Abstract
Modern mass spectrometry (MS) technologies have provided a versatile platform that can be combined with a large number of techniques to analyze protein structure and dynamics. These techniques include the three detailed in this chapter: 1) hydrogen/deuterium exchange (HDX), 2) limited proteolysis, and 3) chemical crosslinking (CX). HDX relies on the change in mass of a protein upon its dilution into deuterated buffer, which results in varied deuterium content within its backbone amides. Structural information on surface exposed, flexible or disordered linker regions of proteins can be achieved through limited proteolysis, using a variety of proteases and only small extents of digestion. CX refers to the covalent coupling of distinct chemical species and has been used to analyze the structure, function and interactions of proteins by identifying crosslinking sites that are formed by small multi-functional reagents, termed crosslinkers. Each of these MS applications is capable of revealing structural information for proteins when used either with or without other typical high resolution techniques, including NMR and X-ray crystallography.
Part 1: Hydrogen/deuterium exchange
1.1 Introduction
Protein functions commonly rely on conformational changes within the protein. In some cases these conformational changes include large sections or entire domains of the protein. In other cases, protein conformational changes are restricted to small specific regions of the protein. Extensive conformational changes are associated with protein folding immediately during or after their synthesis in vivo, when they fold to acquire their native conformational structure. Knowledge of the location of functionally relevant conformational changes within the protein and the magnitude and rates of conformational interconversion among various protein conformations (i.e. dynamics) are of great importance to the understanding of protein function.
Direct or indirect evidence of protein conformational changes have been deduced through the use of several spectroscopic techniques, including circular dichroism, electron paramagnetic resonance, intrinsic protein fluorescence, UV-Vis and IR spectroscopy, and it is not uncommon to use a combination of these techniques to obtain a general description of the structure and dynamics of the protein system under consideration. Measurements of protein dynamics traditionally have been done by determining 15N NMR relaxation times and calculating S2, the average order parameter, a measure of the motion of the N-H vector at peptide amide linkages. Higher order parameters indicate less freedom of movement. Motions measured by these techniques are on the pico- to nano-second time scale but may also indicate if slower motions might be occurring. To fully understand the dynamics of a particular protein, it is desirable to span as wide a time range as possible.
Hydrogen exchange is a well-understood phenomenon, and in conjunction with mass spectrometry (MS) is a useful method for studying protein dynamics and structure. This exchange was first discovered in the early 1950s by Kaj Ulrik Linderstrom-Lang and Aase Hvidt, scientists at the Carlsberg Laboratory in Copenhagen. They discovered that both the polar side chain hydrogens and the peptide group hydrogens undergo continual exchange with the hydrogens from the solvent. Using density gradient tubes, Lang and Hvidt developed a novel method to measure this exchange of amide backbone hydrogens with a heavier isotope, deuterium (1,2). With this method, they were able to show that the newly discovered α-helices and β-sheets in native proteins do indeed have the proposed hydrogen-bonded backbone structures (1,3). Despite having extremely limited resolution and accuracy at this time, Lang and Hvidt were able to derive equations and propose mechanisms that are still being used today in hydrogen/deuterium exchange (HDX) methodologies (1).
During the following 40 years, many developments were made using hydrogen exchange in conjunction with different techniques, including NMR, tritium gel-filtration, and circular dichroism. Some of these advances include showing that the chemical nature of adjacent side chains has a major effect on the exchange rate (4), measuring the rates of both acid- and base-catalyzed exchange (5), developing protein fragmentation and HPLC separation methods (6), and site-resolved HDX (7). Finally, in 1991 Katta and Chait showed that HDX could be used with electrospray ionization mass spectrometry, removing many of the limitations associated with applications of HDX, including the size of the protein that can be studied (8). With the use of MS to analyze the exchange, the use of HDX to study protein structure continues to advance, with the development of faster and more automated software for both analyzing data and running samples (9), and cold boxes for HPLC to maintain low temperatures during injection to avoid back exchange (10). As a result, the size and type of proteins being studied with HDX, as well as the number of people employing this method, continue to grow.
Recently, HDX in combination with MS has been used to characterize protein movements in solution over a time range from milliseconds to several hours. This technique has become increasingly popular to characterize conformational changes and the dynamic transitions between the conformations in proteins. The purpose of this chapter is to describe the procedures and methods for HDX MS to novice users. Thus, we will first describe a basic methodology and a simple experimental set up. Then, we will discuss alternative workflows, caveats and potential problems, and complementary techniques.
HDX MS is a method in which deuterium atoms present in buffer replace hydrogen atoms in the protein (11–16). Of all the hydrogen atoms present in a protein polypeptide, only hydrogen atoms in O-H, N-H and S-H groups can be replaced with deuterium atoms present in the buffer. As a further limitation, only those present in the amide linkages can be measured by HDX MS (all others exchange too rapidly during sample handling to be detected by mass spectrometry). The amino acid sequences of peptides (and thereby their locations in the protein) and their mass (and thereby the identification of which peptides undergo deuterium exchange) are detected by enzymatic (most often pepsin) digestion of the protein into peptides and peptide mass evaluation by MS. The total number of exchangeable protons and the rate of exchange events are both dependent on the equilibrated protein conformational average and the rate of dynamic transitions between conformations. Therefore, HDX MS is a sensitive technique for evaluating both changes in average conformation of the peptide backbone chain and changes in its dynamics.
