Abstract
This paper introduces a statistical estimation framework for magnetic resonance (MR) fingerprinting, a recently proposed quantitative imaging paradigm. Within this framework, we present a maximum likelihood (ML) formalism to estimate multiple parameter maps directly from highly undersampled, noisy k-space data. A novel algorithm, based on variable splitting, the alternating direction method of multipliers, and the variable projection method, is developed to solve the resulting optimization problem. Representative results from both simulations and in vivo experiments demonstrate that the proposed approach yields significantly improved accuracy in parameter estimation, compared to the conventional MR fingerprinting reconstruction. Moreover, the proposed framework provides new theoretical insights into the conventional approach. We show analytically that the conventional approach is an approximation to the ML reconstruction; more precisely, it is exactly equivalent to the first iteration of the proposed algorithm for the ML reconstruction, provided that a gridding reconstruction is used as an initialization.
Index Terms: Maximum likelihood estimation, augmented Lagrangian, variable splitting, variable projection, quantitative magnetic resonance imaging
I. Introduction
Magnetic resonance (MR) fingerprinting [1] provides a new quantitative imaging paradigm for simultaneously obtaining multiple tissue-specific parameters (e.g., T1, T2, spin density) in an efficient imaging experiment. The success of this technique is largely attributed to an ingenious design of incoherent signal excitation and data acquisition schemes. More specifically, it applies a set of randomized flip angles and repetition times to stimulate a sequence of images with randomly varied contrast weightings, contributing to the temporal incoherence; it further uses a set of highly undersampled variable density spiral trajectories to acquire k-space data, leading to the spatial incoherence.
With incoherent data, the conventional MR fingerprinting reconstruction in [1] employs a simple non-iterative procedure: it first performs a gridding reconstruction [2], [3], from which it then estimates unknown parameters by pattern matching. More specifically, given a range of parameters of interest, it constructs a dictionary that contains all possible signal (or magnetization) evolutions simulated from the Bloch equation. Afterwards, voxel-by-voxel pattern matching is performed. This selects a template signal evolution from the dictionary that yields the maximum correlation with the observed signal from the gridding reconstruction. Accordingly, the reconstructed parameters are assigned as those that generate the selected template signal evolution.
Although the conventional reconstruction approach is computationally efficient, it is heuristic in nature, i.e., there is no theoretic optimality associated with the reconstructed parameter maps. Moreover, its practical utility can often be limited by the inherent trade-off between accuracy and acquisition time, especially for applications requiring broad volumetric coverage [4]. Recently, a number of advanced reconstruction approaches, including [5]–[8], have been proposed, resulting in improved accuracy or even higher computational efficiency. For example, in the context of compressed sensing, low-dimensional signal models, including the sparse model [5] and manifold model [6], have been leveraged to enable accurate reconstruction with reduced acquisition time. To enhance the computational efficiency, [7] utilized a low-rank/subspace structure to reduce the dimensionality of the dictionary, whereas [8] developed a group matching strategy for rapid template search.
In this work, we introduce a statistical estimation framework for MR fingerprinting reconstruction, and formulate the reconstruction problem into a maximum likelihood (ML) estimation formalism. To solve the resulting optimization problem, a novel solution algorithm, based on variable splitting (VS) [9]–[11], alternating direction method of multipliers (ADMM) [11]–[13], and variable projection (VARPRO) methods [14]–[18], is described. The proposed algorithm features a unique variable splitting strategy that separates the Bloch equation based physical model from the data model. It allows simultaneous estimation of multiple parameters of interest, in a voxel-by-voxel fashion, through solving a subproblem of the ADMM iteration. This is done by using the VARPRO algorithm within the discrete optimization framework. Results from both simulations and in vivo experiments are shown to demonstrate the improvement in parameter estimation with the proposed approach.
The proposed framework also provides new theoretical insights into the conventional approach. In particular, we bridge the gap between the conventional approach and the statistically principled approach, i.e., ML reconstruction, by showing that with a gridding reconstruction as an initialization, the first iteration of the proposed algorithm for ML estimation exactly produces the conventional approach. With subsequent iterations, the reconstruction can be pushed toward an ML optimum, which has improved estimation performance. A preliminary account of this work was presented in [19].
II. Proposed Approach
A. Formulation
In MR fingerprinting, the contrast-weighted image Im (x) can be parameterized as [6], [20], [21]:
| (1) |
for m = 1, ⋯, M, where ρ(x) denotes the spin density, T1 (x) the longitudinal relaxation time, T2 (x) the transverse relaxation time, and ϕm(·) the contrast-weighting function associated with the mth acquisition parameter (e.g., the flip angle αm and repetition time TRm). Note that ϕm(·) in (1) is determined by the Bloch equation based magnetization dynamics, driven by the acquisition parameters . However, distinct from conventional MR parameter mapping [20], [21], the analytical form of ϕm (·) can be difficult to obtain in MR fingerprinting (though it is feasible to determine ϕm numerically via Bloch simulations).
For simplicity, hereafter we use a discrete image model in which (1) can be represented as
| (2) |
where Im and ρ are N × 1 complex vectors, T1, and T2 are all N × 1 real vectors, and Φm (·) ∈ ℂN×N is a diagonal matrix with [Φm]n,n = ϕm(T1[n], T2[n]). Moreover, under this setting, the imaging equation can be written as
| (3) |
for m = 1, ⋯, M, and c = 1, ⋯, Nc, where dm,c ∈ ℂP, and nm,c ∈ ℂP respectively contain the measured k-space data and measurement noise from the cth receiver coil at TRm, Fm ∈ ℂP×N denotes the undersampled Fourier encoding matrix, Sc ∈ ℂN×N is a diagonal matrix with the diagonal entries containing the coil sensitivities associated with the cth receiver coil, and Nc denotes the total number of receiver coils. Note that (3) includes both single-channel acquisitions (i.e., Nc = 1) and multichannel acquisitions (i.e., Nc > 1) as the special cases. For multichannel acquisitions, we have the following assumptions: 1) is not time-varying; and 2) is known or can be accurately pre-estimated (e.g., via a pilot scan).
