Abstract
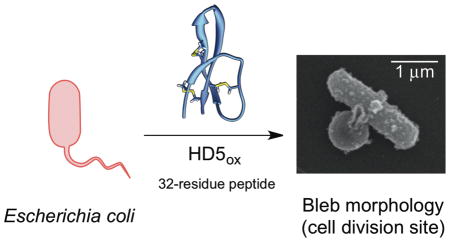
Human α-defensin 5 (HD5) is a 32-residue cysteine-rich host-defense peptide that exhibits broad-spectrum antimicrobial activity and contributes to innate immunity in the human gut and other organ systems. Despite many years of investigation, its antimicrobial mechanism of action remains unclear. In this work, we report that HD5ox, the oxidized form of this peptide that exhibits three regiospecific disulfide bonds, causes distinct morphological changes to Escherichia coli and other Gram-negative microbes. These morphologies include bleb formation, cellular elongation, and clumping. The blebs are up to ~1 μm wide and typically form at the site of cell division or cell poles. Studies with E. coli expressing cytoplasmic GFP reveal that HD5ox treatment causes GFP emission to localize in the bleb. To probe the cellular uptake of HD5ox and subsequent localization, we describe the design and characterization of a fluorophore-HD5 conjugate family. By employing these peptides, we demonstrate that fluorophore-HD5ox conjugates harboring the rhodamine and coumarin fluorophores enter the E. coli cytoplasm. On the basis of the fluorescence profiles, each of these fluorophore-HD5ox conjugates localizes to the site of cell division and cell poles. These studies support the notion that HD5ox, at least in part, exerts its antibacterial activity against E. coli and other Gram-negative microbes in the cytoplasm.
Graphical Abstract
Introduction
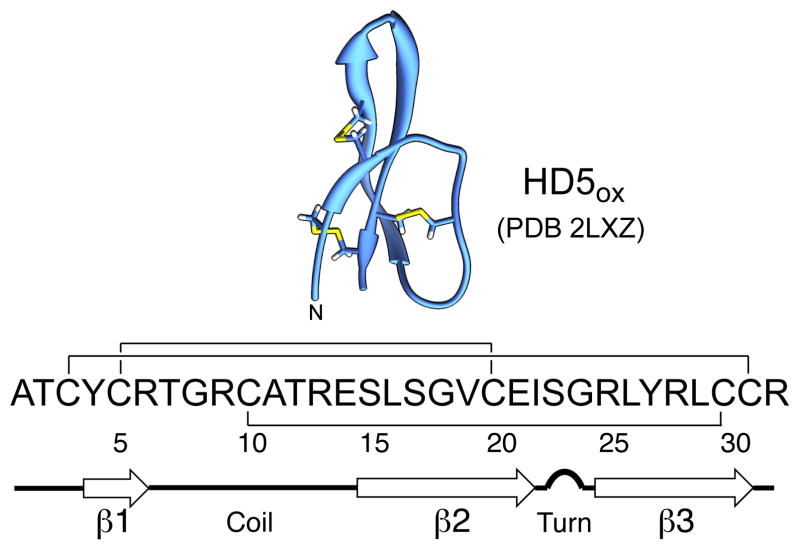
Bacterial infections and the considerable rise in antibiotic resistance in hospital and community settings pose significant problems for global health initiatives.1,2 The dearth of new antibiotics in the drug pipeline as well as an incomplete comprehension of human innate immunity and microbial pathogenesis further confound efforts to address these challenges.3,4 Bacterial pathogens must overcome the innate immune system, which provides first-line defense against microbial invaders, to cause human disease. Fundamental investigations that address the molecular interworking of the innate immune system are critical to further understand the host-microbe interaction and enable the development of new therapeutic strategies for infectious disease. Antimicrobial (AMPs) and/or host-defense peptides are important components of the innate immune system.5–8 In humans, defensins7,9 and the cathelicidin LL-3710,11 are abundant host-defense peptides expressed and utilized by neutrophils and epithelial cells of the gastrointestinal tract, urogenital tract, airway, and skin.12 Mammalian defensins are cysteine-rich peptides that are classified as α-, β-, and θ-defensins on the basis of regiospecific disulfide linkage patterns.12 α-Defensins exhibit three disulfide bonds in the oxidized forms with the linkages CysI—CysVI, CysII—CysIV, CysIII—CysV, and have three-stranded β-sheet structures.7 In humans, six α-defensins are known.7 The enteric α-defensin HD5 (Figure 1), the focus of this work, is an abundant constituent of small intestinal Paneth cell granules.13–16 Although human defensins are established contributors to immunity, and many exhibit broad-spectrum in vitro antimicrobial activity, details pertaining to the physiological function of each peptide are often unclear.12,17,18 In this work, we focus on a fundamental and outstanding question: how does HD5 kill bacteria?
Figure 1.
Structure of the HD5ox monomer determined by solution NMR (PDB 2LXZ)19 and amino acid sequence. The regiospecific disulfide bond linkages and secondary structure are indicated.
How defensins kill bacteria as well as how in vitro antibacterial activity relates to the physiological milieu are questions of current interest and debate.17 The oxidized α-defensins display remarkable similarity in their tertiary structure, and most characterized to date are cationic and amphipathic.20 Moreover, defensins from various organisms have the capacity to disrupt bacterial cell membranes.21–23 On the basis of early investigations, including seminal structural studies of HNP3,24 a working model whereby defensins kill bacteria by non-specific membrane destabilization was presented.21,24 Over the years, this type of model was generalized for many defensins and other antimicrobial peptides.17 Nevertheless, defensins exhibit remarkable diversity in primary sequence, and recent studies support alternative mechanisms of action for some family members. Fungal plectasin,25 oyster defensin,26 and fungal copsin27 bind lipid II and block cell wall biosynthesis. The human defensins human neutrophil peptide 1 (HNP1, α-defensin)28 and human β-defensin 3 (HBD3)29 are also reported to bind lipid II to varying degrees.30 Studies of HBD3 attributed the in vitro antibacterial activity against Staphylococcus aureus to lipid II binding and subsequent cell wall lysis.29 Recently, human β-defensin 2 (HBD2) was found to localize at septal foci of Enterococcus faecalis and disrupt virulence factor assembly.31 Human α-defensin 6 (HD6) lacks antimicrobial activity in vitro and is proposed to serve a host-defense function by self-assembling into a web-like structure termed “nanonet” that captures bacteria in the intestinal lumen.32,33 Taken together, these investigations highlight tremendous variation in defensin mechanism of action despite similar tertiary structures, and demonstrate that membrane permeabilization is only one of the many factors that contribute to the in vitro antimicrobial activities demonstrated by this vast peptide family.
Our laboratory has initiated a research program focused on understanding the biophysical properties and biological functions of human defensins that are produced and released in the small intestine. Paneth cells,16 located in the crypts of Lieberkühn, contain granules that store two α-defensins, HD5 and HD6, as well as other antimicrobial peptides, proteases, and a labile zinc pool of unknown function.34,35 HD5 is the most abundant Paneth cell antimicrobial peptide,36 and it exhibits broad-spectrum antimicrobial activity in vitro.37–40 Moreover, transgenic mice expressing HD5 are more resistant to Salmonella challenge than wild-type mice,41 and studies of the resident intestinal microbiota suggest that HD5 contributes to controlling its composition.41,42 In humans suffering from ileal Crohn’s disease, an inflammatory disorder of the small bowel, a deficiency in Paneth cell defensins is observed.36 Despite these compelling observations from animal models and clinical studies, the antimicrobial mechanism of action of HD5 is not well understood.