A number of attributes of HDX MS make it ideal for evaluating macromolecular systems. 1) Mass spectrometry requires low concentrations of protein. This can remove some of the ambiguity at higher protein concentrations (such as those required for many NMR studies). 2) Deuterium-labeling of a protein results in the introduction of multiple reporting groups (one reporter/protein residue) with minimum structural modification of the protein. This results in higher resolution information compared to many other techniques. 3) The number of exchanging protons can be determined. Each proton that is exchanged for a deuteron adds 1 atomic mass unit (amu) to the average molecular mass of a protein. Thus, the increase in the mass determines the number of deuterium incorporated. 4) By observing the isotopic pattern for a given protein or peptide fragment (discussed in detail below), HDX MS can distinguish between localized unfolding events (referred to as EX2 kinetics and seen as a binomial isotope pattern) and more global, or cooperative unfolding events (referred to as EX1 kinetics and seen as a bimodal isotopic pattern). 5) There is no upper limit to the size of the macromolecule that can be analyzed by HDX MS analysis. This is due to the fact that for detailed analysis of deuterium content in specific regions (peptides) of the protein, the protein is proteolyzed before mass analysis. 6) Measurements are for proteins in solution with no dependency on crystal growth, as is required for X-ray crystallography. 7) As mentioned above, protein dynamics can be probed on a much longer time scale than is accessible with many other techniques (e.g. NMR relaxation). HDX MS can probe dynamics ranging from milliseconds to several hours, and perhaps longer. As a result, HDX MS can increase significantly the overall description of dynamic motions within a protein.
1.2 Theoretical basis and experimental design for HDX MS
The theory and methodology used to study protein conformation and dynamics using HDX MS have been described in several reviews (12,16–20). In the absence of secondary structure restraints, HDX for a specific polypeptide is dependent on the temperature and pH of the reaction. The most common experimental procedure for HDX is continuous labeling. In this method, the exchange is initiated by making a large dilution of a concentrated stock of the protein into deuterated buffer. The progress of the exchange reaction is sampled at different times. Under these conditions, the chemistry and the thermodynamic parameters of HDX are well established (21–23). The rate of HDX at the protein amide linkages is acid or base catalyzed, and can be expressed as follows:
| Equation 1 |
Thus, the rate of HDX for a specific polypeptide is dependent on the pH and temperature of the reaction. This rate, as determined experimentally, has a minimum in the pH range 2.3–2.5. Figure 1 illustrates the theoretical rates of HDX for rat liver mitochondrial aspartate aminotransferase, a 49,000 Da globular protein, in the absence of secondary structure restraints, calculated at 0°C and at both pH 7.5 and 2.3. At pH 7.5 HDX is very fast (t1/2= 0.014 min) and the exchange is completed almost instantly. However, there is a minimum exchange rate at pH 2.4. At this pH, minutes are required before complete exchange occurs. This sensitivity of exchange rates to pH requires careful control of pH during exchange. However, the same pH sensitivity provides a tool to quench exchange by quickly lowering temperature and pH, a step necessary during mass analysis.
Figure 1. Theoretical rates of hydrogen/deuterium exchange of mitochondrial aspartate aminotransferase.
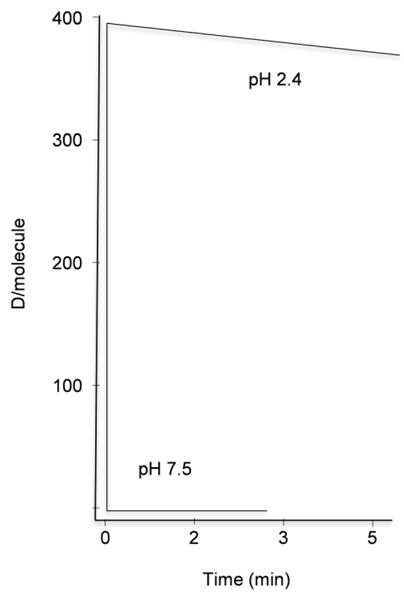
The theoretical rate of HDX at 0°C and pH 7.5 or 2.4 was calculated for mitochondrial aspartate aminotransferase (MW 44,597 Da) according to a previously published algorithm (22) using HXPEP, written and kindly provided by Zhongqi Zhang (Amgen, Thousand Oaks, CA).
In the absence of any structural constraints, the hydrogen atoms of solvent exposed amide linkages exchange at their free, unmodified rates. However, if the amide hydrogen atoms are involved in stable internal hydrogen-bonding, or are not exposed to solvent, they will exchange more slowly. In native proteins, the local differences in these rates are due to the fact that the structure of these molecules is not rigid, but has a certain degree of mobility. This mobility has been called “breathing”, and can be visualized as shown in Figure 2. The kinetics of HDX can be described according to the following kinetic equation:
| Equation 2 |
where kcl, kop and ke are the constants of closing, opening and chemical hydrogen/deuterium exchange, respectively. For proteins in their native state, a common assumption is that kcl >> kop and ke >> kop.
Figure 2. Schematic representation of the mechanism of hydrogen/deuterium exchange.
