ABSTRACT
Aberrant DNA methylation seems to be associated with prostate cancer behavior. We investigated LINE-1 methylation in prostate cancer and non-neoplastic tissue adjacent to tumor (NTAT) in association with mortality from prostate cancer. We selected 157 prostate cancer patients with available NTAT from 2 cohorts of patients diagnosed between 1982–1988 and 1993–1996, followed up until 2010. An association between LINE-1 hypomethylation and prostate cancer mortality in tumor was suggested [hazard ratio per 5% decrease in LINE-1 methylation levels: 1.40, 95% confidence interval (CI): 0.95–2.01]. After stratification of the patients for Gleason score, the association was present only for those with a Gleason score of at least 8. Among these, low (<75%) vs. high (>80%) LINE-1 methylation was associated with a hazard ratio of 4.68 (95% CI: 1.03–21.34). LINE-1 methylation in the NTAT was not associated with prostate cancer mortality. Our results are consistent with the hypothesis that tumor tissue global hypomethylation may be a late event in prostate cancerogenesis and is associated with tumor progression.
KEYWORDS: Global hypomethylation, LINE-1, mortality, methylation, non-neoplastic tissue, prostate cancer
Abbreviations
- NTAT
non-neoplastic tissue adjacent to tumor
- LINE-1
long interspersed nuclear element-1
- PIN
prostatic intraepithelial neoplasia
- APC
adenomatous polyposis coli
- GSTP1
glutathione S-transferase 1
- TURP
transurethral resection of the prostate
- HR
hazard ratio
- CI
confidence interval
- MAR
missing at random
Introduction
Prostate cancer is the most common tumor among men worldwide.1 Clinically, it can be an aggressive or a latent disease and the clinical behavior of prostate cancer is not entirely explained by the tumor's clinical-pathological characteristics. Many molecular markers are being studied in association with tumor aggressiveness, especially gene expression profiling2,3 and epigenetic changes.4,5 Among the latter markers, aberrant DNA methylation seems to play an important role in predicting the tumor behavior.6
Both gene-specific promoter hypermethylation and global hypomethylation may confer aggressiveness to cancer cells.7-9 In prostate cancer, hypermethylation of several genes has been associated with tumor aggressiveness in terms of clinical-pathological characteristics, biochemical recurrence, and/or mortality from prostate cancer.10-16 Global hypomethylation, which may cause reactivation of the transcription of oncogenes and transposable elements and increases chromosomal instability and loss of imprinting,17,18 has been less extensively investigated. We have recently conducted a systematic review of studies on prostatic tumor global hypomethylation and prostate cancer progression:19 although studies were heterogeneous and mostly based on a limited sample size, global hypomethylation was reported to be associated with high Gleason score, advanced tumor stage, high pre-operative PSA, and presence of metastasis.20-26
The genome-wide methylation level of the long interspersed nuclear element-1 (LINE-1) is considered as a good surrogate marker for global methylation,27-29 as LINE-1 is widely interspersed and represents about 15% of human genome.30,31 Thus, in the present study we have analyzed prostate tissue LINE-1 methylation status in a cohort of prostate cancer patients in association with Gleason score, gene-specific hypermethylation, and mortality from prostate cancer. We analyzed both the tumor tissue and the non-neoplastic tissue adjacent to the tumor (NTAT). The study is nested in cohorts of prostate cancer patients that we have previously studied to assess the role of gene-specific promoter hypermethylation in prostate cancer progression.13,32
Results
Selected characteristics of the 157 study patients are reported in Table 1. Most of the patients (82%) were diagnosed in the 1990s and the source of tissue was equally distributed among biopsy (34.4%), transurethral resection of the prostate (TURP) (34.4%), and radical prostatectomy (31.2%). As previously reported,32 among these patients the proportions of promoter methylation of glutathione S-transferase 1 (GSTP1) and adenomatous polyposis coli (APC) were higher in tumor tissue than in NTAT.
Table 1.
Selected characteristics of the study patients.
| Characteristics | Number | % |
|---|---|---|
| Year of diagnosis | ||
| 1982-1988 | 28 | 17.8 |
| 1993-1996 | 129 | 82.2 |
| Range survival time (years) | 0.03–24.11 | |
| Median survival time (years) | 6.79 | |
| Age at diagnosis (years) | ||
| 40-64 | 26 | 16.6 |
| 65-69 | 35 | 22.3 |
| 70-74 | 40 | 25.5 |
| 75+ | 56 | 35.7 |
| Mortality | ||
| Overall | 128 | 81.5 |
| From prostate cancer | 43 | |
| From other causes | 85 | |
| Source of tumor tissue | ||
| Biopsy | 54 | 34.4 |
| TURP | 54 | 34.4 |
| Radical prostatectomy | 49 | 31.2 |
| Gleason score | ||
| <7 | 59 | 37.6 |
| 7 | 41 | 26.1 |
| ≥8 | 57 | 36.3 |
| N genes methylated in tumor tissue (out of APC and GSTP1) | ||
| 0 | 11 | 7.0 |
| 1 | 31 | 19.8 |
| 2 | 115 | 73.2 |
| N genes methylated in NTAT (out of APC and GSTP1) | ||
| 0 | 67 | 42.7 |
| 1 | 45 | 28.7 |
| 2 | 45 | 28.7 |
TURP, transurethral resection of the prostate; NTAT, non-neoplastic tissue adjacent to tumor;
APC, adenomatous polyposis coli; GSTP1, glutathione S-transferase 1.