HD5 is a 32-residue peptide with an overall charge of +4 at neutral pH (Figure 1). Over the past decade, structure-activity relationship studies of HD5ox evaluated the importance of quaternary structure,40,43 disulfide linkages44,45 cationic residues,46 the canonical α-defensin salt bridge between [Arg6] and [Glu14],47 and chirality48 for its in vitro bactericidal activity. Results from several recent studies probing interactions between HD5ox and E. coli support a model whereby (i) the Gram-negative outer membrane serves as a permeability barrier for HD5ox49 and (ii) the inner membrane becomes damaged as a result of HD5ox exposure.44
In this work, we utilize microscopy to investigate the attack of HD5ox on Gram-negative bacteria. We establish that E. coli treated with native HD5ox exhibit distinct morphologies that include clumping, cell elongation, and formation of one or more cellular blebs typically at the cell poles or division site. We demonstrate that similar morphological changes occur for other Gram-negative bacteria, including the opportunistic human pathogens Acinetobacter baumannii and Pseudomonas aeruginosa, following HD5ox exposure. Through the design, characterization and utilization of a fluorophore-HD5 conjugate family, we report that rhodamine- and coumarin-modified HD5ox enter the E. coli cytoplasm. Moreover, these fluorophore-HD5 conjugates preferentially localize at cell poles and cell division sites in E. coli, suggesting locales of a possible intracellular target.
Results and Discussion
HD5 Causes Distinct Morphological Changes in Escherichia coli
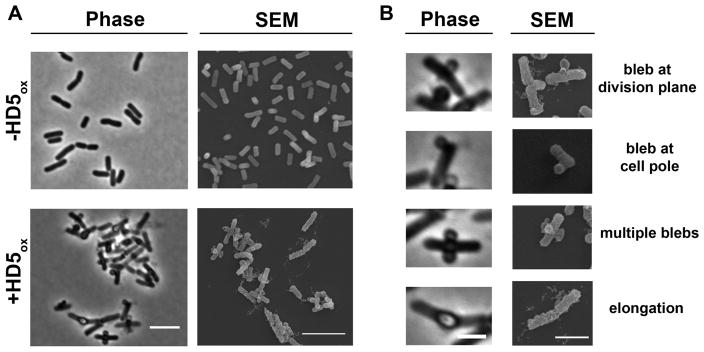
For morphology studies, we selected to image Gram-negative Escherichia coli. This microbe is a commensal microbe of the human gut as well as a pathogen of the gut and urogenital tract, and a number of studies have probed the antibacterial activity of HD5ox against this strain.37,44,47 In AMA assays, the concentration of HD5ox required to kill E. coli (e.g., lethal dose 99.99% or 4-fold log reduction in CFU/mL) depends on the number of colony forming units. Our standard AMA assay for evaluating HD5ox activity employs mid-log phase bacteria at ≈1 × 106 CFU/mL cultured in an AMA buffer (10 mM sodium phosphate buffer pH 7.4, 1% v/v TSB without dextrose). Under these conditions, the concentration of HD5ox required to kill 99.99% of E. coli is ≈4 μM depending on the precise starting CFU/mL. Treatment of mid-log-phase E. coli ATCC 25922 (1 × 106 CFU/mL) with HD5ox (2 and 4 μM) under standard AMA assay conditions resulted in marked changes to the bacterial morphology observable by phase-contrast microscopy that included the formation of large bulges, hereafter called blebs (Figure S1). To facilitate visualizing more cells per experiment, we modified the standard AMA conditions and employed a greater number of cells (1 × 108 CFU/mL) and higher concentrations of HD5ox (0–80 μM). Under these conditions, HD5ox displays AMA, and ≈2-fold log reduction in CFU/mL is observed following treatment of the E. coli with 40 μM HD5ox (Figure S2). Moreover, the E. coli displayed distinct morphological changes as observed in the preliminary experiment (Figure S1). On the basis of this similarity, we employed the modified conditions with greater cell density and higher HD5ox concentration for further imaging experiments. The morphologies observed under these conditions included cellular elongation and the formation of blebs (Figure 2). Clumping of E. coli was also observed. Bacterial cells with lengths of 5 μm or greater were categorized as elongated, and cells with lengths in the 10–15 μm range were periodically observed following HD5ox treatment. The blebs were typically localized at the cell division sites and cell poles; however, some bacteria displayed blebs along the cell body, and some bacteria exhibited multiple blebs per cell. The blebs remained intact during the centrifugation and wash steps required for scanning electron microscopy (SEM) sample preparation. Indeed, the blebs as well as what appeared to be outer membrane vesicles (Figure S3) and cellular debris (vide infra) were markedly apparent in SEM images (Figure 2).
Figure 2.
HD5ox causes distinct morphological changes in E. coli that include bleb formation, cellular elongation, and clumping. (A) Phase-contrast and the SEM images of E. coli ATCC 25922 (1 × 108 CFU/mL) in the absence (−HD5ox) or presence (+HD5ox) of 20 μM HD5ox, scale = 5 μm. (B) Phase-contrast and the SEM images of single cells illustrate the various morphologies caused by HD5ox, scale = 2 μm.
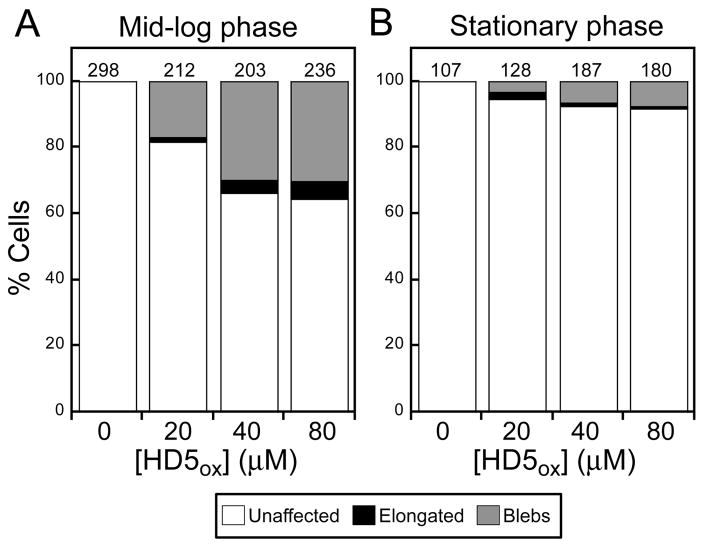
Neither blebs nor elongation were observed for untreated cells, and the number of cells exhibiting these morphological changes increased with increasing HD5ox (0–80 μM) (Figure 3). Following exposure to 20 μM HD5ox, ~17% of cells exhibited blebs and 2% were elongated (n = 212 cells). These values increased to ~30% and ≥4% at ≥40 μM HD5ox, respectively (n = 203 cells). We performed time-course experiments where E. coli were exposed to 20 μM HD5ox on the microscope stage and imaged over time (~2 h). Many of the treated cells displayed blebs, and the blebbing cells did not lyse over the course of this experiment (Figure S4). Propidium iodide (PI) uptake was therefore employed to evaluate the viability of HD5ox-treated cells, and the cells exhibiting blebs were labeled with PI, which indicated that bleb formation correlated with cell death (Figure S5). The morphological changes observed for E. coli ATCC 25922 were comparable to those we recently reported for E. coli K-12 and select mutants from the Keio Collection.50
Figure 3.
Effect of HD5ox treatment on E. coli ATCC 25922 morphology. The cells (1 × 108 CFU/mL) were treated with varying concentrations of HD5ox for 1 h at 37 °C (10 mM sodium phosphate buffer, pH 7.4, 1% (v/v) TSB). (A) Quantification of unaffected as well as elongated and bleb morphologies for mid-log phase E. coli. (B) Quantification for stationary-phase E. coli. The number above each bar indicates the number of cells counted.
The susceptibility of E. coli to HD5ox depends on the growth phase, and stationary phase cultures exhibit resistance to HD5ox relative to mid-log phase cultures as observed for other defensins.21,38 In this work, treatment of mid-log phase E. coli with 4 μM HD5ox resulted in an ~4-fold log-reduction in CFU/mL whereas only ~2-fold log-reduction was observed for stationary phase E. coli (Figure S6). In agreement with this trend, fewer morphological changes were observed for stationary phase cells treated with HD5ox (Figures 3, S6). Taken together, the microscopy and antimicrobial activity assays with HD5ox and E. coli provided a correlation between bacterial susceptibility and altered cellular morphology.
