Abstract Abstract
The banded basslet, Lipogramma evides Robins & Colin, 1979, is shown to comprise two species: Lipogramma evides, which inhabits depths of 133–302 m, and a new species described here as Lipogramma levinsoni, which inhabits depths of 108–154 m and previously was considered to represent the juvenile of Lipogramma evides. A second new species of banded basslet, described here as Lipogramma haberi, inhabits depths of 152–233 m and was previously not reported in the literature. Morphologically, the three species differ in color patterns and modal numbers of gill rakers, whereas various other morphological features distinguish Lipogramma levinsoni from Lipogramma evides and Lipogramma haberi. DNA barcode data and multilocus, coalescent-based, species-delimitation analysis support the recognition of the three species. Phylogenetic analysis of mitochondrial and nuclear genetic data supports a sister-group relationship between the two deepest-living of the three species, Lipogramma evides and Lipogramma haberi, and suggests that the shallower Lipogramma levinsoni is more closely related to Lipogramma anabantoides Böhlke, 1960, which inhabits depths < 120 m. Evolutionary relationships within Lipogramma thus appear to be correlated with species depth ranges, an eco-evolutionary pattern that has been observed in other Caribbean marine teleosts and that warrants further investigation. The new species represent the eleventh and twelfth new fish species described in recent years from exploratory submersible diving in the Caribbean in the globally poorly studied depth zone of 50–300 m. This study suggests that there are at least two additional cryptic species of Lipogramma, which are being analyzed in ongoing investigations of Caribbean deep-reef ecosystems.
Keywords: Manned submersible, cryptic species, integrative taxonomy, phylogeny, ocean exploration, Smithsonian Deep Reef Observation Project (DROP)
Introduction
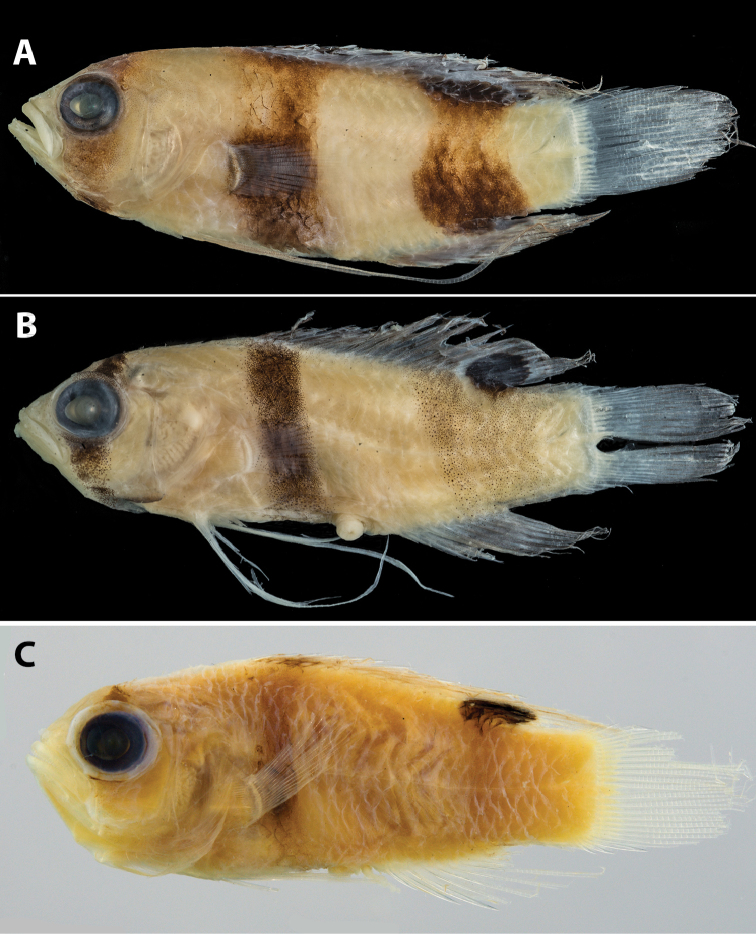
The western Atlantic family Grammatidae comprises small, usually brightly colored fishes in two genera, Gramma with four species and Lipogramma with eight (Robertson and Van Tassell 2015). Among other characters, the two genera are distinguished by the absence of a lateral line and presence of thickened, spinous, outer procurrent rays in Lipogramma (Mooi & Gill, 2002). The Banded Basslet, Lipogramma evides Robins & Colin, 1979, was described based on six specimens collected from Barbuda, Jamaica, Mexico, and Nicaragua. The original description also included observations of the species from Belize by Colin (1974). Subsequently, six additional specimens from the Bahamas were reported by Gilmore and Jones (1988). Robins and Colin (1979) noted differences in pigment pattern between adults and what they thought was a juvenile Lipogramma evides (Fig. 1A, B), in particular the presence of broader and more intense dark bands on the juvenile that completely encircle the body. Gilmore and Jones (1988) further commented on the presumed color differentiation between ontogenetic stages and noted that heavily banded “juveniles” (Fig. 1C) inhabit shallower waters (< 200 m) than adults (as deep as 250 m).
Figure 1.
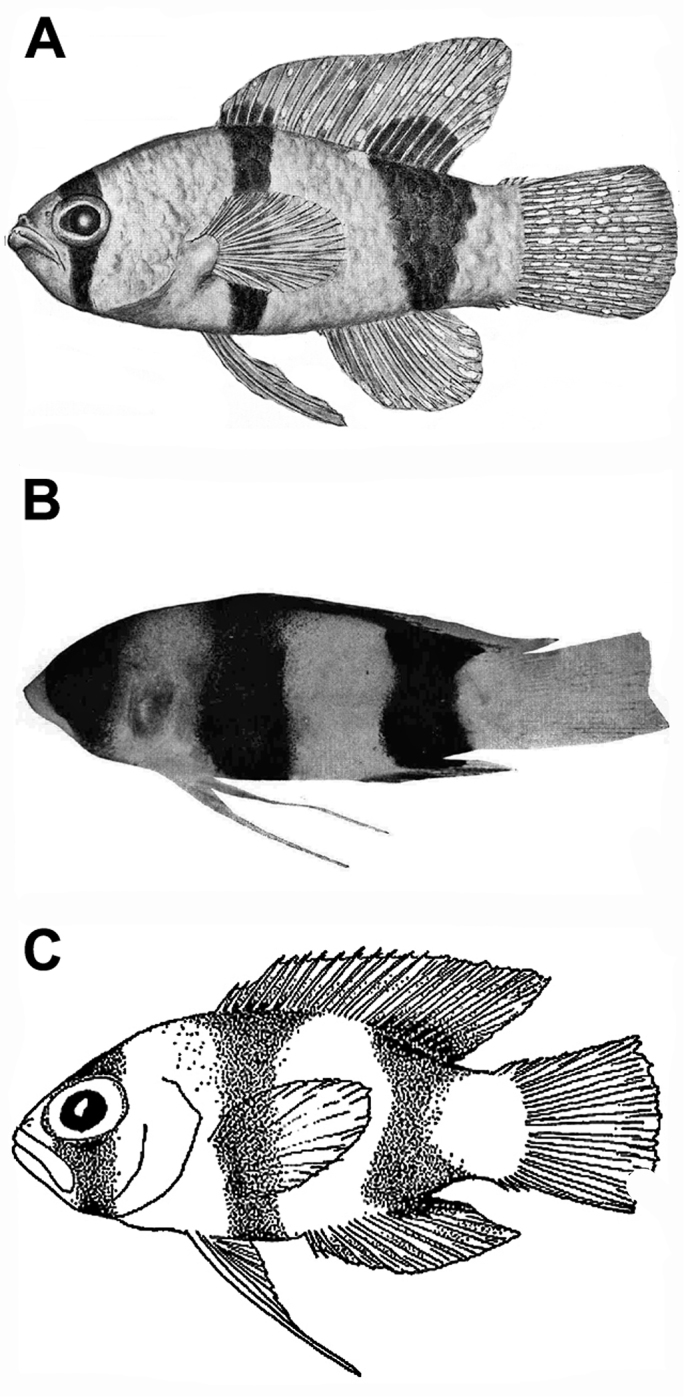
Previously published images of A Lipogramma evides, 34.4 mm SL, ANSP 134329, holotype, from Robins and Colin (1979: fig. 1) B Lipogramma levinsoni sp. n., 12.6 mm SL, ANSP 134332, as juvenile paratype of Lipogramma evides in Robins and Colin (1979: fig. 2) C Lipogramma levinsoni sp. n., 14.1 mm SL, IRCZM 107: 07660, as juvenile of Lipogramma evides in Gilmore and Jones (1988: fig. 3). Images reproduced with permission from Bulletin of Marine Science.
Exploratory submersible diving to 300 m in the southern and eastern Caribbean over the past several years by the (DROP) has resulted in the collection of over 50 specimens of “banded basslets” assignable to Lipogramma based on the absence of a lateral line and presence of spinous procurrent caudal-fin rays. That material includes individuals with both pigment patterns observed by previous authors and a third pigment pattern not previously described. Genetic and morphological analyses of individuals with those three pigment patterns suggested three distinct species and show that the heavily banded pattern is not an ontogenetic feature but diagnostic of a separate species. That species reaches a smaller maximum size than Lipogramma evides and has a shallower depth range. The other new species is similar in size and depth of occurrence to Lipogramma evides. Here we describe those two new species of Lipogramma, morphologically and genetically compare them with Lipogramma evides, and discuss depth distributions and evolutionary relationships of species of the genus.
Methods and materials
Collecting and morphology. Basslets were collected using Substation Curaçao’s manned submersible Curasub (http://www.substation-curacao.com). The sub has two flexible, hydraulic arms, one of which is equipped with a quinaldine/ethanol-ejection system and the other with a suction hose. Anesthetized fish specimens were captured with the suction hose, which empties into a vented plexiglass cylinder attached to the outside of the sub. At the surface, the specimens were photographed, tissue sampled, and fixed in 10% formalin. Measurements were made weeks to months after fixation and subsequent preservation in 75% ethanol and were taken to the nearest 0.1 mm with dial calipers or an ocular micrometer fitted into a Wild stereomicroscope. Selected preserved specimens were later photographed to document preserved pigment pattern and X-rayed with a digital radiography system. Images of supraorbital pores and tooth-like structures on gill rakers were made using a Zeiss Axiocam on a Zeiss Discovery V12 SteREO microscope. Counts and measurements follow Hubbs and Lagler (1947). Specimens were cleared and stained following the protocol of Dingerkus and Uhler (1977). Symbolism for configuration of supraneural bones, anterior neural spines, and anterior dorsal pterygiophores follows Ahlstrom et al. (1976). USNM; ANSP; IRCZM; UF.
Molecular analyses. Tissue samples for 97 specimens assignable to eight species of Lipogramma were used for molecular analyses (Appendix 1). Tissues of Lipogramma rosea Gilbert, 1979 (in Robins and Colin 1979), Lipogramma regia Robins & Colin, 1979, and Lipogramma flavescens Gilmore & Jones, 1988 were not available. Tissues were stored in saturated salt-DMSO (dimethyl sulfoxide) buffer (Seutin et al. 1991). DNA extraction and cytochrome c oxidase subunit I (COI) DNA barcoding were performed for 96 specimens (i.e., for all available specimens except one Lipogramma anabantoides – Appendix 1) as outlined by Weigt et al. (2012). Four nuclear markers were amplified and sequenced—TMO-4C4, Rag1, Rhodopsin, and Histone H3—for 18 specimens of Lipogramma, and one or more of those genes was sequenced for an additional three specimens (Appendix 1). Primers and PCR conditions for the nuclear markers followed Lin and Hastings (2011, 2013). Sequences were assembled and aligned using Geneious v. 9 (Biomatters, Ltd., Aukland). A neighbor-joining (NJ) network was generated for the COI data using the K2P substitution model (Kimura 1980) in the tree-builder application in Geneious. Mean within- and between-species K2P genetic distances were calculated from the COI data in MEGA v. 7 (Kumar et al. 2015). Genetic distances were considered as corroborating morphology-based species delineation if the distances between species were ten or more times the intraspecific differences (Hebert et al. 2004). The alignments of COI and nuclear genes were concatenated and phylogeny was inferred using Bayesian Inference (BI) and Maximum Likelihood (ML), partitioning by gene. For the Bayesian analysis, substitution models and partitioning scheme were chosen using PartitionFinder (Lanfear et al. 2012) according to Bayesian Information Criterion scores. The chosen scheme had the following partitions and models: COI, HKY+I+G; Histone H3 plus Rhodospin, HKY+G; TMO-4C4, K80+G; Rag1, K80+G. All partitions in the ML analysis received a GTR-GAMMA substitution model. The BI phylogeny was inferred in the program MrBayes v. 3.2 (Ronquist et al. 2012) using two Metropolis-coupled Markov Chain Monte Carlo (MCMC) runs, each with four chains. The analysis ran for 10 million generations sampling trees and parameters every 1000 generations. Burn-in, convergence and mixing were assessed using Tracer (Rambaut and Drummond 2007) and by visually inspecting consensus trees from both runs. The ML analysis was done in the program RAxML v.8.2.9 (Stamatakis, 2014), using 20 initial random searches, and topological support was assessed using 1000 bootstrap replicates. Outgroups for the phylogenetic analysis included two species of Gramma and several other genera from the Ovalentaria sensu Wainwright et al. (2012): Acanthemblemaria (Labrisomidae), Helcogramma (Tripterygiidae), Blenniella (Blenniidae), and Tomicodon (Gobiesocidae).
To corroborate the morphologically diagnosed species using our molecular data, we conducted a coalescent-based, Bayesian species-delimitation analysis (Yang and Rannala 2010, 2014). We used the computer program BP&P ver. 3.2 (Bayesian Phylogenetics and Phylogeography – Yang and Rannala [2010], Yang [2015]), which analyzes multi-locus DNA sequence alignments under the multispecies coalescent model (Rannala and Yang 2003). We used the five DNA alignments for the 21 Lipogramma specimens in BP&P, with each sequence in the alignments being assigned to one of eight groups a priori, based on the diagnostic morphological and coloration characters discussed in the ‘Morphological Comparisons’ section below. BP&P was then used to jointly infer a species tree and calculate posterior probabilities of different species-delimitation models containing either eight species, fewer than eight species (i.e. lumping multiple ‘morphological species’), or more than eight species (i.e. splitting ‘morphological species’ into multiple cryptic species).