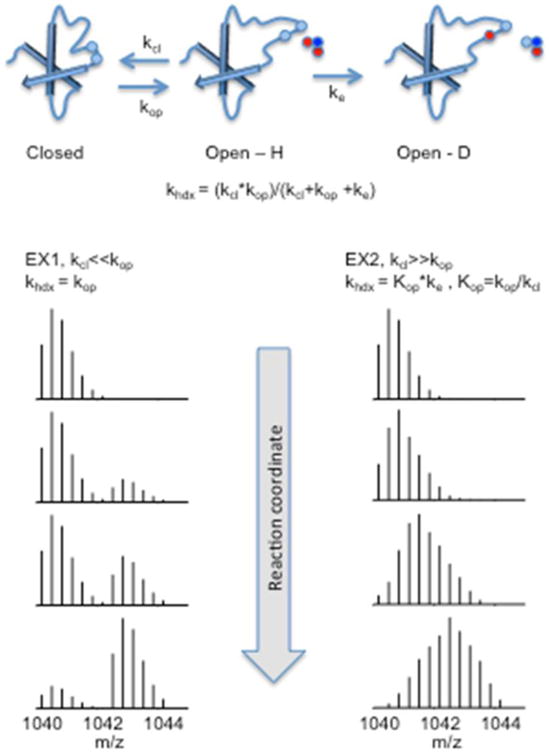
Hydrogen atoms in the peptide backbone (top panel) can exchange with hydrogen (blue) or deuterium (red) atoms in water in a process dependent on accessibility and breathing (opening and closing) of the protein. In the EX1 regime (left panel), opening is faster than closing (kop>>kcl), and the rate of exchange is determined by the rate constant of opening. In the EX2 regime (right panel), closing is faster than opening (kcl>>kop), and the reaction is dependent on the rate of opening and chemical exchange. The isotopic patterns shows the theoretical exchange pattern of a triply charged peptide with m/z = 1040.08 under EX1 or EX2 exchange regimes.
Depending on the relative values of the kinetic constants, two extreme kinetic behaviors can be found. When kop << kcl the exchange rate is determined by the first order rate constant kop. Thus, khdx is dependent exclusively on the conformation of the protein. This first extreme behavior is defined as EX1. EX1 kinetics are rarely observed. However, EX1 exchange can be observed under experimental conditions that favor the unfolded state (24,25) of proteins (high temperature or in the presence of chaotropic or unfolding reagents). On a mass spectrometer, EX1 is characterized by a binomial transition from one mass (i.e. undeuterated) to the final (deuterated) species (Figure 2). In other words, two isotopic envelopes are detected, one for the undeuterated peptide-ion and a second one for the fully deuterated ion. The relative intensity of these two isotopic envelopes changes over time as the exchange reaction proceeds.
In contrast to the conditions that define EX1, when kcl >> kop, the khdx is second order and depends exclusively on the factors determining the chemical hydrogen/deuterium exchange. In this case the rate of exchange is measured by khdx = kopke. This second extreme behavior is defined as EX2. The EX2 mechanism is most commonly observed for proteins in the folded state. EX2 behavior is characterized by a monotonic change of the isotopic envelope with the progress of the exchange reaction (Figure 2). The EX1 kinetic mechanism reflects the activation energy for segmental opening and the EX2 represents the sum of all energies of opening and proton transfer. In EX2, the free energy difference (ΔG0) of the opening event can be described according to the following equation:w
| Equation 3 |
where Kop is the equilibrium constant of the opening/closing reaction (Figure 2).
Based on these concepts, most HDX MS experimental designs rely on two different stages (14): exchange and quenching. In the first stage, reaction conditions (i.e. pH and temperature) are designed to allow HDX while the protein undergoes normal folding/function. In the second stage, the HDX is quenched by rapidly decreasing the temperature (to 0°C or below) and pH (to pH 2.3–2.5).
1.3 Equipment
Cooling HPLC interface. To reduce back exchange during mass analysis of the intact protein or its peptides, all experimental steps after HDX are performed at low pH and temperature. The simplest instrumental set-up consists of immersing the solvents, columns and all parts of an HPLC in an ice bath, or enclosing the entire HPLC set-up in a refrigerated chamber. For better control of temperature, we designed a Semi-Automatic Interface for Deuterium Exchange (SAIDE, Figure 3) (10). This interface consists of a TVC –S2 box (Mecour) equipped with a 6-port valve (Cheminert, N60 SS) and a 4-port valve (Cheminert, C2). The 6-port valve is equipped with a through-the-handle external loop injector and holds the sample loop (10 μL). The sample loop acts as the reaction vessel during protease digestion. The reversed phase column bridges the two valves, and the 4-port valve directs flow to either waste or to the mass spectrometer. Other specialized equipment is available that performs automatic sample pick up, mixing, injection and data acquisition, although at a considerable expense (18).
High performance liquid chromatograph (HPLC). The system should be able to deliver flows between 20 and 50 μL/min. We use a quaternary HPLC MS pump (ThermoFisher Scientific).
