model 7 | < 1 yr | 312 | * | * | 304 | * | * |
| > 1 yr | 2 | * | * | 4 | * | * | |
| ∞ CD4 | 308 | * | * | 306 | * | * | |
| ∞ VL | −5.6 | * | * | −4.98 | * | * | |
| β | −1.12 | 0.014 | −79.3 | −1.03 | 0.003 | −295.6 | |
Estimates for treatment effect (β) are significant at level 10% if the absolute value of Z-stat is greater than 1.64 and significant at level 5% if the absolute value of Z-stat is greater than 1.96.
To be compared with mean treatment effect in treated for (< 1 year; > 1 year; ∞): benchmarks values are (350;12;370) for these simulations.
Simulated delta-method can lead to estimation of these values, but is not implemented here. Indeed, for Model 5, 6 and 7 significance of the treatment effect has to be evaluated through the mechanistic parameters β1,α1 and β.
4. Real data
We used two large cohorts: the ANRS CO3 Aquitaine cohort (Thiébaut et al., 2000) and the Swiss HIV Cohort Study (SHCS) (Sterne et al., 2005; Gran et al., 2016). Like Cole et al. (2005), we took a sub-sample of patients who were alive, HIV positive, yet untreated and under follow-up in April 1996 when HAART became available. All patients taking only one or two antiretroviral drugs (rather than HAART) were excluded. Once a patient was on HAART, we assumed he or she remained on it. For each patient, follow-up began with the first visit after April 1996 and ended with 1) the last visit at which he or she was seen alive, 2) the last visit before patient discontinued the study, or 3) April 2003, whichever comes first. Data were assumed missing completely at random (MCAR); thus we deleted observations where either the viral load or CD4 count was missing. Patients with only one observation were excluded. After exclusions, there were 1591 patients from the Aquitaine cohort and 1726 patients from the SHCS (see Web-Supplementary Material B1 for a description of patient selection). Table 3 gives descriptive statistics. For most patients, follow up ended with administrative censoring and therefore we assumed censoring was not informative.
Table 4 displays the results we obtained for the effect of treatment on CD4 counts. The naive Model 1, not corrected for treatment assignment, indicated a small and non-significant increase in CD4 for SHCS cohort, and a significant negative effect for the Aquitaine Cohort; thus illustrating the need for modeling treatment assignment. This is corrected by the use of a treatment model in MSM Models 2. Models 3 and 4 led to similar results in the SHCS and different results in the Aquitaine cohort. For Model 4, this may be because covariates other than CD4 count are also confounders (such as viral load). For Model 3, some covariates driving the choice of treatment initiation may have been omitted in the treatment model. Estimates from both Models 3 and 4 suggest that treatment causes a significant increase in CD4 within the first year and in subsequent years. The one-year increase, however, was much smaller for the Aquitaine cohort than for the SCHS when using MSM-based models. See Web-Supplementary Material B2 for a discussion of this result in relation with a possible practical violations of the experimental treatment assumption Cole and Hernán (2008). The results of the dynamical models, especially Models 6 and 7, were more consistent between cohorts.
Table 4.
Estimated treatment effect on CD4 counts from real data of the Aquitaine cohort and SHCS: Model 1: Naive regression; Model 2: MSM on ; Model 3: MSM on ; Model 4: Simple LIM; Model 5: autoregressive LIM; Model 6: LIM system; Model 7: mechanistic model.