Treatment of E. coli with Other AMPs Does Not Result in the Bleb Morphology
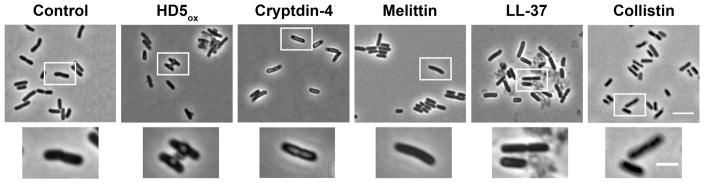
We questioned whether other antimicrobial peptides confer the same morphological changes observed for HD5ox under the experimental conditions used in this work. Prior biophysical studies proposed the ability of the murine Paneth cell defensin cryptdin-4 to induce negative curvature on bacterial membranes, resulting in structures called blebs or pores.51 To visualize the effect of cryptdin-4 on E. coli morphology, we first obtained cryptdin-4 by overexpression in E. coli, and confirmed its antimicrobial activity (Supporting Information, Figure S9). The E. coli treated with cryptdin-4 (20 μM) did not form large blebs as observed for HD5ox (Figure 4). Likewise, E. coli did not exhibit blebs following exposure to the pore-forming antimicrobial peptide melittin52 (20 μM), the LPS-associated membrane-destabilizing peptide colistin53 (20 μM), or the pore-forming antimicrobial peptide human LL-3754 (20 μM) (Figure 4). The bacteria treated with LL-37 were somewhat elongated relative to the untreated control cells. Small membrane protrusions (<100nm wide) and surface roughness observed for bacteria treated with AMPs such as magainin 255 and Bac8c56 have been described as blebs. To the best of our knowledge, larger blebs (~1-μm wide), formed preferentially at poles and cell division sites as observed for HD5ox, have not been reported for a human host-defense peptide. The most similar bleb morphologies we identified in the literature are the bulges that result from β-lactam treatment.57 Moreover, a knockout mutant of elyC,58 an inner-membrane protein involved in peptidoglycan synthesis from the Keio Collection, and some Tol-Pal mutants of Caulobacter crescentus,59 are reported to display large blebs.
Figure 4.
The consequences of various antimicrobial peptides on E. coli morphology. E. coli ATCC 29522 (1 × 108 CFU/mL, mid-log phase) were exposed to each peptide (20 μM) for 1 h at 37 °C (10 mM sodium phosphate buffer, pH 7.4, 1% (v/v) TSB) prior to imaging. Top panels, scale = 5 μm; bottom panels, scale = 2 μm.
On the basis of this modest AMP screen, we concluded that HD5ox affects E. coli differently than the other AMPs considered in this work, including the murine α-defensin cryptdin-4. Although morphology comparisons alone do not provide detailed insight into antibiotic mechanisms, our imaging results suggest that the mechanism of action of HD5ox against E. coli cannot be explained fully by membrane permeabilization.
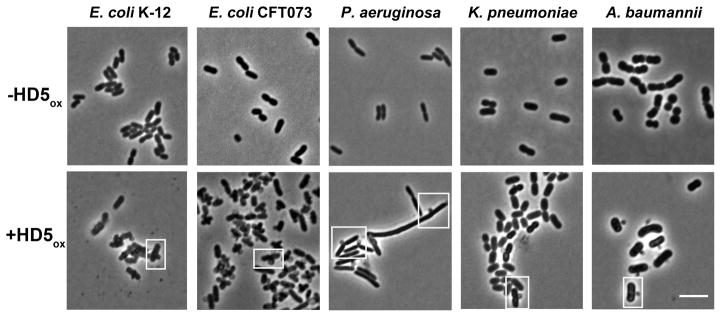
HD5ox Causes Similar Morphological Changes in Other Gram-negative Organisms
HD5ox exhibits broad-spectrum antibacterial activity37,38,40 and whether its mechanism of action is general or strain-specific remains unclear. Several structure/activity relationship studies indicated that HD5ox operates by different mechanisms for Gram-negative and –positive organisms, but these studies were limited to comparisons between E. coli and Staphylococcus aureus.44,45,48 To delineate whether HD5ox perturbs the morphologies of other Gram-negative strains, four human pathogens, Acinetobacter baumannii 17978, Klebsiella pneumoniae 13883, Pseudomonas aeruginosa PAO1, and E. coli CFT073, were evaluated along with the laboratory strain E. coli K-12 (Figure 5). As for non-pathogenic E. coli ATCC 25922 and K-12, bleb formation, elongation, and clumping were observed for the pathogenic strains. We previously reported that A. baumannii exhibits relatively high sensitivity to HD5ox.40 In accordance with this observation, A. baumannii displayed blebs at 10 μM HD5ox (Figure S8). P. aeruginosa is less susceptible to HD5ox killing,40 and blebs were only observed with 40 μM of HD5ox (Figure S8).
Figure 5.
The effect of HD5ox exposure on the morphology of Gram-negative bacteria. Each microbe (1 × 108 CFU/mL, mid-log phase) was exposed to HD5ox (20 μM except for P. aeruginosa where 40 μM was used) for 1 h at 37 °C (10 mM sodium phosphate buffer, pH 7.4, 1% v/v TSB) prior to imaging. Scale = 5 μm. Additional images are provided in Figure S6.
Morphological Changes are Attenuated by Salt and Divalent Metal Ions
The in vitro antimicrobial activity of many defensins is attenuated by the presence of salt and divalent cations.60 This phenomenon is commonly attributed to a disruption of electrostatic interactions between the cationic defensin and anionic bacterial cell membrane.17 Like many defensins, the antibacterial activity of HD5ox is attenuated by millimolar concentrations of sodium chloride.38 We observed that NaCl prevents bleb formation and other morphological changes associated with HD5ox activity. Indeed, E. coli co-treated with HD5ox (40 μM) and NaCl (200 mM) displayed a smooth morphology similar to that of the untreated control (Figure S7). The divalent cations Ca(II), Mg(II) and Zn(II) also blocked HD5ox activity (Figure S5). This effect was most potent for Zn(II) where a 2:1 Zn(II):HD5ox molar ratio resulted in a loss of antibacterial activity and HD5ox- associated morphologies. This observation is of broad interest because the Paneth cell granules, which harbor HD5, also contain a labile zinc pool of unknown function.34
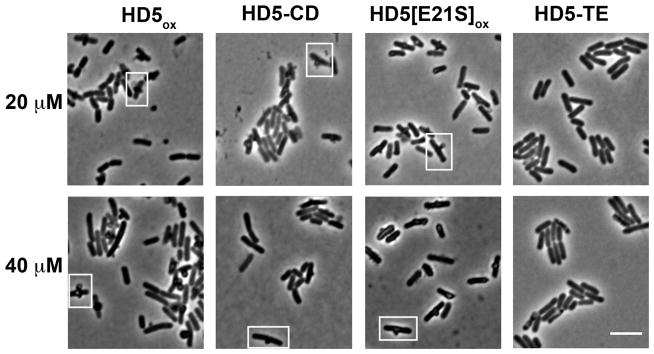
Disulfide Bonds Are Necessary for the Bleb Morphology
The canonical α-defensin disulfide array provides a three-stranded β-sheet fold to each HD5ox monomer. This compact structure orients the positively charged residues on one face of the peptide and the hydrophobic residues on the opposite, rendering HD5 amphipathic.19 To investigate the structural requirements for HD5ox activity, we evaluated the consequences of treating E. coli with three HD5 derivatives, HD5-TE, HD5-CD, and HD5[E21S]ox, selected to probe the disulfide array as well as quaternary structure. HD5-TE is a linear disulfide-null analogue where the six cysteines are carboxymethylated with 2-iodoacetamide.61 Defensins have the propensity to self-associate, and in prior work we reported that HD5ox forms tetramers at neutral pH.19 HD5-CD is a C2-symmetric covalent dimer of HD5ox with a cationic surface that results from intermolecular disulfide exchange between the Cys5–Cys20 disulfide bonds (canonical CysII—CysIV) of two HD5 monomers.40 We also prepared and characterized HD5[E21S]ox, a new HD5ox mutant that forms a non-covalent dimer, but not a tetramer, at neutral pH (Supporting Information, Table S3, Figure S23).
