Abstract
Asthma is a serious and hereditary respiratory disorder affecting all age groups. Interleukin-13 (IL-13) is a central regulator of allergic inflammation. The purpose of the present study was to estimate the relationship between IL-13 +1923C/T polymorphism and asthma susceptibility. Relevant case-control studies published between January 2000 and July 2016 were searched in the online databases. Review Manage (RevMan) 5.3 was used to conduct the statistical analysis. The pooled odds ratio (OR) with its 95% confidence interval (CI) was employed to calculate the strength of association. A total of 26 articles were retrieved, including 17642 asthma patients and 42402 controls. Overall, our results found that IL-13 +1923C/T polymorphism was significantly associated with increased risk of asthma under each genetic model (P<0.00001). Subgroup analysis by ethnicity showed that alleles and genotypes of this variant correlated with asthma among Asians and Caucasians, but only TT genotype under the homozygote model in Africans. When stratified by age group, this variant highly correlated with asthma in children and moderately in adults. Furthermore, the TT, CT and CC genotypes in asthma group were all significantly associated with increased IgE levels in sera of asthma patients when compared with controls. Our results suggested that IL-13 +1923C/T polymorphism contributed to the development of asthma. Further case-control studies with more ethnicities are still needed.
Keywords: Asthma, interleukin-13, meta-analysis, polymorphism
Introduction
Asthma, a heterogeneous disease, is the most common long-term inflammatory disease of the lung airways, remaining a major cause of disability, health resource utilization and poor quality of life worldwide [1,2]. It is characterized by the presence of recurring respiratory symptoms, reversible airflow obstruction and bronchospasm [3]. Its symptoms include episodes of wheezing, chest tightness, shortness of breath and coughing, occurring a few times a day or a week depending on the single individual [4]. According to the World Health Organization, it is estimated that 235 to 330 million people currently suffer from asthma, and approximately 250000 to 345000 people die from this disease per year throughout the world [5]. The risk factors such as tobacco smoking, indoor allergens, obesity, diet, air pollution and social factors might contribute to the development of asthma [6,7]. In addition, an elevation of serum IgE level is considered as a potent predictor of asthma course [8]. Although great advances for the development of asthma controller therapies have been made and death rates due to asthma have reduced greatly over these years, no available therapeutic regimens can cure this disease and its burden will continue to be driven by increasing prevalence [9]. Therefore, there is an urgent need to identify some biomarkers to predict this disease and create a guideline for the therapeutic strategies.
Recent progresses in molecular biology suggest complex interactions of innate and adaptive immune cells, structural cells and their cytokines involved in the process of airway inflammation [10,11]. A growing number of evidence have shown that interleukins (ILs) are critical to mounting inflammation and immune responses [12]. ILs are a multifunctional group of immunomodulators that primarily mediate the leucocyte cross-talk, and mainly regulate the immune cell proliferation, growth, differentiation, survival, activation and functions [13–15]. The interleukin-13 (IL-13) gene, located on human chromosome 5q31–33, is produced by innate lymphoid cells and T-helper type 2 (Th2) cells during allergic inflammation, containing four exons and three introns and encoding an unglycosylated protein composed of 132 amino acids. The most prominent effects of IL-13 include promotion of differentiation and survival of eosinophils and mast cells, activation of fibroblasts, elevation of bronchial hyperresponsiveness and switching of B-cell antibody production from IgM to IgE [16]. Furthermore, IL-13 inhibition may be beneficial in patients who are refractory to existing therapies [17]. Studies have shown that IL-13 might play a role in human diseases such as allergic airway disease [18], fibrosis [19] and renal cell carcinoma [20]. Genetic polymorphisms in IL-13 gene might influence its function, thus involved in the pathogenicity of asthma. One of the most studied single nucleotide polymorphisms (SNPs) was +1923C/T (rs1295686) in third intron at the intron/exon boundary. This variant was shown to be involved in the dysregulation of total IgE [21].
Several studies have identified the role of IL-13 +1923C/T polymorphism in asthma susceptibility; however, the results obtained from different geographical populations were very different. For example, Ramphul et al. [22] found that IL-13 +1923C/T locus had a significant effect predisposed to asthma in Mauritian Indian children, not in Chinese Han population. Moreover, the prevalence rates of asthma vary between countries, ranging from 1 to 18% and it is more common in developed than in developing countries [23]. Also, genetic associations with asthma can differ significantly among different ethnic populations [24,25]. Therefore, we conducted this meta-analysis to reassess the association of IL-13 +1923C/T polymorphism with asthma risk based on all the available case-control studies.
Materials and methods
Study identification
The electronic databases of Medline, Embase, PubMed, Chinese National Knowledge Infrastructure (CNKI) and Wanfang were comprehensively searched to retrieve relevant articles published between January 2000 and July 2016. The following medical subject heading (MeSH) terms: “asthma or asthmatic”, “cytokines or interleukin or interleukin-13 or IL-13” and “polymorphism or variant or SNP” as well as their combinations were employed as the searching keywords. The corresponding Chinese version was used in the Chinese databases. To obtain more data, we manually searched the references of related articles. Our analysis only focused on the studies that were written in English and Chinese. When the same authors or laboratories reported this issue on the same population, only the latest published full-text article was included.
Inclusion and exclusion criteria
The included studies must meet the following criteria: (i) case-control studies evaluating the correlation of IL-13 +1923C/T polymorphism in asthma risk; (ii) patients with asthma were defined according to the Guidelines of the American Thoracic Society [26] or other diagnostic criteria [27]; controls should be unrelated ethnically matched individuals with no symptoms or history of allergy and other pulmonary diseases; (iii) genotype information in patients and controls was available to extract; and (iv) the genotype distribution in controls should be in consistence with Hardy–Weinberg equilibrium (HWE). The exclusion criteria were: (i) without the control group; (ii) conference papers or review reports; (iii) data cannot be extracted; and (iv) with duplicated data.
Data extraction
Two of our authors independently assessed the extracted information of each included study. Any disagreement was resolved by discussion with a third author. Each item should be able to reach a final consensus. The following information was extracted from each article: the name of first author, published year, country, ethnicity, mean age, sample size, genotyping methods, genotype distribution and HWE in controls.
Statistical analysis
The association between IL-13 +1923C/T polymorphism and asthma risk was measured by pooled odds ratio (OR) with 95% corresponding confidence intervals (CIs). The significance of the pooled OR was determined by the Z-test and a P value less than 0.05 was considered significant. The allelic model (T compared with C), homozygote model (TT compared with CC), heterozygote model (CT compared with CC), dominant model (TT + CT compared with CC) and recessive model (TT compared with CT + CC) were calculated. The I2-test and the Q-statistic test were employed to determine the between-study heterogeneity. The fixed-effect model was used when the P-value for the Q-test was more than 0.10 and I2 for the I2 test was <50%; otherwise, the random-effect model was used. Funnel plot was used to assess the publication bias. Analyses were performed using the software Review Manage 5.3 (Oxford, England, U.K.).