| Real Dataset observational studies | |||||||
|---|---|---|---|---|---|---|---|
| SHCS |
Aquitaine Cohort |
||||||
| Model | β treatment | Effect | Sd. | Z-stat† | Effect | Sd. | Z-stat† |
| Model 1 | < 1 yr | 6 | 16 | 0.34 | −94 | 12 | −7.55 |
| > 1 yr | 30 | 6 | 5.42 | 30 | 3 | 9.75 | |
| ∞ | +∞ | - | - | +∞ | - | - | |
| Model 2 | < 1 yr | 208 | 18 | 11.31 | 36 | 19 | 1.87 |
| > 1 yr | 50 | 9 | 5.79 | 53 | 5 | 9.62 | |
| ∞ | +∞ | - | - | +∞ | - | - | |
|
| |||||||
| Model 3 | < 1 yr | 174 | 10 | 17.29 | 27 | 14 | 1.99 |
| > 1 yr | 61 | 5 | 12.37 | 34 | 5 | 6.3 | |
| ∞ | +∞ | - | - | +∞ | - | - | |
|
| |||||||
| Model 4 | < 1 yr | 189 | 11 | 17.33 | 109 | 9 | 12.03 |
| > 1 yr | 73 | 8 | 9.07 | 55 | 6 | 8.99 | |
| ∞ | +∞ | - | - | +∞ | - | - | |
| Model 5 | < 1 yr | 26 | * | * | 45 | * | * |
| > 1 yr | 14 | * | * | 19 | * | * | |
| ∞ | 55 | * | * | 79 | * | * | |
| β 1 | 60 | 4 | 16.04 | 14 | 3 | 4.12 | |
| Model 6 | < 1 yr | 73 | * | * | 92 | * | * |
| > 1 yr | 26 | * | * | 16 | * | * | |
| ∞ CD4 | 104 | * | * | 111 | * | * | |
| ∞ VL | −2.0 | * | * | −2.3 | * | * | |
| β 1 | 80 | 16 | 5 | 28 | 18 | 1.53 | |
| α 1 | −3.29 | 0.09 | −38.4 | −3.19 | 0.1 | −30.55 | |
|
| |||||||
| Model 7 | < 1 yr | 104 | * | * | 71 | * | * |
| > 1 yr | 18 | * | * | 9 | * | * | |
| ∞ CD4 | 127 | * | * | 86 | * | * | |
| ∞ VL | −4.09 | * | * | −3.14 | * | * | |
| β | −1.73 | 0.05 | −34.79 | −0.89 | 0.01 | −85.77 | |
Estimates for treatment effect (β) are significant at level 10% if the Z-stat is greater than 1.64 and significant at level 5% if the Z-stat is greater than 1.96.
Simulated delta-method can lead to estimation of these values, but is not implemented here. Indeed, for Model 5, 6 and 7 significance of the treatment effect has to be evaluated through the mechanistic parameters β1,α1 and β.
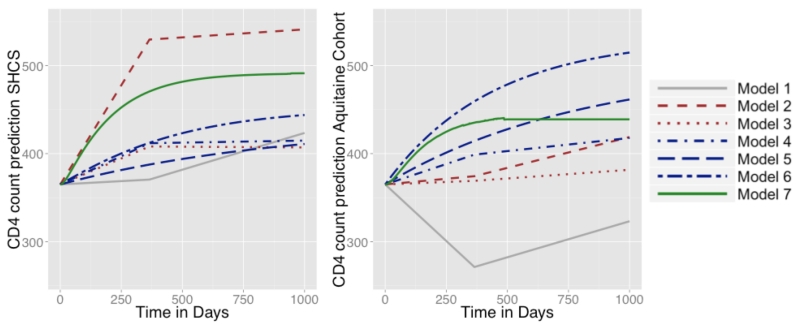
Model 6 is interesting because it dissociates the effect of the treatment on CD4 count from its effects on viral load. In this model, the estimated treatment effect on CD4 count was small in both cohorts and was non-significant for the Aquitaine cohort. In contrast, the effect on viral load was highly significant in both cohorts. This is consistent with the mode of action of antiretroviral treatment: the increase in CD4 count is essentially mediated by the decrease in viral load; the latter is the direct effect of treatment. Such biological knowledge is incorporated in Model 7, where the treatment acts on the infectivity parameter. In view of the Z-statistics obtained by a Wald test of the hypothesis β = 0 in Equation 2.9, the statistical evidence obtained in Model 7 is very high (this is confirmed by a likelihood ratio test). Moreover, Model 7 gives an insight into the value of the biological birth and death rates of cells during the infection (see Web-Supplementary Material B3). Regarding these biological parameters, the estimates from the two data sets are rather consistent in the sense that they have the same order of magnitude, although a formal comparison would show that several parameters are different, potentially due to different characteristics of the patients in the two cohorts. Finally, a simple way to look at these results and to compare them, is to consider the mean evolution in CD4 over time. Figure 2 represents the predicted CD4 counts with Models 1 to 7 for treated patients starting at baseline with CD4 count of 365 and a viral load of 4.4 (which are approximately the mean values at treatment initiation in these cohorts). For Models 1-3, these curves are deterministic, which is not the case for Models 4-7 because these models have random effects. For these latter models, we computed the mean predicted curves depending on the value of the random effect, which have to be set to values compatible with the baseline values of the biomarkers. In order to set them, in both cases, we computed the equilibrium point of the system without treatment and solved the system of equations. Figure 2 shows that the naive Model 1 is not in agreement with medical knowledge. Models 1-3 are unstable, whereas Models 4-7 are more consistent between the two studies. Models 3, 4 and 5 lead to similar trajectories in the SHCS dataset but very different trajectories in the Aquitaine Cohort study. In the latter, because trajectories for Model 3 look unrealistic and too pessimistic, we believe LIM Models 4 or 5 is more realistic. This illustrates the instability of weighted approaches. Finally, Model 5, 6 and 7 have an equilibrium point. Model 7 reaches a steady state in about 1 year which is in line with observed data and is similar in both cohorts.