E. coli treated with HD5-CD or HD5[E21S]ox were indistinguishable from those treated with HD5ox (Figure 6), in agreement with antimicrobial activity assays where both HD5[E21S]ox and HD5-CD killed E. coli (Figure S10). In contrast, the antimicrobial activity of linear and unstructured HD5-TE against E. coli was attenuated (2-fold log reduction of CFU/mL at 16 μM) relative to HD5ox. HD5-TE did not cause bleb formation (Figure 6); however, SEM revealed that the cells treated with HD5-TE were corrugated and frequently elongated (Figure S11). These results highlight the importance of cysteine residues housed in disulfide linkages in the overall antimicrobial activity of HD5 and the induced morphological changes.
Figure 6.
E. coli morphologies in the presence of HD5 derivatives reveal that the disulfide bonds are necessary for bleb formation. E. coli ATCC 25922 (1 × 108 CFU/mL, mid-log phase) was exposed to each peptide for 1 h at 37 °C (10 mM sodium phosphate buffer, pH 7.4, 1% v/v TSB) prior to imaging. Scale = 5 μm.
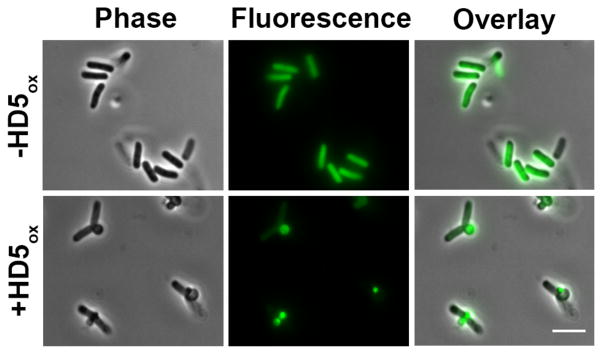
Blebs Accumulate the Cytoplasmic Contents
The blebs observed in this work are reminiscent of the cellular morphologies that result from treatment of E. coli with β-lactams, and E. coli strains expressing GFP have proven to be useful in imaging studies of β-lactam action.57,62 Guided by this work, we investigated the contents as well as time-dependent formation of the blebs by employing an E. coli strain that expresses cytoplasmic GFP (E. coli cyto-GFP). Mid-log phase E. coli cyto-GFP formed blebs following exposure to HD5ox. The blebs displayed GFP emission, and the GFP emission from the cell body was markedly reduced, suggesting that cytoplasmic contents localized to the blebs (Figure 7). Some additional phenotypes were observed for dividing cells. In several instances where a dividing cell exhibited a bleb at the cell division site, the GFP localization differed between the daughter cells (Figure S12). Moreover, for cells with blebs at the cell division site, the GFP intensity in the region between the two daughter cells was relatively weak, indicating that the presence of a membrane at the division plane. In agreement with this observation, labeling HD5ox-treated E. coli ATCC 25922 with the membrane-binding dye FM4-64 confirmed the presence of this membrane (Figure S13).
Figure 7.
Treatment of E. coli cyto-GFP with HD5ox reveals that the cytoplasmic contents leak into the blebs. E. coli cyto-GFP (1 × 107 CFU/mL, mid-log phase) were exposed to 4 μM HD5ox for 1 h at 37 °C (10 mM sodium phosphate buffer, pH 7.4, 1% v/v TSB) prior to imaging. Scale = 5 μm.
Cell viability, as observed by PI uptake, was inversely correlated to the overall GFP fluorescence intensity of the bacteria (Figure S12). When E. coli cyto-GFP (1 × 108 CFU/mL) were treated with HD5ox (20 μM) and subsequently stained as a result of PI uptake (5 μg/mL), the cells with brighter GFP emission and fewer morphological defects exhibited less PI labeling relative to cells that were affected by HD5ox.
In agreement with studies using E. coli ATCC 25922 (Figure 3), negligible changes in morphology and GFP localization were observed for HD5ox-treated E. coli cyto-GFP in stationary phase. To obtain quantitative comparisons, the GFP intensity and morphological changes for both mid-log phase and stationary-phase E. coli cyto-GFP were analyzed (Figure S15). The mean cell length for the mid-log phase cells increased from 2.8 ± 0.8 μm to 3.4 ± 1.0 μm (Student’s t-test: t-value = 6.32, t-probability = <0.0001) with increasing HD5ox (0–80 μM), and the mean GFP fluorescence intensity decreased accordingly (155 ± 48 units to 93 ± 47 units). Only minor changes in both cell lengths (3.0 ± 0.8 μm to 2.9 ± 0.7 μm) (Student’s t-test: t-value = 1.97, t-probability = 0.049) and mean intensities (112 ± 38 units to 139 ± 53 units) were found when stationary phase cells were treated with HD5ox.
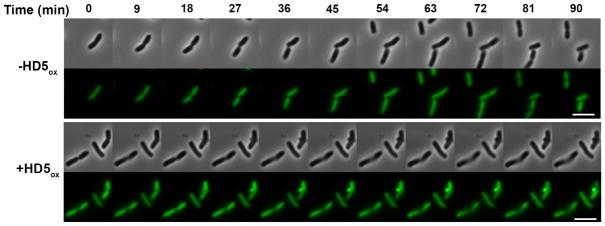
To obtain temporal information on bleb formation and GFP redistribution, we performed time-course experiments where E. coli cyto-GFP were treated with HD5ox on the microscope stage and collected images over a 2 h period (Figure 8). The replication time for E. coli in the standard AMA buffer (10 mM sodium phosphate buffer, pH 7.4, 1% v/v TSB without dextrose) on a MatTek plate ranged from 90–120 min at 37 °C. These cultures were unsynchronized, and bleb formation occurred at different time points depending on the cell. Blebs were observed immediately for some cells whereas others formed blebs after ~30 min exposure to HD5ox. After the appearance of one or more blebs, GFP intensity in the cell body diminished in all cases observed. Although dividing E. coli were observed for untreated cells (Figure 8, top panels), cells that were affected by HD5ox no longer divided (Figure 8, bottom panels).
Figure 8.
Time-lapse imaging of E. coli cyto-GFP (1 × 107 CFU/mL, mid-log phase) treated with 20 μM HD5ox with HD5ox at 37 °C (10 mM sodium phosphate buffer, pH 7.4, 1% v/v TSB). Scale = 5 μm.
Membrane Composition of the Blebs and Observation of Outer Membrane Vesicles
The composition of the membrane surrounding the blebs of HD5ox-treated E. coli was investigated by fluorescence imaging of an E. coli strain harboring a plasmid encoding periplasmic GFP (E. coli peri-GFP) as well as transmission electron microscopy (TEM) of E. coli ATCC 25922. These studies afforded several phenotypes and suggested that two different types of blebs form (Figure S16). Some cells exhibited a ring of GFP emission around the blebs, which indicated that both the outer and inner membranes surrounded the bleb and were in tact. Other cells presented uniform GFP emission throughout the bleb. Possible explanations for this phenotype include (i) only the outer membrane surrounded the blebs or (ii) the inner membrane surrounded the bleb, but was damaged and leaked the periplasmic contents into the bleb. TEM imaging revealed that the blebs are membrane bound; however, it was difficult to confirm whether one or both membranes surround each bleb (Figure S17).
TEM also revealed profound changes to the E. coli cell surface upon HD5ox treatment, including vesicles surrounded by a membrane that often clustered in chain-like arrangements (Figure S17F). During phase-contrast imaging of HD5ox-treated bacteria, we frequently observed surface-appended structures resembling debris. We attribute these structures, at least in part, to outer membrane vesicle (OMVs) on the basis of TEM and SEM studies (Figure S3). Formation of OMVs (~20–200 nm wide) is an important bacterial stress response pathway, and OMVs contribute to pathogenesis.63 Thus, we speculate that E. coli may attempt to evade HD5ox by generating and shedding OMVs, along with HD5ox, into the extracellular space.