Chromatographic columns. A reversed phase C8 (MicroTech Scientific, Zorbax C8 SB Wide Pore Guard Column 2.5 cm × 0.2 cm) is needed to desalt the protein when measuring global rate of the exchange in the intact protein. As an alternative, a reversed phase C4 may be required to desalt highly hydrophobic proteins (MicroTech Scientific, Zorbax C4 SB Wide Pore Guard Column 2.5 cm × 0.2 cm). A reversed phase C18 column (MicroTech Scientific, Zorbax C18 SB Wide Pore Guard Column 2.5 cm × 0.2 cm) is needed to resolve peptic peptides and identify regions with deuterium incorporation.
Mass spectrometer. The mass spectrometers useful for HDX MS characterization of macromolecular complexes are Tandem Mass Spectrometers. That is, those that allow for at least two different stages of mass analysis: one to scan for the peptide-ions (parent ions) present in the sample, and the second to scan for the fragment ions produced after a specific parent ion has undergone a stage of fragmentation (see Section II: Mass Spectrometry). A high resolving power mass spectrometer is recommended, such as an ICR FT or Orbitrap. However, other mass spectrometers with lower resolving power have been used. Because of the high flows used for peptide elution, the ESI tip must be chosen carefully. A 100 μm ID tip with an opening of 30μm has proved to be ideal for our experimental set-up.
Figure 3. Mass spectrometer rigged for HDX MS.

A) The cooling box (SAIDE) is located right before the ESI source of a high resolving mass spectrometer (LTQ FT) and after the HPLC pumps (HPLC). B) Detail of the SAIDE box showing the internal components of the unit: two valves, one reversed phase column, loop and fluid lines. The box is used for temperature control during all stages of protein digestion, peptide desalting and chromatographic elution of peptides.
1.4 Materials
Protein or protein system of interest.
Pepsin. Make a pepsin (Worthington) stock solution by diluting an appropriate weighed amount of pepsin and dilute it in 200 mM ammonium formate, pH 2.3 at a final protein concentration of 1.6 mg/ml. Pepsin concentration can be estimated from its absorbance at 280 nm using a 1% absorptivity coefficient of 1.4.
Protein stock buffer. A buffer appropriate for your particular protein system.
Deuteration buffers. Buffers adequate for your protein system made in D2O (99.9% D2O, ACROS Organics). Note that a correction factor must be introduced when measuring pH of deuterated buffers to account for the differences in activity of protium vs. deuterium: pH = pD + 0.4.
Quench buffer. Quench buffer is 200 mM NH4CH3COOH, pH 2.3, ice cold. Other buffer composition can be used (ammonium phosphate). Note in some cases it might be required to supplement the quench buffer with a low amount of a denaturing or chaotropic agent (i.e., 0.6 M guanidine hydrochloride) to achieve full unfolding of the protein and efficient pepsin digestion.
HPLC solvents. Two solvents are needed to create a gradient. Solvent A is 0.05% trifluoroacetic acid in H2O (TFA, MS grade). Solvent B is 0.05% TFA acid in acetonitrile.
1.5 Experimental procedure
Figure 4A outlines the procedures involved in a continuous labeling experiment. Usually, a stock solution of the protonated protein is diluted into a deuterated buffer and the direction of the exchange is H-> D (on-exchange). Figure 4B outlines also the reverse procedure (off-exchange), when a protein is first fully exchanged with deuterium, and the exchange reaction proceeds in the opposite direction. This method has been used to study the reversible unfolding of a protein. The procedure outlined below describes the steps involved in a continuous labeling, on-exchange procedure. Other experimental procedures are possible, however, and the particular design will depend on the question of interest.
Figure 4. HDX MS general experimental procedure.

The scheme shows the steps to perform continuous labeling HDX on-exchange (A) or off-exchange (B) experimental procedures. HDX is initiated by making a dilution of a concentrated stock of the protein into a deuterated buffer. At different time points the reaction is sampled by taking an aliquot and measuring the mass of the intact protein (global exchange) or of the proteolytic fragments (deuterium level in peptides) with the aid of a mass spectrometer.
1.5.1 Initiate exchange reaction
The exchange reaction is initiated by making a 1:10 (or higher) dilution of a concentrated stock solution of the protein or protein system of interest into a buffer made in D2O.
At different time points, the exchange reaction is sampled by taking an aliquot. Two mass measurements can be made: the global rate of exchange in the intact protein (see 1.5.2. Global rate of exchange) or rate of exchange in pepsin generated peptides (see 1.5.3 Location of deuterium exchange along the peptide backbone).
1.5.2 Global rate of exchange
To obtain a global rate of exchange, the change in mass of the protein at different times following the initiation of the exchange reaction is measured. For the measurement of the mass of the intact protein, mass analysis is performed by direct injection of an aliquot of the labeling reaction mixture on a C4 or C8 nano-column. Following desalting at 0–15% B at high flow, the protein is eluted using a step gradient of acetonitrile (0 to 60% B in B+A in 15 min) and analyzed on-line by mass spectrometry.
1.5.3 Location of deuterium exchange along the peptide backbone
To identify the residues involved in the hydrogen/deuterium exchange reaction, it is first required to identify the peptides resulting from the proteolysis of the protein. This first stage is performed under control conditions; that is, in the absence of deuterium in the buffers but under identical conditions to be used to measure the exchange. This results in a list of peptides of interest. Then, the experiment is repeated under the exchange conditions using deuterated buffers.