Depth distributions. To evaluate depth distributions we searched FishNet2 (www.fishnet2.net) for all Lipogramma specimens that were identified to species and that included data on the depth of capture. For some specimens, capture depth was given as a range of possible depths, and in instances where this range was 50 m or narrower, we took the mean depth as a proxy for a point estimate of the exact depth of capture. Broader depth ranges of capture were excluded. Depth records for Lipogramma evides were only included for specimens whose identifications we confirmed to avoid possible confusion with one of the two new species described here. When combined with depth data from specimens from DROP collections, this search resulted in depth records for 278 identified specimens of Lipogramma. We also included depth records from 83 visual observations from DROP submersible dives, excluding those observations where there was uncertainty regarding identification of the three morphologically similar species (Lipogramma evides and the two species described here).
Accession numbers. GenSeq nomenclature (Chakrabarty et al. 2013) and GenBank accession numbers for DNA sequences derived in this study are presented along with museum catalog numbers for voucher specimens in Appendix 1.
Taxonomy
Hourglass Basslet
Lipogramma levinsoni
Baldwin, Nonaka & Robertson sp. n.
http://zoobank.org/C12172C1-B3BF-48B8-B267-61D845EDCC63
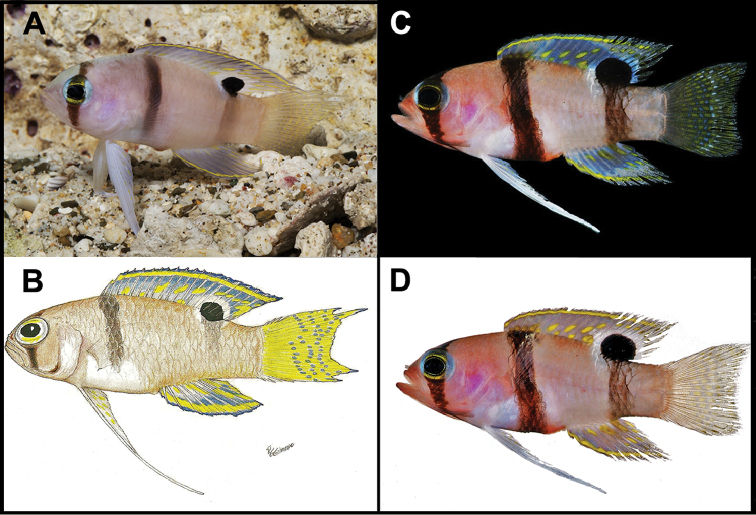
Figure 2.
Lipogramma levinsoni sp. n. A USNM 406139, holotype, 28.3 mm SL, photographed prior to preservation, photo by D. R. Robertson and C. C. Baldwin B USNM 406394, 22.2 mm SL , photographed prior to preservation, photo by D. R. Robertson and C. C. Baldwin C and D Aquarium photos, Curaçao Sea Aquarium, photos by D. Ross Robertson.
Lipogramma evides Robins & Colin, 1979: 43, fig. 2, table 1, ANSP 134332, paratype from Jamaica (photograph, counts, measurements).
Lipogramma evides Robins & Colin, 1979, fig. 3 in Gilmore and Jones (1988: 441), IRCZM 107:07660 from San Salvador, Bahama Islands (illustration, habitat information).
Type locality.
Curaçao, southern Caribbean.
Holotype.
USNM 406139, 28.3 mm SL, tissue no. CUR11139, Curasub submersible, sta. CURASUB11-02, Curaçao, off Substation Curaçao, 12.083197 N, 68.899058 W, 137–146 m depth, 23 May 2011, C. Baldwin, D. Robertson & B. Van Bebber.
Paratypes.
BONAIRE: USNM 426784, 24.2 mm SL, tissue no. CUR13183, Curasub submersible, Bonaire, Bonaire City Dock, Kralendijk, Dive 2, 12.15 N, 68.2829 W, 121-137 m depth, 30 May 2013, B. Van Bebber, A. Schrier, C. Baldwin, T. Christiaan; CURAÇAO: ANSP 201863, 24.0 mm SL, Curasub submersible, Curaçao, off Substation Curaçao, 12.083197 N, 68.899058 W, no depth data available; UF 238589, 25.0 mm SL, tissue no. CUR11018, Curasub submersible, sta. CURASUB11-22, Curaçao, off of Substation Curaçao downline, 12.083197 N, 68.899058 W, no depth data available, 27 February 2011, C. Baldwin & L. Weigt; USNM 406393, 25.7 mm SL, tissue no. CUR11393, Curasub submersible, sta. CURASUB11-06, Curaçao, 132 m depth, 31 May 2011, C. Baldwin, A. Driskell, A. Schrier & B. Van Bebber; USNM 414877, 25.3 mm SL, cleared and stained, tissue no. CUR12159, Curasub submersible, sta. CURASUB12-15, Curaçao, off of Substation Curaçao downline, 12.083197 N, 68.899058 W, 128 m depth, 10 August 2012, A. Schrier, B. Brandt, C. Baldwin, A. Driskell & P. Mace; USNM 440229, 12.7 mm SL, Curasub submersible, sta. CURASUB14-07, Curaçao, in between Porto Marie and Daaibooi beaches, 12.202842 N, 69.089507 W, 123 m depth, 21 March 2014, C. Baldwin et al.; USNM 440230, 13.4 mm SL, Curasub submersible, sta. CUR13-18, Curaçao, Playa Forti, Westpoint, 12.3679 N, 69.1553 W, 127 m, 15 August 2013, C. Baldwin, B. Brandt, A. Schrier, K. Johnson & C. DeForest; USNM 406140, 19.5 mm SL, tissue no. CUR11140, Curasub submersible, sta. CURASUB11-02, Curaçao, 137-146 m depth, 23 May 2011, C. Baldwin, D. Robertson & B. Van Bebber. DOMINICA: USNM 440231, 17.0 mm SL, tissue no. DOM16229, Curasub submersible, off northwest Dominica, no specific collection data available, March 2016, R/V Chapman Crew.
Non-type specimens.
BONAIRE: USNM 426754, 21.2 mm SL, tissue no. CUR13184, Curasub submersible, Bonaire, Bonaire City Dock, Kralendijk, Dive 2, 12.15 N, 68.2829 W, 121–137 m depth, 30 May 2013, B. Van Bebber, A. Schrier, C. Baldwin, T. Christiaan; USNM 426802, 9.4 and 18.3 mm SL, Curasub submersible, Bonaire, Bonaire City Dock, Kralendijk, 12.15 N, 68.2829 W, 114–137 m depth, 30 May 2013, B. Van Bebber, A. Schrier, C. Baldwin, T. Christiaan. CURAÇAO: USNM 426774, 17.6 mm SL, tissue no. CUR13267, Curasub submersible, sta. CURASUB13-18, Curaçao, Playa Forti, Westpoint, 12.3679 N, 69.1553 W, 118 m depth, 15 August 2013, C. Baldwin, B. Brandt, A. Schrier, K. Johnson & C. DeForest; USNM 426730, 12.3 mm SL, tissue no. CUR13268, Curasub submersible, sta. CURASUB13-18, Curaçao, Playa Forti, Westpoint, 12.3679 N, 69.1553 W, 118 m depth, 15 August 2013, C. Baldwin, B. Brandt, A. Schrier, K. Johnson & C. DeForest; USNM 406011, 20.9 mm SL, tissue no. CUR11011, Curasub submersible, sta. CURASUB11-22, Curaçao, off of Substation Curaçao downline, 12.083197 N, 68.899058 W, no depth data available, 27 February 2011, C. Baldwin & L. Weigt; USNM 406012, 18.0 mm SL, tissue no. CUR11012, Curasub submersible, sta. CURASUB11-22, Curaçao, off of Substation Curaçao downline, 12.083197 N, 68.899058 W, no depth data available, 27 February 2011, C. Baldwin & L. Weigt; USNM 406019, 14.0 mm SL, tissue no. CUR11019, Curasub submersible, sta. CURASUB11-22, Curaçao, off of Substation Curaçao downline, 12.083197 N, 68.899058 W, no depth data available, 27 February 2011, C. Baldwin & L. Weigt; USNM 406394, 22.2 mm SL, tissue no. CUR11394, Curasub submersible, sta. CURASUB11-06, Curaçao, 132 m depth, 31 May 2011, C. Baldwin, A. Driskell, A. Schrier & B. Van Bebber. DOMINICA: USNM 438703, 19.0 mm SL, tissue no. DOM16052, Curasub submersible, sta. CURASUB16-07, Toucari Bay, Toucari, Dominica, NW corner of island, 15.608047 N, 61.471788 W, no depth data available, 2 March 2016, A. Schrier, R. Bakmeijer, B. Van Bebber & F. van der Hoeven; JAMAICA: ANSP 134332, 12.6 mm SL, Nekton Gamma dive 141, collection 151-2, Jamaica, Discovery Bay, 145 m depth, 15 August 1972, L. Land & S. Hastings.
Diagnosis.
A species of Lipogramma distinguishable from congeners by the following combination of characters: pectoral-fin rays 16–18 (modally 17), gill rakers 17–20 (modally 19); three supraorbital pores present along dorsal margin of orbit, no pore present between pore at mid orbit and one at posterodorsal corner of orbit; caudal fin truncate, tips of lobes rounded; body with three broad blackish bars (one on head, two on trunk) on white background, width of bar on head sufficient to encompass entire eye, width just ventral to eye averaging 26.4% head length; trunk bars sometimes hourglass shaped, with narrower and less intensely colored central regions; anterior trunk bar covering pectoral-fin base; posterior trunk bar extending onto dorsal and anal fins as large oval blotches bordered in part by white or blue pigment to form partial ocelli; dorsal and anal fins with thin orange sub-marginal stripe. The new species is further differentiated from congeners for which molecular data are available in mitochondrial COI and nuclear Histone 3, Rhodopsin, TMO-4C4, and RAG1.
Description.
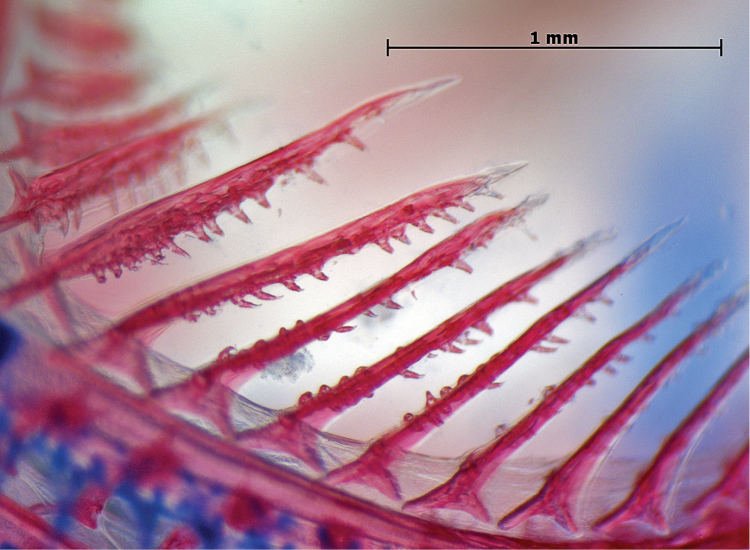
Counts and measurements of type specimens given in Table 1. Frequency distributions of pectoral-fin rays and gill rakers on the first arch are given in Table 2. Twenty specimens examined, 9.4 to 28.3 mm SL. Dorsal-fin rays XII, 9 (last ray composite); anal-fin rays III, 8 (last ray composite); pectoral-fin rays 16–18, modally 17, 17 on both sides in holotype; pelvic-fin rays I,5; total caudal-fin rays 25 (13 + 12), principal rays 17 (9 + 8), spinous procurrent rays 6 (III + III), and 2 additional rays (i + i) between principal and procurrent rays that are neither spinous nor typically segmented; vertebrae 25 (10 + 15); pattern of supraneural bones, anterior dorsal-fin pterygiophores and dorsal-fin spines 0/0/0+2/1+1/1/; ribs on vertebrae 3–10; epineural bones present on vertebrae 1-16 in holotype and cleared and stained paratype (difficult to assess in radiographs of most other specimens); gill rakers on first arch 17–20 (5-6 + 12–14), modally 19 (6 + 13), 19 (6 + 13) in holotype; uppermost four and lowermost one or two rakers very small or present only as nubs, all other gill rakers elongate and slender with tooth-like secondary rakers as in Lipogramma evides (Fig. 3); pseudobranchial filaments 5–7 (7 in holotype), filaments fat and fluffy; branchiostegals 6.
Table 1.