Substituting (2) into (3), we have the following data model:
| (4) |
which describes the data generating process, from the underlying tissue MR parameters, to magnetization dynamics, to measured data. Equation (4) is a nonlinear model, due to the dependency of magnetization dynamics on {T1,T2} and the multiplicative coupling between Φm (T1,T2) and ρ. With respect to the noise, we assume: 1) for a fixed m and c, nm,c can be modeled as additive complex white Gaussian noise; 2) nm,c1 is correlated with nm,c2, if c1 ≠ c2; and 3) nm1,c1 is independent of nm2,c2, whenever m1 ≠ m2. For multichannel acquisitions, we further assume that the noise covariance matrix is known or can be accurately pre-estimated (e.g., via a dummy scan), with which we can pre-whiten the colored Gaussian noise [22].
The goal of MR fingerprinting reconstruction is to estimate {T1,T2, ρ} from highly undersampled, noisy k-space data . Given the above data model and noise characteristics, the reconstruction problem can be formulated as the following ML estimation problem:
| (5) |
where d̃m,c and S̃c respectively denote the measured data and sensitivity maps after noise prewhitening. Note that (5) enables direct parameter estimation from highly undersampled k-space data, as opposed to the conventional approach in which unknown parameters are estimated from severely aliased images generated by the gridding reconstruction. For notational simplicity, hereafter we use θ = {T1,T2} to denote the unknown parameters in Φm (·).
B. Solution Algorithm
Equation (5) results in a large-scale nonlinear least-squares problem. There are a number of key challenges associated with solving this problem:
The cost function in (5) is a nonconvex, implying that the success of an iterative algorithm can critically depend on its initialization.
Equation (5) optimizes over a large set of variables that are at very distinct scales. This leads to difficulty of applying a generic nonlinear optimization method, such as the nonlinear conjugate gradient (CG) method [23], which often needs careful variable rescaling (e.g., [21], [24]).
The lack of an analytical expression for ϕm(·) makes it cumbersome to evaluate the cost function value and gradient in iterative algorithms, since each iteration involves Bloch simulations for all voxels, which can be computationally expensive.
Here we present a novel solution algorithm, integrating a variable splitting strategy, the alternating direction method of multipliers, and the variable projection method, to combat these difficulties. The main idea is to employ a special VS scheme and the ADMM method to convert (5) into a series of well-understood optimization problems that admit efficient solutions. In the following, we describe the proposed algorithm in detail.
First, we introduce a set of auxiliary variables to split the parametric image model in (2) from the data consistency term in (5). For multichannel acquisitions, we also introduce another set of auxiliary variables, , to split the coil sensitivities [11]. As a consequence, (5) can be converted into the following equivalent constrained optimization problem:
| (6) |
for m = 1, ⋯, M, and c = 1, ⋯, Nc.
Second, we apply the ADMM method [11]–[13] to solve (6). Specifically, the associated augmented Lagrangian function can be formed as follows:
| (7) |
where xm,c and ym are the Lagrangian multipliers, μg, μh > 0 are the penalty parameters, and Re takes the real part of a complex number. We iteratively minimize (7) by solving the following subproblems:
| (8) |
| (9) |
| (10) |
and updating the Lagrangian multipliers:
until the solution change between consecutive iterations is less than the tolerance ε, or the maximum number of iterations Kmax is reached. Next, we describe the detailed solutions to (8)–(10).
1) Solution to (8)
The subproblem in (8) can be written as
| (11) |
This is a nonlinear least-squares fitting problem, which is decoupled for each voxel, i.e.,
| (12) |
for n = 1, ⋯,N, where ρn and θn respectively denote the nth element of ρ and θ, z(θn) = [ϕ1(θn), ⋯, ϕM(θn)]T, , and gm,n and ym,n denote the nth element of gm and ym, respectively.
Equation (12) has a separable structure, i.e., with a fixed value of θ, it reduces to a linear least-squares problem with respect to ρ. A class of algorithms based on variable projection (VARPRO) [14]–[18] can efficiently solve this type of problems. The VARPRO method expresses the update for { } as follows1:
| (13) |
and
| (14) |
Note that it can be shown that solving (13) and (14) together can yield the same set of optimal solutions as (12) (see [15] for more details). With VARPRO, ρn is separated from the optimization problem (12), resulting in a simpler problem in (13). This problem basically determines a set of parameters for the nth voxel, which generates magnetization dynamics having the maximum correlation with . Although (13) is more amenable to generic nonlinear optimization methods, applying these methods still involves Bloch simulations, which can be time consuming. Inspired by [16]–[18] and the idea from MR fingerprinting [1], here we bypass this issue by reformulating (13) as a discrete optimization problem. More specifically, we discretize the feasible search space of θn into a finite set of parameters , with which we use Bloch simulations to generate a dictionary that contains all possible signal evolutions, i.e., . After that, a global optimization of (13) can be performed via a grid search, i.e.,
| (15) |
Note that the above scheme requires constructing the dictionary only once, which avoids repeating Bloch simulations for every voxel at each iteration.
2) Solution to (9)
The subproblem in (9) is a linear least-squares problem, which is decoupled for each gm. It is equivalent to solving the following normal equation:
| (16) |
where
Noting that is a diagonal matrix, (16) can be efficiently solved by an entry-by-entry inversion.
Algorithm 1.
Proposed algorithm for (5).
3) Solution to (10)
The subproblem in (10) is also a linear least-squares problem, which is decoupled for each hm,c. Its solution can be obtained by solving
| (17) |
where . Note that for arbitrary k-space trajectories, the Gram matrix , or the coefficient matrix of (17), has a block Toeplitz structure [25]. A number of off-the-shelf numerical solvers taking advantage of this structure enables very efficient solution of (17), including the pre-conditioned CG solvers (e.g., [26], [27]) and hierarchically semiseparable (HSS) solvers (e.g., [28], [29]).