Design and Synthesis of Fluorophore-HD5 Conjugates for Visualizing Peptide Localization
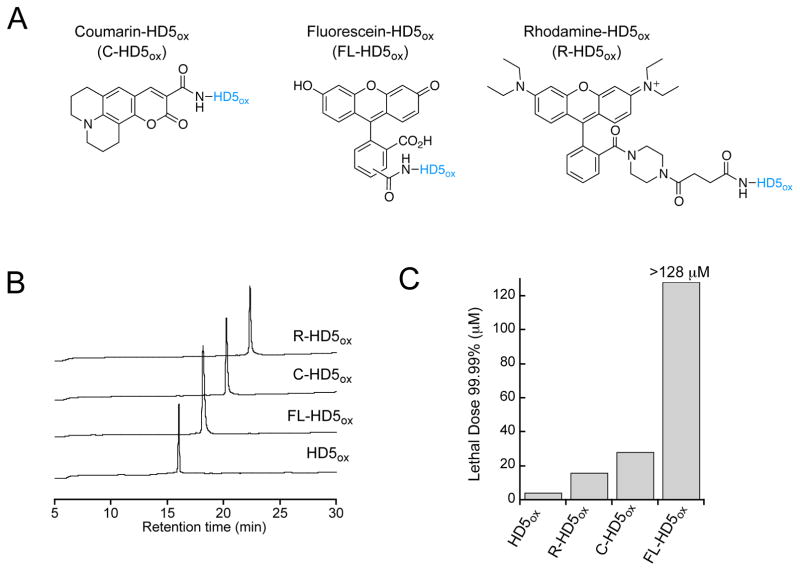
To probe the cellular localization of HD5ox, we designed, prepared and characterized a seven-member family of fluorophore-HD5 conjugates (Figure 9A, B, Figure S16 and Supporting Information). Because addition of a fluorophore constitutes a substantial modification to a 32-residue peptide that may perturb the function and cellular localization of the native peptide, we evaluated the behavior of fluorophore-HD5 conjugates harboring different fluorophores as well as peptides harboring rhodamine attached to different positions (Table S1). First, we modified the N-terminus of HD5 with three different fluorophores that afford variable photophysical properties as well as overall charge. We selected coumarin 343 (C), fluorescein (FL), and rhodamine B 4-(3-carboxypropionyl)piperazine amide (R)64 as fluorophores to achieve emission properties spanning the ~490- to ~590-nm range. Moreover, we reasoned that the overall charge of the fluorophore-modified peptide may influence its antimicrobial activity. Coumarin 343 is neutral following coupling to an α-amino group, fluorescein is anionic, and rhodamine B is cationic. This selection allowed us to probe the effect of fluorophore charge on the antimicrobial activity and cellular localization of fluorophore-HD5ox conjugates. We prepared R-HD5[R9K] and R-HD5[R13K] to evaluate the consequences of fluorophore positioning within the peptide sequence. We selected these positions because (i) prior structure-activity relationship studies reported that R9K and R13K mutants retained some antimicrobial activity,46, and (ii) the X-ray crystal structure (PDB: 1ZMP)20 and NMR solution structure (PDB: 2LXZ)19 of HD5ox revealed that side-chains of R9 and R13 are solvent exposed and directed away from the dimer interface of native HD5ox.
Figure 9.
Functionalization of the HD5ox N-terminus affords three fluorophore-HD5ox conjugates. (A) Structures of coumarin-, fluorescein-, and rhodamine-derivatized HD5ox. The HD5ox primary sequence is presented in Figure 1. (B) Analytical HPLC traces (220 nm absorption, 10–60% B over 30 min, 1 mL/min) of purified peptides. (C) Antimicrobial activity of the peptides against E. coli ATCC 25922 (n = 3).
The syntheses of the fluorophore-HD5 conjugates were achieved using Fmoc-based solid-phase peptide synthesis (SPPS) as described previously.40 Analytical and photophysical characterization of the peptides is detailed in the Supporting Information (Tables S1 and S2, Figure S18). To determine whether the absence of disulfide bonds influences cellular localization, two disulfide-null mutants, RHD5-TE and FL-HD5-TE, were synthesized by capping the cysteines of the reduced peptides, R-HD5red and FL-HD5red, with 2-iodoacetamide (Supporting Information).
Fluorophore Modifications Influence the Antibacterial Activity of HD5
To assess the impact of fluorophore modification on HD5ox function, the AMAs of the conjugates against E. coli ATCC 25922 (1 × 106 CFU/mL) were first evaluated and compared to that of native HD5ox (Figures 9C, S18C). Fluorophore modification attenuated the cell killing ability of HD5ox to varying degrees. The antibacterial activity (reported as the lethal dose for 99.99% killing, LD99.99) of HD5ox and the analogues harboring N-terminal fluorophores followed a trend where HD5ox (8 μM) > R-HD5ox (16 μM) > C-HD5ox (28 μM) > FL-HD5ox (>128 μM) (Figure 9C). This trend suggests that the charge of the fluorophore may influence antimicrobial activity. Additional factors, such as the physiochemical properties of the fluorophores and linkers (e.g. succinic acid spacer for R-HD5ox) may also contribute to such changes in the antimicrobial activity of modified HD5ox. R-HD5[R9K] and R-HD5[R13K] were active against E. coli and the linear derivatives were less active compared to their respective folded peptides, as expected (Figure S18C).
HD5ox Enters the E. coli Cytoplasm and Localizes to the Cell Poles and Division Site
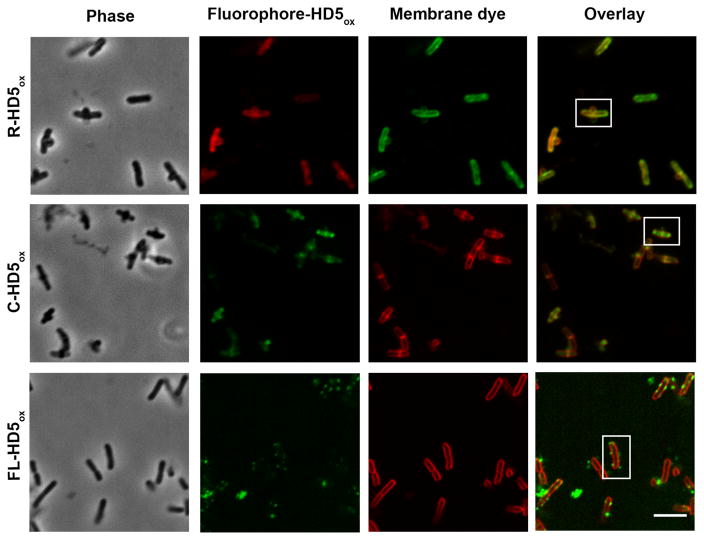
On the basis of the AMA assay results described above, we first investigated the cellular localization of R-HD5ox. We observed HD5ox-like morphological changes, including bleb formation, for E. coli ATCC 25922 (Figure 10) and E. coli CFT073 (Figure S19). Intracellular fluorescence was also observed. Co-labeling studies with the membrane-binding dye FM1-43 revealed that the FM1-43 emission profile enclosed a significant portion of the R-HD5ox fluorescence intensity profile (Figures 10,11), which indicates that the peptide penetrated the outer and inner membranes and entered the cytoplasm. In most of the cells examined by fluorescence microscopy, the rhodamine emission was most intense at the poles and division plane. This type of labeling pattern was not observed for R-HD5-TE or rhodamine-modified LL-37 (Figure S20), both of which provided uniform cytoplasmic fluorescence.
Figure 10.