Tumor tissue LINE-1 methylation
Analyses were performed in 152 patients, as 5 patients were excluded due to non-detectable LINE-1 methylation. LINE-1 methylation was higher in TURPs than biopsies and radical prostatectomies and was not associated with Gleason score or hypermethylation of APC and GSTP1, although the association with gene hypermethylation was, if anything, inverse (Table 2). Results stratified by Gleason score showed that an inverse association between LINE-1 hypomethylation and gene-specific hypermethylation was evident only among patients with a Gleason score of at least 8 [adjusted coefficient = −2.06, 95% confidence interval (CI): −5.17–1.06] (data not shown).
Table 2.
Selected characteristics in association with LINE-1 methylation in the tumor tissue and in the non-neoplastic tissue adjacent to tumor.
| Tumor tissue patients (n=152) |
NTAT patients (n=105) |
||||||
|---|---|---|---|---|---|---|---|
| Characteristic | Mean LINE-1 (sd) | Coefficient1 | 95% CI | Mean LINE-1 (sd) | Coefficient1 | 95% CI | |
| 78.6 (4.29) | — | — | 79.3 (3.87) | — | — | ||
| Source of tumor tissue | |||||||
| Biopsy | 77.5 (4.32) | Ref | 77.4 (3.32) | Ref | |||
| TURP | 80.7 (3.42) | 2.91 | 1.23, 4.59 | 81.1 (3.48) | 3.71 | 1.73-5.69 | |
| Radical prostatectomy | 77.3 (4.26) | −0.64 | −2.38-1.11 | ||||
| Gleason score | |||||||
| <7 | 77.9 (3.63) | Ref | 78.0 (3.58) | Ref | |||
| 7 | 78.3 (4.89) | 0.37 | −1.34-2.08 | 79.7 (3.98) | 1.19 | -1.27-3.65 | |
| ≥8 | 79.4 (4.29) | 0.41 | −1.21-2.04 | 80.0 (3.89) | 0.34 | -1.75-2.43 | |
| Tumor tissue methylation in APC and GSTP1 | |||||||
| Continuous: 0, 1, 2 methylated genes | 78.6 (4.29) | −0.37 | −1.54-0.80 | 79.2 (4.13) | 0.33 | -0.89-1.56 | |
sd, standard deviation; CI, confidence interval; TURP, transurethral resection of the prostate; Ref, reference; APC, adenomatous polyposis coli; GSTP1, glutathione S-transferase 1; NTAT, non-neoplastic tissue adjacent to tumor.
Coefficient adjusted for age at diagnosis, year of diagnosis, source of tumor tissue, Gleason score, and tumor tissue methylation in APC and GSTP1.
There was some evidence of an association between LINE-1 hypomethylation in tumor tissue and mortality from prostate cancer, which remained after adjustment for Gleason score and gene-specific hypermethylation [hazard ratio (HR) per 5% decrease in LINE-1 methylation levels: 1.40, 95% CI: 0.95–2.01] and was evident also when LINE-1 methylation was treated as a categorical variable (Table 3). Analyses on mortality stratified for Gleason score revealed that the association between LINE-1 hypomethylation and mortality from prostate cancer was present only in patients with a Gleason score of at least 8 (adjusted HR per 5% decrease in LINE-1 methylation levels: 1.76, 95% CI: 1.00–2.93) (Table 3). In these patients, a low LINE-1 methylation level (< 75%) compared with a high methylation level (at least 80%) was associated with a HR of dying from prostate cancer of 4.68 (95% CI: 1.03–21.34). We also performed analyses by source of tissue, finding homogeneous results among TURPs, radical prostatectomies, and biopsies (Supplementary Table 1).
Table 3.
LINE-1 methylation in the tumor tissue and in the non-neoplastic tissue adjacent to tumor in association with mortality from prostate cancer.
| Tumor tissue patients (n=152) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All patients |
Patients with Gleason score<8 (n=96) |
Patients with Gleason score ≥8 (n=56) |
NTAT patients (N=105) |
|||||||||
| LINE-1 methylation | n | HR11 (95% CI) | HR22 (95% CI) | n | HR11 (95% CI) | HR22 (95% CI) | n | HR11 (95% CI) | HR22 (95% CI) | n | HR1 (95% CI) | HR22 (95% CI) |
| Decrease in 5% methylation | 41 | 1.28 (0.86-1.92) | 1.40 (0.95-2.01) | 13 | 0.90 (0.44-1.84) | 0.95 (0.47-1.84) | 28 | 1.68 (1.00-2.81) | 1.76 (1.00-2.93) | 37 | 0.81 (0.39-1.76) | 0.86 (0.42-1.76) |
| Categorical | N3 | |||||||||||
| <75 | 6 | 2.07 (0.71-6.02) | 2.22 (0.75-6.58) | 2 | 0.91 (0.15-5.38) | 0.83 (0.14-4.96) | 4 | 4.33 (1.02-8.39) | 4.68 (1.03-21.34) | 3.45 | 0.57 (0.05-6.60) | 0.71 (0.06-7.89) |
| 75-79,9 | 18 | 1.47 (0.73-2.95) | 1.60 (0.81-3.18) | 5 | 0.74 (0.20-1.72) | 0.67 (0.17-2.59) | 13 | 1.98 (0.85-4.64) | 2.03 (0.85-4.86) | 17.4 | 0.91 (0.33-2.47) | 0.79 (0.30-2.09) |
| ≥80 | 17 | Ref | Ref | 6 | Ref | Ref | 11 | Ref | Ref | 16.15 | Ref | Ref |
CI, confidence interval; n, number of prostate cancer deaths; Ref, reference; HR, hazard ratio; NTAT, non-neoplastic tissue adjacent to tumor.