4) Algorithm initialization
Considering that (5) is a non-convex optimization problem, the solution of the algorithm generally depends on the initialization. Here, we initialize with the gridding reconstruction (or coil-combined gridding reconstruction for the multichannel case), , and . Note that this initialization scheme is always feasible, and also very efficient to implement [3]. As will be demonstrated later, it consistently leads to good performance for the algorithm.
From a modeling/formulation standpoint, this initialization scheme eventually bridges the gap between the conventional MR fingerprinting reconstruction and ML reconstruction. In fact, the derivations in (11)–(15) for VARPRO has already yielded a matched filter based solution similar to the conventional approach. Here, given the above initialization (i.e., an appropriate ), the proposed algorithm for ML estimation will exactly produce the conventional approach at its first iteration. Generally, the use of additional iterations can drive the solution toward an ML optimum, which has a number of desirable properties [30, Ch.7], such as asymptotic unbiasedness and asymptotic efficiency. In the next section, we will illustrate some of the benefits with concrete examples.
5) Computational complexity
It is worth analyzing the computational complexity of the proposed algorithm. For clarity, we summarize the overall algorithm in Algorithm 1. With the VS strategy and ADMM method, the original problem in (5) boils down to iteratively solving one parameter fitting problem and two structured linear least-squares problems. The parameter fitting problem in (8) is solved by the discrete VARPRO-based method on a voxel-by-voxel basis. For each voxel, this can be done in 𝒪(MQ) operations. Note that higher computational efficiency can be achieved by further integrating with the dictionary compression [7] or group search strategy [8]. The linear least-squares problem in (9) is decoupled for each time frame. Solving each subproblem only involves inverting a diagonal matrix, which can be done in 𝒪(N) operations. Regarding the linear least-squares problem in (10), it is decoupled with respect to each time frame and each coil, and each subproblem can be solved by the HSS solver in 𝒪(N log2N) operations [28].
III. Results
In this section, representative results from both simulations and in vivo experiments are shown to illustrate the performance of the proposed approach.
A. Simulation Results
A numerical brain phantom was designed to simulate single-channel MR fingerprinting experiments, in which the T1, T2, and spin density maps were taken from the Brainweb database [31], as shown in Fig. 1. The Bloch simulations were performed to generate a sequence of contrast-weighted images using the inversion recovery fast imaging with steady state precession (IR-FISP) sequence2 in which the same set of flip angles and repetition times as in [32] were applied. For simplicity, we assume that the flip angle is uniform over the field of view. We set the acquisition matrix size as 256×256, and the field-of-view (FOV) as 300 × 300 mm2. The same set of spiral trajectories as in [32] was used, and the k-space data were synthesized with the nonuniform fast Fourier transforms [33]. One spiral interleaf was acquired for each image frame (the fully sampled data consists of 48 interleaves). Complex white Gaussian noise was added to the measured data with the signal-to-noise ratio (SNR) defined as 20 log10(s/σ), where s denotes the average signal intensity within a region of the white matter in the first contrast-weighted image, and σ denotes the standard deviation of the noise. For convenience, we use the number of TRs, as denoted by M, to measure the length of a MR fingerprinting acquisition, which corresponds to the total acquisition time .
Fig. 1.
Ground truth for the numerical phantom: (a) T1 map, (b) T2 map, and (c) spin density map.
We performed reconstructions using the conventional approach and the proposed ML approach. The gridding reconstruction involved in both approaches were performed on a 2X oversampled Cartesian grid, with an optimized Kaiser-Bessel convolution kernel [3] and Voronoi density compensation. Moreover, we used the same dictionary for both approaches based on the following parameter discretization scheme: 1) the possible T1 values were set within [100, 5000] ms, with an increment of 10 ms for the range [100, 3000] ms and an increment of 200 ms for the range [3001, 5000] ms; and 2) the possible T2 values were set within [1, 500] ms, with an increment of 5 ms. Additionally, we manually selected the penalty parameter to optimize the performance of the proposed ML reconstruction. Lastly, to assess the reconstruction accuracy, the following error metrics are used: 1) voxelwise relative error = |θn −θ̂n|/θn, where θn and θ̂n denote the true and reconstructed parameter at the nth voxel, respectively; and 2) overall error = ||θ − θ̂||2/||θ||2.3
1) General evaluation
We first demonstrate the benefit of the ML reconstruction in terms of improving the accuracy of parameter estimation. Specifically, we simulated a MR fingerprinting experiment with the acquisition length M = 300 and SNR = 40 dB, and performed reconstructions using the above two approaches. Fig. 2 (a), (c), and (e) respectively show the plots of overall error versus the number of iterations for the T1, T2, and spin density reconstructions using the ML reconstruction (the first iteration of the ML reconstruction produces the conventional approach). As can be seen, with the iterative procedure, the ML reconstruction leads to better accuracy than the conventional approach for all the parameter maps, where the improvements for the T2 and spin density reconstructions are more pronounced. Furthermore, we show the reconstructed parameter maps and associated error maps produced at different number of iterations of the ML reconstruction in Fig. 2 (b), (d), and (f). This not only confirms the quantitative improvements shown before, but also illustrates how the iterative procedure refines the results qualitatively. In particular, notice that as the iteration progresses, the aliasing-like artifacts appearing in the T2 and spin density maps are significantly reduced.
Fig. 2.
Evaluation of the ML reconstruction at different number of iterations with the acquisition length M = 300. (a) Overall error versus iteration number for T1. (b) Reconstructed T1 maps and associated relative error maps. (c) Overall error versus iteration number for T2. (d) Reconstructed T2 maps and associated relative error maps. (e) Overall error versus iteration number for spin density. (f) Reconstructed spin density maps and associated relative error maps. For each error map, the overall error is labeled at the lower right corner, and the regions associated with the background, skull, and scalp are set to be zero.