Results
Main characteristics of selected studies
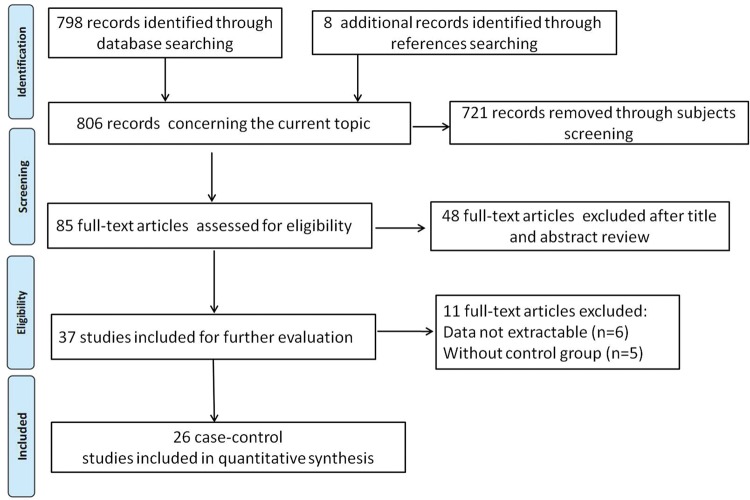
Figure 1 outlined the study process of selection. Briefly, we first identified 798 articles. After applying the inclusion and exclusion criteria, a total of 26 articles including 17642 asthma patients and 42402 controls were screened out. Of the 26 articles, 12 were written in Chinese [28–39] and 14 in English [40–53]. Among them, 17 were conducted in Asian populations, five in Caucasian populations and four in African populations. One article contained two study populations. The IL-13 +1923C/T polymorphism was measured by 11 different methods. The genotypes of IL-13 +1923C/T polymorphism in controls were all in accordance with HWE (P>0.05). Table 1 listed the main characteristics of included studies. Table 2 exhibited the distribution information of alleles and genotypes of IL-13 +1923C/T polymorphism.
Figure 1.
Flow chart of selection process in this meta-analysis
Table 1.
Main characteristics of included studies in this meta-analysis
| First author | Year | Country | Ethnicity | Mean age | Sample size | Genotyping methods | ||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |||||
| Hákonarson, H. | 2001 | Iceland | Caucasian | 38 (12–59) | 38 (12–59) | 94 | 94 | PCR |
| Chen, J.Q. | 2004 | China | Asian | 2.59 ± 1.44 | 2.90 ± 1.45 | 96 | 53 | PCR-RFLP |
| Donfack, I, J. | 2005 | U.S.A. | Caucasian | NA | NA | 126 | 205 | LAS |
| Donfack, II, J. | 2005 | U.S.A. | African | NA | NA | 205 | 183 | LAS |
| Song, Z.Q. | 2005 | China | Asian | 14–67 | 18–70 | 100 | 100 | PCR-RFLP |
| Battle, N.C. | 2007 | U.S.A. | African | 19.4 (7.3–40.9) | 29.8 (8.2–41.2) | 264 | 176 | PCR-RFLP |
| Shi, X.H. | 2008 | China | Asian | 34 (14–66) | 18–56 | 48 | 48 | PCR-RFLP |
| Daley, D. | 2009 | Australia | Caucasian | NA | NA | 644 | 751 | Illumina Bead Array System |
| Wang, X.H. | 2009 | China | Asian | 39 ± 11 | 48 ± 12 | 150 | 160 | PCR-RFLP |
| Li, X.N. | 2010 | U.S.A. | Caucasian | 46.9 ± 18.4 | 31.4 ± 21.9 | 473 | 1892 | Illumina HumanCNV370 BeadChip |
| Moffatt, M.F. | 2010 | Mixed | Caucasian | NA | NA | 10365 | 16110 | Illumina Human610 quad array |
| Wu, X.H. | 2010 | China | Asian | 8.8 ± 3.2 | 9.2 ± 2.8 | 252 | 227 | PCR-RFLP |
| Noguchi, E. | 2011 | Japan | Asian | 8.0 ± 4.4 | 60.3 ± 14.2 | 938 | 2376 | TaqMan |
| El-Behady, E.M. | 2012 | Egypt | African | 9.3 ± 1.8 | 9.2 ± 1.6 | 50 | 30 | PCR-RFLP |
| Yoon, D.K. | 2012 | Korea | Asian | 52.2 ± 8.9 | 52.2 ± 8.9 | 237 | 16095 | Afymetrix Genome-Wide Human SNP array 5.0 |
| Jia, C.M. | 2013 | China | Asian | 4.27 ± 2.52 | 4.15 ± 2.91 | 77 | 50 | PCR-RFLP |
| Kelibiena, T. | 2013 | China | Asian | 38.35 ± 9.17 | 38.12 ± 8.23 | 76 | 89 | PCR-RFLP |
| Liu, Q.H. | 2013 | China | Asian | 3–12 | NA | 384 | 384 | TaqMan |
| Pu, H.P. | 2013 | China | Asian | 5.8 ± 2.9 | 5.6 ± 2.6 | 96 | 96 | PCR-RFLP |
| Wang, Y. | 2014 | China | Asian | 3–12 | 3–12 | 435 | 601 | SNaPshot assay |
| Xia, M.Q. | 2014 | China | Asian | 6.30 ± 3.39 | 4.96 ± 3.61 | 305 | 200 | PCR-RFLP |
| Xu, J.X. | 2014 | China | Asian | 7.5 ± 8.2 | 8.4 ± 8.8 | 230 | 220 | Sequenom |
| Ramphul, K. | 2015 | Mauritius | African | 3–12 | 18–22 | 193 | 189 | TaqMan |
| Xi, S.Y. | 2015 | China | Asian | 20–65 | 18–66 | 100 | 100 | PCR-RFLP |
| Li, T.X. | 2016 | China | Asian | 5.06 (4.0–8.0) | 5.00 (4.0–8.0) | 652 | 752 | SnaPshot assay |
| Tang, M.F. | 2016 | China | Asian | 2–7 | 2–7 | 903 | 1205 | TaqMan |
| Wang, H.L. | 2016 | China | Asian | 8 ± 1.7 | 6.6 ± 2.0 | 173 | 56 | PCR-RFLP |
LAS, multiplex PCR and an immobilized linear array system; NA, not available; PCR-RFLP, PCR-restriction fragment length polymorphism.
Table 2.