HR1 adjusted for age and year of diagnosis and source of tumor tissue.
HR2 adjusted as HR1, Gleason score and APC and GSTP1 methylation in the tumor tissue.
Failure average among imputed datasets.
LINE-1 methylation in non-neoplastic tissue adjacent to the tumor
As described in the Methods, analyses of LINE-1 methylation in NTAT were restricted to the 105 patients who underwent a biopsy or a TURP. Consistently with the findings in the tumor tissue, LINE-1 methylation in NTAT was higher in TURPs than in biopsies (adjusted coefficient = 3.71, 95% CI: 1.73- 5.69), while it was not associated with Gleason score or hypermethylation of APC and GSTP1 (Table 2).
As reported in Table 3, LINE-1 methylation in NTAT was not associated with prostate cancer mortality (HR per 5% decrease in LINE-1 methylation levels: 0.86, 95% CI: 0.42–1.76). When analyses were stratified by source of tumor tissue or Gleason score, all HRs remained close to 1.0 (Supplementary Table 2).
Results from a sensitivity analysis restricted to patients with molecular data from newly bisulfite modified DNA from NTAT samples were consistent with those obtained from the full sample after multiple imputation (Supplementary Tables 3 and 4).
For 100 patients, we had information on LINE-1 methylation both in the tumor tissue and in the NTAT. Although there was no correlation between LINE-1 methylation values of the tumor and the NTAT, most of the lowest values were observed in the tumor tissue (Fig. 1). Average methylation levels were similar in the two tissues (difference = 0.1%, P = 0.29) but there was no evidence of correlation (r = 0.01). No correlation (data not shown) was found either when analyses were stratified by source of tumor tissue (TURPs or biopsies).
Figure 1.
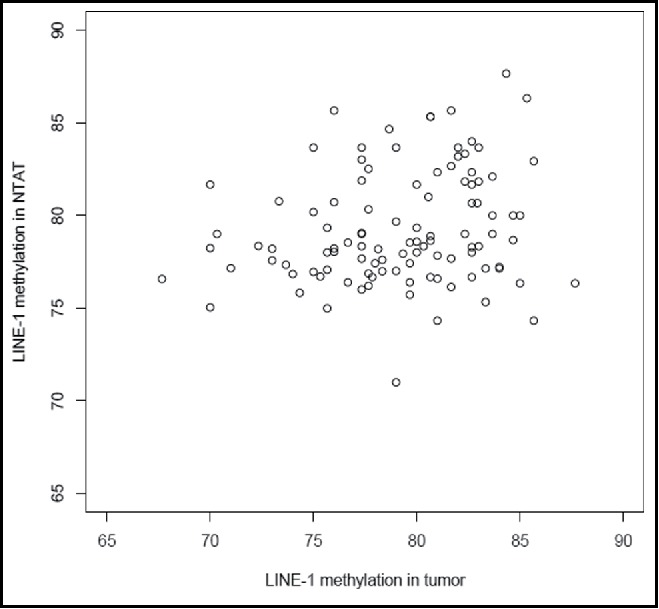
Correlation between LINE-1 methylation values of the tumor and the NTAT.
Discussion
We studied LINE-1 methylation status as a marker of global methylation and found that LINE-1 hypomethylation in the tumor tissue could be associated with mortality from prostate cancer, especially in patients with an aggressive disease (i.e., Gleason score of at least 8). These associations were not replicated when LINE-1 methylation was analyzed in NTAT.
In addition, our findings of higher LINE-1 methylation levels in TURPs than in biopsies or radical prostatectomies, both in the tumor tissue and in the NTAT, suggest that prostate tumors arising from the transition zone could have a different methylation pattern than tumors arising from the peripheral zone. This is consistent with data suggesting that tumors of the transition zone, which are 20–25% of all prostate tumors, show a specific morphological,33 clinical,34,35 biological,36 and molecular37-39 behavior. Furthermore, findings of the current study are similar to those of a recent study on tumor-negative prostate samplings, in which we found higher LINE-1 methylation levels in TURPs than in biopsies.40
Although our study does not have a large sample size, to our knowledge it is the first to explore the relationship between LINE-1 methylation status and mortality from prostate cancer. Furthermore, it presented a broad approach, as we analyzed both the tumor tissue and the NTAT and we used information on gene-specific hypermethylation. A potential limitation of our study is related to the fact that we included heterogeneous sources of tissue (biopsies, TURPs, and radical prostatectomies). We have, however, always adjusted for source of tumor tissue in the analyses, as well as performed stratified analyses in which we did not find evidence of effect modification of the association between LINE-1 methylation and mortality from prostate cancer. Interestingly, lack of heterogeneity by source of tissue implies also internal consistency among heterogeneous groups of patients, e.g., in regards to the treatment approach, which provides support to the robustness of our results. In our study, we did not have information on treatments after the tissue sampling, but it should be considered that treatment decisions after the sampling were taken also on the basis of the morphological/molecular characteristics of the tumor tissue. Thus, adjustment for post-sampling treatments could induce bias rather than control for it. Multifocality of the prostate tumor could represent a further critical point, as we did not always had the whole prostate tissue available (e.g., in biopsies and TURPs) and, in any case, we analyzed only one randomly chosen block for each patient. However, multifocal differences in LINE-1 methylation would likely introduce non-differential misclassification that is expected, if anything, to underestimate the association between LINE-1 methylation and prostate cancer mortality. It is thus reassuring that we found a stronger association for the tumor tissue, in which multifocality is an issue, than for the NTAT. Finally, we had to use multiple imputation to include cases whose DNA could not be newly modified. This method imputes missing data while acknowledging the uncertainty associated with the imputed values and provides valid statistical inferences under the missing at random (MAR) assumption.41,42,43 Indeed, when data are MAR, analyses based on complete cases may be biased because cases with missing values may differ systematically from the completely observed cases. In our study, results obtained using multiple imputations or a complete case approach were similar.