-
Peptic mass maps
The first step is to make a dilution (1:10 to 1:20) of the protein stock in the protonated buffer. This dilution is equivalent to the dilution that will be made later in deuterated buffer to initiate the exchange reaction.
Peptic digestions of the protein are performed by making a second 1:10 dilution of an aliquot of the protein in ice cold 200 mM ammonium formate (pH 2.3) containing pepsin at a final protein:protease ratio of 1:1 (w:w). Note that the ratio protein:protease must be optimized experimentally.
Inject the reaction sample immediately into the loop of the 6-port valve on the SAIDE interface.
Allow pepsin digestion to proceed for 2–5 min (time of digestion must be optimized for each protein).
Switch the 6-port valve, start HPLC gradient. The resulting enzyme digest is desalted on a C18 nanocolumn at 75–100 μL/min for 2 min while the flow on the 4-port valve is diverted to waste.
Following desalting, switch the 4-port valve to direct the flow to the mass spectrometer ESI source for peptide detection.
Elute peptides using a 2 to 40% gradient of 0.05% TFA acetonitrile in 0.05% TFA in 15 min. The peptides are detected on-line using a high resolving power mass spectrometer. Figure 5 shows a representative elution profile of a peptide digest using our chromatographic system. The MS settings must have been optimized for detection of peptides using high flow mobile phase. Data are acquired under automatic control to perform MS followed by tandem mass scans of the four to six most intense ions, using an exclusion list of 2–4 min, depending on the capabilities of your mass spectrometer and chromatographic system.
-
Measurement of deuterium content in peptides
The exchange reaction is initiated as indicated above, using deuterated buffer instead of the protonated buffer.
At different times during the exchange reaction, remove an aliquot and dilute it in the quench buffer in the presence of pepsin, as before (see sections 1.5.3.A.b–g).
MS analysis is performed as above with the exception that the mass spectrometer is operated to perform mass analysis only (no data dependence).
Figure 5. Representative chromatographic profile and data analysis.
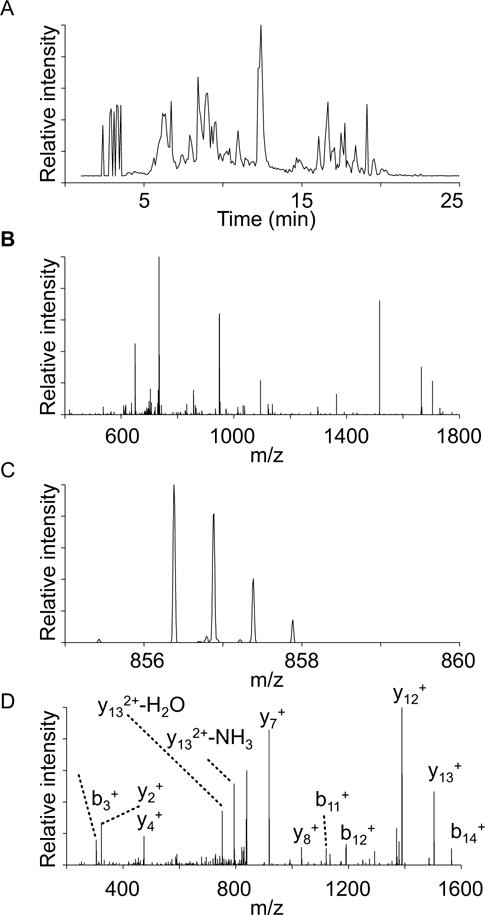
A) Base line chromatographic profile of a peptide digest of mitochondrial aspartate aminotransferase. B) Mass spectrum at 8.5 min of elution. C) A magnification of the mass spectrum of panel B, showing the doubly charged ion with m/z of 856.5 corresponding to the peptic peptide AHNPTGTDPTEEEWK. D) Tandem mass spectrum of the same peptide; to simplify the figure only the most prominent b- and y- ions are indicated.
1.6 Data analysis
1.6.1 Peptide identification
When working with pure proteins, as is the case in HDX MS, statistical tools for False Discovery Rate (FDR) and peptide/protein probabilities calculation are, as a general rule, not useful. Instead, peptide identification is based on parameters that rely on the quality of the tandem mass spectra. When data are acquired on a high resolving power mass spectrometer and Proteome Discoverer is used to analyze them, peptide identifications are made using an in-house protein database. This database includes the protein of interest, pepsin and common contaminant protein sequences. The databased is made assuming that pepsin has no specificity, using a fragment ion mass tolerance of 20 ppm, and a parent ion tolerance of 0.30 Da. Peptide identifications are accepted if they can be established at Xcorr score of at least 1.5, 2.0 or 2.5 for peptides with 1, 2 or 3 charges, respectively, with a ΔCorrelation score larger than 0.08. Note, manual inspection and validation of some tandem mass spectra may be required. See Chapter 8 for more information on tandem mass spectrometry peptide/protein sequencing and identification.