Distribution information of alleles and genotypes in IL-13 +1923C/T polymorphism among asthma patients and controls
| First author | Cases | Controls | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | CC | CT | TT | C | T | HWE | |
| Hákonarson, H. | 65 | 27 | 2 | 157 | 31 | 64 | 27 | 3 | 155 | 33 | 0.997 |
| Chen, J.Q. | 41 | 43 | 12 | 125 | 67 | 39 | 14 | 0 | 92 | 14 | 0.541 |
| Donfack, I, J. | 72 | 45 | 9 | 189 | 63 | 120 | 77 | 8 | 317 | 93 | 0.598 |
| Donfack, II, J. | 18 | 101 | 86 | 137 | 273 | 25 | 75 | 83 | 125 | 241 | 0.486 |
| Song, Z.Q. | 24 | 55 | 21 | 103 | 97 | 43 | 47 | 10 | 133 | 67 | 0.860 |
| Battle, N.C. | 31 | 117 | 113 | 179 | 343 | 21 | 77 | 72 | 119 | 221 | 0.998 |
| Shi, X.H. | 12 | 26 | 10 | 50 | 46 | 30 | 16 | 2 | 76 | 20 | 0.997 |
| Daley, D. | 422 | 199 | 23 | 1043 | 245 | 516 | 213 | 22 | 1245 | 257 | 0.939 |
| Wang, X.H. | 31 | 57 | 61 | 119 | 179 | 66 | 68 | 26 | 200 | 120 | 0.498 |
| Li, X.N. | 278 | 167 | 28 | 723 | 223 | 1247 | 578 | 67 | 3072 | 712 | 0.998 |
| Moffatt, M.F. | 6306 | 3558 | 501 | 16170 | 4560 | 10310 | 5156 | 644 | 25776 | 6444 | 0.984 |
| Wu, X.H. | 106 | 114 | 32 | 326 | 178 | 126 | 85 | 16 | 337 | 117 | 0.949 |
| Noguchi, E. | 387 | 439 | 112 | 1213 | 663 | 1125 | 1025 | 226 | 3275 | 1477 | 0.944 |
| El-Behady, E.M. | 20 | 20 | 10 | 60 | 40 | 25 | 5 | 0 | 55 | 5 | 0.883 |
| Yoon, D.K. | 110 | 99 | 28 | 319 | 155 | 7729 | 6768 | 1580 | 22226 | 9928 | 0.081 |
| Jia, C.M. | 22 | 42 | 13 | 86 | 68 | 25 | 22 | 3 | 72 | 28 | 0.812 |
| Kelibiena, T. | 37 | 26 | 13 | 100 | 52 | 66 | 19 | 4 | 151 | 27 | 0.274 |
| Liu, Q.H. | 174 | 164 | 46 | 512 | 256 | 179 | 169 | 36 | 527 | 241 | 0.912 |
| Pu, H.P. | 39 | 45 | 12 | 123 | 69 | 67 | 24 | 5 | 158 | 34 | 0.379 |
| Wang, Y. | 182 | 191 | 62 | 555 | 315 | 291 | 246 | 64 | 828 | 374 | 0.542 |
| Xia, M.Q. | 84 | 128 | 93 | 296 | 314 | 95 | 73 | 32 | 263 | 137 | 0.997 |
| Xu, J.X. | 150 | 71 | 9 | 371 | 89 | 151 | 62 | 7 | 364 | 76 | 0.979 |
| Ramphul, K. | 78 | 79 | 25 | 235 | 129 | 77 | 96 | 13 | 250 | 122 | 0.066 |
| Xi, S.Y. | 21 | 41 | 38 | 83 | 117 | 41 | 44 | 15 | 126 | 74 | 0.854 |
| Li, T.X. | 304 | 290 | 58 | 898 | 406 | 355 | 316 | 81 | 1026 | 478 | 0.698 |
| Tang, M.F. | 345 | 439 | 110 | 1129 | 659 | 512 | 530 | 150 | 1554 | 830 | 0.781 |
| Wang, H.L. | 38 | 58 | 77 | 134 | 212 | 24 | 23 | 9 | 71 | 41 | 0.690 |
Association of IL-13 +1923C/T polymorphism in asthma risk
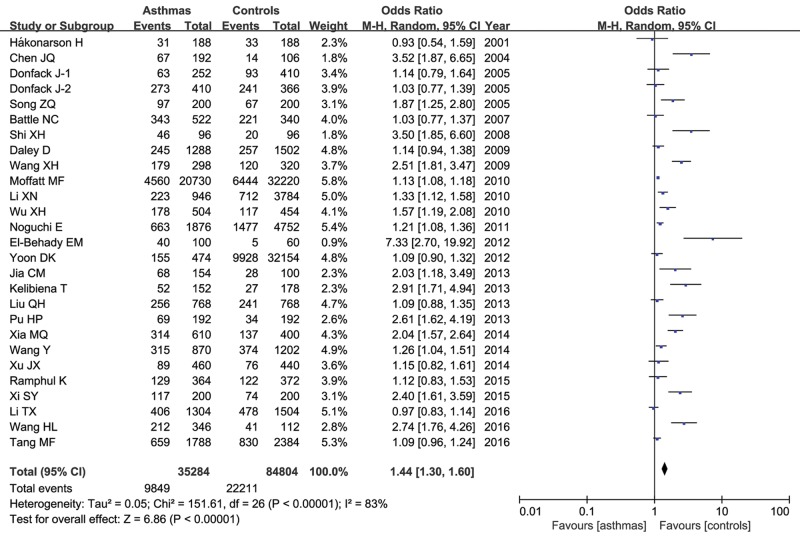
Table 3 presented the genetic effect of IL-13 +1923C/T variant on asthma risk. The between-study heterogeneity was detected (P<0.01 and I2>50%), and the random-effect model was employed. Overall, the frequency of T allele of IL-13 +1923C/T polymorphism was found to be a little higher in asthma patients than that in controls (27.9% compared with 26.2%), and the statistical analysis demonstrated that this allele was significantly related with increased asthma susceptibility (T compared with C: OR=1.44, 95% CI= 1.30–1.60, P<0.00001) as shown in Figure 2. This significant association was obtained in other genetic models as well (TT compared with CC: OR=1.93, 95% CI=1.57–2.37, P<0.00001; CT compared with CC: OR=0.32, 95% CI=1.20–1.46, P<0.00001; TT + CT compared with CC: OR=1.49, 95% CI=1.32–1.68, P<0.00001; TT compared with CT + CC: OR=1.59, 95% CI=1.34–1.89, P<0.00001).
Table 3.