Figure 2.
Mean evolution in CD4 predicted by Model 1 (plain line, simple regression), Model 2 (dashed line, MSM on ), Model 3 (dotted line, MSM on ), Model 4 (dotted line, simple LIM), Model 5 (dotted line, autoregressive LIM), Model 6 (dashed-dotted line, LIM system), and Model 7 (long-dashed line, mechanistic model) for treated patients starting with 365 CD4 cells/mL and a viral load of 4.4 log10 copies/mm3: (left) estimates from the SHCS data (right) estimates from the Aquitaine cohort. This figure appears in color in the electronic version of this article.
5. Conclusion
In this paper we estimated the effect of HAART on CD4 count using four dynamic models and compared estimates with those from a naive regression model, a variant of a previously proposed MSM and a MSM based on linear increments. This is an empirical comparison rather than a theoretical comparison. We discussed assumptions and validity of the various approaches together with possible bias in estimates, statistical evidence and practicability of use. The naive regression model (Model 1) strongly underestimated the effect of the treatment. The MSMs (Models 2 and 3) corrected this misleading result but sometimes failed to reach significance or were unstable across cohorts. The discrete-time dynamic models (Models 4, 5 and 6) based on LIM gave rather good estimates and show higher statistical evidence, although they may sometimes be too rigid. All the discrete-time models are easily fit without specialist software. The continuous-time dynamic model based on ODE-NLME (Model 7) gave good results. Models 6 and 7, which jointly model CD4 and viral load gave the most consistent results, with a richer interpretation since they explicitly model the effect of HAART on CD4 via a direct effect on viral load.
We have used a linear MSM with two slopes similar to that proposed by Cole et al. (2005). This model adequately represents the short term (few years) effect of treatment but not the long-term effect because the effect in subsequent years implies a biologically implausible indefinite increase over time. As pointed out by the Associate Editor, because MSM can specify any reasonable outcome regression model, ranging from a very simple model which posits that only the most recent exposures affect the outcome to something very complex - e.g. a spline-weighted sequence of exposures (Xiao et al., 2014), it would be possible to define an MSM in which the effect would be bounded. However, this would be at the cost of additional non-linear parametrization. Also, it would be possible to use more recent methods such as the history adjusted MSM (Petersen et al., 2007); these are most suited for dynamic treatment regimes whereas we assessed the effect of a static treatment regimes in this work. In contrast most dynamical models (although not Model 4) have an equilibrium point. We showed that a MSM could be fitted for the increment rather than for . Thus, the MSM approach could complement the dynamic approach in the sense that less stringent assumptions would be needed for causal inference. However, the dynamic models already do a good job and the need to correct them (at the price of more complex procedure and loss of statistical evidence) is not obvious. In this paper we assumed MCAR observations for GEE, which is appropriate because most patients were administratively censored. MAR observations can be treated by using inverse probability of censorship weights (see Web-Supplementary Material Section B4). One advantage of likelihood-based dynamic models is that they are still valid given MAR observations.