Our data suggest increased mortality from prostate cancer associated with lower levels of LINE-1 methylation in the tumor tissue. It has been suggested that global hypomethylation is a late epigenetic mechanism that could play a role in progression of advanced prostate cancer.24 Moreover, it has been suggested that a high de-repression, via hypomethylation, of LINE-1 promoter could induce a general dysregulation that involves chromosomal instability, reactivation of repeated sequences, and oncogenes.24,22 Our results are consistent with these suggestions, as the association between LINE-1 hypomethylation and mortality from prostate cancer was limited to men with an aggressive tumor (Gleason score 8+). On the other hand, there was no association between LINE-1 methylation and Gleason score. Although this latter finding is not completely inconsistent with previous studies (a clear association between LINE-1 hypomethylation and Gleason score has been found only in one out of 3 previous studies22,23,26), if global methylation was a late event in prostate cancerogenesis, we would have expected higher hypomethylation among patients with a Gleason score of at least 8. It could be speculated that global methylation is more related to the metastatic potential of aggressive prostate cancers than to the Gleason score, and it is of interest that previous studies strongly indicate a decreased global methylation in the prostate cancer tissue among prostate cancer patients with systemic metastases.21,24 In addition, some studies have found widespread genome-wide hypomethylation as well as focal hypermethylation in prostate cancer metastases from different sites (e.g., bone, liver). These studies confirm that the hypomethylation in metastasis showed a pattern that induces genetic instability rather than the regulation of specific genes. Hypermethylation at specific genes is less common but strongly maintained among different metastasis from a patient.44,45
Gene-specific hypermethylation and global hypomethylation are often considered to coexist in the process of carcinogenesis.7,9,17 We did not find clear evidence of an association of LINE-1 hypomethylation with GSTP1 and APC hypermethylation, although the inverse association became stronger in more advanced tumors. These data give some support to (or at least do not contradict) the concept of an “epigenetic catastrophe”46,9 occurring with prostate tumor development and progression.
In a previous study, based on the same patients included in the current study, we found that hypermethylation in APC and GSTP1 in the NTAT was associated with a strongly increased risk of dying from prostate cancer.32 Those results supported the concept of field cancerization, i.e., that molecular dysregulations may occur in the NTAT before the tumor becomes morphologically evident. Our current null results on LINE-1 methylation in NTAT do not provide further support to the concept of field cancerization; however, they are consistent with the hypothesis that global hypomethylation is a late event in cancerogenesis that, accordingly, would not be detectable in NTAT.
In conclusion, our data suggest that global hypomethylation may be a late event in cancerogenesis that is associated with tumor progression and mortality for tumors with high Gleason score.
Materials and methods
Patients and samples
The study is nested in 2 cohorts of, in total, 459 patients with prostate cancer diagnosed between 1982–1988 (1980s cohort) and 1993–1996 (1990s cohort) at the San Giovanni Battista Hospital of the city of Turin, Italy. These patients were followed up for cause-specific mortality until August 2010. A detailed description of the 2 cohorts has been published previously.13 Briefly, for all 459 patients we collected demographic and clinical information (age, residence, source of prostatic tumor tissue-biopsy, radical prostatectomy or transurethral resection of the prostate, and tumor grade) from the pathology reports and traced the corresponding blocks of formalin fixed tumor tissue. All diagnostic slides were traced and reviewed by an uropathologist to assign a uniform Gleason score. In addition, as previously described,32 diagnostic slides were analyzed to identify areas of (NTAT). We identified a subcohort of 157 patients with NTAT relying on restrictive criteria, including absence of areas of prostatic intraepithelial neoplasia (PIN), availability of tissue (all biopsies from the 1980s cohort were a priori excluded as areas of NTAT that were too limited) and possibility to identify an area of NTAT. If more than one area of NTAT was present, the farthest area from the tumor was chosen. The portions of NTAT collected had at least 1.5 mm of distance from tumor cells. The selected NTAT were manually dissected from slices overlapping the area highlighted from the uropathologist.
The current study is conducted in this cohort of 157 patients, for whom we potentially had access on both the tumor tissue and the NTAT. From previous studies we had qualitative information on promoter hypermethylation of adenomatous polyposis coli (APC) and glutathione S-transferase 1 (GSTP1) in the tumor tissue and in the NTAT.13,32 We also had stored (at −20°C) DNA extracted from the tumor tissue (for all patients) and from NTAT (for 91 patients) as well as stored (at -80°C) bisulfite-modified DNA from NTAT (for 49 patients). For 17 patients, stored DNA (modified or not) from NTAT was not available.
The study was approved by the Ethical Committee of the San Giovanni Battista Hospital - CTO/CRF/Maria Adelaide Hospital of Turin.
Molecular analysis
Available stored genomic DNA from the 157 tumor tissue samples and from the 91 NTAT samples (248 samples in total for 157 patients) underwent bisulfite modification using the Epitect Bisulfite Kit (Qiagen, Hilden, Germany). Controls fully methylated and unmethylated (CpGenomeTM universal methylated DNA and CpGenomeTM universal unmethylated DNA, Chemicon Co. Billerica, MA) were included in the modification set. We conducted molecular analyses on LINE- 1 methylation both on these 248 newly modified samples and on the 49 stored bisulfite-modified DNA samples.