2) Influence of acquisition length
We have also evaluated the performance of the ML reconstruction with different acquisition lengths. Fig. 3 shows the reconstructed T1, T2, and spin density maps by the two approaches at M = 250, 500, and 1000. As expected, both approaches improve with the increase of acquisition length. However, note that the ML reconstruction not only outperforms the conventional approach for all three acquisition windows, but also exhibits better robustness to the change of acquisition length. It is worth noting that the ML reconstruction with M = 500 yields similar accuracy to the conventional approach with M = 1000, demonstrating its capability of reducing acquisition time. In Fig. 4, we plot the overall error versus acquisition length, which further confirms the benefit offered by the proposed ML approach.
Fig. 3.
Evaluation of the conventional approach and ML reconstruction with different acquisition lengths. (a)–(c): Reconstructed T1 maps and associated relative error maps. (d)–(f): Reconstructed T2 maps and associated relative error maps. (h)–(j): Reconstructed spin density maps and associated relative error maps. For each error map, the overall error is labeled at the lower right corner, and the regions associated with the background, skull, and scalp are set to be zero.
Fig. 4.
Overall error versus acquisition length for (a) T1, (b) T2, and (c) spin density maps from the conventional approach and ML reconstruction.
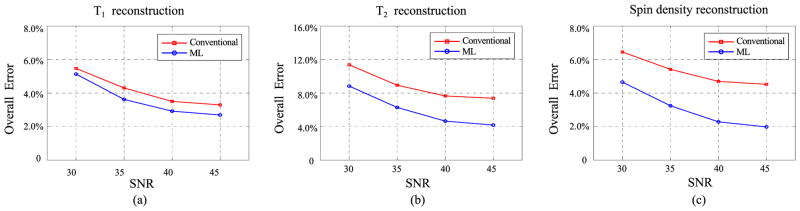
3) Influence of SNR
We have also evaluated the performance of the ML reconstruction at different noise levels. Specifically, we simulated the experiments at SNR = 30, 35, 40, and 45 dB, all of which were with M = 500. We performed reconstructions using the conventional approach and ML reconstruction. Fig. 5 shows the plots of overall error versus SNR for the reconstructed T1, T2, and spin density maps. It can be seen that the ML reconstruction consistently yields better accuracy than the conventional approach for all tested SNR levels.
Fig. 5.
Overall error versus SNR for (a) T1, (b) T2, and (c) spin density maps from the conventional approach and ML reconstruction.
4) Bias-variance analysis
We have also investigated the statistical properties of the proposed approach. Specifically, we conducted Monte Carlo (MC) simulations (with 100 trials) for the conventional approach and ML reconstruction at SNR = 35dB and M = 500. To compare the bias, variance, and mean square error at the same scale, the following normalized quantities are calculated for the nth voxel:
-
1)normalized bias:
-
2)normalized variance:and 3) normalized root-mean-square error:
where 𝔼̂(·) denotes the empirical mean evaluated for the MC simulations. Note that . Fig. 6 shows the maps of NBias, NVar, and NRMSE from the two reconstruction approaches. As can be seen, the ML reconstruction has significantly reduced bias compared to the conventional approach, which is consistent with the theoretical property that the ML estimation is asymptotically unbiased [30]. Moreover, it results in a similar level of variance to the conventional approach, except for the region of cerebrospinal fluid in the T2 map and spin density map. But note that these regions are generally not of interest for neuroimaging. Lastly, by taking into account both the bias and variance, the ML reconstruction leads to improved NRMSE over the conventional approach.
Fig. 6.
Normalized bias, variance, and root-mean-square error for (a) T1, (b) T2, and (c) spin density estimation using the conventional approach and ML reconstruction at SNR = 35 dB and M = 500. Note that the regions associated with the background, skull, and scalp are set to be zero.
B. In Vivo Results
In this subsection, we show representative results of applying the proposed approach to in vivo MR fingerprinting experiments. Specifically, the experiments were performed on a 3T Siemens Tim Trio scanner (Siemens Medical Solutions, Erlangen, Germany) equipped with a 32-channel head coil. One healthy human subject was scanned using the IR-FISP sequence with the approval of the Institutional Review Board. The same sets of acquisition parameters and spiral trajectories as in [32] were used in the protocol, and one spiral interleaf was acquired for each acquisition parameter. Other relevant imaging parameters include: FOV = 300 × 300 mm2, matrix size = 256×256, and slice thickness = 5 mm. Two slices were acquired with the acquisition lengths of 1400 and 1000 TRs, respectively. For each slice, we further collected a set of fully sampled data as reference by repeating the above acquisition multiples times, all with the same set of acquisition parameters, . But, in each repetition, we switched to a different spiral interleaf to acquire data for {(αm, TRm)} until all the interleaves had been collected.4 Here, to ensure that the magnetization returns to thermal equilibrium, a short delay was given before consecutive repetitions. Subsequently, we combined the measured data from different repetitions to form a set of fully sampled data for each {(αm, TRm)}. Additionally, we performed pilot scans with a rapid gradient echo (GRE) sequence, from which the coil sensitivity maps were estimated using the subspace method [34]. We also measured the actual spiral trajectories [35], rather than using the theoretical ones, to avoid the potential trajectory distortion caused by eddy currents and other hardware imperfections.
We performed the conventional MR fingerprinting reconstruction and ML reconstruction using the same set of data from a single acquisition. The same dictionary, as described in the simulations, was used for both approaches. We initialized the ML reconstruction with the coil combined gridding reconstruction, and manually selected the penalty parameters μg and μh for the optimized performance. Additionally, we reconstructed the contrast-weighted images from the fully sampled data, and then estimated the parameter maps as the underlying references.