Fluorescence imaging of E. coli treated with fluorophore-HD5ox conjugates and membrane dyes. E. coli ATCC 25922 (1 × 108 CFU/mL, mid-log phase) treated with R-HD5ox (20 μM), C-HD5ox (20 μM), and FL-HD5ox (8 μM) at 37 °C (10 mM sodium phosphate buffer, pH 7.4, 1% v/v TSB) and incubated with 2 μg/mL of FM1-43 for R-HD5ox sample and 2 μg/mL of FM 4–64 for both C-HD5ox and FL-HD5ox samples prior to imaging. The boxed cells are depicted in Figure 11. Scale = 5 μm.
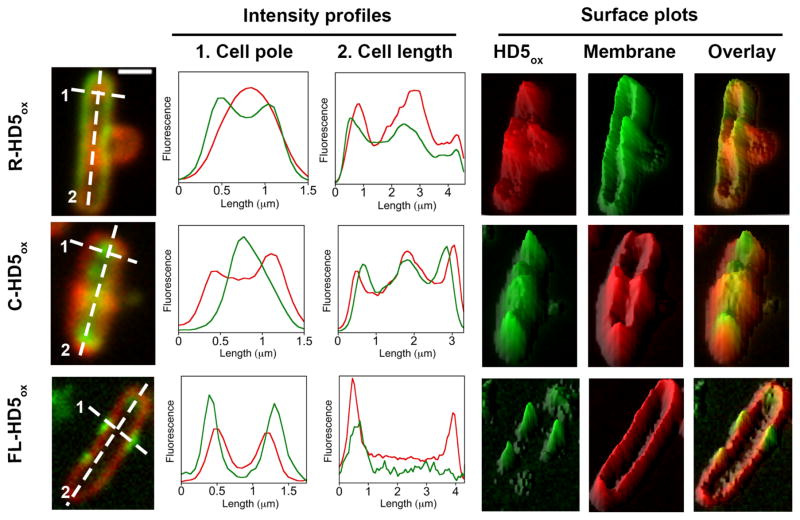
Figure 11.
Intensity profiles and surface plots of the boxed cells depicted in Figure 10. Fluorescence intensities along the cell poles (dashed line 1) and across cell (dashed line 2) are plotted. Surface plots were generated using ImageJ software. Scale = 1 μm.
When E. coli were treated with R-HD5ox under conditions that result in attenuated HD5ox activity (vide supra), different labeling patterns were observed. For instance, the presence of NaCl (200 mM) resulted in rhodamine emission only at the cell surface, indicating that R-HD5ox did not enter E. coli (Figure S22). Stationary-phase E. coli treated with R-HD5ox exhibited diminished intracellular fluorescence relative to mid-log phase cells (Figure S22). Taken together, these observations support a model whereby HD5ox must overcome the outer membrane permeability barrier and enter the cytosol to exert its full capacity to kill bacteria.
Co-incubation of E. coli with a 1:1 molar ratio of rhodamine B and HD5ox resulted in uniform cytoplasmic staining whereas treatment of E. coli with rhodamine B alone resulted in negligible cellular fluorescence (Figure S21). This result suggested that HD5ox treatment allowed for rhodamine B entry as a result of membrane permeabilization, and confirmed that covalent attachment of rhodamine to HD5ox is essential for observing fluorescence localized to the cell poles and division site. The R-HD5[R13K]ox conjugate provided a similar labeling pattern as R-HD5ox and thereby indicated that localization of the rhodamine-HD5ox conjugate is not an artifact resulting from a particular site of rhodamine attachment (Figure S20). Moreover, C-HD5ox preferentially labeled the cell poles and cell division sites, and co-labeling with the membrane-binding dye FM 4-64 confirmed its cytoplasmic localization (Figures 10,11). The fact that the same labeling pattern was observed for HD5ox modified with either rhodamine or coumarin, taken with these modified peptides causing the same morphological changes as observed for unmodified HD5ox, provides a strong indication that the localization is a result of HD5ox, and not the fluorophore.
It should be noted that FL-HD5ox, which provided no in vitro antibacterial activity against E. coli at the highest concentration evaluated (128 μM), did not enter E. coli. Rather, the peptide afforded a punctate labeling pattern on and around the bacterial surface (Figures 10,11). We attribute this phenomenon to the overall negative charge of the fluorescein moiety.
The fluorescence imaging studies of HD5ox may be considered in the context of other AMPs that have been examined by similar approaches to probe uptake and mechanism of action. For instance, fluorescence microscopy revealed that buforin II entered the bacterial cytoplasm without permeabilizing the inner membrane.65 A rhodamine derivative of the only human cathelicidin, LL-37, attacked septating E. coli more readily than non-septating cells and permeabilized the bacteria via a carpet-model of membrane destabilization.66,67 A fluorophore-labeled lantibiotic was shown to interact with lipid II of B. subtilis 168.68 Recent imaging studies of Cy3-derivaitzed HBD2 indicated that HBD2 preferentially localizes to the nascent poles and cell division site of Enterococcus faecalis.31 This focal labeling pattern was attributed to the binding of the HBD2 to anionic lipids in these regions. The cell poles and division site of E. coli are also enriched in negatively charged phospholipids that include cardiolipin and phosphatidylglycerol.69 Whether HD5ox also binds to anionic lipids at the cell poles of E. coli and/or another target in these locales is currently unknown and a topic for future investigation.
Summary and Outlook
In this study, we defined how the antimicrobial peptide HD5ox affects the morphology of E. coli and other Gram-negative bacteria. We established that HD5ox causes distinct morphological changes to E. coli as well as K. pneumoniae, A. baumannii, and P. aeruginosa that include bleb formation, cellular clumping, and elongation. We also demonstrated that other AMPs, including human LL-37 and murine α-defensin cryptdin-4, do not cause such morphologies. Taken together, these observations indicate that HD5ox kills E. coli by a different mechanism than the other peptides examined in this work. Our results highlight the importance of treating host-defense peptides individually, and not generalizing cell-killing mechanisms.
Defensins are often described as membrane-disrupting peptides. Our prior investigations indicate that HD5ox traverses the outer membrane and subsequently damages the inner membrane of E. coli.44,50 In this work, the insights gained from utilizing fluorophore-HD5ox conjugates modified with rhodamine or coumarin support a model that HD5ox enters the E. coli cytoplasm. Because the cytoplasm is a reducing environment, intracellular reduction of the HD5ox disulfide array to liberate free cysteine residues may occur. Deciphering whether such redox chemistry contributes to altered cellular morphologies and cell killing warrants exploration. The cytoplasmic localization also supports the possibility that HD5ox (or the reduced form) has an as-yet undiscovered intracellular target. Indeed, the intracellular staining pattern at the cell poles and cell division site routinely observed for E. coli treated with R/C-HD5ox suggests the locale of a possible target. On the basis of this localization, coupled with the elongation phenotype observed for E. coli and other Gram-negative microbes, we reason that HD5ox may exert antimicrobial activity by affecting cell division. Efforts are underway to further investigate this notion.
Materials and Methods
Complete Materials and Methods are provided as Supporting Information.
Supplementary Material
Acknowledgments
We thank Prof. Daniel Kahne for insightful discussions; Prof. Bradley Pentelute and members of the Pentelute Lab for expertise in flow-based peptide synthesis; Yunfei Zhang for performing initial Zn(II) experiments; Jill Tomaras for microbiology assistance; Nicki Watson at of the Whitehead Institute for Biomedical Research for preparing the TEM samples and imaging; Prof. K. Ribbeck for providing the E. coli cyto-GFP strain.
Funding. This work was supported by the NIH (Grant DP2OD007045 from the Office of the Director). HRC was a recipient of a 2014 Richard R. Schrock summer graduate fellowship; PC is a recipient of a Royal Thai Government Fellowship; SAL and ILC received research funding from the MIT UROP Program. The Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-007031) is gratefully acknowledged. The Institute for Soldier Nanotechnologies is supported in part by the U. S. Army Research Laboratory and the U. S. Army Research Office through the Institute for Soldier Nanotechnologies, under contract number W911NF-13-D-0001.