LINE-1 (GenBank accession number X58075) promoter methylation was analyzed using PyroMark Q24 MDx (Qiagen). Primers (forward 5′-TTTGAGTTAGGTGTGGGATATAGTT-3′, reverse 5′-Biot-CACCTAAAAAATCCAATCACTCC-3′ and sequencing 5′-TTAGGTGTGGGATATAGTTT-3′), not covering CpG sites, produce a 98 bp amplicon containing 3 CpG sites (positions 819, 826, and 829) and were designed according to PyroMark Assay Design software version 2.0 (Qiagen). We performed PCR reaction in a total volume of 35 µl containing 1X buffer (KCl), 2 mM MgCl2, 0.8 mM dNTPs, 0.5 µM of each primer, 0.05 U Taq polymerase and 6 µl of bisulfite-modified DNA with the following cycling profile: 95°C for 10 min followed by 45 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 1 min, extension at 72°C for 1 min and final extension at 72°C for 10 min. The sequence to analyze was C/TGTGGTGC/TGTC/TG and the dispensation order was GTCGTAGATAGTCAGATC. Positive controls for methylated and unmethylated status were included in each pyrosequencing run. Methylation quantification was expressed for each CpG site selected as percentage of methylated cytosine divided by the sum of methylated and unmethylated cytosine.
Statistical analysis
As measure of LINE-1 methylation status, we calculated the mean methylation levels of the 3 CpG sites analyzed in the LINE-1 promoter (all pairways correlation coefficients were >0.83). LINE-1 methylation was treated as a continuous variable, but we also created a categorical variable using 3 levels of methylation: <75%, 75–79.9%, and ≥80%.
LINE-1 methylation in the tumor tissue
We used multivariable linear regression to estimate the association between selected characteristics (age, year of diagnosis, source of tumor tissue, Gleason score, and tumor tissue methylation in APC and GSTP1) and level of LINE-1 methylation in the tumor tissue. Normality was satisfied as indicated by tests based on skewness and kurtosis (P>0.15).
We used a Cox proportional hazard regression model to estimate the hazard ratio (HR) of mortality from prostate cancer in association with LINE-1 methylation in the tumor tissue. Time from diagnosis was used as the time axis, while age and year of diagnosis were introduced as continuous variables. The proportional hazard assumption was met as indicated by a test based on Schoenfeld residuals (P = 0.34). Models were always adjusted for the source of tumor tissue and progressively adjusted for tumor tissue methylation in APC and GSTP1 and Gleason score (variables categorized as shown in Table 1). We adjusted for gene-specific methylation as it has been suggested that global hypomethylation may coexist with gene-specific hypermethylation, and the latter has been associated with mortality from prostate cancer.13 Additional analyses were performed stratifying by source of tumor tissue and Gleason score (<8, ≥8).
LINE-1 methylation in the NTAT
Analyses of LINE-1 methylation in NTAT were restricted to biopsies and transurethral resections of the prostate (TURPs) (105 patients out of 157). Through the preliminary results of this study and the results of a parallel study of our group,40 we found that the distinction between transition and peripheral zone of prostate was important as these two tissues have strong differences in LINE-1 methylation levels. In our study, the NTAT was sampled without considering whether it came from the peripheral or the transition zone. To identify a posteriori the tissue of origin (i.e., peripheral or transition) of the NTAT samples obtained from the radical prostatectomies, we reviewed the slides selected for the DNA extraction: in most of the cases (30) the tissue was of a mixed nature, while we could identify a clear peripheral origin for 9 cases and a clear transition origin for 10 cases. When we analyzed LINE-1 methylation in these 3 groups, we found a mean LINE-1 methylation of 77.8% in the peripheral tissues, 78.8% in the mixed tissues, and 79.5% in the transition tissues, thus further supporting the notion that the transition tissue has a higher methylation compared with the peripheral tissue. These patients were however excluded from the mortality analyses as NTAT of mixed origin could bias the estimates, while there were too few events among the other 2 groups (2 events among patients with NTAT of peripheral origin and 1 event among patients with NTAT of transition origin) to be added to the analyses.
Since the long-time storage of bisulfite-modified DNA could alter the molecular results and data on LINE-1 methylation level in NTAT from newly modified DNA were available only for 52 patients (48% of the 105 patients with a TURP or a biopsy), we performed multiple imputation to obtain complete and more valid information on LINE-1 methylation for all 105 patients. Specifically, a multivariate imputation by chained equations (MICE) approach was adopted assuming the data were missing at random (MAR).47,48 The imputation model involved regression of the LINE-1 methylation level in the NTAT measured on the newly modified DNA on: year of diagnosis, age at diagnosis, source of tissue, Gleason score, number of methylated genes out of APC and GSTP1 in the tumor tissue and in NTAT, LINE-1 tumor methylation level, LINE-1 NTAT methylation level measured on the stored modified DNA, and survival data, including the event indicator, the cumulative hazard at the time of entry for the event actually experienced, and the difference between the estimated cumulative hazard at the exit time and that at entry.49 After fitting the imputation model, imputed values for the missing data were created using predictive mean matching.