Counts and measurements of type specimens of Lipogramma levinsoni sp. n.. Measurements are in percent SL except width of bar ventral to eye, which is in percent head length. C&S = cleared and stained; CP = caudal peduncle; PFO = pelvic-fin origin; P1 = pectoral fin; P2 = pelvic fin; DXII = twelfth dorsal-fin spine. “Other Caudal” rays include “i” – a slender, flexible, non-spinous, and typically non-segmented ray and “I” – a spinous procurrent ray.
| USNM 406139 | USNM 406393 | USNM 406140 | USNM 414877 | USNM 426784 | USNM 440230 | USNM 440229 | USNM 440231 | ANSP 201863 | UF 238589 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Holotype | Paratype | Paratype | Paratype (C&S) | Paratype | Paratype | Paratype | Paratype | Paratype | Paratype | |
| SL | 28.3 | 25.7 | 19.5 | 25.3 | 24.2 | 13.4 | 26.3 | 17.0 | 24.0 | 25.0 |
| Dorsal-fin rays | XII, 9 | XII, 9 | XII, 9 | XII, 9 | XII, 9 | XII, 9 | XII, 9 | XII, 9 | XII, 9 | XII, 9 |
| Anal-fin rays | III, 8 | III, 8 | III, 8 | III, 8 | III, 8 | III, 8 | III, 8 | III, 8 | III, 8 | III, 8 |
| Principal caudal | 9+8 | 9+8 | 9+8 | 9+8 | 9+8 | 9+8 | 9+8 | Broken | 9+8 | 9+8 |
| Other caudal | IIIi+iIII | IIIi+iIII | IIIi+iIII | IIIi+iIII | IIIi+iIII | IIIi+iIII | IIIi+iIII | Broken | IIIi+iIII | IIIi+iIII |
| Pectoral-fin rays | 17, 17 | 17, 16 | 17, 17 | 17, 17 | 17*, 17 | 17, 17 | 17, 18 | 16, - | 17, 17 | 17, 17 |
| Gill rakers | 19 | 20 | 19 | 19 | 19 | 19 | 18 | 18 | 19 | 18 |
| Head length | 33.2 | 36.2 | 35.4 | - | 38.0 | 37.3 | 34.6 | 39.4 | 35.8 | 37.0 |
| Eye diameter | 12.0 | 12.1 | 14.9 | - | 11.6 | 15.7 | 11.4 | 13.5 | 12.1 | 13.0 |
| Snout length | 7.1 | 7.4 | 6.2 | - | 7.4 | 6.7 | 5.7 | 5.9 | 7.1 | 5.5 |
| Depth at CP | 20.1 | 18.7 | 17.4 | - | 19.8 | 18.7 | 16.3 | 17.6 | 17.1 | 17.2 |
| Depth at PFO | 33.9 | 36.6 | 34.4 | - | 35.5 | 35.1 | 35.4 | 31.2 | 40.0 | 33.6 |
| Length P1 | 25.8 | 24.5 | 23.1 | - | 21.9 | 26.1 | 24.7 | 22.4 | 28.8 | 25.6 |
| Length P2 | 72.4 | 45.5 | Broken | - | 47.1 | 42.5 | 62.7 | Broken | 83.3 | Broken |
| Length DXII | 18.7 | 20.2 | 20.5 | - | 16.5 | 17.2 | 19.0 | 15.9 | 22.2 | 21.0 |
| Width of bar ventral to eye | 25.5 | 28.0 | 21.5 | - | 26.1 | 26.0 | 23.1 | 25.4 | 25.6 | 28.4 |
6th and 7th rays (counting from dorsalmost ray) separate proximally but joined distally within same sheath and appearing as a single fat ray.
Table 2.
Frequency distributions of gill rakers on first arch and left and right pectoral-fin rays in Lipogramma levinsoni sp. n., Lipogramma evides, and Lipogramma haberi sp. n. Counts for the holotype and three paratypes of Lipogramma evides are included from Robins and Colins (1979). Counts of gill rakers and pectoral-fin rays for a fourth paratype of Lipogramma evides, ANSP 134330, were not given in the original description. The fifth and smallest paratype, ANSP 134332, is a specimen of Lipogramma levinsoni, and counts of that specimen made in this study are included. An asterisk indicates count of gill rakers or left pectoral-fin rays in holotype.
| Gill Rakers | Pectoral-fin Rays | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 15 | 16 | 17 | 18 | 19 | |
| Lipogramma levinsoni | 1 | 5 | 9* | 1 | 5 | 26* | 3 | ||||||
| Lipogramma evides | 3 | 14* | 11 | 1 | 1 | 45* | 9 | ||||||
| Lipogramma haberi | 1* | 2 | 1 | 5* | |||||||||
Figure 3.
Tooth-like, secondary rakers on the first gill arch in Lipogramma evides, USNM 34771, cleared and stained paratype. Photo by L. Tornabene.
Spinous and soft dorsal fins confluent, several soft rays at rear of fin forming elevated lobe that extends posteriorly beyond base of caudal fin. Pelvic fin, when depressed, extending posteriorly to point between anterior base of anal fin and beyond base of caudal fin, elongate first pelvic-fin ray broken in most preserved specimens. Dorsal profile from snout to origin of dorsal fin convex. Diameter of eye of holotype contained 2.8 times in head length. Pupil slightly tear shaped, with small aphakic space anteriorly. Scales extending anteriorly onto posterior portion of head, ending short of coronal pore. Scales present on cheeks, opercle, preopercle, interopercle, and isthmus. Scales lacking on top of head, snout, jaws, and branchiostegals. Scales large and deciduous, too many scales missing in most specimens to make accurate scale counts. In holotype, approximately 23 lateral scales between shoulder and base of caudal fin, approximately 4 scale rows on cheek, and approximately 9 scale rows across body above anal-fin origin. Scales on head and nape without cteni, scales on rest of body ctenoid. Fins naked except small scales present at bases of soft dorsal and anal fins.
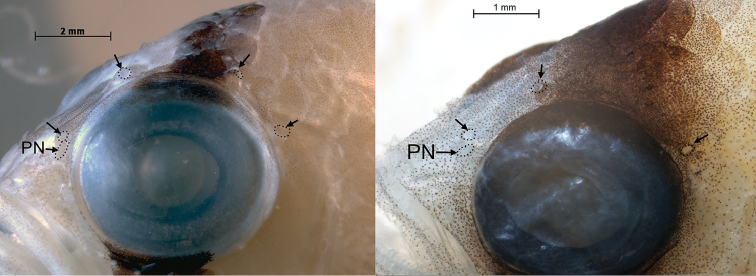
Margins of bones of opercular series smooth, opercle without spines. Single row of teeth on premaxilla posteriorly, broadening to 2-3 rows anteriorly, teeth in innermost row smallest, some teeth in outer row enlarged into small canines. Dentary similar, holotype with 3 enlarged teeth in outer row near symphysis. Vomer with chevron-shaped patch of teeth, palatine with long series of small teeth. Several canals and pores visible on head, but most pores inconspicuous. Conspicuous pores present in infraorbital canal (2 pores) and portion of supraorbital canal bordering dorsal portion of orbit (3); less conspicuous pores present on top of head (1 median coronal pore), preopercle (7), and lateral-line canal in the posttemporal region (3). Anteriormost of the 3 supraorbital pores situated at anterodorsal corner of orbit, middle supraorbital pore situated above mid orbit, and posteriormost supraorbital pore situated at posterodorsal corner of orbit (Fig. 4). This pore with fleshy rim in holotype, and mid-orbit supraorbital pore with smaller fleshy rim. Posterior nostril situated just ventral to anteriormost supraorbital pore, nostril a single large opening with ventral portion of rim slightly elevated. Anterior nostril in tube with anterior flap and situated just posterior to upper lip. No lateral line present on body.
Figure 4.
Supraorbital pore patterns in Lipogramma evides, UF 238591, 34.5 mm SL (left) and Lipogramma levinsoni sp. n., UF 238589, 25.0 mm SL (right). Arrows point to pores, which have been outlined with tiny dots for emphasis. PN – posterior nostril.
Coloration: In life (Fig. 2), ground color of head and trunk white to tan dorsally grading to white below. Head: dark brown to black bar encompassing orbit and extending ventrally to ventral midline; above orbit, bar narrowing across dorsal midline; eye with dark brown outer ring, yellowish to bluish iris. Trunk: two broad, dark brown to blackish bars present beneath dorsal fin, bars sometimes hourglass shaped, with narrower and less intensely colored central regions (central regions losing almost all dark color in some freshly dead specimens); anterior bar extending ventrally from anterior third of spinous dorsal fin to ventral midline, its anterior border extending forward to encompass base of pectoral fin; posterior bar extending ventrally from base of soft dorsal fin to posterior half of anal fin. Dorsal fin: dark trunk bars extending onto base of fin as two blotches, anterior blotch short, low, less conspicuous (than posterior blotch) and sometimes with faint orange upper border. Posterior blotch an intense, dark, longitudinal oval spanning lower half of soft dorsal and bordered posteriorly by white to bluish-white pigment. Base of fin between trunk bars whitish, central portion of fin brown to grey, and distal third of fin with bluish tint and thin, orange, submarginal stripe; this stripe breaking into spots along the rear third of fin. Anal fin: posterior trunk bar extending onto proximal portion of posterior half of fin as a strong, horizontally elongate, black blotch edged distally with bluish white line; base of fin with thin, white stripe, fin color grading into blackish to bluish-black distally. A thin, orange, sub-marginal stripe breaks into spots along posterior portion of fin. Caudal fin: basal half translucent pale orange, grading into translucent bluish distally, sometimes with indistinct, very narrow, submarginal orange band around entire edge. Pectoral fins: base blackish, fin translucent, rays translucent or tinted with orange. Pelvic fins: translucent white to bluish white, with orange tint medially on basal half of fin. In preservative (Fig. 5A), barred color pattern retained, but orange, yellow, and bluish pigments absent.
Figure 5.
Preserved specimens of A Lipogramma levinsoni sp. n., holotype, USNM 406139, 28.3 mm SL B Lipogramma haberi sp. n., holotype, USNM 422679, 40.1 mm SL C Lipogramma evides, paratype, ANSP 134330, 30.5 mm SL Photos A and B by Sandra Raredon, C by Mark Sabaj.
Distribution.
Known from specimens collected from the Bahamas, Bonaire, Curaçao, Dominica, and Jamaica. This species was also clearly observed in October 2016 by DRR and LT from the mini-submarine “Idabel” at 140 m depth adjacent to Half Moon Bay, Roatan, Honduras.
Habitat.
Lives in or hovers above small rocky rubble on gradual slopes at depths of 108-154 m. When approached by the submersible, Lipogramma levinsoni disappears into the rubble. We observed them often in pairs.
Etymology.
Named Lipogramma levinsoni in recognition of the generous, continuing support of research on neotropical biology at the Smithsonian Tropical Research Institute (Panamá) made by Frank Levinson.
Common name.
We propose “Hourglass basslet” (Cabrilleta hierba-horaria as the Spanish equivalent) to differentiate this species from the Banded Basslet, Lipogramma evides, and the Yellow-banded Basslet, Lipogramma haberi (see description below), both of which have narrower, straight-sided bars on the trunk.
Genetic comparisons.
Table 3 shows average inter- and intraspecific divergences in COI among species of Lipogramma analyzed genetically in this study. With the exception of a single substitution in one specimen, the 15 specimens of Lipogramma levinsoni exhibit no intraspecific genetic variation at this locus and differ from other Lipogramma species by 15.4–26.0%. Lipogramma levinsoni differs from Lipogramma evides by 17.1% and Lipogramma haberi by 19.0%.
Table 3.
Average Kimura two-parameter distance summary for species of Lipogramma based on cytochrome c oxidase I (COI) sequences analyzed in this study. Intraspecific averages are in bold.
| “robinsi1” | “robinsi2” | levinsoni | haberi | anabantoides | trilineata | klayi | evides | |
|---|---|---|---|---|---|---|---|---|
| “robinsi1” (n=6) | 0.003 | |||||||
| “robinsi2” (n=7) | 0.119 | 0.002 | ||||||
| levinsoni (n=15) | 0.162 | 0.169 | 0 | |||||
| haberi (n=3) | 0.111 | 0.132 | 0.19 | 0.002 | ||||
| anabantoides (n=2) | 0.195 | 0.184 | 0.154 | 0.202 | 0.005 | |||
| trilineata (n=12) | 0.217 | 0.251 | 0.227 | 0.236 | 0.258 | 0.005 | ||
| klayi (n=21) | 0.266 | 0.259 | 0.26 | 0.279 | 0.246 | 0.242 | 0.003 | |
| evides (n=30) | 0.103 | 0.128 | 0.171 | 0.11 | 0.22 | 0.249 | 0.263 | 0.001 |
Comments.
The smallest paratype of Lipogramma evides, ANSP 134332 (Fig. 1B), 12.6 mm SL, is a specimen of Lipogramma levinsoni. Although Robins and Colin (1979) indicated 15 pectoral-fin rays on both sides of this specimen, we count 17 on the right and find the left side too bent to make an accurate count. Lipogramma levinsoni typically has 17–18 pectoral-fin rays, modally 17. The gill-raker count of 19 given by Robins and Colin (1979) was confirmed by our examination, and is the typical count for Lipogramma levinsoni. Counts of pectoral-fin rays (15–16, usually 16) and gill rakers on the first arch (19–21, usually 20 or 21) given by Robins and Colin (1979) for the remaining paratypes of Lipogramma evides support their identification as specimens of Lipogramma evides. As noted, previous authors have mistakenly identified the broad-banded Lipogramma levinsoni as the juvenile form of the more narrow-banded Lipogramma evides. Our material includes juvenile specimens of both Lipogramma levinsoni and Lipogramma evides, which in each case have the adult configuration of dark bands (Fig. 6).
Figure 6.
Comparison of juveniles of A Lipogramma levinsoni sp. n., USNM 440230, paratype, 13.4 mm SL and B Lipogramma evides, USNM 431410, 12.7 mm SL.
Yellow-banded Basslet
Lipogramma haberi
Baldwin, Nonaka & Robertson sp. n.
http://zoobank.org/4A8447E9-205C-4639-9209-428D8DCDAC1F
Figure 7.
Lipogramma haberi sp. n., USNM 422679, holotype, 40.1 mm SL, photographed prior to preservation against white (top) and black (bottom) backgrounds. Photos by D. R. Robertson and C. C. Baldwin.
Type locality.
Curaçao, southern Caribbean
Holotype.
USNM 422679, 40.1 mm SL, tissue no. CUR13171, Curasub submersible, sta. CURASUB13-09, Curaçao, southwest tip of Klein Curaçao, 11.975783 N, 68.646192 W, 152 m depth, 27 May 2013, M. Harasewych, L. Weigt, B. Van Bebber & A. Schrier.
Paratypes.
USNM 434772, 26.4 mm SL, tissue no. CUR15092, Curasub submersible, sta. CURASUB15-12, northwest corner of Klein Curaçao, 11.998453 N, 68.651308 W, 187 m depth, 27 August 2015, B. Brandt, A. Schrier, S. Haber & T. Haber; USNM 422670, 23.0 mm SL, tissue no. CUR13158, Curasub submersible, sta. CURASUB13-08, Curaçao, southwest tip of Klein Curaçao, 11.975783 N, 68.646192 W, 233 m depth, 27 May 2013, C. Baldwin, D. Robertson, B. Brandt, A. Schrier & L. Weigt.
Diagnosis.