Abbreviations
- AMA
Antimicrobial activity
- C
Coumarin 343
- CFU
Colony forming unit
- Crp-4
Cryptdin-4 (a murine α-defensin)
- FL
5(6)-Carboxyfluorescein
- FM1-43
(N-(3-Triethylammoniumpropyl)-4-(4-(dibutylamino) styryl)pyridiniumdibromide)
- FM4-64
(N-(3-Triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridiniumdibromide)
- HD5
Human α-defensin 5
- HD5red
Reduced form of human α-defensin 5
- HD5ox
Oxidized form of human α-defensin 5
- HD5-TE
Linear form of human α-defensin 5 (iodoacetamide-capped)
- HD5-CD
Human α-defensin 5 covalent dimer
- PI
Propidium iodide
- R
Rhodamine B 4-(3-carboxypropionyl)piperazine amide
- SEM
Scanning electron microscopy
- TEM
Transmission electron microscopy
- TSB
Trypticase soy broth
Footnotes
Supporting Information. Complete experimental methods, additional data.
References
- 1.WHO. Antimicrobial resistance: global report on surveillance. 2014. [Google Scholar]
- 2.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 5.Jenssen H, Hamill P, Hancock REW. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi KY, Chow LNY, Mookherjee N. Cationic host defence peptides: multifaceted role in immune modulation and inflammation. J Innate Immun. 2012;4:361–370. doi: 10.1159/000336630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehrer RI, Lu W. α-Defensins in human innate immunity. Immunol Rev. 2012;245:84–112. doi: 10.1111/j.1600-065X.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 8.Cruz J, Ortiz C, Guzmán F, Fernández-Lafuente R, Torres R. Antimicrobial peptides: promising compounds against pathogenic microorganisms. Curr Med Chem. 2014;21:2299–2321. doi: 10.2174/0929867321666140217110155. [DOI] [PubMed] [Google Scholar]
- 9.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 10.Ramanathan B, Davis EG, Ross CR, Blecha F. Cathelicidins: microbicidal activity, mechanisms of action, and roles in innate immunity. Microbes Infect. 2002;4:361–372. doi: 10.1016/s1286-4579(02)01549-6. [DOI] [PubMed] [Google Scholar]
- 11.Gennaro R, Zanetti M. Structural features and biological activities of the cathelicidin-derived antimicrobial peptides. Biopolymers. 2000;55:31–49. doi: 10.1002/1097-0282(2000)55:1<31::AID-BIP40>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 12.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 13.Jones DE, Bevins CL. Paneth cells of the human small intestine express an antimicrobial peptide gene. J Biol Chem. 1992;267:23216–23225. [PubMed] [Google Scholar]
- 14.Porter EM, Liu L, Oren A, Anton PA, Ganz T. Localization of human intestinal defensin 5 in Paneth cell granules. Infect Immun. 1997;65:2389–2395. doi: 10.1128/iai.65.6.2389-2395.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayabe T, Ashida T, Kohgo Y, Kono T. The role of Paneth cells and their antimicrobial peptides in innate host defense. Trends Microbiol. 2004;12:394–398. doi: 10.1016/j.tim.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Clevers HC, Bevins CL. Paneth cells: maestros of the small intestinal crypts. Annu Rev Physiol. 2013;75:289–311. doi: 10.1146/annurev-physiol-030212-183744. [DOI] [PubMed] [Google Scholar]
- 17.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 18.Hilchie AL, Wuerth K, Hancock REW. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat Chem Biol. 2013;9:761–768. doi: 10.1038/nchembio.1393. [DOI] [PubMed] [Google Scholar]
- 19.Wommack AJ, Robson SA, Wanniarachchi YA, Wan A, Turner CJ, Wagner G, Nolan EM. NMR solution structure and condition-dependent oligomerization of the antimicrobial peptide human defensin 5. Biochemistry. 2012;51:9624–9637. doi: 10.1021/bi301255u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szyk A, Wu Z, Tucker K, Yang D, Lu W, Lubkowski J. Crystal structures of human alpha-defensins HNP4, HD5, and HD6. Protein Sci. 2006;15:2749–2760. doi: 10.1110/ps.062336606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehrer RI, Barton A, Daher KA, Harwig SS, Ganz T, Selsted ME. Interaction of human defensins with Escherichia coli Mechanism of bactericidal activity. J Clin Invet. 1989;84:553–561. doi: 10.1172/JCI114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 23.Figueredo SM, Weeks CS, Young SK, Ouellette AJ. Anionic amino acids near the pro-alpha-defensin N terminus mediate inhibition of bactericidal activity in mouse pro-cryptdin-4. J Biol Chem. 2009;284:6826–6831. doi: 10.1074/jbc.M807024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill CP, Yee J, Selsted ME, Eisenberg D. Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization. Science. 1991;251:1481–1485. doi: 10.1126/science.2006422. [DOI] [PubMed] [Google Scholar]
- 25.Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, Jansen A, Nielsen AK, Mygind PH, Raventós DS, Neve S, Ravn B, Bonvin AMJJ, De Maria L, Andersen AS, Gammelgaard LK, Sahl HG, Kristensen HH. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science. 2010;328:1168–1172. doi: 10.1126/science.1185723. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt P, Wilmes M, Pugnière M, Aumelas A, Bachère E, Sahl HG, Schneider T, Destoumieux-Garzón D. Insight into invertebrate defensin mechanism of action: oyster defensins inhibit peptidoglycan biosynthesis by binding to lipid II. J Biol Chem. 2010;285:29208–29216. doi: 10.1074/jbc.M110.143388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Essig A, Hofmann D, Münch D, Gayathri S, Künzler M, Kallio PT, Sahl HG, Wider G, Schneider T, Aebi M. Copsin, a novel peptide-based fungal antibiotic interfering with the peptidoglycan synthesis. J Biol Chem. 2014;289:34953–34964. doi: 10.1074/jbc.M114.599878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Leeuw E, Li C, Zeng P, Li C, Diepeveen-de Buin M, Lu WY, Breukink E, Lu W. Functional interaction of human neutrophil peptide-1 with the cell wall precursor lipid II. FEBS Lett. 2010;584:1543–1548. doi: 10.1016/j.febslet.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sass V, Schneider T, Wilmes M, Körner C, Tossi A, Novikova N, Shamova O, Sahl HG. Human beta-defensin 3 inhibits cell wall biosynthesis in Staphylococci. Infect Immun. 2010;78:2793–2800. doi: 10.1128/IAI.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilmes M, Sahl HG. Defensin-based anti-infective strategies. Int J Med Microbiol. 2014;304:93–99. doi: 10.1016/j.ijmm.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Kandaswamy K, Liew TH, Wang CY, Huston-Warren E, Meyer-Hoffert U, Hultenby K, Schröder JM, Caparon MG, Normark S, Henriques-Normark B, Hultgren SJ, Kline KA. Focal targeting by human β-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc Natl Acad Sci U S A. 2013;110:20230–20235. doi: 10.1073/pnas.1319066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu H, Pazgier M, Jung G, Nuccio SP, Castillo PA, de Jong MF, Winter MG, Winter SE, Wehkamp J, Shen B, Salzman NH, Underwood MA, Tsolis RM, Young GM, Lu W, Lehrer RI, Bäumler AJ, Bevins CL. Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science. 2012;337:477–481. doi: 10.1126/science.1218831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chairatana P, Nolan EM. Molecular basis for self-assembly of a human host-defense Peptide that entraps bacterial pathogens. J Am Chem Soc. 2014;136:13267–13276. doi: 10.1021/ja5057906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giblin LJ, Chang CJ, Bentley AF, Frederickson C, Lippard SJ, Frederickson CJ. Zinc-secreting Paneth cells studied by ZP fluorescence. J Histochem Cytochem. 2006;54:311–316. doi: 10.1369/jhc.5A6724.2005. [DOI] [PubMed] [Google Scholar]
- 35.Dinsdale D. Ultrastructural localization of zinc and calcium within the granules of rat Paneth cells. J Histochem Cytochem. 1984;32:139–145. doi: 10.1177/32.2.6693753. [DOI] [PubMed] [Google Scholar]