For each imputed data set (n = 20) we performed a multivariable linear regression to estimate the association between selected characteristics and level of LINE-1 methylation in NTAT and a Cox regression to estimate the HR of mortality from prostate cancer for NTAT LINE-1 methylation. Analyses stratified by Gleason score (<8, ≥8) were also conducted. Estimates obtained in each data set were then combined into overall estimates, with standard errors, confidence intervals and P-values calculated using Rubin's rule.50
We also compared the mean LINE-1 global methylation levels between the NTAT and the tumor tissue, using the Wilcoxon test for paired data and estimating the Spearman correlation coefficient. This analysis was further stratified by source of tumor tissue (TURP or biopsy).
As a sensitivity analysis, we repeated the analyses on LINE-1 methylation status in the NTAT restricting to patients for whom newly modified DNA was available (n = 52).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
The study was partially supported by the Italian Association for Cancer Research (AIRC) and the Ministry of University and Research (ex-60%). The work of Lorenzo Richiardi has been partially supported by a Fulbright Research Scholar grant.
Author Contributions
V Fiano developed protocols of molecular biology, performed and interpreted molecular analysis and wrote the manuscript. D Zugna performed and interpreted statistical analysis and contributed to the manuscript revision. C Grasso performed and interpreted molecular analysis and contributed to the manuscript revision. M Trevisan performed molecular analysis. L Delsedime and L Molinaro reviewed histo-pathological slices. A Gillio-Tos supervised molecular analyses and interpreted molecular results. F Merletti contributed to the conception of study design and to the revision of the manuscript. L Richiardi conceived study design, interpreted statistical analysis and reviewed the manuscript.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer 2013; PMID:25220842; http://dx.doi.org/ 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Mohammed AA. Biomarkers in prostate cancer: new era and prospective. Med Oncol 2014; 31:140; PMID:25048724; http://dx.doi.org/ 10.1007/s12032-014-0140-3 [DOI] [PubMed] [Google Scholar]
- 3.Davis JW. Novel commercially available genomic tests for prostate cancer: a roadmap to understanding their clinical impact. BJU Int 2014; 114:320-2; PMID:25146527; http://dx.doi.org/ 10.1111/bju.12695 [DOI] [PubMed] [Google Scholar]
- 4.Valdés-Mora F, Clark SJ. Prostate cancer epigenetic biomarkers: next-generation technologies. Oncogene 2015; 34:1609-18; PMID:24837368; http://dx.doi.org/ 10.1038/onc.2014.111 [DOI] [PubMed] [Google Scholar]
- 5.Blute ML Jr, Damaschke NA, Jarrard DF. The epigenetics of prostate cancer diagnosis and prognosis: update on clinical applications. Curr Opin Urol 2015; 25:83-8; PMID:25405932; http://dx.doi.org/ 10.1097/MOU.0000000000000132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strand SH, Orntoft TF, Sorensen KD. Prognostic DNA methylation markers for prostate cancer. Int J Mol Sci 2014; 15:16544-76; PMID:25238417; http://dx.doi.org/ 10.3390/ijms150916544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szyf M. DNA methylation and cancer therapy. Drug Resist Updat 2003; 6:341-53; PMID:14744498; http://dx.doi.org/ 10.1016/j.drup.2003.10.002 [DOI] [PubMed] [Google Scholar]
- 8.Szyf M, Pakneshan P, Rabbani SA. DNA demethylation and cancer: therapeutic implications. Cancer Lett 2004; 211:133-43; PMID:15219937; http://dx.doi.org/ 10.1016/j.canlet.2004.04.009 [DOI] [PubMed] [Google Scholar]
- 9.Perry AS, Watson RW, Lawler M, Hollywood D. The epigenome as a therapeutic target in prostate cancer. Nat Rev Urol 2010; 7:668-80; PMID:21060342; http://dx.doi.org/ 10.1038/nrurol.2010.185 [DOI] [PubMed] [Google Scholar]
- 10.Woodson K, O'Reilly KJ, Ward DE, Walter J, Hanson J, Walk EL, Tangrea JA. CD44 and PTGS2 methylation are independent prognostic markers for biochemical recurrence among prostate cancer patients with clinically localized disease. Epigenetics 2006; 1:183-6; PMID:17998819 [DOI] [PubMed] [Google Scholar]
- 11.Henrique R, Ribeiro FR, Fonseca D, Hoque MO, Carvalho AL, Costa VL, Pinto M, Oliveira J, Teixeira MR, Sidransky D, et al.. High promoter methylation levels of APC predict poor prognosis in sextant biopsies from prostate cancer patients. Clin Cancer Res 2007; 13:6122-9; PMID:17947477; http://dx.doi.org/ 10.1158/1078-0432.CCR-07-1042 [DOI] [PubMed] [Google Scholar]
- 12.Weiss G, Cottrell S, Distler J, Schatz P, Kristiansen G, Ittmann M, Haefliger C, Lesche R, Hartmann A, Corman J, et al.. DNA methylation of the PITX2 gene promoter region is a strong independent prognostic marker of biochemical recurrence in patients with prostate cancer after radical prostatectomy. J Urol 2009; 181:1678-85; PMID:19233404; http://dx.doi.org/ 10.1016/j.juro.2008.11.120 [DOI] [PubMed] [Google Scholar]