1.6.2 Deuterium content
The change in deuterium content is measured as the change in mass of the deuterated and undeuterated averaged masses of the protein. Many software packages can be used, and usually the instrument manufacturer will provide a program to obtain this measurement. Specialized software is recommended. HDExaminer (Sierra Analytics) is a commercial software that performs automatic isotopic envelope isolation, measurement of the average mass and deuterium content of the peptides, and can plot the results in a variety of formats, including the comparison of multiple states of a protein. There are, however, several free tools for the same purpose: HDXFinder (26), HD desktop (27) and its successor HDX Workbench (9), HX Express (28), Hexicon (29,30) and MagTran (31), among others.
1.6.3 Mathematical analysis
- Curve fitting – Equation 2 describes the exchange reaction for a single amide linkage. In theory, one could expect one phase per amide linkage. However, in practice, multiple protons in the peptide might exchange and individual rate constants of exchange cannot be measured. In practice, the exchange reaction is fitted to an exponential rise (on-exchange) or decay (off-exchange):
Where Dt is the deuterium content at time t, Ai and ki are the amplitude and the rate constant for the ith phase. In practice, multiple HDX reactions are grouped into fast, medium and slow phases (n=3).Equation 4 Use of overlapping peptides – Because pepsin has low selectivity for cleavage site, pepsin digestion results in the production of multiple overlapping peptides. Statistical and logical analysis of the deuterium content of these overlapping peptides can provide higher spatial resolution than that obtained at the peptide level. Some of the programs mentioned above will apply logical restrictions and will provide a value for the amount of deuterium incorporated/retained in smaller units than obtained at the peptide level.
Additional considerations – When calculating the total number of exchanged H/D, one must keep several things in mind. 1) Any HDX at the N-terminal end of the peptide is lost during proteolysis. 2) Previous studies have demonstrated that any HDX at the second amide linkage is also lost very quickly during the chromatographic step (22,32). 3) Proline in peptic bonds does not have an exchangeable proton at its amide linkage.
1.7 Alternative workflows
As mentioned above, the generic experimental protocol outlined in Figure 4 can be modified to fit specific questions. In most cases these require additional equipment. For example, manual mixing, as indicated in the protocol outlined above, allows the measurement of deuterium content after the first few seconds of exchange (10 s), but exchange reactions that occur below that threshold cannot be measured. For rapid mixing and quenching of the reaction in the time range below seconds, a quench flow instrument is required. In this situation, quench flow in combination with HDX MS has been used to access these very fast rates of exchange of enzymes during catalysis (33). In pulse labeling experiments, an additional pump is used to expose briefly the protein sample to a pulse of deuterium and quench it quickly. This method has been used to study intermediates of folding pathways of proteins (34–36).
Most HDX MS studies make use of in-solution pepsin digestion. However, immobilized pepsin columns have been shown to improve digestion efficiency (37,38). In some cases there is too much back exchange, rendering the data unusable. Care must be taken on the choice of support used to conjugate the protein (39).
1.8 Complementary methodology
It has been observed that ESI of proteins in an unfolded state will produce higher charged envelopes than those produced by ESI of proteins in native conditions. This indicates that the protein ions in gas phase retain some of the structure that the protein had in solution, thus the charge distribution of the protein ions is an indication of the global structure of the protein. This is thought to be a consequence of the higher exposure of potentially charged residues that are otherwise protected in the core of the protein in the native state.
To obtain higher spatial resolution it would be necessary to interpret the tandem mass spectrum. However, due to the low energy of fragmentation of CID, this fragmentation method results in scrambling of deuterium among the resulting fragment ions (40–46). Thus, the CID mass spectrum of these peptides cannot be used to determine the position of the deuterium in amide linkages. The development of ETD, a more energetic method of fragmentation, results in the efficient fragmentation of peptides with little or no scrambling and interpretation of the tandem mass spectra of these peptides results in amino acid resolution.
In hydroxyl radical labeling (47), a protein solution is exposed briefly to oxidative conditions. This results in oxidative modifications of solvent exposed amino acid side chains. This can be achieved by either chemical reaction using Fenton chemistry (48) or by UV cleavage of hydrogen peroxide in fast photo oxidation of proteins (FPOP) (49). The appearance of covalently modified amino acid residues with oxygen can be identified by tandem mass spectrometry following trypsin digestion. When interpreting these data, it is important to keep in mind that reactivity of individual amino acid residues is determined not only by their accessibility to solvent but also by their individual reactivity. The reactivity of amino acid side chains is as follows: Cys > Met > Trp > Tyr > Phe > Cystine > His > Leu ~ Ile > Arg ~ Lys ~ Val > Ser ~ Thr ~ Pro > Gln ~ Glu > Asp ~ Asn > Ala > Gly (47). For detailed discussion of this methodology see the reviews by Chance (50) and Konermann (51).
1.9 Problems and caveats
1.9.1 Back exchange
A primary concern in mass analysis is the loss of deuterium during sample handling for mass analysis. Reducing pH to quench exchange requires the addition of acid. This quenching results in a reduced exchange rate, not a complete absence of exchange. Furthermore, reduced pH also exposes the now deuterated protein to additional protons. Also, the deuterated protein is further exposed to protonated buffer during the HPLC stage of desalting/peptide separation. Therefore, deuterons can be replaced with buffer protons during data analysis in a process known as back-exchange. In order to minimize loss of deuterium, mass measurement must be taken quickly, usually within the first few minutes following quenching. Despite efforts to work quickly, the back exchange of side chains is too rapid to be assessed with normal mass spectrometry methodologies and is the reason that HDX MS is limited to detecting information about the peptide backbone.