- 36.Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, Shen B, Schaeffeler E, Schwab M, Linzmeier R, Feathers RW, Chu H, Lima H, Fellermann K, Ganz T, Stange EF, Bevins CL. Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc Natl Acad Sci U S A. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ericksen B, Wu Z, Lu W, Lehrer RI. Antibacterial activity and specificity of the six human {alpha}-defensins. Antimicrob Agents Chemother. 2005;49:269–275. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter EM, van Dam E, Valore EV, Ganz T. Broad-spectrum antimicrobial activity of human intestinal defensin 5. Infect Immun. 1997;65:2396–2401. doi: 10.1128/iai.65.6.2396-2401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nuding S, Zabel LT, Enders C, Porter E, Fellermann K, Wehkamp J, Mueller HAG, Stange EF. Antibacterial activity of human defensins on anaerobic intestinal bacterial species: a major role of HBD-3. Microbes Infect. 2009;11:384–393. doi: 10.1016/j.micinf.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Wommack AJ, Ziarek JJ, Tomaras J, Chileveru HR, Zhang Y, Wagner G, Nolan EM. Discovery and characterization of a disulfide-locked C2-symmetric defensin Peptide. J Am Chem Soc. 2014;136:13494–13497. doi: 10.1021/ja505957w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 42.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjöberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajabi M, Ericksen B, Wu X, de Leeuw E, Zhao L, Pazgier M, Lu W. Functional determinants of human enteric α-defensin HD5: crucial role for hydrophobicity at dimer interface. J Biol Chem. 2012;287:21615–21627. doi: 10.1074/jbc.M112.367995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wanniarachchi YA, Kaczmarek P, Wan A, Nolan EM. Human defensin 5 disulfide array mutants: disulfide bond deletion attenuates antibacterial activity against Staphylococcus aureus. Biochemistry. 2011;50:8005–8017. doi: 10.1021/bi201043j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Leeuw E, Burks SR, Li X, Kao JPY, Lu W. Structure-dependent functional properties of human defensin 5. FEBS Lett. 2007;581:515–520. doi: 10.1016/j.febslet.2006.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Leeuw E, Rajabi M, Zou G, Pazgier M, Lu W. Selective arginines are important for the antibacterial activity and host cell interaction of human alpha-defensin 5. FEBS Lett. 2009;583:2507–2512. doi: 10.1016/j.febslet.2009.06.051. [DOI] [PubMed] [Google Scholar]
- 47.Rajabi M, de Leeuw E, Pazgier M, Li J, Lubkowski J, Lu W. The conserved salt bridge in human alpha-defensin 5 is required for its precursor processing and proteolytic stability. J Biol Chem. 2008;283:21509–21518. doi: 10.1074/jbc.M801851200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei G, de Leeuw E, Pazgier M, Yuan W, Zou G, Wang J, Ericksen B, Lu WY, Lehrer RI, Lu W. Through the looking glass, mechanistic insights from enantiomeric human defensins. J Biol Chem. 2009;284:29180–29192. doi: 10.1074/jbc.M109.018085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomassin JL, Lee MJ, Brannon JR, Sheppard DC, Gruenheid S, Le Moual H. Both group 4 capsule and lipopolysaccharide O-antigen contribute to enteropathogenic Escherichia coli resistance to human α-defensin 5. PLoS One. 2013;8:e82475. doi: 10.1371/journal.pone.0082475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moser S, Chileveru HR, Nolan EM, Tomaras J. A Bacterial Mutant Library as a Tool to Study the Attack of a Defensin Peptide. Chembiochem. 2014;15:2684–2688. doi: 10.1002/cbic.201402354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt NW, Mishra A, Lai GH, Davis M, Sanders LK, Tran D, Garcia A, Tai KP, McCray PB, Ouellette AJ, Selsted ME, Wong GCL. Criterion for amino acid composition of defensins and antimicrobial peptides based on geometry of membrane destabilization. J Am Chem Soc. 2011;133:6720–6727. doi: 10.1021/ja200079a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van den Bogaart G, Guzmán JV, Mika JT, Poolman B. On the mechanism of pore formation by melittin. J Biol Chem. 2008;283:33854–33857. doi: 10.1074/jbc.M805171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falagas ME, Kasiakou SK. Colistin: the revival of polymyxins for the management of multidrug-resistant Gram-negative bacterial infections. Clin Infect Dis. 2005;40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 54.Sochacki Ka, Barns KJ, Bucki R, Weisshaar JC. Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc Natl Acad Sci U S A. 2011;108:E77–81. doi: 10.1073/pnas.1101130108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsuzaki K, Sugishita K, Harada M, Fujii N, Miyajima K. Interactions of an antimicrobial peptide, magainin 2, with outer and inner membranes of Gram-negative bacteria. Biochim Biophys Acta. 1997;1327:119–130. doi: 10.1016/s0005-2736(97)00051-5. [DOI] [PubMed] [Google Scholar]
- 56.Spindler EC, Hale JDF, Giddings TH, Hancock REW, Gill RT. Deciphering the mode of action of the synthetic antimicrobial peptide Bac8c. Antimicrob Agents Chemother. 2011;55:1706–1716. doi: 10.1128/AAC.01053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao Z, Kahne D, Kishony R. Distinct single-cell morphological dynamics under beta-lactam antibiotics. Mol Cell. 2012;48:705–712. doi: 10.1016/j.molcel.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paradis-Bleau C, Kritikos G, Orlova K, Typas A, Bernhardt TG. A genome-wide screen for bacterial envelope biogenesis mutants identifies a novel factor involved in cell wall precursor metabolism. PLoS Genet. 2014;10:e1004056. doi: 10.1371/journal.pgen.1004056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeh YC, Comolli LR, Downing KH, Shapiro L, McAdams HH. The caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J Bacteriol. 2010;192:4847–4858. doi: 10.1128/JB.00607-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldman MJ, Anderson GM, Stolzenberg ED, Kari UP, Zasloff M, Wilson JM. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, Cougnon FBL, Wanniarachchi YA, Hayden JA, Nolan EM. Reduction of human defensin 5 affords a high-affinity zinc-chelating peptide. ACS Chem Biol. 2013;8:1907–1911. doi: 10.1021/cb400340k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chung HS, Yao Z, Goehring NW, Kishony R, Beckwith J, Kahne D. Rapid beta-lactam-induced lysis requires successful assembly of the cell division machinery. Proc Natl Acad Sci U S A. 2009;106:21872–21877. doi: 10.1073/pnas.0911674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen T, Francis MB. Practical synthetic route to functionalized rhodamine dyes. Org Lett. 2003;5:3245–3248. doi: 10.1021/ol035135z. [DOI] [PubMed] [Google Scholar]
- 65.Park CB, Yi KS, Matsuzaki K, Kim MS, Kim SC. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: the proline hinge is responsible for the cell-penetrating ability of buforin II. Proc Natl Acad Sci U S A. 2000;97:8245–8250. doi: 10.1073/pnas.150518097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barns KJ, Weisshaar JC. Real-time attack of LL-37 on single Bacillus subtilis cells. Biochim Biophys Acta. 2013;1828:1511–1520. doi: 10.1016/j.bbamem.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding B, Soblosky L, Nguyen K, Geng J, Yu X, Ramamoorthy A, Chen Z. Physiologically-relevant modes of membrane interactions by the human antimicrobial peptide, LL-37, revealed by SFG experiments. Sci Rep. 2013;3:1854. doi: 10.1038/srep01854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bindman NA, van der Donk WA. A general method for fluorescent labeling of the N-termini of lanthipeptides and its application to visualize their cellular localization. J Am Chem Soc. 2013;135:10362–10371. doi: 10.1021/ja4010706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oliver PM, Crooks JA, Leidl M, Yoon EJ, Saghatelian A, Weibel DB. Localization of Anionic Phospholipids in Escherichia coli Cells. J Bacteriol. 2014;196:3386–3398. doi: 10.1128/JB.01877-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.