A species of Lipogramma distinguishable from congeners by the following combination of characters: pectoral-fin rays 15–16 (modally 16), gill rakers 15–16 (modally 16); four supraorbital pores along dorsal portion of orbit, a pore present between pore at mid orbit and one at posterodorsal corner or orbit; caudal fin truncate, tips of lobes rounded; body with three dusky bars (one on head, two on trunk) on yellow/white background; width of bar on head sufficient to encompass pupil but not entire eye, width just ventral to eye averaging 17.6% head length; anterior trunk bar narrow and not extending forward to cover pectoral-fin base, bar lighter and less conspicuous ventrally; posterior trunk bar a broad, yellow/tan triangle that is wider dorsally than ventrally; this triangle extending onto soft dorsal fin as large, round, well-defined ocellus; posterior trunk bar not extending onto anal fin; dorsal fin with thin yellow sub-marginal stripe; no yellow submarginal stripe on anal fin; dorsal, anal, and caudal fins with numerous yellow spots. The new species is further differentiated from congeners for which molecular data are available in COI and RAG1.
Description.
Counts and measurements of type specimens given in Table 4. Frequency distributions of pectoral-fin rays and gill rakers on the first arch are given in Table 2. Three specimens examined, 23.0–40.1 mm SL. Dorsal-fin rays XII, 9 (last ray composite); anal-fin rays III, 8 (last ray composite); pectoral-fin rays 15–16, modally 16, 16 on both sides in holotype; pelvic-fin rays I,5; total caudal-fin rays 25 (13 + 12), principal rays 17 (9 + 8), spinous procurrent rays 6 (III + III), and 2 additional rays (i + i) between principal and procurrent rays that are neither spinous nor typically segmented; vertebrae 25 (10 + 15); pattern of supraneural bones, anterior dorsal-fin pterygiophores and dorsal-fin spines 0/0/0+2/1+1/1/; ribs on vertebrae 3-10; epineural bones present on vertebrae 1–15 in one paratype, difficult to assess in other specimens; gill rakers on first arch 15–16 (4-5 + 11), 15 (4 + 11) in holotype, both paratypes with 16 (5 + 11); lowermost two rakers very small, all other gill rakers elongate and slender with tooth-like secondary rakers as in Lipogramma evides (Fig. 3); pseudobranchial filaments 6, filaments fat and fluffy; branchiostegals 6.
Table 4.
Counts and measurements of type specimens of Lipogramma haberi sp. n. Measurements are in percent SL except width of bar ventral to eye, which is in percent head length. CP = caudal peduncle; PFO = pelvic-fin origin; P1 = pectoral fin; P2 = pelvic fin; DXII = twelfth dorsal-fin spine. “Other Caudal” rays include “i” – a slender, flexible, non-spinous, and typically non-segmented ray and “I” – a spinous procurrent ray.
| USNM 422679 | USNM 434772 | USNM 422670 | |
|---|---|---|---|
| Holotype | Paratype | Paratype | |
| SL | 40.1 | 26.4 | 23.0 |
| Dorsal-fin Rays | XII, 9 | XII, 9 | XII, 9 |
| Anal-fin Rays | III, 8 | III, 8 | III, 8 |
| Principal Caudal | 9+8 | 9+8 | Broken |
| Other Caudal | IIIi+iIII | IIIi+iIII | Broken |
| Pectoral-fin Rays | 16, 16 | 16, 15 | 16, 16 |
| Gill Rakers | 15 | 16 | 16 |
| Head Length | 35.2 | 39.0 | 34.8 |
| Eye Diameter | 11.2 | 14.0 | 13.0 |
| Snout Length | 6.7 | 5.7 | 6.1 |
| Depth at CP | 18.7 | 20.1 | 17.8 |
| Depth at PFO | 32.4 | 34.1 | 27.0 |
| Length P1 Fin | 22.2 | 27.7 | 24.3 |
| Length P2 Fin | 62.3 | 54.5 | 46.1 |
| Length DXII | 22.4 | 23.1 | 17.4 |
| Width of Bar Ventral to Eye | 14.9 | 20.4 | 17.5 |
Spinous and soft dorsal fins confluent, several soft rays in posterior portion of fin forming elevated lobe that extends posteriorly beyond base of caudal fin. Pelvic fin extending posteriorly to anterior third of caudal peduncle in holotype when depressed, longest pelvic-fin rays broken in preserved specimens. Dorsal profile from snout to origin of dorsal fin convex. Diameter of eye of holotype contained 2.7 times in head length. Pupil slightly tear shaped, with small aphakic space anteriorly. Scales extending anteriorly onto top of head, ending short of coronal pore. Scales present on cheeks, opercle, preopercle, interopercle, and isthmus. Scales lacking on frontal region, snout, jaws, and branchiostegals. Scales large and deciduous, too many missing in paratypes to make counts, holotype with approximately 24 lateral scales between shoulder and base of caudal fin, 5 cheek rows, and 11 rows across body above anal-fin origin. Scales on head and nape without cteni, scales on rest of body ctenoid. Fins naked except small scales present at bases of soft dorsal and anal fins.
Margins of bones of opercular series smooth, opercle without spines. Premaxilla with band of small conical teeth, band widest at symphysis, outer row with largest teeth, 3 or 4 near symphysis enlarged into canines. Dentary similar except 4-6 anterior teeth enlarged into canines. Vomer with chevron-shaped patch of teeth, palatine with long series of small teeth. Several canals and pores visible on head, but most pores inconspicuous. Conspicuous pores present in infraorbital canal (2) and in supraorbital canal bordering dorsal portion of orbit (4); less conspicuous pores present on top of head (1 median coronal pore), preopercle (8), and lateral-line canal in posttemporal region (3). An additional 4 tiny pores present beneath orbit in holotype in infraorbital canal. Supraorbital pore pattern as in Lipogramma evides (Fig. 4): anteriormost of 4 supraorbital pores situated at anterodorsal corner of orbit, second supraorbital pore situated above mid orbit, and posteriormost supraorbital pore situated at posterodorsal corner of orbit. Between second and posteriormost supraorbital pores, another pore present and situated closer to latter. Posterior nostril situated just ventral to anteriormost supraorbital pore, nostril a single large opening with ventral portion of rim slightly elevated. Anterior nostril in tube with anterior flap and situated just posterior to upper lip. No lateral line present on body.
Coloration: In life, ground color of head and trunk pale yellow to tan dorsally, white ventrally. Head: mostly pale yellow-tan with white blotch on operculum; a brown to black C-shaped bar with yellow-brown edges originating on top of head, widening ventrally above orbit to width of pupil and passing over orbit at that width, then narrowing ventrally and continuing as dark line along lower edge of operculum; iris dark brown above and below where bar passes through, yellowish-white anteriorly and posteriorly, a thin gold ring circling pupil. Trunk: two dark bars beneath dorsal fin, anterior one brown to blackish (edged with yellow-brown) originating below anterior dorsal spines and descending obliquely behind pectoral-fin base to ventral midline; bar fading below pectoral-fin base; posterior bar much broader than anterior bar but paler and less conspicuous, bar spanning dorsal and ventral body margins and covering anterior half of caudal peduncle; bar narrowing ventrally. Dorsal fin: grey with a bluish tint (when photographed against black background – Fig. 7, bottom), with thin, submarginal yellow stripe; spinous dorsal fin with row of round to oblong yellow spots along base, 1–2 rows of obliquely oriented, oval, yellow spots above that; soft dorsal with large, conspicuous, circular, black ocellus covering lower half of fin and extending onto dorsal portion of trunk; thin, white, outer ring surrounding ocellus on both fin and trunk complete in holotype (Fig. 7), absent along underside of ocellus in both paratypes; above ocellus, soft dorsal fin with approximately three rows of rounded yellow spots; grey spaces between yellow spots appearing as well-defined grey to blue spots posteriorly. Anal fin: grey with bluish tint (when photographed against black background), each ray with 3-6 elongate yellow spots from base to fin edge; grey spaces between yellow spots appearing as well-defined grey to blue spots posteriorly. Caudal fin: base of fin mostly yellow, remainder of fin with rows of yellow spots along fin rays; grey spaces between yellow spots appearing as well-defined grey or blue spots. Pectoral fins: base yellowish with black dots, fin translucent. Pelvic fins: bright white, inner 2-3 rays with series of small yellow-brown dots. In preservative (Fig. 5B), barred color pattern retained, posterior trunk bar faint, and yellow and bluish pigments absent.
Distribution.
Known only from Klein Curaçao, a 1.7 km2 island 11 km southeast of Curaçao.
Habitat.
No specific habitat information recorded.
Etymology.
Named in honor of Spencer and Tomoko Haber, who funded and participated in a submersible dive by the Smithsonian’s Deep Reef Observation Project (DROP) that resulted in the collection of USNM 434772, a paratype of the new species.
Common name.
We propose “Yellow Banded Basslet” (“Cabrilleta cinta-amarilla” as the spanish equivalent) to distinguish Lipogramma haberi from Lipogramma evides and Lipogramma levinsoni. Although Lipogramma evides has a submarginal yellow stripe along the dorsal and anal fins, it lacks the overall yellow body color of Lipogramma haberi.
Genetic comparisons.
Table 3 shows average inter- and intraspecific divergences in COI among species of Lipogramma analyzed genetically in this study. Lipogramma haberi exhibits 0.2% intraspecific genetic variation and 11.0–27.9% divergence from other Lipogramma species. It differs from Lipogramma evides by 11.0% and from levinsoni by 19.0%.
Comments.
Relative to Lipogramma levinsoni and Lipogramma evides, which are known from multiple localities within the Caribbean Sea, Lipogramma haberi is an uncommon species on deep reefs and may have a more restricted geographic distribution. Although both Lipogramma levinsoni and Lipogramma evides are frequently observed and collected off the southern coast of Curaçao, in more than one hundred submersible dives there we have not collected Lipogramma haberi. Rather, we have only collected Lipogramma haberi on infrequent trips to Klein Curaçao, a small island, as noted above, 11 km southeast of Curaçao.
Discussion
Comments on Lipogramma evides. The type series of Lipogramma evides includes the holotype and five paratypes (Robins and Colin 1979). We examined specimens or photographs of specimens of the type series from ANSP and FMNH and conclude that all except one, ANSP 134332, 12.6 mm SL, represent Lipogramma evides. We also examined 31 specimens of Lipogramma evides that we recently collected at Curaçao and that range in size from 12.7–45.4 mm SL. Frequency distributions of pectoral-fin rays and gill rakers on the first arch are given in Table 2, an illustration of the holotype that was included in the original description of the species is shown in Fig. 1A, color patterns of live and recently deceased individuals are shown in Fig. 8, a photographed of a preserved paratype (ANSP 134330) is provided in Fig. 5C, secondary spines on gill rakers of the first arch are shown in Fig. 3, supraorbital pore pattern is shown in Fig. 4, and a photograph of a preserved juvenile is featured in Fig. 6.
Figure 8.
Lipogramma evides A Aquarium photograph by Barry Brown, Substation Curacao B USNM 276560, 45.3 mm SL, illustration by Grant Gilmore in Gilmore and Jones (1988: fig. 1) C and D USNM 414885, 24.4 mm SL, photos by D. R. Robertson and C. C. Baldwin against black (C) and white (D) backgrounds.
The illustration of the holotype (Fig. 1A) shows a triangular-shaped bar on the posterior portion of the trunk that more closely resembles the shape of that bar in Lipogramma haberi (Fig. 7) than Lipogramma evides (Fig. 8). The pigment is too faded in the preserved holotype now to determine the shape of that bar, but we note that in specimens or photographs of the four paratypes of the type series that are actually specimens of Lipogramma evides, the posterior trunk bar is narrow (e.g., Fig. 5C). This suggests that the shape of the posterior trunk bar illustrated in the holotype of Lipogramma evides is either incorrectly drawn or represents an anomaly for the species. Lipogramma evides and Lipogramma haberi are easily distinguished by numbers of gill rakers on the first arch—15 or 16 in Lipogramma haberi, 19-22 (modally 20) in Lipogramma evides (Table 2). Our examination of the holotype confirms the count by Robins and Colin (1979) of 20 gill rakers in the holotype of Lipogramma evides. Furthermore, the triangular-shaped posterior trunk bar in Lipogramma haberi is very pale relative to the anterior trunk and head bars. Robins and Colin’s (1979) illustration of the holotype shows three body bars of equal intensity.
Colin’s (1974) observation of “Lipogramma sp.” at Glover’s Reef, Belize, was cited as Lipogramma evides by Robins and Colin (1979), and based on the recorded depths of observations, 165–180 m, we tentatively agree with this identification, as Belize is between Arrowsmith Bank and Nicaragua, where Lipogramma evides does occur. Lipogramma evides inhabits depths of 133-302 m, whereas Lipogramma levinsoni occurs from 103 to 154 m. However, Colin’s (1974) observed fish could have been Lipogramma haberi, which occurs from 152–233 m. Polanco et al. (2012) recorded several specimens of Lipogramma evides from the Coralinos Archipelago off Colombia, and based on the stated counts of 15–16 pectoral-fin rays and 20–21 gill rakers, those specimens are correctly identified. In addition to Colombia and the tentative Belize location, Lipogramma evides is known definitively from the type locality of Arrowsmith Bank in the Yucatan Peninsula, Nicaragua, southeast of Barbuda, and Curaçao (including Klein Curaçao). It was also observed but not collected in October 2016 by DRR and LT from the mini-submarine “Idabel” at 232–250 m depth adjacent to Half Moon Bay, Roatan, Honduras. It was not collected or observed on DROP submersible dives at Bonaire or Dominica. A list of Lipogramma evides material examined in this study is given in Appendix 2.