Summary of the genetic effect of IL-13 +1923C/T variant on asthma risk and subgroup analyses
| Groups | Comparisons | N | Test of association | Test of heterogeneity | |||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | I2 | Ph | Model | |||
| Total | T compared with C | 26 | 1.44 (1.30, 1.60) | <0.00001 | 83% | <0.00001 | R |
| TT compared with CC | 1.93 (1.57, 2.37) | <0.00001 | 73% | <0.00001 | R | ||
| CT compared with CC | 1.32 (1.20, 1.46) | <0.00001 | 59% | <0.0001 | R | ||
| TT + CT compared with CC | 1.49 (1.32, 1.68) | <0.00001 | 76% | <0.0001 | R | ||
| TT compared with CT + CC | 1.59 (1.34, 1.89) | <0.00001 | 67% | <0.0001 | R | ||
| Asian | T compared with C | 17 | 1.66 (1.41, 1.95) | <0.00001 | 86% | <0.00001 | R |
| TT compared with CC | 2.34 (1.72, 3.18) | <0.00001 | 79% | <0.00001 | R | ||
| CT compared with CC | 1.46 (1.27, 1.69) | <0.00001 | 60% | 0.0005 | R | ||
| TT + CT compared with CC | 1.74 (1.45, 2.08) | <0.00001 | 78% | <0.00001 | R | ||
| TT compared with CT + CC | 1.85 (1.43, 2.39) | <0.00001 | 73% | <0.00001 | R | ||
| Caucasian | T compared with C | 5 | 1.14 (1.09, 1.18) | <0.00001 | 0% | 0.41 | F |
| TT compared with CC | 1.30 (1.16, 1.46) | <0.00001 | 0% | 0.46 | F | ||
| CT compared with CC | 1.31 (1.08, 1.19) | <0.00001 | 0% | 0.72 | F | ||
| TT+CT compared with CC | 1.15 (1.10, 1.21) | <0.00001 | 0% | 0.56 | F | ||
| TT compared with CT + CC | 1.25 (1.11, 1.40) | 0.0001 | 0% | 0.52 | F | ||
| African | T compared with C | 4 | 1.32 (0.88, 1.98) | 0.18 | 79% | 0.003 | R |
| TT compared with CC | 1.59 (1.09, 2.31) | 0.02 | 45% | 0.14 | F | ||
| CT compared with CC | 1.45 (0.77, 2.74) | 0.25 | 73% | 0.01 | R | ||
| TT + CT compared with CC | 1.60 (0.83, 3.08) | 0.16 | 77% | 0.005 | R | ||
| TT compared with CT + CC | 1.26 (0.76, 2.09) | 0.38 | 63% | 0.04 | R | ||
| Adult | T compared with C | 6 | 1.66 (1.20, 2.31) | 0.002 | 86% | <0.00001 | R |
| TT compared with CC | 2.61 (1.42, 4.79) | 0.002 | 78% | 0.0004 | R | ||
| CT compared with CC | 1.27 (1.10, 1.48) | 0.001 | 42% | 0.12 | F | ||
| TT + CT compared with CC | 1.66 (1.18, 2.34) | 0.004 | 77% | 0.0007 | R | ||
| TT compared with CT + CC | 2.19 (1.35, 3.56) | 0.002 | 70% | 0.006 | R | ||
| Children | T compared with C | 14 | 1.50 (1.27, 1.77) | <0.00001 | 84% | <0.00001 | R |
| TT compared with CC | 1.90 (1.40, 2.57) | <0.0001 | 73% | <0.00001 | R | ||
| CT compared with CC | 1.38 (1.17, 1.62) | <0.0001 | 64% | 0.0006 | R | ||
| TT + CT compared with CC | 1.56 (1.29, 1.89) | <0.00001 | 78% | <0.00001 | R | ||
| TT compared with CT + CC | 1.59 (1.23, 2.07) | 0.0004 | 67% | 0.0002 | R | ||
F, fixed-effect model; N, number of included studies; R, random-effect model.
Figure 2.
Meta-analysis of the correlation between the IL-13 +1923C/T polymorphism and asthma risk under the allelic model
Subgroup analysis by ethnicity and age group of the association between IL-13 +1923C/T polymorphism and asthma risk
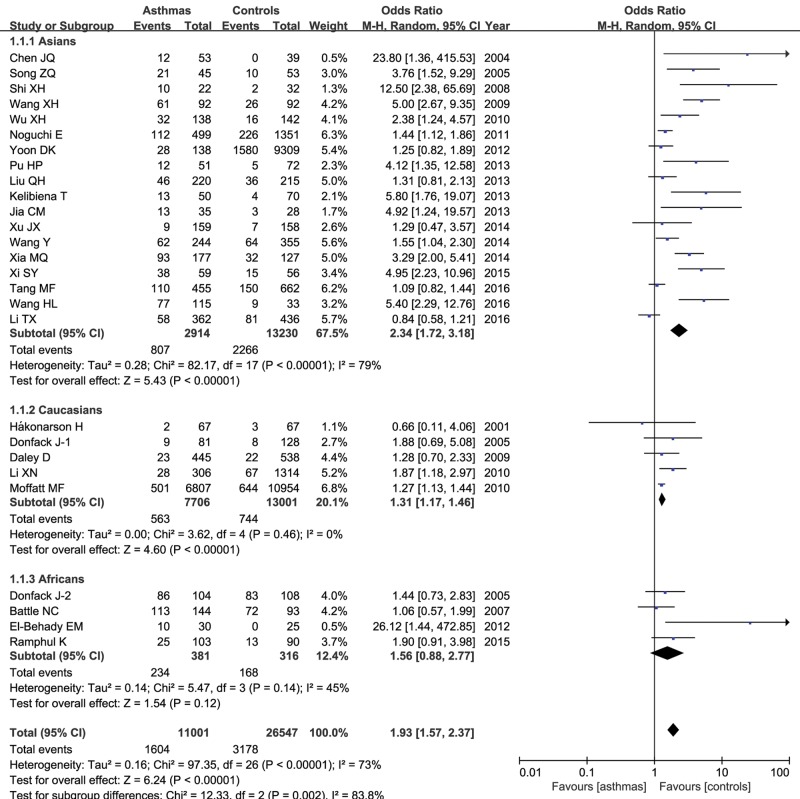
In the stratified analysis by ethnicity, 17 articles including 5242 asthma patients and 22781 controls were conducted in Asians, five articles including 11702 patients and 19052 controls in Caucasians, and four articles including 698 patients and 569 controls in Africans. Overall, we found that IL-13 +1923C/T variant was associated with increased asthma susceptibility among Asians under each genetic model (T compared with C: OR=1.66, 95% CI=1.41–1.95, P<0.00001; TT compared with CC: OR=2.34, 95% CI=1.72–3.18, P<0.00001; CT compared with CC: OR=1.46, 95% CI=1.27–1.69, P<0.00001; TT + CT compared with CC: OR=1.74, 95% CI=1.45–2.08, P<0.00001; TT compared with CT + CC: OR=1.85, 95% CI=1.43–2.39, P<0.00001). This statistical significance was detected in Caucasians as well (T compared with C: OR=1.14, 95% CI=1.09–1.18, P<0.00001; TT compared with CC: OR=1.30, 95% CI=1.16–1.46, P<0.00001; CT compared with CC: OR=1.31, 95% CI=1.08–1.19, P<0.00001; TT + CT compared with CC: OR=1.15, 95% CI=1.10–1.21, P<0.00001; TT compared with CT + CC: OR=1.25, 95% CI=1.11–1.40, P=0.0001). However, only TT genotype under the homozygote model was related with increased risk of asthma in Africans (TT compared with CC: OR=1.59, 95% CI=1.09–2.31, P=0.02) in the fixed-effect model. Figure 3 showed the relationship between TT genotype of +1923C/T variant and asthma risk under the homozygote model among Asians, Caucasians and Africans respectively.
Figure 3.