In most cases, two states of the protein are compared (control and experimental condition). Thus, assuming that the experimental conditions are maintained constant for each state, the differences in both total deuterium content and/or rate constants in identical peptides are used to describe different states of the protein. However, if a fully deuterated form of the protein is available, the following equation can be used to correct for the loss of deuterium during the analytical stages (13):
| Equation 5 |
where Dt is the content of deuterium at time t, mt is the average mass at time t, mH the mass of the undeuterated peptide and mD the mass of the fully deuterated peptide.
1.9.2 Overlapping peptides
To reduce back exchange, peptides are eluted using sharp gradients. In most cases there are only 30 minutes for data collection after quenching of the exchange reaction, which includes protease digestion, desalting and peptide separation. Moreover, the use of an enzyme with low selectivity results in the co-elution of multiple peptides. The isotopic envelopes of these peptides are changing in shape and average mass as the exchange reaction proceeds. This often results in the overlapping of peptide isotopic envelopes. Most software applications resolve this problem by either extracting ion chromatograms (HDFinder) or by curve fitting a theoretical envelope to the experimental data (HDExaminer, HD Desktop). The use of high resolving power mass spectrometer alleviates this problem. However, each peptide assignment must be validated individually.
1.9.3 Spatial resolution
The spatial resolution of HDX MS detected with simple mass measurements is at the peptide level. Most HDX MS studies published to-date have been made using this mode of operation. As a result, such experimental designs provide medium resolution, i.e. deuterium content is measured at the peptide level. To gain more spatial resolution using this experimental design, multiple overlapping peptides are required and deuterium assignment content is provided by logical analysis of these multiple overlapping peptides. However, this is not always possible, since certain regions of the protein may not produce the necessary number of overlapping peptides to obtain the degree of resolution desired.
Part 2: Limited proteolysis
2.1 Introduction
The development of the concept of “limited proteolysis” is widely attributed to work from the Linderstrom-Lang laboratory in the 1940’s (3). Among other studies, his laboratory demonstrated that proteins could be “enzymatically modified without serious degradation” by restricting proteolysis (52). Subsequently, the Neurath laboratory also made extensive use of this technique to study the structure of proteins (53,54). Unlike the complete proteolysis that is normally used for mass spectrometry, limited proteolysis refers to proteolysis that is halted by some means, so that complete degradation of the protein does not occur (see 2.3.3 for details on quenching proteolysis). Limited, controlled, in vitro proteolysis is a simple, but powerful, tool to study the conformation of proteins.
Proteases have a variety of specificities, i.e., residues at which they preferentially cleave. This specificity controls the sites of cleavage based on the primary structure of proteins not showing higher order structure. With the added dimension of folding, however, the normal specificities of proteases are no longer the only factor dictating cleavage location. Secondary structure will obscure sites from proteases, regardless of exposure, as will any additional structure that hides regions within folds or causes stereochemical constraints (55). Accessibility to the protease active site by the protein target becomes more restricted upon folding, thus the structure of the substrate contributes to the selectivity of the protease.
As an experimental technique, limited proteolysis was initially used to cleave larger proteins or complexes into separate domains to study them individually. It was first used to probe protein structure by Neurath in 1980, when he observed that most globular proteins were relatively resistant to proteolysis until they were denatured (53). He proposed that, as with other enzymatic reactions, optimal proteolytic activity occurred when there was complete complementarity between the substrate structure and the active site of the protease. The ability of the protease to cleave the substrate also depends on the location of a potential cleavage site within the structure, as only solvent exposed regions will be accessible in a tightly folded protein. Neurath proposed a model in which functional domains of proteins are tightly packed, and therefore relatively protease resistant, whereas linker regions or loops are surface exposed and more susceptible to proteolysis (53,54). Using crystal structures and limited proteolysis to confirm correlations between flexibility and cleavage, this model became the basis of limited proteolysis theory: that is, limited proteolysis occurs only at sites on a protein’s structure that are solvent accessible and flexible enough to conform and fit within the active site of the protease (56–59). Generally, this solvent accessibility and flexibility occurs at specific region(s) of a protein; so that even when multiple proteases with different specificities are used, the cleavage sites are clustered together, although not necessarily with cleavage at the same residue (55).
Because protease specificity still plays a role in determining cleavage sites, it is important to use proteases with broad specificities, along with multiple proteases with differing specificities. This will ensure that the regions being targeted reflect their exposure in the tertiary structure, rather than their primary structure. Therefore, it is also imperative to maintain the protein’s higher order structure. When planning and executing an experiment, it is essential to keep in mind the basic premise of limited proteolysis: brief proteolysis of surface exposed regions while maintaining the protein core. Because proteolysis of a protein can cause conformational changes, it should not be allowed to proceed for too long, as regions that were not originally surface exposed may become so, causing results to be skewed. If the protein core becomes compromised, information about the structure is no longer reliable.