Morphological comparisons. Lipogramma levinsoni, Lipogramma evides, and Lipogramma haberi can be readily distinguished from all congeners in having three dark bars (one on the head, two on the trunk) on a white background vs. a brown body with a reddish head in Lipogramma anabantoides; a yellow body with one black bar on the head in Lipogramma flavescens; a purple head and yellow trunk in Lipogramma klayi Randall, 1963; a brown body with one broad white bar and multiple narrow orange bars in Lipogramma regia; a brownish body with about 12 thin dark bars in Lipogramma robinsi Gilmore, 1997; a pink head and trunk with a yellow stripe along the dorsal profile of the head in Lipogramma rosea; and a yellow head and trunk with three long iridescent blue stripes on the head in Lipogramma trilineata Randall, 1963. The major differences among Lipogramma levinsoni, Lipogramma evides, and Lipogramma haberi are summarized in Table 5. Lipogramma evides and Lipogramma haberi are morphologically similar and reach a similar maximum size (45.5 and 40.1 in our material, respectively). They are easy to distinguish from one another on the basis of number of gill rakers on the first arch (usually 20-21 in Lipogramma evides, 15-16 in Lipogramma haberi – Table 2) and by live and preserved color pattern (Figs 5, 7, 8). In life, Lipogramma haberi has a considerable amount of yellow as ground color and associated with the dark bars, whereas the ground color of Lipogramma evides is mostly white. In fresh and preserved specimens, the posterior trunk bar in Lipogramma haberi is broad and much wider at the top than the bottom, whereas in Lipogramma evides it is narrower and of uniform width. There is also a difference in the shape of the caudal fin of the two species, with Lipogramma haberi having a truncate fin with rounded lobe tips and Lipogramma evides having a slightly emarginate fin with pointed lobe tips.
Table 5.
Comparisons among Lipogramma levinsoni sp. n., Lipogramma haberi sp. n., and Lipogramma evides.
| Lipogramma levinsoni | Lipogramma haberi | Lipogramma evides | |
|---|---|---|---|
| Standard length (mm) | 9.4–28.3 | 23.0–40.1 | 13.7–45.5 |
| Gill rakers on 1st arch | Usually 19 | 15–16 | Usually 20–21 |
| Pectoral-fin rays | Usually 17 | Usually 16 | Usually 16 |
| Supraorbital pores/pore present between pore at mid orbit and one at posterodorsal corner of orbit (Fig. 4) | 3/Absent | 4/Present | 4/Present |
| Ground color | White, grey on nape & snout | Yellow above, white below | White |
| Dark bar on head | Black; relatively wide, widens to encompasses entire eye No rearward extension along lower edge of opercle |
Brown, edged with yellow; relatively narrow, widens to encompass pupil Narrow rearward extension along lower edge of opercle |
Black; relatively narrow, widens to encompass pupil Narrow rearward extension along lower edge of opercle |
| Width of dark bar on head (measured immediately ventral to eye) in % HL) | 21.5–34.8 (x = 26.4) | 14.9–20.4 (x = 17.6) | 8.7–19.4 (x = 14.7) |
| Anterior trunk bar | Black, wide, vertical, center often narrower & paler Covers pectoral base Extension onto dorsal fin large, intense |
Brown, edged with yellow; narrow, slightly oblique, uniform width, paler on belly Behind pectoral base No extension onto dorsal fin |
Black; narrow, slightly oblique, uniform width, paler on belly Behind pectoral base Extension onto dorsal fin small, weak |
| Posterior trunk bar | Same form and color as anterior bar Extension onto dorsal fin = oval partial ocellus Extension onto anal fin = elongate, partial ocellus |
Yellow-brown; broad dorsally, thinning ventrally, triangular in shape Extension onto dorsal fin = round, well defined ocellus No extension onto anal fin |
Same form and color as anterior bar but usually paler than anterior bar Extension onto dorsal fin = round, well defined ocellus Extension onto anal fin absent or a small, weak smudge |
| Dorsal-fin pigment | Submarginal stripe orange Remainder of fin without pale spots |
Submarginal stripe yellow Remainder of fin with 2–3 rows of yellow spots |
Submarginal stripe yellow Remainder of fin with 1–2 rows of yellow spots |
| Anal-fin pigment | Submarginal stripe orange No pale spots on remainder of fin |
No pale submarginal stripe Remainder of fin with 1–6 rows of yellow spots |
Submarginal stripe yellow Remainder of fin with 1–3 rows of yellow spots near base |
| Caudal-fin shape | Truncate, lobe tips rounded | Truncate, lobe tips rounded | Slightly emarginate, lobe tips pointed |
| Depth range (m) | 108–154 | 152–233 | 133–302 |
| Geographical distribution | Bahamas, Bonaire, Curaçao, Dominica, and Jamaica | Klein Curaçao | Barbuda, Belize(?),Colombia, Curaçao, Klein Curaçao, Mexico (Caribbean), and Nicaragua |
Lipogramma levinsoni reaches a smaller maximum size than Lipogramma haberi and Lipogramma evides (largest specimen examined 28.3 mm SL) and differs in modal numbers of gill rakers on first arch and pectoral-fin rays (Table 2), supraorbital pore pattern (Fig. 4), and numerous aspects of color pattern (Figs 2, 5, 7, 8). In life, Lipogramma levinsoni is easily distinguished from Lipogramma haberi and Lipogramma evides by having an orange submarginal stripe on the dorsal fin (vs. yellow) and an orange submarginal stripe on the anal fin (vs. no stripe in Lipogramma haberi, a yellow submarginal stripe in Lipogramma evides). In preservative, Lipogramma levinsoni is easily distinguished from Lipogramma haberi and Lipogramma evides by the shape, size, and configuration of the dark body bars (Table 5).
Species delimitation and phylogeny. The neighbor-joining network (Suppl. material 1) shows eight distinct genetic lineages with an average within-lineage genetic distance of 0.002 substitutions/site and an average between-lineage genetic distance of 0.20 substitutions/site (Table 3). Considering between-lineage distances that are 10 or more times greater than within-lineage distances as indicative of distinct species (Hebert et al. 2014), genetic distances corroborate the recognition of the Lipogramma levinsoni and Lipogramma haberi lineages as species. Average between-lineage divergence for Lipogramma levinsoni is 19% (18% between Lipogramma levinsoni and the other two banded basslets, Lipogramma haberi and Lipogramma evides), whereas average within-lineage divergence is 0%. For Lipogramma haberi, average between-lineage divergence is 18% (11% between Lipogramma haberi and Lipogramma levinsoni/Lipogramma evides), whereas average within-lineage divergence is 0.2%.
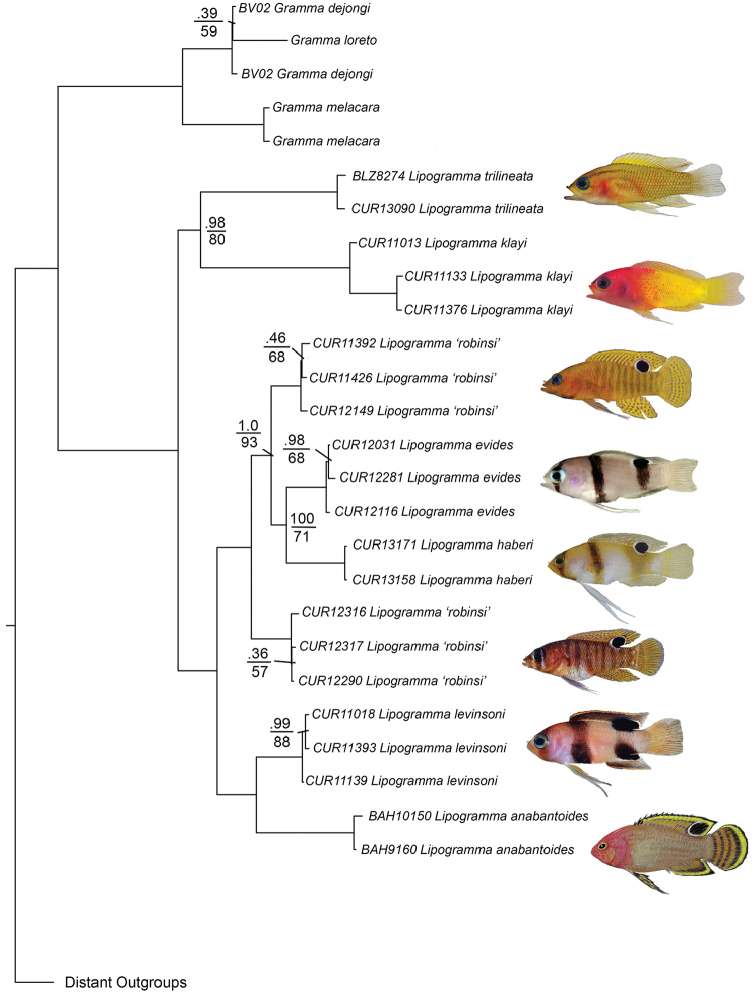
The ML and BI analyses resulted in identical topologies, with most relationships supported by 1.0 posterior probability and 100% bootstrap support (Fig. 9). The BP&P analysis inferred a coalescent-based species-tree that was identical in topology to the ML and BI trees. In addition, the BP&P analysis provided overwhelming support for the presence of eight species of Lipogramma in our phylogeny (posterior probability 0.99981), including three distinct species of banded basslets (Lipogramma evides, Lipogramma haberi and Lipogramma levinsoni), indicating perfect congruence between molecular and morphology-based species delimitation criteria. Lipogramma trilineata and Lipogramma klayi, which have two of the shallowest depth ranges among Lipogramma species (Fig. 10), were recovered as sister species. This pair is sister to a larger clade comprising Lipogramma anabantoides + Lipogramma levinsoni (as sister species) and Lipogramma evides + Lipogramma haberi + two putative new species superficially resembling Lipogramma robinsi. Not surprising given their morphological similarity, Lipogramma evides, and Lipogramma haberi resolve as sister species. There is strong support for a clade comprising the four deepest-known species in our phylogeny—Lipogramma evides, Lipogramma haberi, and the two “Lipogramma robinsi” species (Figs 9, 10). Baldwin and Robertson (2014) found a similar evolutionary grouping of deep-water species in the serranid genus Liopropoma, and Tornabene et al. (2016a) found repeated invasions of deep-reef depths in the family Gobiidae with subsequent species radiations entirely within the deep-reef zone. Lipogramma flavescens, which also inhabits deep water (200–300 m, Fig. 10), may be part of this clade. A dark ocellus on the base of the soft dorsal fin appears to be a synapomorphy of the clade comprising Lipogramma anabantoides, Lipogramma levinsoni, Lipogramma evides, Lipogramma haberi, and the two “Lipogramma robinsi” species. Presence of this ocellus in Lipogramma flavescens and Lipogramma regia suggests that they may also belong to this group, but genetic samples of both are needed for phylogenetic analysis. Lipogramma flavescens may be closely related to Lipogramma haberi, as they share a narrow dark bar through the eye, yellow coloration, and low gill-raker count (15–16), and they inhabit similar deep-reef depths (152–233 m for Lipogramma haberi, 200–300 m for Lipogramma flavescens). If the evolutionary relationships of Lipogramma species are correlated with depth as our data suggest, and if Lipogramma regia, which is known only from depths < 100 m is a member of the clade diagnosed by a dark ocellus on the soft dorsal fin, it may be most closely related to Lipogramma anabantoides and Lipogramma levinsoni, which are known only from depths < 120 m (Lipogramma anabantoides) and < 154 m (Lipogramma levinsoni). Those three are the only known Lipogramma species with a modal pectoral-fin count of 17 (Gilmore and Jones 1988: Table 2, this study). We note that the addition to our molecular phylogeny of the four known species of Lipogramma that are currently unavailable for analysis (Lipogramma flavescens, Lipogramma regia, Lipogramma rosea, and Lipogramma robinsi) could change our hypotheses of relationships within the genus.
Figure 9.
Bayesian Inference molecular phylogeny of Lipogramma based on combined mitochondrial and nuclear genes. Numbers of individuals analyzed for each species are given in Appendix 1, along with the genes sequenced for each individual. Topology is identical to that from Maximum Likelihood analysis. Support values are Bayesian posterior probabilities (above) and bootstrap values (below). Nodes without labels have 1.0 posterior probability and 100 bootstrap values. Photos or illustrations by C. C. Baldwin, D. R. Robertson, R. G. Gilmore, and C. R. Robins.
Figure 10.
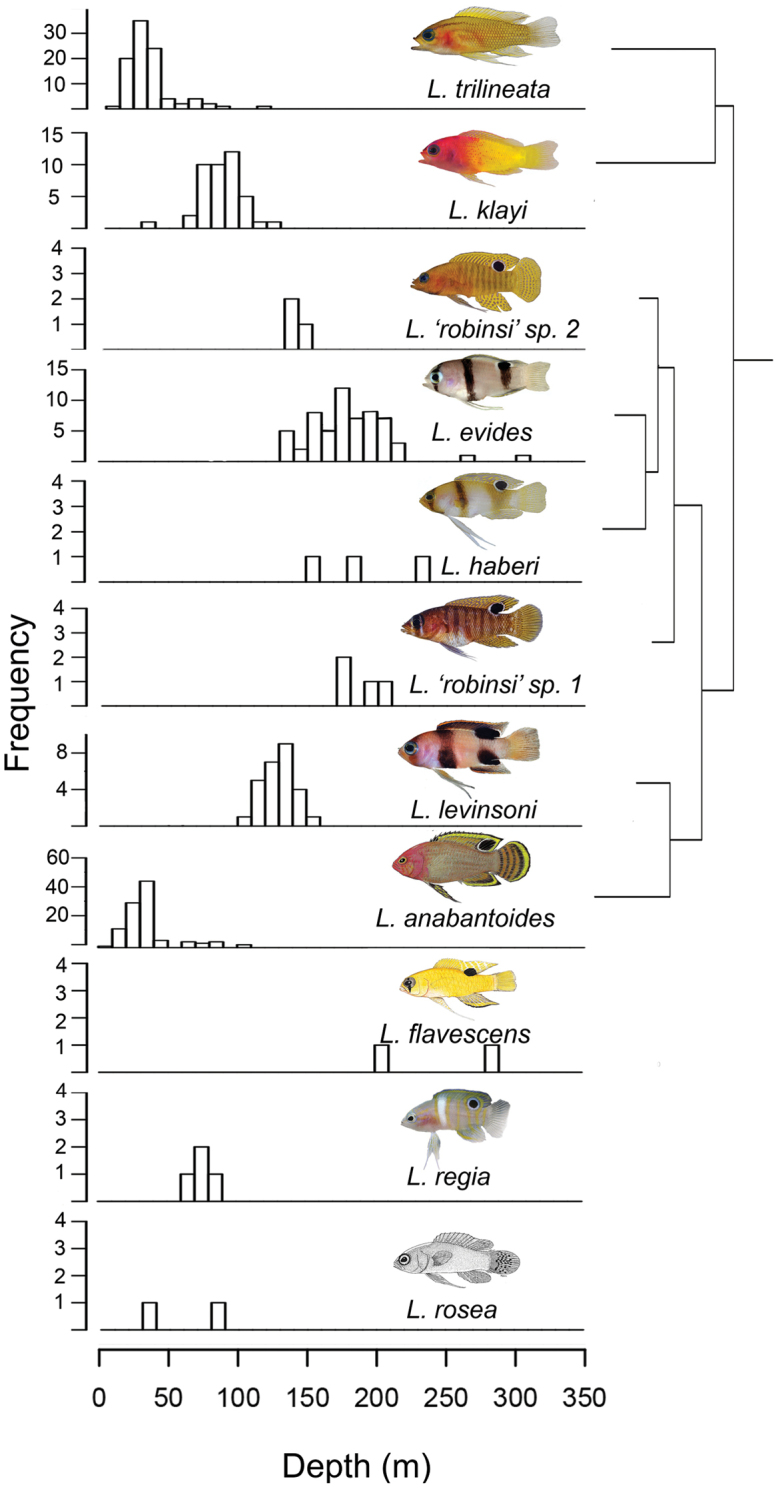
Depth distributions for species of Lipogramma. Photos or illustrations by C. C. Baldwin, D. R. Robertson, R. G. Gilmore, and Mooi and Gill (2002: fig. 9).