Forest plot of the relative strength of the association between IL-13 +1923C/T polymorphism and asthma risk under the homozygote model among Asians, Caucasians and Africans
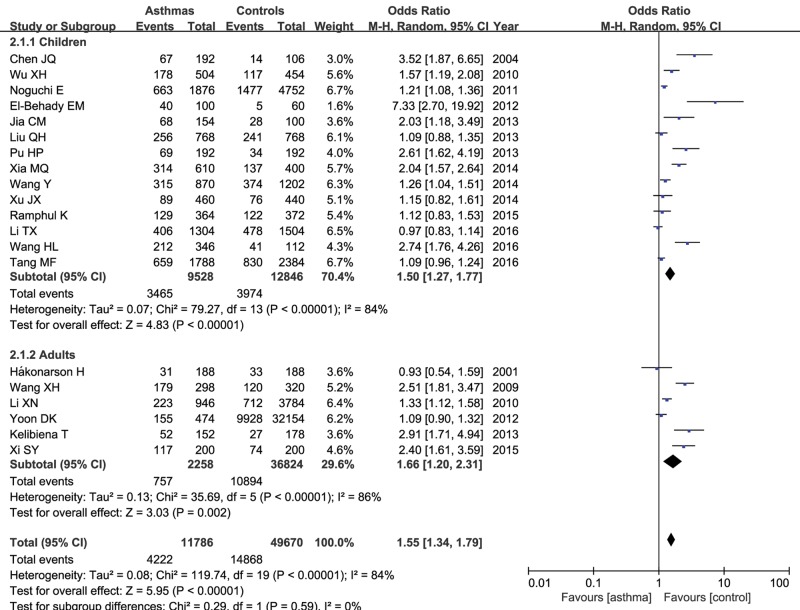
In the stratified analysis by age group, 14 articles containing 4764 cases and 6423 controls were performed in children (<18 years old), six articles containing 1129 cases and 18412 controls were in adults (≥18 years old) and the other six were mixed age group. Our result identified that IL-13 +1923C/T variant correlated with increased risk of asthma in both children and adult groups under each genetic model (Table 3). The significant effect was higher in children group than that in adult group. Figure 4 showed the relationship between T allele of IL-13 +1923C/T polymorphism and asthma risk in children and adults respectively.
Figure 4.
Meta-analysis of correlation of IL-13 +1923C/T polymorphism in asthma in children and adults
Association of IL-13 +1923C/T polymorphism in IgE levels (k-units/l)
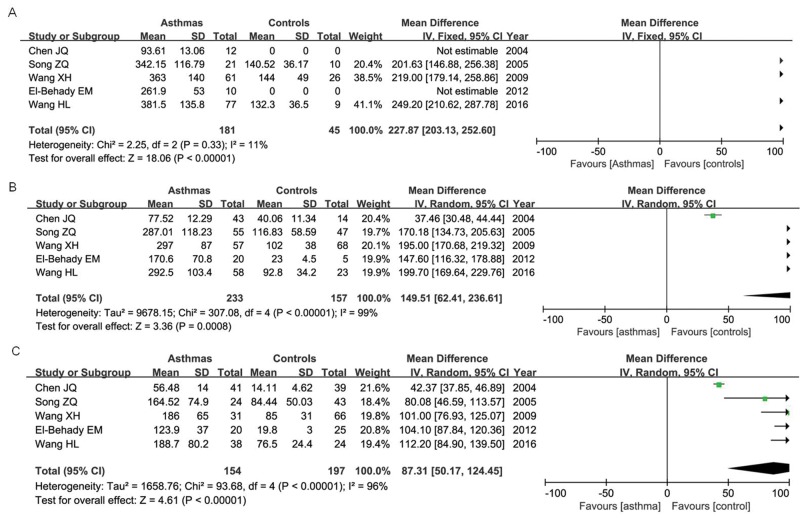
Eight articles reported the association between IL-13 +1923C/T polymorphism and IgE levels in serum; however, the relevant data could only be extracted from five of them, including 569 asthma patients and 399 controls. The statistical analysis found that the TT, CT and CC genotypes in asthma group were all significantly associated with increased IgE levels in serum when compared with controls as shown in Figure 5.
Figure 5.
Forest plot of the association between TT (A), CT (B) and CC (C) genotypes of IL-13 +1923C/T polymorphism and IgE level (k-units/l) of patients with asthma
Sensitivity analysis and publication bias
We omitted each particular study to verify whether our results were influenced by each included study or not. The pooled ORs were not materially altered. The funnel plot was used to evaluate the publication bias. All the plots were found to be roughly symmetrical, indicating no publication bias presented as shown in Figure 6.
Figure 6.
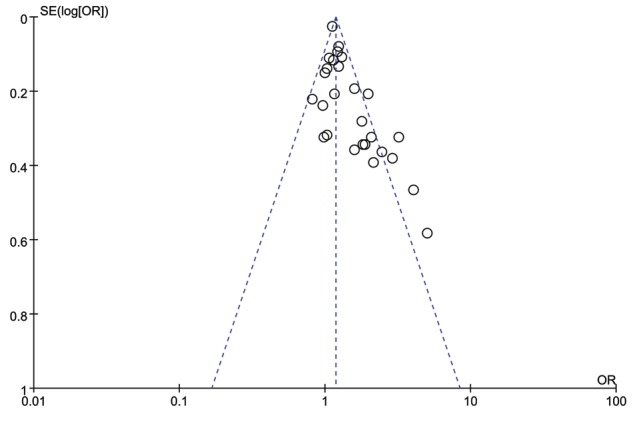
Funnel plot of IL-13 +1923C/T polymorphism in asthma risk under the heterozygote model
Discussion
In this meta-analysis, we retrieved a total of 26 relevant articles. Our results found that IL-13 +1923C/T polymorphism was significantly associated with increased risk of asthma under each genetic model. Subgroup analysis by ethnicity showed that the alleles and genotypes of this genetic variant correlated with asthma susceptibility among Asians and Caucasians, but only TT genotype under the homozygote model in Africans. This variant was related with increased risk of asthma in both children group and adult group under each genetic model as well. Furthermore, the TT, CT and CC genotypes in asthma group were all significantly associated with increased IgE levels in sera of asthma patients when compared with controls. Our result was consistent with previous meta-analysis conducted by Liu et al. [54], which contained ten included studies and suggested that IL-13 +1923C/T polymorphism was a risk factor for asthma.
Asthma is a hereditary disorder that is caused by a combination of intrinsic factors and environmental exposure [55]. Exposure to allergens is one of the environmental factors. In response to allergen presentation by airway DCs, T-helper lymphocytes of the adaptive immune system control many aspects of the disease through secretion of IL-4, IL-5, IL-13, IL-17 and IL-22, and these are counterbalanced by cytokines produced by Treg cells [56]. IL-13 is a key Th2 cytokine that directs many of the important features of airway inflammation and remodelling in patients with allergic asthma [57]. The IL-13 transcriptional “signature” can be used to identify individuals with “Th2 high” and “Th2 low” asthma [58]. IL-13 induces characteristic changes in mRNA [59] and miRNA [60] expression patterns in airway epithelial cells, and it induced protein periostin that is secreted basally from airway epithelial cells and can be used as a biomarker for Th2 high asthma [61]. Furthermore, sputum IL-13 levels could serve as a useful biomarker for asthma control assessment [62]. Current studies have identified that target IL-13 pathway is a promising therapeutic approach for asthma [63,64]. The association of IL-13 with asthma pathology and reduced corticosteroid sensitivity suggests a potential benefit of anti-IL-13 therapy in refractory asthma [65,66].