- 13.Richiardi L, Fiano V, Vizzini L, De Marco L, Delsedime L, Akre O, Tos AG, Merletti F. Promoter methylation in APC, RUNX3, and GSTP1 and mortality in prostate cancer patients. J Clin Oncol 2009; 27:3161-8; PMID:19470943; http://dx.doi.org/ 10.1200/JCO.2008.18.2485 [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Kron KJ, Pethe VV, Demetrashvili N, Nesbitt ME, Trachtenberg J, Ozcelik H, Fleshner NE, Briollais L, van der Kwast TH, et al.. Association of tissue promoter methylation levels of APC, TGFβ2, HOXD3 and RASSF1A with prostate cancer progression. Int J Cancer 2011; 129:2454-62; PMID:21207416; http://dx.doi.org/ 10.1002/ijc.25908 [DOI] [PubMed] [Google Scholar]
- 15.Haldrup C, Mundbjerg K, Vestergaard EM, Lamy P, Wild P, Schulz WA, Arsov C, Visakorpi T, Borre M, Høyer S, et al.. DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer. J Clin Oncol 2013; 31:3250-8; PMID:23918943; http://dx.doi.org/ 10.1200/JCO.2012.47.1847 [DOI] [PubMed] [Google Scholar]
- 16.Litovkin K, Joniau S, Lerut E, Laenen A, Gevaert O, Spahn M, Kneitz B, Isebaert S, Haustermans K, Beullens M, et al.. Methylation of PITX2, HOXD3, RASSF1 and TDRD1 predicts biochemical recurrence in high-risk prostate cancer. J Cancer Res Clin Oncol 2014; 140:1849-61; PMID:24938434; http://dx.doi.org/ 10.1007/s00432-014-1738-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics 2009; 1:239-59; PMID:20495664; http://dx.doi.org/ 10.2217/epi.09.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehrlich M, Lacey M. DNA hypomethylation and hemimethylation in cancer. Adv Exp Med Biol 2013; 754:31-56; PMID:22956495; http://dx.doi.org/ 10.1007/978-1-4419-9967-2_2 [DOI] [PubMed] [Google Scholar]
- 19.Zelic R, Fiano V, Grasso C, Zugna D, Pettersson A, Gillio-Tos A, Merletti F, Richiardi L. Global DNA hypomethylation in prostate cancer development and progression: a systematic review. Prostate Cancer Prostatic Dis 2015; 18:1-12; PMID:25384337; http://dx.doi.org/ 10.1038/pcan.2014.45 [DOI] [PubMed] [Google Scholar]
- 20.Santourlidis S, Florl A, Ackermann R, Wirtz HC, Schulz WA. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. Prostate 1999; 39:166-174; PMID:10334105; http://dx.doi.org/ 10.1002/(SICI)1097-0045(19990515)39:3%3c166::AID-PROS4%3e3.0.CO;2-J [DOI] [PubMed] [Google Scholar]
- 21.Schulz WA, Elo JP, Florl AR, Pennanen S, Santourlidis S, Engers R, Buchardt M, Seifert HH, Visakorpi T. Genomewide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes Chromosomes Cancer 2002; 35:58-65; PMID:12203790; http://dx.doi.org/ 10.1002/gcc.10092 [DOI] [PubMed] [Google Scholar]
- 22.Florl AR, Steinhoff C, Muller M, Seifert HH, Hader C, Engers R, Ackermann R, Schulz WA. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br J Cancer 2004; 91:985-994; PMID:15292941; http://dx.doi.org/ 10.1038/sj.bjc.6602030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, Kim D, Kang GH. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol 2007; 211:269-277; PMID:17139617; http://dx.doi.org/ 10.1002/path.2106 [DOI] [PubMed] [Google Scholar]
- 24.Yegnasubramanian S, Haffner MC, Zhang Y, Gurel B, Cornish TC, Wu Z, Irizarry RA, Morgan J, Hicks J, DeWeese TL, et al.. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res 2008; 68:8954-8967; PMID:18974140; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-6088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanzer S, Balic M, Strutz J, Heitzer E, Obermair F, Hauser-Kronberger C, Samonigg H, Dandachi N. Rapid and reliable detection of LINE-1 hypomethylation using high-resolution melting analysis. Clin Biochem 2010; 13:1443-1448; PMID:20883681; http://dx.doi.org/ 10.1016/j.clinbiochem.2010.09.013 [DOI] [PubMed] [Google Scholar]
- 26.Delgado-Cruzata L, Hruby GW, Gonzalez K, McKiernan J, Benson MC, Santella RM, Shen J. DNA methylation changes correlate with Gleason score and tumor stage in prostate cancer. DNA Cell Biol 2012; 31:187-192; PMID:21830905; http://dx.doi.org/ 10.1089/dna.2011.1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 2004; 32:e38; PMID:14973332; http://dx.doi.org/ 10.1093/nar/gnh032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 2005; 33:6823-36; PMID:16326863; http://dx.doi.org/ 10.1093/nar/gki987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Estécio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, et al.. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One 2007; 2:e399; PMID:17476321; http://dx.doi.org/ 10.1371/journal.pone.0000399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phokaew C, Kowudtitham S, Subbalekha K, Shuangshoti S, Mutirangura A. LINE-1 methylation patterns of different loci in normal and cancerous cells. Nucleic Acids Res 2008; 36:5704-12; PMID:18776216; http://dx.doi.org/ 10.1093/nar/gkn571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wild L, Flanagan JM. Genome-wide hypomethylation in cancer may be a passive consequence of transformation. Biochim Biophys Acta 2010; 1806:50-7; PMID:20398739; http://dx.doi.org/ 10.1016/j.bbcan.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 32.Richiardi L, Fiano V, Grasso C, Zugna D, Delsedime L, Gillio-Tos A, Merletti F. Methylation of APC and GSTP1 in non-neoplastic tissue adjacent to prostate tumour and mortality from prostate cancer. PLoS One 2013; 8:e68162; PMID:23874531; http://dx.doi.org/ 10.1371/journal.pone.0068162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greene DR, Wheeler TM, Egawa S, Dunn JK, Scardino PT. A Comparison of the morphological features of cancer arising in the transition zone and in the peripheral zone of the prostate. J Urol 1991; 146:1069-1076; PMID:1895423 [DOI] [PubMed] [Google Scholar]