The two “Lipogramma robinsi” included here (Table 3, Figs 9, 10) are genetically distinct and superficially different from one another and from Lipogramma robinsi Gilmore, 1997. A more thorough investigation of those three taxa is in progress, after which a key to all Lipogramma species will be constructed.
Lipogramma is currently classified along with Gramma in the family Grammatidae based on a single synapomorphy in the arrangement of cheek musculature (Gill and Mooi 1993). Molecular data have failed to corroborate the monophyly of the Grammatidae (Betancur-R et al. 2013, Mirande 2016); rather, those data suggest that Lipogramma and Gramma are each related to different taxa within the diverse Ovalentaria. Relationships within the Ovalentaria have proven difficult to resolve with traditional molecular markers (Betancur-R et al. 2013), molecular markers plus some morphological characters (Mirande 2016), and phylogenomic data (Eytan et al. 2015). Potential close relatives of Lipogramma based on molecular data include blennioids, cichlids, plesiopids, pseudochromids, and Pholidichthys. Some of these groups have been previously linked to either Lipogramma, Gramma, or both, based on shared morphological characters, but the homologies of many of these characters have been questioned (Gill and Mooi 1993). At present, the phylogenetic position of Lipogramma is unresolved.
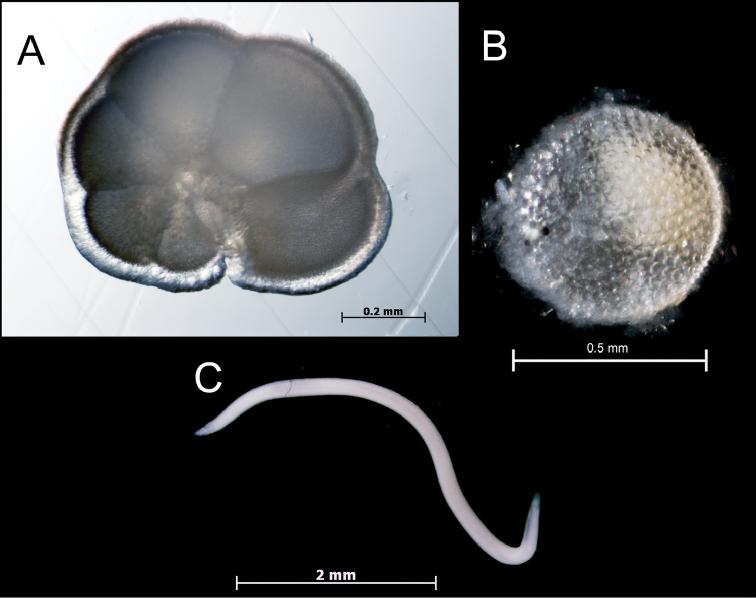
Ecology and life history. Little is known about community structure on deep reefs, including food networks. Although an analysis of the diet of banded basslets based on stomach contents is beyond the scope of this study, the gastrointestinal tract of the cleared and stained Lipogramma evides (USNM 434771) contained numerous individuals of a planktonic foraminiferan that was tentatively identified by Smithsonian Curator of Planktic Foraminifera Brian Huber as Globorotalia manardii (d’Orbigny) – Fig. 11A. Two items found in the gastrointestinal tract of Lipogramma levinsoni (USNM 406140) appear to be a diatom (possibly Coscinodiscus eccentricus Ehrenberg, Huber pers. comm, Fig. 11B) and a parasitic nematode (identification by Smithsonian Curator of Invertebrate Zoology Jon Norenberg and Assistant Professor of Biology at Virginia Military Institute Ashleigh Smythe, Fig. 11C). Future investigations of diets of deep-reef fish species are planned.
Figure 11.
Items from stomachs of deep-reef Lipogramma: A Planktonic foraminiferan, possibly Globorotalia manardii, from Lipogramma evides, USNM 434771, collected at 174 m B Diatom, possibly Coscinodiscus eccentricus, from Lipogramma levinsoni sp. n., USNM 406140, collected between 137 and 146 m C Parasitic Nematoda from same specimen as B. Photos by A. Nonaka and L. Tornabene.
The broad Caribbean distributions of Lipogramma levinsoni and Lipogramma evides (Table 5) suggest a pelagic larval stage capable of dispersal, so it is perplexing that there are no records of Lipogramma or Gramma larvae from plankton tows (Asoh and Yoshikawa 1996, Hardy 2006). Thresher (1980) noted that Lipogramma trilineata constructs nests within algae in aquaria settings, and Asoh and Yoshikawa (1996) described similar nesting behavior in Gramma loreto Poey, 1868. The apparent restricted geographic distribution of Lipogramma haberi (Table 5) could indicate that some species have limited dispersal capabilities; however, the paucity of faunal investigations of deep-reef ecosystems may mask a larger geographic distribution for that species. Interestingly, Leis et al. (2012) calculated swimming speed for reared Gramma loreto larvae and found that the actual and relative critical speed (Ucrit) were so low that for most of the pelagic larval duration their ability to influence their dispersal by horizontal swimming would be much less than that of many other tropical fish species. Further study of the early life history of Lipogramma is needed, including exploring the possibility that planktonic dispersal in the genus may be limited.
Conclusions
Adults and juveniles of the banded basslet, Lipogramma evides, were previously recognized as different ontogenetic color patterns of a single species. This study shows that the juvenile color pattern belongs to a cryptic species, described here as Lipogramma levinsoni. This study also documents the first known juveniles of Lipogramma evides, which share the color pattern of adults. A second new species that is morphologically similar to Lipogramma evides, Lipogramma haberi, is also described. These three basslet species are confined to deep-reef depths, but they stratify such that Lipogramma levinsoni occurs at shallower depths than Lipogramma evides and Lipogramma haberi. A molecular analysis of evolutionary relationships among available Lipogramma species reveals correlations between depth of occurrence and phylogeny, an eco-evolutionary pattern observed in other deep-reef Caribbean fishes that warrants further investigation. The two new basslets represent the eleventh and twelfth new fish species described in recent years from exploratory submersible diving by the Smithsonian’s Deep Reef Observation Project (DROP) to Caribbean depths of 300 m (Baldwin and Robertson 2013, 2014, 2015; Baldwin and Johnson 2014; Baldwin et al. 2016; Tornabene et al. 2016b, 2016c). Numerous other new fish and invertebrate species discovered by DROP await description, including the two putative new species identified in this study as morphologically similar to but distinct from Lipogramma robinsi. Considerably more effort is needed to adequately explore tropical deep reefs, diverse but largely overlooked global ecosystems.
Supplementary Material
Acknowledgments
We thank Laura Albini, Bruce Brandt, Barry Brown, Mary Brown, Cristina Castillo, Loretta Cooper, Tico Christiaan, Tommy Devine, Grant Gilmore, Brian Horne, Brian Huber, Dave Johnson, Rob Loendersloot, Caleb McMahon, Dan Mulcahy, Jon Norenberg, Diane Pitassy, Sandra Raredon, Rob Robins, Laureen Schenk, Adriane “Dutch” Schrier, Ian Silver-Gorges, Ashleigh Smythe and Barbara van Bebber for assistance in various ways with this study. Funding for the Smithsonian Institution’s Deep Reef Observation Project was provided internally by the Consortium for Understanding and Sustaining a Biodiverse Planet to CCB, the Competitive Grants for the Promotion of Science program to CCB and DRR, the Herbert R. and Evelyn Axelrod Endowment Fund for systematic ichthyology to CCB, and STRI funds to DRR. Externally the research was funded by National Geographic Society’s Committee for Research and Exploration to CCB (Grant #9102-12). This is Ocean Heritage Foundation/Curacao Sea Aquarium/Substation Curacao (OHF/SCA/SC) contribution number 27.
Appendix 1.
Links between DNA voucher specimens, GenBank accession numbers, and DNA sequences of Lipogramma derived for use in this study.
| Catalog number | Tissue number | Species | GenBank COI | GenBank H3 | GenBank TMO-4C4 | GenBank Rag1 | GenBank Rhodopsin | GenSeq designation |
|---|---|---|---|---|---|---|---|---|
| Photo Voucher Only | BAH10150 | Lipogramma anabantoides | KX713732 | KX713823 | KX713880 | KX713842 | KX713862 | genseq-5 |
| USNM 413759 | BAH9160 | Lipogramma anabantoides | KX713824 | KX713881 | KX713843 | KX713863 | genseq-4 | |
| USNM 420334 | BLZ5340 | Lipogramma anabantoides | KX713733 | – | – | – | – | genseq-4 |
| USNM 414886 | CUR12013 | Lipogramma evides | KX713750 | – | – | – | – | genseq-4 |
| USNM 414889 | CUR12031 | Lipogramma evides | KX713751 | KX713834 | KX713891 | KX713852 | KX713872 | genseq-4 |
| USNM 414883 | CUR12044 | Lipogramma evides | KX713752 | – | – | – | – | genseq-4 |
| USNM 414884 | CUR12050 | Lipogramma evides | KX713753 | – | – | – | – | genseq-4 |
| USNM 414887 | CUR12078 | Lipogramma evides | KX713754 | – | – | – | – | genseq-4 |
| USNM 414890 | CUR12084 | Lipogramma evides | KX713755 | – | – | – | – | genseq-4 |
| USNM 414888 | CUR12116 | Lipogramma evides | KX713757 | KX713835 | KX713892 | KX713853 | KX713873 | genseq-4 |
| USNM 414882 | CUR12118 | Lipogramma evides | KX713758 | – | – | – | – | genseq-4 |
| USNM 414878 | CUR12276 | Lipogramma evides | KX713760 | – | – | – | – | genseq-4 |
| USNM 414881 | CUR12280 | Lipogramma evides | KX713761 | – | – | – | – | genseq-4 |
| USNM 414885 | CUR12281 | Lipogramma evides | KX713762 | KX713837 | KX713894 | KX713855 | KX713875 | genseq-4 |
| USNM 414879 | CUR12288 | Lipogramma evides | KX713763 | – | – | – | – | genseq-4 |
| USNM 414876 | CUR12353 | Lipogramma evides | KX713767 | – | – | – | – | genseq-4 |
| USNM 421602 | CUR13100 | Lipogramma evides | KX713771 | – | – | – | – | genseq-4 |
| USNM 426769 | CUR13233 | Lipogramma evides | KX713779 | – | – | – | – | genseq-4 |
| USNM 426770 | CUR13234 | Lipogramma evides | KX713780 | – | – | – | – | genseq-4 |
| USNM 426771 | CUR13265 | Lipogramma evides | KX713781 | – | – | – | – | genseq-4 |
| USNM 426737 | CUR13266 | Lipogramma evides | KX713782 | – | – | – | – | genseq-4 |
| USNM 426746 | CUR13279 | Lipogramma evides | KX713785 | – | – | – | – | genseq-4 |
| USNM 426709 | CUR13286 | Lipogramma evides | KX713786 | – | – | – | – | genseq-4 |
| USNM 426722 | CUR13294 | Lipogramma evides | KX713787 | – | – | – | – | genseq-4 |
| Photo Voucher Only | CUR15032 | Lipogramma evides | KX713793 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15055 | Lipogramma evides | KX713795 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15057 | Lipogramma evides | KX713796 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15060 | Lipogramma evides | KX713798 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15061 | Lipogramma evides | KX713799 | – | – | – | – | genseq-5 |
| USNM 434771 | CUR15091 | Lipogramma evides | KX713811 | – | – | – | – | genseq-4 |
| USNM 434783 | CUR15103 | Lipogramma evides | KX713813 | – | – | – | – | genseq-4 |
| USNM 434784 | CUR15104 | Lipogramma evides | KX713814 | – | – | – | – | genseq-4 |
| USNM 431313 | TIK003 | Lipogramma evides | KX713822 | – | – | – | – | genseq-4 |
| USNM 422670, paratype | CUR13158 | Lipogramma haberi | KX713775 | – | – | KX713860 | – | genseq-2 |
| USNM 422679, holotype | CUR13171 | Lipogramma haberi | KX713776 | – | – | KX713861 | – | genseq-1 |
| USNM 434772, paratype | CUR15092 | Lipogramma haberi | KX713812 | – | – | – | – | genseq-2 |
| USNM 406013 | CUR11013 | Lipogramma klayi | KX713737 | KX713826 | KX713883 | KX713845 | KX713865 | genseq-3 |
| USNM 406133 | CUR11133 | Lipogramma klayi | KX713740 | KX713828 | KX713885 | KX713847 | KX713867 | genseq-3 |
| USNM 406134 | CUR11134 | Lipogramma klayi | KX713741 | – | – | – | – | genseq-3 |
| USNM 406375 | CUR11375 | Lipogramma klayi | KX713744 | – | – | – | – | genseq-3 |
| USNM 406376 | CUR11376 | Lipogramma klayi | KX713745 | KX713830 | KX713887 | KX713849 | KX713869 | genseq-3 |
| USNM 422669 | CUR13112 | Lipogramma klayi | KX713772 | – | – | – | – | genseq-3 |
| USNM 422676 | CUR13113 | Lipogramma klayi | KX713773 | – | – | – | – | genseq-3 |
| USNM 422690 | CUR13114 | Lipogramma klayi | KX713774 | – | – | – | – | genseq-3 |
| Photo Voucher Only | CUR15064 | Lipogramma klayi | KX713800 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15066 | Lipogramma klayi | KX713801 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15068 | Lipogramma klayi | KX713802 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15069 | Lipogramma klayi | KX713803 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15070 | Lipogramma klayi | KX713804 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15075 | Lipogramma klayi | KX713805 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15076 | Lipogramma klayi | KX713806 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15077 | Lipogramma klayi | KX713807 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15084 | Lipogramma klayi | KX713810 | – | – | – | – | genseq-5 |