For slice 1, we performed reconstructions with two acquisition lengths, i.e., M = 700 and 1400. Note that for M = 700, the data were retrospectively taken from the first half of the entire acquisition with 1400 TRs. Fig. 7 shows the T1, T2, and spin density maps obtained from the reference, conventional reconstruction, and ML reconstruction. Here, we also show the relative error map and overall error associated with each reconstruction for quantitative assessment. Clearly, both approaches yield improved performance, as the number of measurements doubles. However, the ML reconstruction outperforms the conventional approach for both acquisition lengths. Also, note that the ML reconstruction with M = 700 results in even higher accuracy than the conventional approach with M = 1400. In Fig. 8, we further show the relative error maps associated with the ML approach at different iterations with M = 700, illustrating how the accuracy was progressively improved. With respect to slice 2, we performed reconstructions at the original acquisition length, i.e., M = 1000. The related results are shown in the supplementary material, which further illustrates the performance of the proposed approach.
Fig. 7.
In vivo reconstructions for slice 1 with two acquisition lengths. (a) Reference T1 map. (b)–(c): Reconstructed T1 maps and associated relative error maps. (d) Reference T2 map. (e)–(f): Reconstructed T2 maps and associated relative error maps. (g) Reference spin density map. (g)–(h): Reconstructed spin density maps and associated relative error maps. For each error map, the overall error is labeled at the lower right corner, and the regions associated with the background, skull, and scalp are set to be zero.
Fig. 8.
Relative error maps associated with the ML reconstruction at different number of iterations. Here the acquisition length is M = 700. The three rows respectively show the relative error maps associated with the T1, T2, and spin density reconstructions. For each error map, the overall error is labeled at the lower right corner, and the regions associated with the background, skull, and scalp are set to be zero.
We also report the computational efficiency of the two reconstruction approaches, which were ran on a Linux workstation with 24 Intel Xeon E5-2643, 3.40 GHz processors and 128 GB RAM using Matlab R2013b. For both approaches, the dictionary for IR-FISP was pre-generated, which took about 14 min based on the extended phase graph formalism [32], [36]. Note that the dictionary only needs to be generated once, and then it can be saved for subsequent experiments. In terms of reconstruction, for the above in vivo data set, the conventional approach (including the gridding reconstruction and dictionary pattern matching) took about 1.5 min, whereas the ML reconstruction took about 32 min. Given the highly separable algorithmic structure, we expect that the computation time of the ML reconstruction could be substantially reduced with an optimized implementation on GPU. This is beyond the scope of this paper, but is worthwhile to explore in future work.
IV. Discussion
The effectiveness of the proposed ML reconstruction has been demonstrated for MR fingerprinting. It is worth making further comments on some points. First of all, it has been observed that both the conventional approach and ML reconstruction yield relatively accurate T1 estimates, even when the acquisition time is short. This can be attributed to the design of the imaging sequence that applies an inversion preparation pulse at the beginning of acquisition, stimulating the T1 recovery-like dynamics that dominate the early portion of signal evolutions (e.g., the first 200 TRs). Such type of magnetization dynamics typically exhibits higher sensitivity to the change of T1 values, thus resulting in more accurate T1 estimation.
The proposed formulation is based on the data model in (4), the fidelity of which impacts the accuracy of parameter estimation. In this work, we used the simple data model to illustrate the effectiveness of the ML-based formalism, although more sophisticated models could be adopted. For example, we could model the partial volume effect occurring at tissue interfaces by including more complex signal evolutions into the dictionary (e.g., [37]). Additionally, for certain applications, it may be desirable to incorporate an appropriate motion model (e.g., [38]) into (4) to compensate motion during acquisitions. However, these extensions generally lead to complex data models with more degrees of freedom, which often incurs higher SNR penalty for parameter estimation. Thus, their effectiveness depends on application-specific tradeoffs between model mismatch and noise contamination, which should be evaluated on a case-by-case basis.
Similar to the conventional approach [1], [32], this work assumes for simplicity that the transmit radiofrequency field (i.e., ) is homogeneous over the field of view. For in vivo brain imaging experiments at 3T, in which inhomogeneity is often not severe, such an assumption generally does not incur significant bias in parameter estimation. But for body imaging [39] or ultra-high field imaging [40], [41], strong B1 inhomogeneity can result in severe bias for estimated parameter maps (especially for T2 maps). In these scenarios, it is necessary to incorporate a spatially dependent into the signal model (or signal evolution dictionary). To obtain , we can either conduct auxiliary scans (e.g., [42]) or perform joint estimation with other parameters of interest, although the latter may suffer from amplified variance in parameter estimation.
The proposed ADMM-based algorithm requires the selection of the penalty parameters μg and μh. For the nonconvex optimization problem in (5), we empirically observed that the solution of the algorithm did depend on the choice of penalty parameters5. In this work, we manually chose these parameters, with the reference to the fully sampled data, to optimize the performance of the algorithm. For practical applications, a useful way is to perform parameter selection based on a set of fully sampled calibration data acquired from a physical phantom (with a similar range of T1 and T2 values to the study), and then apply the selected parameters to subsequent experiments using the same imaging protocol and hardware configurations. Some preliminary investigations indicate the effectiveness of this approach, although it is worth studying the optimal parameter selection from a theoretical standpoint.
Statistical estimation provides a powerful framework to address the MR fingerprinting reconstruction problem. Within this framework, this paper described an ML approach for parameter estimation, and also proved that the conventional approach is equivalent to the first iteration of the proposed algorithm for ML estimation. There are a number of useful extensions that can be further pursued. For example, we can derive the estimation-theoretic bounds (e.g., [20], [21]) to characterize the performance of ML estimation. Also, these bounds can be utilized to optimally design MR fingerprinting experiments. Such an extension is under investigation, and will be reported in a subsequent work. Furthermore, we can incorporate a prior information on and/or {ρ,θ} (e.g., [43]–[45]) to derive a penalized ML formulation for improved performance. A preliminary investigation along this line is presented in [46].