The mechanistic Model 7 directly incorporates biological knowledge. This leads to a more significant test for the parameter of interest. The simulated data in the paper are obtained with a dynamic model, so one might think that this favors dynamic models; however the true generation process comes itself from a complex dynamic system. Thus, the simulated model is much more complex than Models 4-7 which are highly misspecified. Of course, misspecification is not only a binary feature but can be quantified, in principle, for instance by the Kullback-Leibler risk between the generating distribution and the best distribution in the model (Commenges, 2015). Both simulated and real data are more complex than the models used to analyse that data: in both cases, we know that the model is misspecified but we hope that the structure of the model captures essential features of the dynamics of the system. The comparison between dynamic and MSM approach remains empirical: we compare estimates from plausible MSM and dynamic models. Finally, mechanistic models, once estimated, open the possibility of designing optimal control of the therapy, as has been proposed on simulations by Adams et al. (2004); Ernst et al. (2006), and also Prague et al. (2012). The issue of “optimal treatment regime” has also been tackled outside of the context of mechanistic models (Petersen et al., 2007; Orellana et al., 2010; Saarela et al., 2015). The drawback of the continuous-time approach is that it is numerically challenging and requires special software running on cluster computers.
Supplementary Material
Acknowledgement
The authors thank the investigators of the Aquitaine Cohort and the Swiss HIV Cohort Study. Parts of this work was founded by NIH grants R37 AI 51164. Parallel computing was used thanks to the MCIA (Mésocentre de Calcul Intensif Aquitain) of the Université de Bordeaux and of the Université de Pau et des Pays de l’Adour.
APPENDIX: Correspondence between parameters of Model 2, 3 and 4
The question of how parameters from a MSM and a dynamical model relate is difficult for two reasons: the models are constructed differently; the philosophical approach to causality is different. We will make this exercise for comparing the MSM Model 2 and the LIM Model 4. One can reconcile the two philosophical approaches by saying that the “causal” interpretation (in a interventional point of view) is that for a given treatment trajectory , we expect under the MSM: . Model 4 is formulated in terms of the increments: , which by summation gives , where is a martingale. This is the Doob decomposition of the process Y. With the assumption of a “perfect” system or a NUC system (see Commenges and Gégout-Petit (2015) and Chapter 9 of Commenges and Jacqmin-Gadda (2015)), if we apply treatment trajectory , this define a new probability P a under which the Doob decomposition, which is: , from which we deduce: . Thus, Model 4 yields the same expectation under an intervention imposing treatment trajectory as Models 2 if β4 = 1, and the parameters giving the effects of and correspond. The same correspondence holds for Model 3, which is equivalent to Model 2 if the observations are equally spaced. Whereas Models 2 and 3 make the assumption that all confounders between A and Z are included in the computation of the inverse probability of treatment, Model 4 assumes that there is no confounder between A and Z. For instance the viral load Vt−1 might be a confounder which is taken into account in inverse probability of treatment in Models 2 and 3; in the dynamic approach one must use more complex models describing the dynamics of the viral load (such as Models 5 and 6). For Models 5, 6 and 7, marginal effects can still be computed (analytically or by simulation), but this may lead to complex forms, while generally MSM assume simple mathematical structures.
Footnotes
Web-Supplementary Material referenced in Sections 3, 4 and 5, the simulated data analyzed in Section 3 and a R program implementing Models 1-6 are available with this paper at the Biometrics website on Wiley Online Library. Programs to estimate the parameters with Model 7 are available on a dedicated website: http://www.isped.u-bordeaux.fr/NIMROD.