| USNM 438687 | DOM16036 | Lipogramma klayi | KX713817 | – | – | – | – | genseq-4 |
| USNM 438741 | DOM16090 | Lipogramma klayi | KX713819 | – | – | – | – | genseq-4 |
| USNM 438742 | DOM16091 | Lipogramma klayi | KX713820 | – | – | – | – | genseq-4 |
| USNM 438803 | DOM16152 | Lipogramma klayi | KX713821 | – | – | – | – | genseq-4 |
| USNM 406011 | CUR11011 | Lipogramma levinsoni | KX713735 | – | – | – | – | genseq-3 |
| USNM 406012 | CUR11012 | Lipogramma levinsoni | KX713736 | – | – | – | – | genseq-3 |
| USNM 406018, paratype | CUR11018 | Lipogramma levinsoni | KX713738 | KX713827 | KX713884 | KX713846 | KX713866 | genseq-2 |
| USNM 406019 | CUR11019 | Lipogramma levinsoni | KX713739 | – | – | – | – | genseq-3 |
| USNM 406139, holotype | CUR11139 | Lipogramma levinsoni | KX713742 | KX713829 | KX713886 | KX713848 | KX713868 | genseq-1 |
| USNM 406140, paratype | CUR11140 | Lipogramma levinsoni | KX713743 | – | – | – | – | genseq-2 |
| USNM 406393, paratype | CUR11393 | Lipogramma levinsoni | KX713747 | KX713832 | KX713889 | KX713851 | KX713871 | genseq-2 |
| USNM 406394 | CUR11394 | Lipogramma levinsoni | KX713748 | – | – | – | – | genseq-3 |
| USNM 426784, paratype | CUR13183 | Lipogramma levinsoni | KX713777 | – | – | – | – | genseq-2 |
| USNM 426754 | CUR13184 | Lipogramma levinsoni | KX713778 | – | – | – | – | genseq-3 |
| USNM 426774 | CUR13267 | Lipogramma levinsoni | KX713783 | – | – | – | – | genseq-3 |
| USNM 426730 | CUR13268 | Lipogramma levinsoni | KX713784 | – | – | – | – | genseq-3 |
| Photo Voucher Only | CUR15031 | Lipogramma levinsoni | KX713792 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15058 | Lipogramma levinsoni | KX713797 | – | – | – | – | genseq-5 |
| USNM 438703 | DOM16052 | Lipogramma levinsoni | KX713818 | – | – | – | – | genseq-4 |
| USNM 406392 | CUR11392 | Lipogramma “robinsi” | KX713746 | KX713831 | KX713888 | KX713850 | KX713870 | genseq-4 |
| USNM 406426 | CUR11426 | Lipogramma “robinsi” | KX713749 | KX713833 | KX713890 | – | – | genseq-4 |
| USNM 414913 | CUR12101 | Lipogramma “robinsi” | KX713756 | – | – | – | – | genseq-4 |
| USNM 414914 | CUR12149 | Lipogramma “robinsi” | KX713759 | KX713836 | KX713893 | KX713854 | KX713874 | genseq-4 |
| USNM 413864 | CUR12290 | Lipogramma “robinsi” | KX713764 | KX713838 | KX713895 | KX713856 | KX713876 | genseq-4 |
| USNM 414911 | CUR12316 | Lipogramma “robinsi” | KX713765 | KX713839 | KX713896 | KX713857 | KX713877 | genseq-4 |
| USNM 414912 | CUR12317 | Lipogramma “robinsi” | KX713766 | KX713840 | KX713897 | KX713858 | KX713878 | genseq-4 |
| USNM 430035 | CUR13329 | Lipogramma “robinsi” | KX713788 | – | – | – | – | genseq-4 |
| USNM 431687 | CUR14079 | Lipogramma “robinsi” | KX713789 | – | – | – | – | genseq-4 |
| USNM 431722 | CUR14114 | Lipogramma “robinsi” | KX713790 | – | – | – | – | genseq-4 |
| USNM 435299 | CUR15012 | Lipogramma “robinsi” | KX713791 | – | – | – | – | genseq-4 |
| USNM 436460 | CUR15125 | Lipogramma “robinsi” | KX713815 | – | – | – | – | genseq-4 |
| USNM 436474 | CUR15139 | Lipogramma “robinsi” | KX713816 | – | – | – | – | genseq-4 |
| Photo Voucher Only | BLZ8127 | Lipogramma trilineata | JQ841643 | – | – | – | – | genseq-5 |
| Photo Voucher Only | BLZ8128 | Lipogramma trilineata | JQ841642 | – | – | – | – | genseq-5 |
| USNM 415245 | BLZ8168 | Lipogramma trilineata | JQ841645 | – | – | – | – | genseq-4 |
| USNM 415298 | BLZ8274 | Lipogramma trilineata | JQ841646 | KX713825 | KX713882 | KX713844 | KX713864 | genseq-4 |
| Photo Voucher Only | BLZ8343 | Lipogramma trilineata | JQ841644 | – | – | – | – | genseq-5 |
| USNM 404204 | BLZWF204 | Lipogramma trilineata | KX713734 | – | – | – | – | genseq-4 |
| USNM 414989 | CUR13082 | Lipogramma trilineata | KX713768 | – | – | – | – | genseq-3 |
| USNM 414990 | CUR13089 | Lipogramma trilineata | KX713769 | – | – | – | – | genseq-3 |
| USNM 414991 | CUR13090 | Lipogramma trilineata | KX713770 | KX713841 | KX713898 | KX713859 | KX713879 | genseq-3 |
| Photo Voucher Only | CUR15034 | Lipogramma trilineata | KX713794 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15078 | Lipogramma trilineata | KX713808 | – | – | – | – | genseq-5 |
| Photo Voucher Only | CUR15079 | Lipogramma trilineata | KX713809 | – | – | – | – | genseq-5 |
Appendix 2.
Specimens of Lipogramma evides examined in this study.
ANSP 134329, holotype, 34.4 mm SL, R/V Pillsbury Sta. 581, Mexico, Arrowsmith Bank, 21°05'N, 86°23'W, 146–265 m depth, 22 May 1967; ANSP 134330, n=2, paratypes, 28.0–32.0 mm SL, R/V Pillsbury Sta. 581, Mexico, Arrowsmith Bank, 21 05'N, 86 23'W, 146–265 m depth, 22 May 1967; ANSP 134331, paratype, 17.2 mm SL, R/V Pillsbury Sta. 969, southeast of Barbuda, 17°27.8'N, 61°41.1'W, 68–216 m depth, 20 July 1969; FMNH 82583, paratype, 34.5 mm SL, Nicaragua, 12°32'N, 82°25 ‘ W, 155 m depth, 23 May 1692; USNM 426801 25.1 mm SL, Curasub submersible, sta. CURASUB13-18, Curaçao, Playa Forti, Westpoint, 12.3679 N, 69.1553 W, no depth data, 15 August 2013, C. Baldwin, B. Brandt, A. Schrier, K. Johnson & C. DeForest; USNM 431410, 12.7 mm SL , Curasub submersible, sta. CURASUB14-07, Curaçao, in between Porto Marie and Daaibooi beaches, 12.202842 N, 69.089507 W, 123 m depth, 21 March 2014, C. Baldwin et al.; USNM 426746, 45.4 mm SL, tissue no. CUR13279, Curasub submersible, sta. CURASUB13-19, Curaçao, Playa Forti, Westpoint, 12.3679 N, 69.1553 W, 179 m depth, 15 August 2013, B. Van Bebber, N. Knowlton, A. Schrier & R. Sant; USNM 431408, 35.5 mm SL, Curasub submersible, sta. CURASUB14-02, Curaçao, off Substation Curaçao downline., 12.083197 N, 68.899058 W, no depth data available, 17 March 2014, B. Brooks et al.; USNM 410992, 43.0 mm SL, Curasub submersible, sta. CURASUB13-33, Caracas Baii and back to Substation Curaçao downline, 12.068 N, 68.873367 W, 215 m depth, 5 November 2013, C. Baldwin, B. Brandt, A. Schrier & C. Castillo; UF 238591, 34.5 mm SL, Curasub submersible, sta. CURASUB15-13, Northwest corner of Klein Curaçao, 11.998453 N, 68.651308 W, 182 m depth, 28 August 2015, C. Baldwin & B. Van Bebber; USNM 434771, 33.3 mm SL, cleared and stained, tissue no. CUR15091, Curasub submersible, sta. CURASUB15-12, northwest corner of Klein Curaçao, 11.998453 N, 68.651308 W, 174 m depth, 27 August 2015, B. Brandt & A. Schrier; USNM 434783, 17.9 mm SL, tissue no. CUR15103, Curasub submersible, sta. CURASUB15-13, Northwest corner of Klein Curaçao, 11.998453 N, 68.651308 W, 171 m depth, 28 August 2015, C. Baldwin & B. Van Bebber; UF 238590, 27.7 mm SL, tissue no. CUR15104, Curasub submersible, sta. CURASUB15-13, Northwest corner of Klein Curaçao, 11.998453 N, 68.651308 W, 172 m depth, 28 August 2015, C. Baldwin & B. Van Bebber; USNM 431313, 39.2 mm SL, tissue no. T1K003, Curasub submersible, sta. CURASUB14-03, Curaçao, west of Substation Curaçao downline, 12.083197 N, 68.899058 W, 177 m depth, 18 March 2014, C. Baldwin et al.; USNM 414886, 24.9 mm SL, tissue no. CUR12013, Curasub submersible, sta. CURASUB12-01, Curaçao, off of Substation Curaçao downline, 12.083197 N, 68.899058 W, 171 m depth, 21 May 2012, C. Baldwin, A. Schrier & B. Brandt; USNM 414889, 31.3 mm SL, tissue no. CUR12031, Curasub submersible, Curaçao, off of Substation Curaçao downline, 12.083197 N, 68.899058 W, no depth data available, 21 May 2012, C. Baldwin et al.; USNM 414883, 40.0 mm SL, tissue CUR12044, Curasub submersible, Curaçao, off of Substation Curaçao downline, 12.083197 N, 68.899058 W, no depth data available, 21 May 2012, C. Baldwin et al.; USNM 414884, 32.0 mm SL, tissue no. CUR12050, Curasub submersible, sta. CURASUB12-11, Curaçao, off of Substation Curaçao downline, 12.083197 N, 68.899058 W, 164 m depth, 6 August 2012, B. Brandt, C. Baldwin, A. Schrier & A. Driskell; USNM 414887, 31.3 mm SL, tissue CUR12078, Curasub submersible, Curaçao, off of Substation Curaçao downline, 12.083197 N, 68.899058 W, no depth data available, 21 May 2012, C. Baldwin et al.; USNM 414890, 40.3 mm SL, tissue no. CUR12084, Curasub submersible, Curaçao, off of Substation Curaçao downline, 12.083197 N, 68.899058 W, no depth data available, 21 May 2012, C. Baldwin et al.; USNM 414888, 39.8 mm SL, tissue no. CUR12116, Curasub submersible, sta. CURASUB12-14, Curaçao, east of downline off Substation dock, 12.083197 N, 68.899058 W, 133 m depth, 9 August 2012, A. Schrier, B. Brandt, C. Castillo, A. Driskell & D. Robertson; USNM 414880, 29.0 mm SL, tissue no. CUR12117, Curasub submersible, sta. CURASUB12-14, Curaçao, east of downline off Substation dock, 12.083197 N, 68.899058 W, 134 m depth, 9 August 2012, A. Schrier, B. Brandt, C. Castillo, A. Driskell & D. Robertson; USNM 414882, 20.1 mm SL, tissue CUR12118, Curasub submersible, sta. CURASUB12-14, Curaçao, east of downline off Substation dock, 12.083197 N, 68.899058 W, 134 m depth, 9 August 2012, A. Schrier, B. Brandt, C. Castillo, A. Driskell & D. Robertson; USNM 414878, 24.1 mm SL, tissue no. CUR12276, Curasub submersible, sta. CURASUB12-16, Curaçao, west to Stella Maris and down, 154 m depth, 13 August 2012, A. Schrier, C. Baldwin & B. Van Bebber; USNM 414881, 21.1 mm SL, tissue no. CUR12280, 21.2 mm SL, Curasub submersible, sta. CURASUB12-17, Curaçao, East of downline off Substation Curaçao dock, 12.083197 N, 68.899058 W, 161 m depth, 13 August 2012, A. Schrier, B. Brandt, C. Castillo & D. Robertson; USNM 414885, 24.4 mm SL, tissue no. CUR12281, Curasub submersible, sta. CURASUB12-17, Curaçao, East of downline off Substation Curaçao dock, 12.083197 N, 68.899058 W, 161 m depth, 13 August 2012, A. Schrier, B. Brandt, C. Castillo & D. Robertson; USNM 414879, 25.0 mm SL, tissue no. CUR12288, Curasub submersible, sta. CURASUB12-16, Curaçao, west to Stella Maris and down, 154 m depth, 13 August 2012, A. Schrier, C. Baldwin & B. Van Bebber; USNM 414876, 41.7 mm SL, tissue no. CUR12353, Curasub submersible, no station data, off Substation Curaçao dock, 12.083197 N, 68.899058 W, no depth data available, 2012, Substation Curaçao crew; USNM 421602, 43.6 mm SL, tissue CUR13100, Curasub submersible, Curaçao, off Substation Curaçao, 12.083197 N, 68.899058 W, no depth data available; USNM 426769, 19.0 mm SL, tissue CUR13233, Curasub submersible, sta. CURASUB13-12, Curaçao, off downline at Substation Curaçao, 12.083197 N, 68.899058 W, 137–173 m depth, 7 August 2013, C. Baldwin, D. Robertson, C. Castillo & B. Van Bebber; USNM 426770, 13.7 mm SL, tissue no. CUR13234, Curasub submersible, sta. CURASUB13-12, Curaçao, off downline at Substation Curaçao, 12.083197 N, 68.899058 W, 137–173 m depth, 7 August 2013, C. Baldwin, D. Robertson, C. Castillo & B. Van Bebber; USNM 426771, 24.9 mm SL, tissue CUR13265, Curasub submersible, sta. CURASUB13-18, Curaçao, Playa Forti, westpoint, 12.3679 N, 69.1553 W, 137–164 m depth, 15 August 2013, C. Baldwin, B. Brandt, A. Schrier, K. Johnson & C. DeForest; USNM 426737, 19.3 mm SL, tissue CUR13266, Curasub submersible, sta. CURASUB13-18, Curaçao, Playa Forti, westpoint, 12.3679 N, 69.1553 W, 137–173 m depth, 15 August 2013, C. Baldwin, B. Brandt, A. Schrier, K. Johnson & C. DeForest; USNM 426709, 40.2 mm SL, tissue no. CUR13286, Curasub submersible, sta. CURASUB13-21, Curaçao, off Substation Curaçao downline, 12.083197 N, 68.899058 W, 171–201 m depth, 17 August 2013, C. Baldwin, A. Schrier, B. Brandt & A. Driskell; USNM 426722, 32.5 mm SL, tissue no. CUR13294, Curasub submersible, sta. CURASUB13-21, Curaçao, off Substation Curaçao downline, 12.083197 N, 68.899058 W, 171–201 m depth, 17 August 2013, C. Baldwin, A. Schrier, B. Brandt & A. Driskell.