Asthma is primarily an inflammatory disorder of the airways associated with Th2 cell-dependent promotion of IgE production and recruitment of mast cells [67]. The elevated level of total IgE and allergy-specific IgE may function as independent risk factors for asthma [68–70]. IL-13 is known to be a key regulator in IgE synthesis. IgE production in allergic asthma patients is more dependent on IL-13 than in non-atopics, due to enhanced IL-13 production and to enhance IgE production in response to IL-13 [71]. SNPs in IL-13 were shown to be associated with allergic phenotypes in several ethnically diverse populations and might affect IgE level [72,73,76]. Allelic variation in the IL-13 gene was robustly confirmed as a contributor to the variance of IgE levels [74]. T allele of IL-13 +1923C/T was highly significant associated with total serum IgE (P=0.00022) [51]. Li et al. [75] found this variant was significantly associated with asthma risk in Chinese children and adults. The potential mechanism might be that: IL-13 +1923C/T variant is located in the third intron of IL-13 gene. The +1923C was easy to be DNA methylated, thus inhibiting the transcription of IL-13 gene; while the +1923T variant might suppress this inhibitive effect, thus promoting high expression of IL-13 and serum IgE level. In addition, other IL-13 polymorphisms might be associated with asthma risk as well. IL-13 rs20541 and rs1800925 were risk factors for asthma and rs1800925 was significantly associated with total serum IgE levels [77]. SNP rs848 in the IL-13 gene region was significantly associated with a continuous measure of symptom severity in adult subjects with severe asthma [78].
Several limitations were presented in our meta-analysis. Firstly, the between-study heterogeneity in any genetic models was high, which might influence the result. Secondly, most of the included studies were conducted in Asian and Caucasian populations, although other ethnicities should be considered. Thirdly, different genotyping methods were used in the respective studies, which may be associated with different call rates. Lastly, the interaction of gene–gene and gene–environment should be considered.
In conclusion, our results suggested that IL-13 +1923C/T polymorphism was a risk factor for asthma susceptibility, especially in Asians and Caucasians. Future large-scale and well-designed studies with more ethnicities are still required to validate the relationship between this genetic polymorphism and asthma risk.
Abbreviations
- CI
confidence interval
- HWE
Hardy–Weinberg equilibrium
- IL
interleukin
- IL-13
interleukin-13
- OR
odds ratio
- SNP
single nucleotide polymorphism
- Th2
T-helper type 2
Author contribution
W.T. conceived and designed the study. Y.X., J.L. and W.T. performed literature research and analysed the data. Z.D. and J.L. performed statistical analysis. B.L. and Z.Y. contributed materials/analysis tools. B.L., Z.Y., W.T., Y.X. and J.L. wrote the manuscript. All authors read and agreed with the final version of this manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Alpers J.H. (1968) The definition of asthma. Lancet 2, 876. [DOI] [PubMed] [Google Scholar]
- 2.Moorman J.E., Akinbami L.J., Bailey C.M., Zahran H.S., King M.E., Johnson C.A. et al. (2012) National surveillance of asthma: United States, 2001-2010. Vital Health Stat 3. , 35 1–58 [PubMed] [Google Scholar]
- 3.Burrows B., Barbee R.A., Cline M.G., Knudson R.J. and Lebowitz M.D. (1991) Characteristics of asthma among elderly adults in a sample of the general population. Chest 100, 935–942 [DOI] [PubMed] [Google Scholar]
- 4.Global Initiative for Asthma (2002) Global strategy for asthma management and prevention. NIH Publication 3602–3659 [Google Scholar]
- 5.Lozano R., Naghavi M., Foreman K., Lim S., Shibuya K., Aboyans V. et al. (2012) Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beasley R., Semprini A. and Mitchell E.A. (2015) Risk factors for asthma: is prevention possible? Lancet 386, 1075–1085 [DOI] [PubMed] [Google Scholar]
- 7.Anto J.M. (2012) Recent advances in the epidemiologic investigation of risk factors for asthma: a review of the 2011 literature. Curr. Allergy Asthma Rep. 12, 192–200 [DOI] [PubMed] [Google Scholar]
- 8.Lama M., Chatterjee M. and Chaudhuri T.K. (2013) Total serum immunoglobulin e in children with asthma. Indian J. Clin. Biochem. 28, 197–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roche N., Pribil C., Devillier P., Van Ganse E., Schuck S., Doussaint J. et al. (2016) Limited treatment adaptation despite poor asthma control in asthma patients treated with inhaled corticosteroids. J. Asthma 53, 76–85 [DOI] [PubMed] [Google Scholar]
- 10.Lambrecht B.N. and Hammad H. (2015) The immunology of asthma. Nat. Immunol. 16, 45–56 [DOI] [PubMed] [Google Scholar]
- 11.Erle D.J. and Sheppard D. (2014) The cell biology of asthma. J. Cell. Biol. 205, 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wenzel S.E. (2012) Asthma phenotypes: the evolution from clinical to molecular approaches. Nat. Med. 18, 716–725 [DOI] [PubMed] [Google Scholar]
- 13.Fietta P., Costa E. and Delsante G. (2015) Interleukins (ILs), a fascinating family of cytokines. Part II: ILs from IL-20 to IL-38. Theor. Biol. Forum 108, 19–40 [PubMed] [Google Scholar]
- 14.Fietta P., Costa E. and Delsante G. (2014) Interleukins (ILs), a fascinating family of cytokines. Part I: ILs from IL-1 to IL-19. Theor. Biol. Forum 107, 13–45 [PubMed] [Google Scholar]
- 15.Chen Q., Carroll H.P. and Gadina M. (2006) The newest interleukins: recent additions to the ever-growing cytokine family. Vitam. Horm. 74, 207–228 [DOI] [PubMed] [Google Scholar]
- 16.Vercelli D. (2002) Genetics of IL-13 and functional relevance of IL-13 variants. Curr. Opin. Allergy Clin. Immunol. 2, 389–393 [DOI] [PubMed] [Google Scholar]
- 17.Corren J. (2013) Role of interleukin-13 in asthma. Curr. Allergy Asthma Rep. 13, 415–420 [DOI] [PubMed] [Google Scholar]
- 18.Gour N. and Wills-Karp M. (2015) IL-4 and IL-13 signaling in allergic airway disease. Cytokine 75, 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gieseck R.L. III, Ramalingam T.R., Hart K.M., Vannella K.M., Cantu D.A., Lu W.Y. et al. (2016) Interleukin-13 activates distinct cellular pathways leading to ductular reaction, steatosis, and fibrosis. Immunity 45, 145–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang Y., Xu L., An H., Fu Q., Chen L., Lin Z. et al. (2015) Expression of IL-4 and IL-13 predicts recurrence and survival in localized clear-cell renal cell carcinoma. Int. J. Clin. Exp. Pathol. 8, 1594–1603 [PMC free article] [PubMed] [Google Scholar]