- 34.Augustin H, Erbersdobler A, Hammerer PG, Graefen M, Huland H. Prostate cancers in the transition zone: Part 2; clinical aspects. BJU Int 2004; 94:1226-9; PMID:15610094; http://dx.doi.org/ 10.1111/j.1464-410X.2004.05147.x [DOI] [PubMed] [Google Scholar]
- 35.Erbersdobler A, Augustin H, Schlomm T, Henke RP. Prostate cancers in the transition zone: Part 1; pathological aspects. BJU Int 2004; 94:1221-5; PMID:15610093; http://dx.doi.org/ 10.1111/j.1464-410X.2004.05146.x [DOI] [PubMed] [Google Scholar]
- 36.Lee JJ, Thomas IC, Nolley R, Ferrari M, Brooks JD, Leppert JT. Biologic differences between peripheral and transition zone prostate cancer. Prostate 2015; 75:183-90; PMID:25327466; http://dx.doi.org/ 10.1002/pros.22903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noel EE, Ragavan N, Walsh MJ, James SY, Matanhelia SS, Nicholson CM, Lu YJ, Martin FL. Differential gene expression in the peripheral zone compared to the transition zone of the human prostate gland. Prostate Cancer Prostatic Dis 2008; 11:173-80; PMID:17646851; http://dx.doi.org/ 10.1038/sj.pcan.4500997 [DOI] [PubMed] [Google Scholar]
- 38.Carlsson J, Helenius G, Karlsson MG, Andrén O, Klinga-Levan K, Olsson B. Differences in microRNA expression during tumor development in the transition and peripheral zones of the prostate. BMC Cancer 2013; 13:362; PMID:23890084; http://dx.doi.org/ 10.1186/1471-2407-13-362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinnott JA, Rider JR, Carlsson J, Gerke T, Tyekucheva S, Penney KL, Sesso HD, Loda M, Fall K, Stampfer MJ, et al.. Molecular differences in transition zone and peripheral zone prostate tumors. Carcinogenesis 2015; 36:632-8; PMID:25870172; http://dx.doi.org/ 10.1093/carcin/bgv051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zelic R, Fiano V, Zugna D, Grasso C, Delsedime L, Daniele L, Galliano D, Pettersson A, Gillio-Tos A, Merletti F, et al.. Gene-specific hypermethylation (GSTP1) and global hypomethylation (LINE-1) on initial negative prostate biopsy as markers of prostate cancer on a rebiopsy. Clin Cancer Res 2016; 22:984-92; PMID:26475336; http://dx.doi.org/ 10.1158/1078-0432.CCR-15-0606 [DOI] [PubMed] [Google Scholar]
- 41.Ali AM, Dawson SJ, Blows FM, Provenzano E, Ellis IO, Baglietto L, Huntsman D, Caldas C, Pharoah PD. Comparison of methods for handling missing data on immunohistochemical markers in survival analysis of breast cancer. Br J Cancer 2011; 104:693-9; PMID:21266980; http://dx.doi.org/ 10.1038/sj.bjc.6606078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phipps AI, Limburg PJ, Baron JA, Burnett-Hartman AN, Weisenberger DJ, Laird PW, Sinicrope FA, Rosty C, Buchanan DD, Potter JD, et al.. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 2015; 148:77-87; PMID:25280443; http://dx.doi.org/ 10.1053/j.gastro.2014.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Heijden GJ, Donders AR, Stijnen T, Moons KG. Imputation of missing values is superior to complete case analysis and the missing-indicator method in multivariable diagnostic research: a clinical example. J Clin Epidemiol 2006; 59:1102-9; PMID:16980151; http://dx.doi.org/ 10.1016/j.jclinepi.2006.01.015 [DOI] [PubMed] [Google Scholar]
- 44.Aryee MJ, Liu W, Engelmann JC, Nuhn P, Gurel M, Haffner MC, Esopi D, Irizarry RA, Getzenberg RH, Nelson WG, et al.. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med 2013; 5:169ra10; PMID:23345608; http://dx.doi.org/ 10.1126/scitranslmed.3005211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, et al.. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016; 22:298-305; PMID:26855148; http://dx.doi.org/ 10.1038/nm.4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, Bova GS, De Marzo AM, Isaacs WB, Nelson WG. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res 2004; 64:1975-86; PMID:15026333; http://dx.doi.org/ 10.1158/0008-5472.CAN-03-3972 [DOI] [PubMed] [Google Scholar]
- 47.Rubin DB. Inference and missing data. Biometrika 1976; 63:581-592; http://dx.doi.org/ 10.1093/biomet/63.3.581 [DOI] [Google Scholar]
- 48.Raghunatan T, Bondarenko I. Diagnostics for Multiple Imputations. SSRN 2007URLhttp://ssrn.com; http://dx.doi.org/ 10.2139/ssrn.1031750 [DOI] [Google Scholar]
- 49.White IR. Imputing missing covariate values for the Cox model. Statistics in medicine 2009; 28:1982-1998; PMID:19452569; http://dx.doi.org/ 10.1002/sim.3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rubin DB. Multiple imputation for non-response in surveys. New Jersey: J Wiley & Sons; 1987; http://dx.doi.org/ 10.1002/9780470316696 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