The proposed solution algorithm effectively tackles several key challenges of the ML reconstruction. It also provides a new way to address the computational problem associated with direct parameter estimation from k-space data arising in a variety of quantitative imaging applications, including the conventional MR parameter mapping6 (e.g., [20], [24]), quantitative diffusion imaging (e.g., [53], [54]), water-fat imaging (e.g., [55]), and contrast-enhanced imaging (e.g., [56]), which are worthwhile to explore in future research. Despite the empirical success, little is known about the convergence property of the proposed algorithm for solving the nonconvex optimization problem in (5), although the behavior of ADMM is well understood in the context of convex optimization [12]. Recently, there has been active research on analyzing alternating minimization algorithms for solving structured nonconvex optimization problems (e.g., [57]–[60]), which may provide useful insights into the theoretical characterization of the convergence of the proposed algorithm.
V. Conclusion
This work presented a maximum likelihood approach to MR fingerprinting reconstruction. A novel algorithm, integrating VS, the ADMM method, and the VARPRO method, was described to efficiently solve the resulting optimization problem. The proposed algorithm for the ML reconstruction includes the conventional approach as its first iteration, while rapidly improving reconstruction quality with additional iterations. The advantages offered by the proposed approach, including improving parameter estimation accuracy and reducing acquisition time, have been demonstrated with both simulations and in vivo experiments.
Acknowledgments
This work was supported in part by the following research grants: NIH-R01-EB017219, NIH-R01-EB017337, NIH-R01-EB013695, NIH-P41-EB015896, NIH-U01-MH093765, NIH-R00-EB012107, and NIH-R24-MH106096.
The authors thank Drs. Mark Griswold, Dan Ma, and Yun Jiang at Case Western Reserve University for their help on the implementation of MR fingerprinting acquisition methods.
Footnotes
Unlike the original inversion recovery balanced steady state free precession sequence in [1], IR-FISP is not sensitive to the off-resonance effect [32], making it immune to banding artifacts.
The regions associated with the background, skull, scalp, and cerebrospinal fluid were not included into the overall error calculation, given that they are not regions of interest for our study.
For example, to collect a set of fully sampled data consisting of 48 spiral interleaves, the standard MR fingerprinting acquisition with needs to be performed 48 times. For the in vivo experiment with M = 1000, the standard acquisition took 13.1 sec, whereas the fully sampled acquisition took 18.3 min. Here, a delay of 10 sec was given between consecutive acquisitions.
In contrast, for convex optimization problems, the ADMM penalty parameters only affect the convergence speed rather than the optimal solution, as long as certain mild conditions are satisfied (see [12], [13] and references therein for more details).
In this context, the proposed approach has key differences from the existing ones (e.g., [20], [21], [24], [45], [47]–[51]). From a formulation viewpoint, it performs parameter estimation directly from undersampled k-space data. This is distinct from the two-step approach (e.g., [45], [47], [50], [52]), which first reconstructs contrast-weighted images with various constraints (e.g., sparsity and/or low-rank constraint) and then performs parameter estimation in the spatial domain. From an algorithmic viewpoint, it features a novel primal-dual based alternating minimization algorithm, as opposed to the generic nonlinear programming based algorithms (e.g., [20], [21], [24], [48]) and standard alternating minimization algorithms (e.g., [49], [51]).
Contributor Information
Bo Zhao, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA 02129 USA, and also with the Department of Radiology, Harvard Medical School, Boston, MA 02115 USA. The initial part of this work was done while he was with the Department of Electrical and Computer Engineering and Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL 61801 USA.
Kawin Setsompop, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA 02129 USA, and also with the Department of Radiology, Harvard Medical School, Boston, MA 02115 USA.
Huihui Ye, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA 02129 USA, and also with the Department of Radiology, Harvard Medical School, Boston, MA 02115 USA.
Stephen Cauley, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA 02129 USA, and also with the Department of Radiology, Harvard Medical School, Boston, MA 02115 USA.
Lawrence L. Wald, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA 02129 USA, and also with the Department of Radiology, Harvard Medical School, Boston, MA 02115 USA, and also with the Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139 USA.
References
- 1.Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature. 2013;495:187–192. doi: 10.1038/nature11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Sullivan JD. A fast sinc function gridding algorithm for Fourier inversion in computer tomography. IEEE Trans Med Imaging. 1985;4(4):200–207. doi: 10.1109/TMI.1985.4307723. [DOI] [PubMed] [Google Scholar]
- 3.Jackson JI, Meyer CH, Nishimura DG, Macovski A. Selection of a convolution function for Fourier inversion using gridding [computerised tomography application] IEEE Trans Med Imaging. 1991;10(3):473–478. doi: 10.1109/42.97598. [DOI] [PubMed] [Google Scholar]