- 21.Granada M., Wilk J.B., Tuzova M., Strachan D.P., Weidinger S., Albrecht E. et al. (2012) A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J. Allergy Clin. Immunol. 129, 840.e21–845.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramphul K., Lv J., Hua L., Liu Q.H., Fang D.Z., Ji R.X. et al. (2014) Single nucleotide polymorphisms predisposing to asthma in children of Mauritian Indian and Chinese Han ethnicity. Braz. J. Med. Biol. Res. 47, 394–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.To T., Stanojevic S., Moores G., Gershon A.S., Bateman E.D., Cruz A.A. et al. (2012) Global asthma prevalence in adults: findings from the cross-sectional world health survey. BMC Public Health 12, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathias R.A., Grant A.V., Rafaels N., Hand T., Gao L., Vergara C. et al. (2010) A genome-wide association study on African-ancestry populations for asthma. J. Allergy Clin Immunol. 125, 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torgerson D.G., Ampleford E.J., Chiu G.Y., Gauderman W.J., Gignoux C.R., Graves P.E. et al. (2011) Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat. Genet. 43, 887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.American Thoracic Society . (1987) Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease (COPD) and asthma. Am. Rev. Respir. Dis. 136, 225–244 [DOI] [PubMed] [Google Scholar]
- 27.Groneberg D.A., Welte T. (2006) Asthma and COPD. Exp. Toxicol. Pathol. 57, 35–40 [DOI] [PubMed] [Google Scholar]
- 28.Wang H. (2016) The influence of IL-4 and IL-13 gene polymorphisms on IgE level in children with asthma. Er Ke Yao Xue Za Zhi 22, 7–9 [Google Scholar]
- 29.Xi S., Zhang L., Zhang X. and Wang S. (2015) Correlation analysis between +1923C/T and +2044G/A polymorphism in IL-13 gene and asthma. Clin. Focus 30, 1421–1425 [Google Scholar]
- 30.Wang Y., Tian-Xiao L.I., Deng Y., Song J.J., Zhang H.Y. and Zhou H.B. (2014) Association study of rs1295686 polymorphisms in IL13 gene with pediatric asthma in a northeastern Han Chinese population. J. Harbin Med. Univ. 48, 1–4 [Google Scholar]
- 31.Xia M. and Zhu X. (2014) Study on relationship between specific IgE and polymorphisms of IL-4 -33C/T and IL-13 +1923C/T in 305 asthmatic children of Guiyang area [Master’s thesis]. Guiyang Medical College, China [Google Scholar]
- 32.Xu J. (2014) The relationship between IL-13 genetic polymorphisms and pediatric asthma in Hubei, China. Zhongguo Fu You Bao Jian 29, 551–553 [Google Scholar]
- 33.Pu H., Liu H., Lv Z. and Sun X. (2013) Study on the correlation between asthma and IL-13 gene polymorphism. Shi Yong Yi Yao Za Zhi 30, 289–292 [Google Scholar]
- 34.Jia C., Liu X., Jiang C. and Wang Y. (2013) Relationship between polymorphisms of IL-13 gene intron 3 +1923 and bronchial asthma in children. Zhonghua Shi Yong Er Ke Lin Chuang Za Zhi 28, 682–685 [Google Scholar]
- 35.Kelibiena T., Mihereguli S., Rinaguli A. and Dilinuer W. (2013) Relationship between asthma patients with abnormal savda and the gene polymorphism of IL-13. Keji Daobao/Sci. Technol. Rev. 31, 87–91 [Google Scholar]
- 36.Wang X.H., Zhao W., Liu S.G. and Feng X.P. (2009) Correlation of IL-4 and IL-13 gene polymorphisms with asthma and total serum IgE levels. Zhonghua Jie He He Hu Xi Za Zhi 32, 161–164 [PubMed] [Google Scholar]
- 37.Shi X. and Zhou J. (2008) Association between IL-13 and β2-AR polymorphisms and asthma. Shandong Yi Yao 48, 119–121 [Google Scholar]
- 38.Song Z.Q., Bin W.U., Wen L.I., Liu J.L. and Zhang W.Z. (2005) Association between IL-13 gene polymorphism and level of IL-13 and TIgE in patients with asthma. Chinese J. Immunol. 6, 469–471 [Google Scholar]
- 39.Chen J., Sun H. and Guo X. (2004) Effect of IL-13 gene polymorphism on the levels of serum IL-13 and total-IgE in asthmatic children. Zhongguo Shi Yong Er Ke Za Zhi 19, 209–211 [Google Scholar]
- 40.Tang M.F., Sy H.Y., Kong A.P., Ko F.W., Wang S.S., Liu T.C. et al. (2016) Genetic effects of multiple asthma loci identified by genomewide association studies on asthma and spirometric indices. Pediatr. Allergy Immunol. 27, 185–194 [DOI] [PubMed] [Google Scholar]
- 41.Li T., Ren Z., Deng Y., Wang Y. and Zhou H. (2016) Lack of association between RAD50-IL13 polymorphisms and pediatric asthma susceptibility in Northeastern Han Chinese. J. Asthma 53, 114–118 [DOI] [PubMed] [Google Scholar]
- 42.Ramphul K., Hua L., Bao Y.X., Li J.Y., Liu Q.H., Ji R.X. et al. (2015) Identification of IL13 C1923T as a single nucleotide polymorphism for asthma in children from Mauritius. Pediatr. Allergy Immunol. Pulmonol. 28, 92–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q., Hua L., Fang D., Lin Q., Zhu Y., Gan X. et al. (2013) Interleukin-13 and RANTES polymorphisms in relation to asthma in children of Chinese Han nationality. Asian. Pac. J. Allergy Immunol. 31, 247–252 [DOI] [PubMed] [Google Scholar]
- 44.Yoon D., Ban H.J., Kim Y.J., Kim E.J., Kim H.C., Han B.G. et al. (2012) Replication of genome-wide association studies on asthma and allergic diseases in Korean adult population. BMB Rep. 45, 305–310 [DOI] [PubMed] [Google Scholar]
- 45.Noguchi E., Sakamoto H., Hirota T., Ochiai K., Imoto Y., Sakashita M. et al. (2011) Genome-wide association study identifies HLA-DP as a susceptibility gene for pediatric asthma in Asian populations. PLoS Genet. 7, e1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moffatt M.F., Gut I.G., Demenais F., Strachan D.P., Bouzigon E., Heath S. et al. (2010) A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 363, 1211–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X., Howard T.D., Zheng S.L., Haselkorn T., Peters S.P., Meyers D.A. et al. (2010) Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J. Allergy Clin. Immunol. 125, 328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X., Li Y., Chen Q., Chen F., Cai P., Wang L. et al. (2010) Association and gene-gene interactions of eight common single-nucleotide polymorphisms with pediatric asthma in middle China. J. Asthma 47, 238–244 [DOI] [PubMed] [Google Scholar]
- 49.Daley D., Lemire M., Akhabir L., Chan-Yeung M., He J.Q., McDonald T. et al. (2009) Analyses of associations with asthma in four asthma population samples from Canada and Australia. Hum. Genet. 125, 445–459 [DOI] [PubMed] [Google Scholar]