References
- Aalen O, RØysland K, Gran J, Ledergerber B. Causality, mediation and time: a dynamic viewpoint. J. R. stat. Soc. A. 2012;175:831–861. doi: 10.1111/j.1467-985X.2011.01030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams B, Banks H, Davidian M, Kwon H, Tran H, Wynne S, Rosenberg E. HIV dynamics: modeling, data analysis, and optimal treatment protocols. J Comput. Appl. Math. 2005;184:10–49. [Google Scholar]
- Adams B, Banks H, Kwon H-D, Tran HT. Dynamic multidrug therapies for HIV: Optimal and STI control approaches. Math. Biosc. Eng. 2004;1:223–241. doi: 10.3934/mbe.2004.1.223. [DOI] [PubMed] [Google Scholar]
- Arjas E, Parner J. Causal reasoning from longitudinal data. Scand. J. Stat. 2004;31:171–187. [Google Scholar]
- Cole S, Hernán M. Constructing inverse probability weights for marginal structural models. Am. J. Epidemiol. 2008;168:656–664. doi: 10.1093/aje/kwn164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S, Hernán M, Anastos K, Jamieson B, Robins J. Determining the effect of highly active antiretroviral therapy on changes in human immunodeficiency virus type-1 RNA viral load using a marginal structural left-censored mean model. Am. J. Epidemiol. 2007;166:219–227. doi: 10.1093/aje/kwm047. [DOI] [PubMed] [Google Scholar]
- Cole S, Hernán M, Margolick J, Cohen M, Robins J. Marginal structural models for estimating the effect of highly active antiretroviral therapy initiation on CD4 cell count. Am. J. Epidemiol. 2005;162:471–478. doi: 10.1093/aje/kwi216. [DOI] [PubMed] [Google Scholar]
- Commenges D. Information theory and statistics: an overview. 2015. arXiv:1511.00860. [Google Scholar]
- Commenges D, Gégout-Petit A. A general dynamical statistical model with causal interpretation. J. R. stat. Soc. B. 2009;71:719–736. [Google Scholar]
- Commenges D, Gégout-Petit A. The stochastic system approach for estimating dynamic treatments effect. Lifetime Data Anal. 2015;21:561–578. doi: 10.1007/s10985-015-9322-3. [DOI] [PubMed] [Google Scholar]
- Commenges D, Jacqmin-Gadda H. Dynamical Biostatistical Models. Chapman & Hall; 2015. [Google Scholar]
- Dawid AP. Causal inference without counterfactuals. J. Am. Stat. Assoc. 2000;95:407–424. [Google Scholar]
- Didelez V. Graphical models for marked point processes based on local independence. J. R. stat. Soc. B. 2008;70:245–264. [Google Scholar]
- Diggle P, Farewell D, Henderson R. Analysis of longitudinal data with drop-out: objectives, assumptions and a proposal. J. R. stat. Soc. C. 2007;56:499–550. [Google Scholar]
- Eichler M, Didelez V. On granger causality and the effect of interventions in time series. Lifetime data anal. 2010;16:3–32. doi: 10.1007/s10985-009-9143-3. [DOI] [PubMed] [Google Scholar]
- Ernst D, Stan G-B, Goncalves J, Wehenkel L. Clinical data based optimal STI strategies for HIV; Decision and Control, 2006 45th; IEEE. 2006.pp. 667–672. [Google Scholar]
- Gégout-Petit A, Commenges D. A general definition of influence between stochastic processes. Lifetime data anal. 2010;16:33–44. doi: 10.1007/s10985-009-9131-7. [DOI] [PubMed] [Google Scholar]
- Gran J, Ho R, Roysland K, Ledergerber B, Young J, Aalen O. Estimating the treatment effect of the treated under time-dependent confounding applied to simulated data and to data from the swiss HIV cohort study. J. R. stat. Soc. C. 2016 [Google Scholar]
- Granger C. Investigating causal relations by econometric models and cross-spectral methods. Econometrica. 1969;37:424–438. [Google Scholar]
- Guedj J, Thiébaut R, Commenges D. Maximum likelihood estimation in dynamical models of HIV. Biometrics. 2007;63:1198–1206. doi: 10.1111/j.1541-0420.2007.00812.x. [DOI] [PubMed] [Google Scholar]
- Hernán M. a., Brumback B, Robins J. Estimating the causal effect of zidovudine on CD4 count with a marginal structural model for repeated measures. Stat. Med. 2002;21:1689–709. doi: 10.1002/sim.1144. [DOI] [PubMed] [Google Scholar]
- Ho R, Gran J, Farewell D. Farewell’s linear increments model for missing data: The FLIM package. The R Journal. 2014;6:137–150. [Google Scholar]
- Ko H, Hogan JW, Mayer KH. Estimating causal treatment effects from longitudinal HIV natural history studies using marginal structural models. Biometrics. 2003;59:152–162. doi: 10.1111/1541-0420.00018. [DOI] [PubMed] [Google Scholar]