- 4.Ye H, Ma D, Jiang Y, Cauley SF, Du Y, Wald LL, Griswold MA, Setsompop K. Accelerating magnetic resonance fingerprinting (MRF) using t-blipped simultaneous multislice (SMS) acquisition. Magn Reson Med. doi: 10.1002/mrm.25799. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Zhang Q, Yuan J, Wang X. MRF denoising with compressed sensing and adaptive filtering. Proc IEEE Int Symp Biomed Imaging. 2014:870–873. [Google Scholar]
- 6.Davies M, Puy G, Vandergheynst P, Wiaux Y. A compressed sensing framework for magnetic resonance fingerprinting. SIAM J Imaging Sciences. 2014;7:2623–2656. [Google Scholar]
- 7.McGivney D, Pierre E, Ma D, Jiang Y, Saybasili H, Gulani V, Griswold MA. SVD compression for magnetic resonance fingerprinting in the time domain. IEEE Trans Med Imaging. 2014;33(12):2311–2322. doi: 10.1109/TMI.2014.2337321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cauley SF, Setsompop K, Ma D, Jiang Y, Ye H, Adalsteinsson E, Griswold MA, Wald LL. Fast group matching for MR fingerprinting reconstruction. Magn Reson Med. 2015;74:523–528. doi: 10.1002/mrm.25439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afonso MV, Bioucas-Dias JM, Figueiredo MAT. Fast image recovery using variable splitting and constrained optimization. IEEE Trans Image Process. 2010;19(9):2345–2356. doi: 10.1109/TIP.2010.2047910. [DOI] [PubMed] [Google Scholar]
- 10.Lingala SG, DiBella E, Jacob M. Deformation corrected compressed sensing (DC-CS): A novel framework for accelerated dynamic MRI. IEEE Trans Med Imaging. 2015;34(1):72–85. doi: 10.1109/TMI.2014.2343953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramani S, Fessler JA. Parallel MR image reconstruction using augmented Lagrangian methods. IEEE Trans Med Imaging. 2011;30(3):694–706. doi: 10.1109/TMI.2010.2093536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyd S, Parikh N, Chu E, Peleato B, Eckstein J. Distributed optimization and statistical learning via the alternating direction method of multipliers. Found Trends Mach Learn. 2011;3:1–122. [Google Scholar]
- 13.Yang J, Zhang Y. Alternating direction algorithms for ℓ1-problems in compressive sensing. SIAM J Sci Comput. 2011;33:250–278. [Google Scholar]
- 14.Golub GH, Pereyra V. The differentiation of pseudo-inverses and nonlinear least squares problems whose variables separate. SIAM J Numer Anal. 1973;10:413–432. [Google Scholar]
- 15.Golub GH, Pereyra V. Separable nonlinear least squares: the variable projection method and its applications. Inverse Probl. 2003;19:R1–R26. [Google Scholar]
- 16.Haldar JP, Anderson J, Sun SW. Maximum likelihood estimation of T1 relaxation parameters using VARPRO. Proc Int Symp Magn Reson Med. 2007:41. [Google Scholar]
- 17.Hernando D, Haldar JP, Sutton BP, Ma J, Kellman P, Liang Z-P. Joint estimation of water/fat images and field inhomogeneity map. Magn Reson Med. 2008;59:571–580. doi: 10.1002/mrm.21522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trzasko JD, Mostardi PM, Riederer SJ, Manduca A. Estimating T1 from multichannel variable flip angle SPGR sequences. Magn Reson Med. 2013;69:1787–1794. doi: 10.1002/mrm.24401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao B, Lam F, Bilgic B, Ye H, Setsompop K. Maximum likelihood reconstruction for magnetic resonance fingerprinting. Proc IEEE Int Symp Biomed Imaging. 2015:905–909. [Google Scholar]
- 20.Haldar JP, Hernando D, Liang Z-P. Super-resolution reconstruction of MR image sequences with contrast modeling. Proc IEEE Int Symp Biomed Imaging. 2009:266–269. [Google Scholar]
- 21.Zhao B, Lam F, Liang ZP. Model-based MR parameter mapping with sparsity constraints: Parameter estimation and performance bounds. IEEE Trans Med Imaging. 2014;33(9):1832–1844. doi: 10.1109/TMI.2014.2322815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: Sensitivity encoding for fast MRI. Magn Reson Med. 1999;42:952–962. [PubMed] [Google Scholar]
- 23.Nocedal J, Wright SJ. Numerical Optimization. 2. New York, NY: Springer; 2006. [Google Scholar]
- 24.Sumpf TJ, Uecker M, Boretius S, Frahm J. Model-based nonlinear inverse reconstruction for T2 mapping using highly undersampled spin-echo MRI. J Magn Reson Imaging. 2011;34:420–428. doi: 10.1002/jmri.22634. [DOI] [PubMed] [Google Scholar]
- 25.Wajer F, Pruessmann KP. Major speedup of reconstruction for sensitivity encoding with arbitrary trajectories. Proc Int Symp Magn Reson Med. 2001:767. doi: 10.1002/mrm.1241. [DOI] [PubMed] [Google Scholar]
- 26.Chan TF. An optimal circulant preconditioner for Toeplitz systems. SIAM J Sci Stat Comp. 1988;9:766–771. [Google Scholar]
- 27.Chan RH, Ng MK. Conjugate gradient methods for Toeplitz systems. SIAM Rev. 1996;38:427–482. [Google Scholar]
- 28.Xi Y, Xia J, Cauley SF, Balakrishnan V. Superfast and stable structured solvers for Toeplitz least squares via randomized sampling. SIAM J Matrix Anal Appl. 2014;35:44–72. [Google Scholar]
- 29.Cauley SF, Xi Y, Bilgic B, Xia J, Adalsteinsson E, Balakrishnan V, Wald LL, Setsompop K. Fast reconstruction for multichannel compressed sensing using a hierarchically semiseparable solver. Magn Reson Med. 2015;73:1034–1040. doi: 10.1002/mrm.25222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kay SM. Fundamentals of Statistical Signal Processing: Estimation Theory. I Upper Saddle River, NJ: Printice Hall; 1993. [Google Scholar]
- 31.Collins DL, Zijdenbos AP, Kollokian V, Sled JG, Kabani NJ, Holmes CJ, Evans AC. Design and construction of a realistic digital brain phantom. IEEE Trans Med Imaging. 1998;17(3):463–468. doi: 10.1109/42.712135. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y, Ma D, Seiberlich N, Gulani V, Griswold MA. MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout. Magn Reson Med. 2015;74:1621–1631. doi: 10.1002/mrm.25559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fessler JA, Sutton BP. Nonuniform fast Fourier transforms using min-max interpolation. IEEE Trans Signal Process. 2003;51(2):560–574. [Google Scholar]