- 50.Battle N.C., Choudhry S., Tsai H.J., Eng C., Kumar G., Beckman K.B. et al. (2007) Ethnicity-specific gene-gene interaction between IL-13 and IL-4Rα among African Americans with asthma. Am. J. Respir. Crit. Care Med. 175, 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donfack J., Schneider D.H., Tan Z., Kurz T., Dubchak I., Frazer K.A. et al. (2005) Variation in conserved non-coding sequences on chromosome 5q and susceptibility to asthma and atopy. Respir. Res. 6, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hakonarson H., Bjornsdottir U.S., Ostermann E., Arnason T., Adalsteinsdottir A., Halapi E. et al. (2001) Allelic frequencies and patterns of single-nucleotide polymorphisms in candidate genes for asthma and atopy in Iceland. Am. J. Respir. Crit. Care Med. 164, 2036–2044 [DOI] [PubMed] [Google Scholar]
- 53.El-Behady E.M. and El-Behady R.M. (2012) Identification of Il-13 1923 C/T as risk locus for asthma in children. Egyptian Journal of Medical Microbiology. 21, 101–107 [Google Scholar]
- 54.Liu Y., Liu T., Nie W., Lai G. and Xiu Q. (2013) Interleukin-13 +1923C/T polymorphism is associated with asthma risk: a meta-analysis. Biomed. Res. Int. 2013, 394316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siroux V., Gonzalez J.R., Bouzigon E., Curjuric I., Boudier A., Imboden M. et al. (2014) Genetic heterogeneity of asthma phenotypes identified by a clustering approach. Eur. Respir. J. 43, 439–452 [DOI] [PubMed] [Google Scholar]
- 56.Lambrecht B.N. and Hammad H. (2013) Asthma: the importance of dysregulated barrier immunity. Eur. J. Immunol. 43, 3125–3137 [DOI] [PubMed] [Google Scholar]
- 57.Brombacher F. (2000) The role of interleukin-13 in infectious diseases and allergy. Bioessays 22, 646–656 [DOI] [PubMed] [Google Scholar]
- 58.Woodruff P.G., Modrek B., Choy D.F., Jia G., Abbas A.R., Ellwanger A. et al. (2009) T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am. J. Respir. Crit. Care Med. 180, 388–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woodruff P.G., Boushey H.A., Dolganov G.M., Barker C.S., Yang Y.H., Donnelly S. et al. (2007) Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc. Natl. Acad. Sci. U.S.A. 104, 15858–15863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Solberg O.D., Ostrin E.J., Love M.I., Peng J.C., Bhakta N.R., Hou L. et al. (2012) Airway epithelial miRNA expression is altered in asthma. Am. J. Respir. Crit. Care Med. 186, 965–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parulekar A.D., Atik M.A. and Hanania N.A. (2014) Periostin, a novel biomarker of TH2-driven asthma. Curr. Opin. Pulm. Med. 20, 60–65 [DOI] [PubMed] [Google Scholar]
- 62.Tsilogianni Z., Hillas G., Bakakos P., Aggelakis L., Konstantellou E., Papaioannou A.I. et al. (2016) Sputum interleukin-13 as a biomarker for the evaluation of asthma control. Clin. Exp. Allergy 46, 1498. [DOI] [PubMed] [Google Scholar]
- 63.Ingram J.L. and Kraft M. (2012) IL-13 in asthma and allergic disease: asthma phenotypes and targeted therapies. J. Allergy Clin. Immunol. 130, 829–842 [DOI] [PubMed] [Google Scholar]
- 64.Wynn T.A. (2015) Type 2 cytokines: mechanisms and therapeutic strategies. Nat. Rev. Immunol. 15, 271–282 [DOI] [PubMed] [Google Scholar]
- 65.De Boever E.H., Ashman C., Cahn A.P., Locantore N.W., Overend P., Pouliquen I.J. et al. (2014) Efficacy and safety of an anti-IL-13 mAb in patients with severe asthma: a randomized trial. J. Allergy Clin. Immunol. 133, 989–996 [DOI] [PubMed] [Google Scholar]
- 66.Brightling C.E., Nordenmark L.H., Jain M., Piper E., She D., Braddock M. et al. (2016) Effect of anti-IL-13 treatment on airway dimensions in severe asthma. Am. J. Respir. Crit. Care Med. 194, 118–120 [DOI] [PubMed] [Google Scholar]
- 67.Holgate S.T. (2012) Innate and adaptive immune responses in asthma. Nat. Med. 18, 673–683 [DOI] [PubMed] [Google Scholar]
- 68.Oettgen H.C. and Geha R.S. (2001) IgE regulation and roles in asthma pathogenesis. J. Allergy Clin. Immunol. 107, 429–440 [DOI] [PubMed] [Google Scholar]
- 69.Bachert C., van Steen K., Zhang N., Holtappels G., Cattaert T., Maus B. et al. (2012) Specific IgE against Staphylococcus aureus enterotoxins: an independent risk factor for asthma. J. Allergy Clin. Immunol. 130, 376–381 [DOI] [PubMed] [Google Scholar]
- 70.Irani A.M. (2014) The relationship between a specific IgE level and asthma outcomes: results from the 2005-2006 National Health and Nutrition Examination Survey. Pediatrics 134, S168. [DOI] [PubMed] [Google Scholar]
- 71.Van der Pouw Kraan T.C., Van der Zee J.S., Boeije L.C., De Groot E.R., Stapel S.O. and Aarden L.A. (1998) The role of IL-13 in IgE synthesis by allergic asthma patients. Clin. Exp. Immunol. 111, 129–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fahy J.V. (2015) Type 2 inflammation in asthma–present in most, absent in many. Nat. Rev. Immunol. 15, 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jung Y.H., Seo J.H., Kim H.Y., Kwon J.W., Kim B.J., Kim H.B. et al. (2015) The relationship between asthma and bronchiolitis is modified by TLR4, CD14, and IL-13 polymorphisms. Pediatr. Pulmonol. 50, 8–16 [DOI] [PubMed] [Google Scholar]
- 74.Maier L.M., Howson J.M., Walker N., Spickett G.P., Jones R.W., Ring S.M. et al. (2006) Association of IL13 with total IgE: evidence against an inverse association of atopy and diabetes. J. Allergy Clin. Immunol. 117, 1306–1313 [DOI] [PubMed] [Google Scholar]
- 75.Li X., Zhang Y., Zhang J., Xiao Y., Huang J., Tian C. et al. (2010) Asthma susceptible genes in Chinese population: a meta-analysis. Respir. Res. 11, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doong S.H., Dhruva A., Dunn L.B., West C., Paul S.M., Cooper B.A. et al. (2015) Associations between cytokine genes and a symptom cluster of pain, fatigue, sleep disturbance, and depression in patients prior to breast cancer surgery. Biol. Res. Nurs. 17, 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cui L., Jia J., Ma C.F., Li S.Y., Wang Y.P., Guo X.M. et al. (2012) IL-13 polymorphisms contribute to the risk of asthma: a meta-analysis. Clin. Biochem. 45, 285–288 [DOI] [PubMed] [Google Scholar]
- 78.Accordini S., Calciano L., Bombieri C., Malerba G., Belpinati F., Lo Presti A.R. et al. (2016) An interleukin 13 polymorphism is associated with symptom severity in adult subjects with ever asthma. PLoS ONE 11, e0151292. [DOI] [PMC free article] [PubMed] [Google Scholar]