- Lavielle M, Samson A, Karina Fermin A, Mentré F. Maximum likelihood estimation of long-term HIV dynamic models and antiviral response. Biometrics. 2011;67:250–259. doi: 10.1111/j.1541-0420.2010.01422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Zeger S. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Orellana L, Rotnitzky A, Robins JM. Dynamic regime marginal structural mean models for estimation of optimal dynamic treatment regimes, part I: main content. Int. J. Biostat. 2010;6:1557–4679. [PubMed] [Google Scholar]
- Pepe M, Anderson G. A cautionary note on inference for marginal regression models with longitudinal data and general correlated response data. Commun. Stat. Simulat. 1994;23:939–951. [Google Scholar]
- Perelson A. Modelling viral and immune system dynamics. Nat. Rev. Immunol. 2002;2:28–36. doi: 10.1038/nri700. [DOI] [PubMed] [Google Scholar]
- Petersen ML, Deeks SG, Martin JN, van der Laan MJ. History-adjusted marginal structural models for estimating time-varying effect modification. Am. J. Epidemiol. 2007;166:985–993. doi: 10.1093/aje/kwm232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen ML, Deeks SG, van der Laan MJ. Individualized treatment rules: Generating candidate clinical trials. Stat. Med. 2007;26:4578–4601. doi: 10.1002/sim.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen ML, Wang Y, van der Laan MJ, Bangsberg DR. Assessing the effectiveness of antiretroviral adherence interventions: using marginal structural models to replicate the findings of randomized controlled trials. J AIDS. 2006;43:S96–S103. doi: 10.1097/01.qai.0000248344.95135.8d. [DOI] [PubMed] [Google Scholar]
- Prague M, Commenges D, Drylewicz J, Thiébaut R. Treatment monitoring of HIV-infected patients based on mechanistic models. Biometrics. 2012;68:902–911. doi: 10.1111/j.1541-0420.2012.01749.x. [DOI] [PubMed] [Google Scholar]
- Prague M, Commenges D, Guedj J, Drylewicz J, Thiébaut R. NIMROD: a program for inference via a normal approximation of the posterior in models with random effects based on ordinary differential equations. Comput. Meth. Prog. Bio. 2013;111:447–458. doi: 10.1016/j.cmpb.2013.04.014. [DOI] [PubMed] [Google Scholar]
- Prague M, Commenges D, Thiébaut R. Dynamical models of biomarkers and clinical progression for personalized medicine: The HIV context. Adv. Drug. Deliver. Rev. 2013;65:954–965. doi: 10.1016/j.addr.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Robins J, Hernán M, Brumback B. Marginal structural models and causal inference in epidemiology. Epidemiology. 2000;11:550–60. doi: 10.1097/00001648-200009000-00011. [DOI] [PubMed] [Google Scholar]
- Saarela O, Stephens DA, Moodie EE, Klein MB. On bayesian estimation of marginal structural models. Biometrics. 2015;71:279–288. doi: 10.1111/biom.12269. [DOI] [PubMed] [Google Scholar]
- Sterne J, Hernán M, Ledergerber B, Tilling K, Weber R, Sendi P, Rickenbach M, Robins J, Egger M. Long-term effectiveness of potent antiretroviral therapy in preventing AIDS and death: a prospective cohort study. The Lancet. 2005;366:378–384. doi: 10.1016/S0140-6736(05)67022-5. [DOI] [PubMed] [Google Scholar]
- Thiébaut R, Morlat P, Jacqmin-Gadda H, Neau D, Mercié P, Dabis F, Chêne G, the Groupe d’Epidmiologie du SIDA en Aquitaine (GECSA) Clinical progression of HIV-1 infection according to the viral response during the first year of antiretroviral treatment. J AIDS. 2000;14:971–978. doi: 10.1097/00002030-200005260-00008. [DOI] [PubMed] [Google Scholar]
- Walker A. Confounding by indication. Epidemiology. 1996;7:335–336. [PubMed] [Google Scholar]
- Wu H. Statistical methods for HIV dynamic studies in AIDS clinical trials. Stat. Methods Med. Res. 2005;14:171–192. doi: 10.1191/0962280205sm390oa. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Abrahamowicz M, Moodie EE, Weber R, Young J. Flexible marginal structural models for estimating the cumulative effect of a time-dependent treatment on the hazard: reassessing the cardiovascular risks of didanosine treatment in the swiss HIV cohort study. J. Am. Stat. Assoc. 2014;109:455–464. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.