- 34.Uecker M, Lai P, Murphy MJ, Virtue P, Elad M, Pauly JM, Vasanawala SS, Lustig M. ESPIRiT-an eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA. Magn Reson Med. 2014;71:990–1001. doi: 10.1002/mrm.24751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan H, Meyer CH. Estimation of k-space trajectories in spiral MRI. Magn Reson Med. 2009;61:1396–1404. doi: 10.1002/mrm.21813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hennig J. Multiecho imaging sequences with low refocusing flip angles. J Magn Reson. 1988;78:397–407. [Google Scholar]
- 37.Hamilton JI, Deshmane A, Hougen S, Griswold M, Seiberlich N. Magnetic resonance fingerprinting with chemical exchange (MRF-X) for quantification of subvoxel T1, T2, volume fraction, and exchange rate. Proc Int Symp Magn Reson Med. 2015:239. [Google Scholar]
- 38.Du H, Lam F. Compressed sensing MR image reconstruction using a motion-compensated reference. Magn Reson Imaging. 2012;30:954–963. doi: 10.1016/j.mri.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, Jiang Y, Ma D, Wright KL, Seiberlich N, Griswold MA, Gulani V. Magnetic resonance fingerprinting (MRF) for rapid quantitative abdominal imaging. Proc Int Symp Magn Reson Med. 2014:561. [Google Scholar]
- 40.Jiang Y, Ye H, Bilgic B, Ma D, Witzel T, Cauley SF, Adalsteinsson E, Setsompop K, Griswold MA, Wald LL. Simultaneous T1 and T2 quantification of the human brain at 7 Tesla by MR fingerprinting. Proc Int Symp Magn Reson Med. 2015:3199. [Google Scholar]
- 41.Cloos MA, Wiggins C, Wiggins G, Sodickson D. Plug and play parallel transmission at 7 and 9.4 Tesla based on principles from MR fingerprinting. Proc Int Symp Magn Reson Med. 2014:542. [Google Scholar]
- 42.Yarnykh VL. Actual flip-angle imaging in the pulsed steady state: A method for rapid three-dimensional mapping of the transmitted radiofrequency field. Magn Reson Med. 2007;57:192–200. doi: 10.1002/mrm.21120. [DOI] [PubMed] [Google Scholar]
- 43.Zhao B, Haldar JP, Brinegar C, Liang Z-P. Low rank matrix recovery for real-time cardiac MRI. Proc IEEE Int Symp Biomed Imaging. 2010:996–999. [Google Scholar]
- 44.Zhao B, Haldar JP, Christodoulou AG, Liang ZP. Image reconstruction from highly undersampled (k, t)-space data with joint partial separability and sparsity constraints. IEEE Trans Med Imaging. 2012;31(9):1809–1820. doi: 10.1109/TMI.2012.2203921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao B, Lu W, Hitchens TK, Lam F, Ho C, Liang Z-P. Accelerated MR parameter mapping with low-rank and sparsity constraints. Magn Reson Med. 2015:489–498. doi: 10.1002/mrm.25421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao B. Model-based iterative reconstruction for magnetic resonance fingerprinting. Proc IEEE Int Conf Image Process. 2015:3392–3396. [Google Scholar]
- 47.Doneva M, Börnert P, Eggers H, Stehning C, Sénégas J, Mertins A. Compressed sensing reconstruction for magnetic resonance parameter mapping. Magn Reson Med. 2010;64:1114–1120. doi: 10.1002/mrm.22483. [DOI] [PubMed] [Google Scholar]
- 48.Sumpf TJ, Petrovic A, Uecker M, Knoll F, Frahm J. Fast T2 mapping with improved accuracy using undersampled spin-echo MRI and model-based reconstructions with a generating function. IEEE Trans Med Imaging. 2014;33(12):2213–2222. doi: 10.1109/TMI.2014.2333370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tran-Gia J, Stäb D, Wech T, Hahn D, Köstler H. Model-based acceleration of parameter mapping (MAP) for saturation prepared radially acquired data. Magn Reson Med. 2013;70:1524–1534. doi: 10.1002/mrm.24600. [DOI] [PubMed] [Google Scholar]
- 50.Zhang T, Pauly JM, Levesque IR. Accelerating parameter mapping with a locally low rank constraint. Magn Reson Med. 2015;73:655–661. doi: 10.1002/mrm.25161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi C, Zhou Y, Wang Y, Lyu J, Liang D, Ying L. Recovery of parametric manifold from reduced measurements: Aapplication to magnetic resonance parameter mapping. Proc IEEE Int Symp Biomed Imaging. 2015:901–904. [Google Scholar]
- 52.Feng L, Otazo R, Jung H, Jensen JH, Ye JC, Sodickson DK, Kim D. Accelerated cardiac T2 mapping using breath-hold multiecho fast spin-echo pulse sequence with k-t FOCUSS. Magn Reson Med. 2011;65:1661–1669. doi: 10.1002/mrm.22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu Y, Wu Y, Zheng Y, Wu EX, Ying L, Liang D. A model-based method with joint sparsity constraint for direct diffusion tensor estimation. Proc IEEE Int Symp Biomed Imaging. 2012:510–513. [Google Scholar]
- 54.Welsh CL, DiBella EV, Adluru G, Hsu EW. Model-based reconstruction of undersampled diffusion tensor k-space data. Magn Reson Med. 2013;70:429–440. doi: 10.1002/mrm.24486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doneva M, Börnert P, Eggers H, Mertins A, Pauly J, Lustig M. Compressed sensing for chemical shift-based water-fat separation. Magn Reson Med. 2010;64:1749–1759. doi: 10.1002/mrm.22563. [DOI] [PubMed] [Google Scholar]
- 56.Guo Y, Zhu Y, Lingala SG, Lebel RM, Nayak K. Highly accelerated brain DCE MRI with direct estimation of pharmacokinetic parameter maps. Proc Int Symp Magn Reson Med. 2015:573. [Google Scholar]
- 57.Netrapalli P, Jain P, Sanghavi S. Phase retrival using alternating minimization. Adv Neural Inf Process Syst. 2013:2796–2804. [Google Scholar]
- 58.Hardt M. Understanding alternating minimization for matrix completion. Proc IEEE 55th Annu Symp Found Comput Sci. 2013:665–674. [Google Scholar]
- 59.Qu Q, Sun J, Wright J. Finding a sparse vector in a subspace: Linear sparsity using alternating directions. Adv Neural Inf Process Syst. 2014:3401–3409. [Google Scholar]
- 60.Wang Y, Yin W, Zeng J. Global convergence of ADMM in nonconvex nonsmooth optimization. UCLA CAM Report. 2015:15–62. [Google Scholar]