Citation
Baldwin CC, Robertson RD, Nonaka A, Tornabene L (2016) Two new deep-reef basslets (Teleostei, Grammatidae, Lipogramma), with comments on the eco-evolutionary relationships of the genus. ZooKeys 638: 45–82. https://doi.org/10.3897/zookeys.638.10455
Supplementary materials
Figure S1
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Carole C. Baldwin, D. Ross Robertson, Ai Nonaka, Luke Tornabene
Data type: Tif image file
Explanation note: Neighbor-joining network based on COI sequences of Lipogramma species investigated in this study. Scale-bar units are substitutions per site.
References
- Ahlstrom EH, Butler JL, Sumida BY. (1976) Pelagic stromateoid fishes (Pisces, Perciformes) of the eastern Pacific: Kinds, distributions, and early life histories and observations on five of these from the northwest Atlantic. Bulletin of Marine Science 26: 285–402. [Google Scholar]
- Asoh K, Yoshikawa T. (1996) Nesting behavior, male parental care, and embryonic development in the fairy basslet, Gramma loreto. Copeia 1996: 1–8. https://doi.org/10.2307/1446936 [Google Scholar]
- Baldwin CC, Johnson GD. (2014) Connectivity across the Caribbean Sea: DNA barcoding and morphology unite an enigmatic fish larva from the Florida Straits with a new species of sea bass from deep reefs off Curaçao. PLoS ONE 9(5): e97661 https://doi.org/10.1371/journal.pone.0097661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin CC, Robertson DR. (2013) A new Haptoclinus blenny (Teleostei, Labrisomidae) from deep reefs off Curacao, southern Caribbean, with comments on relationships of the genus. ZooKeys 306: 71–81. https://doi.org/10.3897/zookeys.306.5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin CC, Robertson DR. (2014) A new Liopropoma sea bass (Serranidae: Epinephelinae: Liopropomini) from deep reefs off Curaçao, southern Caribbean, with comments on depth distributions of western Atlantic liopropomins. ZooKeys 409: 71–92. https://doi.org/10.3897/zookeys.409.7249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin CC, Robertson DR. (2015) A new, mesophotic Coryphopterus goby (Teleostei: Gobiidae) form the southern Caribbean, with comments on relationships and depth distributions within the genus. ZooKeys 513: 123–142. doi: 10.3897/zookeys.513.9998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin CC, Pitassy DE, Robertson DR. (2016) A new deep-reef scorpionfish (Teleostei: Scorpaenidae: Scorpaenodes) from the southern Caribbean with comments on depth distributions and relationships of western Atlantic members of the genus. ZooKeys 606: 141–158. https://doi.org/10.3897/zookeys.606.8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur-R R, Broughton RE, Wiley EO, Carpenter K, López JA, Li C, Holcroft NI, Arcila D, Sanciangco M, Cureton JC, II, Zhang F, Buser T, Campbell MA, Ballesteros JA, Roa-Varon A, Willis S, Borden WC, Rowley T, Reneau PC, Hough DJ, Lu G, Grande T, Arratia G, Ortí G. (2013) The tree of life and a new classification of bony fishes. PLoS Currents Tree of Life. 2013 Apr 18 [last modified: 2013 Apr 18]. Edition 1. https://doi.org/10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288 [DOI] [PMC free article] [PubMed]
- Böhlke JE. (1960) Comments on serranoid fishes with disjunct lateral lines, with the description of a new one from the Bahamas. Notulae Naturae (Philadelphia) 330: 1–11. [Google Scholar]
- Chakrabarty P, Warren M, Page LM, Baldwin CC. (2013) GenSeq: An updated nomenclature and ranking for genetic sequences from type and non-type sources. ZooKeys 346: 29–41. https://doi.org/10.3897/zookeys.346.5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin PL. (1974) Observation and collection of deep-reef fishes off the coasts of Jamaica and British Honduras (Belize). Marine Biology 24: 29–38. https://doi.org/10.1007/BF00402844 [Google Scholar]
- Dingerkus G, Uhler LD. (1977) Enzyme clearing of alcian blue stained whole small vertebrates for demonstration of cartilage. Stain Technol. 52: 229–232. https://doi.org/10.3109/10520297709116780 [DOI] [PubMed] [Google Scholar]
- Eytan RI, Evans BR, Dornburg A, Lemmon AR, Lemmon EM, Wainright PC, Near TJ. (2015) Are 100 enough? Inferring Acanthomorph teleost phylogeny using Anchored Hybrid Enrichment. BMC Evolutionary Biology 15: 113 https://doi.org/10.1186/s12862-015-0415-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore RG. (1997) Lipogramma robinsi, a new basslet from the tropical western Atlantic, with descriptive and distributional notes on L. flavescens and L. anabantoides (Perciformes: Grammatidae). Bulletin of Marine Science 60: 782–788. [Google Scholar]
- Gilmore RG, Jones RS. (1988) Lipogramma flavescens, a new grammid fish from the Bahama Islands, with descriptive and distributional notes on L. evides and L. anabantoides. Bulletin of Marine Science 42: 435–445. [Google Scholar]
- Gill AC, Mooi (1993) Monophyly of the Grammatidae and of the Notograptidae, with evidence for their phylogenetic positions among perciforms. Bulletin of Marine Science 25: 327–350. [Google Scholar]
- Hardy JD. (2006) Chapter 121 – Grammatidae. In: Richards WJ. (Ed.) Early stages of Atlantic fishes: an identification guide for western central North Atlantic, Vol. 2 CRC Press, Boca Raton, FL. [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B 270: 313–322. https://doi.org/10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. (2004) Identification of birds through DNA barcodes. PLoS Biology 2: 1657–1663. https://doi.org/10.1371/journal.pbio.0020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbs CL, Lagler KF. (1947) Fishes of the Great Lakes region. Cranbrook Institute of Science Bulletin No. 26.
- Kimura M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120. https://doi.org/10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2015) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. https://doi.org/10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S. (2012) PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29: 1695–1701. https://doi.org/10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- Leis JM, Bullock S, Duday A, Guion C, Galzin R. (2012) Development of morphology and swimming in larvae of a coral-reef fish, the royal gramma, Gramma loreto (Grammatidae: Teleostei). Scienta Marina 76: 281–288. https://doi.org/10.3989/scimar.03409.03A [Google Scholar]
- Lin H-C, Hastings PA. (2011) Evolution of a Neotropical marine fish lineage (Subfamily Chaenopsinae, Suborder Blennioidei) based on phylogenetic analysis of combined molecular and morphological data. Molecular Phylogenetics and Evolution 60: 236–248. https://doi.org/10.1016/j.ympev.2011.04.018 [DOI] [PubMed] [Google Scholar]
- Lin H-C, Hastings PA. (2013) Phylogeny and biogeography of a shallow water fish clade (Teleostei: Blenniiformes). BMC Evolutionary Biology 13: 210 https://doi.org/10.1186/1471-2148-13-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirande JM. (2016) Combined phylogeny of ray-finned fishes (Actinopterygii) and the use of morphological characters in large-scale analyses. Cladistics 2016: 1–18. https://doi.org/10.1111/cla/12171 [DOI] [PubMed] [Google Scholar]
- Mooi RD, Gill AC. (2002) Grammatidae. In: Carpenter KE. (Ed.) The living marine resources of the western Central Atlantic. Volume 2 Bony fishes part 1 (Acipenseridae to Grammatidae). FAO Species Identification Guide for Fishery Purposes, 1370–1373. [Google Scholar]
- Poey F. (1868) Synopsis piscium cubensium. Catalogo Razonado de los peces de la isla de Cuba. Repertorio Fisico-Natural de la Isla de Cuba v. 2: 279–484. [Google Scholar]
- Polanco A, Acero PA, Mejía-Ladino LM, Mejía LS. (2012) Nuevos registros de peces de las familias Serranidae, Grammatidae y Labridae (Actinopterygii: Perciformes) para el Caribe Colombiano. Boletín de Investigaciones Marinas y Costeras 41: 287–298. [Google Scholar]
- Rambaut A, Drummond AJ. (2007) Tracer. http://beast.bio.ed.ac.uk/Tracer
- Randall JE. (1963) Three new species and six new records of small serranoid fishes from Curaçao and Puerto Rico. Studies on the Fauna of Curacao and other Caribbean Islands 19: 77–110, Pls. 1–3. [Google Scholar]
- Rannala B, Yang Z. (2003) Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164: 1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DR, Van Tassell J. (2015) Shorefishes of the Greater Caribbean: online information system. Version 1.0 Smithsonian Tropical Research Institute, Balboa, Panamá: http://biogeodb.stri.si.edu/caribbean/en/pages [accessed 7.IX.2016] [Google Scholar]
- Robins CR, Colin PL. (1979) Three new grammid fishes from the Caribbean Sea. Bulletin of marine Science 29: 41–52. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DP, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seutin G, White BN, Boag PT. (1991) Preservation of avian blood and tissue samples for DNA analyses. Canadian Journal of Zoology 69(1): 82–90. https://doi.org/10.1139/z91-013 [Google Scholar]
- Stamatakis A. (2014) RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. doi: https://doi.org/10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thresher RE. (1980) Reef fish: behavior and ecology on the reef and in the aquarium. Palmetto Publishing Company, St. Petersburg, FL. [Google Scholar]
- Tornabene L, Van Tassel JL, Robertson DR, Baldwin CC. (2016a) Repeated invasions into the twilight zone: evolutionary origins of a novel assemblage of fishes from deep Caribbean reefs. Molecular Ecology 25: 3662–3682. https://doi.org/10.1111/mec.13704 [DOI] [PubMed] [Google Scholar]
- Tornabene L, Robertson DR, Baldwin CC. (2016b) Varicus lacerta, a new species of goby (Teleostei, Gobiidae, Gobiosomatini, Nes subgroup) from mesophotic reefs in the southern Caribbean. ZooKeys 596: 143–156. https://doi.org/10.3897/zookeys.596.8217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornabene L, Van Tassell JL, Gilmore RG, Robertson DR, Young F, Baldwin CC. (2016c) Molecular phylogeny, analysis of character evolution, and submersible collections enable a new classification of a diverse group of gobies (Teleostei: Gobiidae: Nes subgroup) including nine new species and four new genera. Zoological Journal of the Linnean Society 177: 764–812. https://doi.org/10.1111/zoj.12394 [Google Scholar]
- Wainwright PC, Smith WL, Price SA, Tang KL, Sparks JS, Ferry LA, Kuhn KL, Eytan RI, Near TJ. (2012) The evolution of pharyngognathy: a phylogenetic and functional appraisal of the pharyngeal jaw key innovation in labroid fishes and beyond. Systematic Biology 61: 1001–1027. Systematic biology 61(6): 1001–1027. https://doi.org/10.1093/sysbio/sys060 [DOI] [PubMed] [Google Scholar]
- Weigt LA, Driskell AC, Baldwin CC, Ormos A. (2012) DNA barcoding fishes. Chapter 6 In: Kress WJ, Erickson DL. (Eds) DNA Barcodes: Methods and Protocols, Methods in Molecular Biology 858: 109–126. https://doi.org/10.1007/978-1-61779-591-6_6 [DOI] [PubMed]
- Yang Z. (2015) The BPP program for species tree estimation and species delimitation. Current Zoology 61(5): 854–865. https://doi.org/10.1093/czoolo/61.5.854 [Google Scholar]
- Yang Z, Rannala B. (2010) Bayesian species delimitation using multilocus sequence data. Proceedings of the National Academy of Sciences of United States of America 107: 9264–9269. https://doi.org/10.1073/pnas.0913022107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B. (2014) Unguided species delimitation using DNA sequence data from multiple loci. Molecular Biology and Evolution 31: 3125–3135. https://doi.org/10.1093/molbev/msu279 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Carole C. Baldwin, D. Ross Robertson, Ai Nonaka, Luke Tornabene
Data type: Tif image file
Explanation note: Neighbor-joining network based on COI sequences of Lipogramma species investigated in this study. Scale-bar units are substitutions per site.