Limited proteolysis was initially analyzed using SDS-PAGE and Edman degradation; however, with the development of MS to study proteins in the late 1980’s (60,61), it became the preferred method of analysis for limited proteolysis. MS has allowed the applications and capabilities of limited proteolysis to greatly increase. With the use of MS, it is now possible to easily identify the exact sites where proteolysis occurs, providing a map of the regions cleaved by the brief proteolysis, allowing for detailed identification of the flexible and surface exposed regions. Unlike NMR or crystallography, MS requires only a minimal amount of protein to obtain structural information, and the ratio of protein to protease is key, rather than the absolute amount of either. Limited proteolysis and MS can also be used on proteins of any size, as there are no minimum or maximum protein size restrictions. It can be used on single-domain proteins, multi-domain proteins, multi-subunit proteins, etc. Another advantage of limited proteolysis/MS is the ability of MS to analyze complex mixtures (62,63).
2.2 Limited proteolysis applications
Limited proteolysis can be used to study different aspects of protein structure, several of which are described below. Because surface accessibility and flexibility are required for proteolysis to occur, the most obvious application of limited proteolysis is the identification of exposed loops and disordered regions. By employing proteases of different specificities and limiting proteolysis, while maintaining the protein core, it is possible to map exposed loops and identify regions of disorder. This can be used to complement NMR or crystallography data (64,65), or even to replace these techniques if they cannot be used on the protein of interest. Crystallography can be especially difficult for disordered or dynamic regions, as it results in low resolution. Limited proteolysis can be used to confirm the disorder and dynamic properties of these regions (66,67).
Likewise, as first proposed by Neurath, multi-domain proteins often have flexible and disordered linker segments joining the domains, and these will be preferentially cleaved during partial proteolysis (57,68). Therefore, identification of domains and their exact boundaries is possible. This separation of domains was one of the first applications of limited proteolysis, as seen in several early papers (54,69,70). More recently, this application has been used in conjunction with MS for the specific identification of linker regions. For example, applying these techniques to the E. coli transcriptional activator protein NtrC, a protein with three separate domains, Bantscheff et al. (57) developed a system combining limited proteolysis, MS, and SDS-PAGE to identify two flexible linker regions. Limited proteolysis can also be used to cleave flexible linker regions to produce separate domains, making feasible the study of single domains and potential folding intermediates (71).
Another application of limited proteolysis is the study of complexes formed between proteins and their targets. This is possible because the interface regions of the complex will initially be solvent accessible on the surface of the protein, but become protected when the complex forms. Therefore, by first performing limited proteolysis on an isolated protein and then on the protein in complex, it is possible to identify the interface regions, although regions affected by conformational changes prompted by the interaction may also show changes in the level of protection. Different peptide maps for the two protein states, free and in complex, will be observed by MS following the limited proteolysis. An example of this approach is the study of the calmodulin-melittin complex (72). The authors performed limited proteolysis on free calmodulin, free melittin, and the calmodulin-melittin complex, observing different peptide maps for the free proteins vs. the proteins in complex. From the regions that changed, they designed a model showing the sites of interactions between melittin and calmodulin. A similar application of limited proteolysis to study protein complexes is to identify regions of protein-DNA, protein-RNA interactions, and antibody epitope identification (73–75).
Regardless of the experimental design – identifying domain linkers, mapping exposed loops, or interactions – another use of limited proteolysis is comparing changes in those regions upon protein activation, mutagenesis, or ligand binding. In these cases, the limited proteolysis of the protein in its basal state is compared to that of the altered protein. If there are conformational changes occurring on the surface of the protein, the resultant peptide maps can show regions of differential proteolysis, indicating they are more or less flexible or exposed.
2.3 Methodology
2.3.1 Optimization
The most basic rule to keep in mind when designing and executing a limited proteolysis experiment is that the protein core must remain intact, or it is no longer “limited” proteolysis, and information about the protein structure may no longer be valid. Because this is so essential, experiments must be performed under conditions that maintain the stability and structure of the protein being studied, regardless of the optimal conditions for the proteases being used.
Because ensuring that the higher order structure dictates the sites of cleavage is so important, as opposed to the protease specificity with respect to primary structure, it is advisable to use multiple proteases with varying specificities, as well as various proteases with broad specificities. This means, however, that the individual proteases will most likely not be cleaving under their optimal conditions (pH, temperature, etc.). Given that maintaining target protein stability is the most important factor, one must first identify conditions that are optimal for that stability. This will include such things as buffer, pH, temperature, and duration that proteolysis can occur. Once these conditions are determined, the concentration of proteases required for sufficient, yet limited proteolysis, can be optimized. Because sub-optimal conditions will undoubtedly be used for some of the proteases, it will likely be necessary to use different ratios of protein to protease for each protease in order to ensure similar levels of proteolysis with minimal cleavage. Examples of this are shown in Table 1.
Table 1.
Protease specificities and final concentrationsa
| Protease | Specificity | Kinase: protease ratio |
|---|---|---|
| Thermolysin | Hydrophobic | 15:1 |
| Chymotrypsin | Aromatic | 2,000:1 |
| Protease V8 (Glu C) | Asp and Glu | 150:1 |
| Trypsin | Arg and Lys | 5,000:1 |
| Ficin | Nonspecific | 10,000:1 |
| Arg C (Clostripain) | Arg | 10:1 |
| Lys C | Lys | 50:1 |
| Papain | Nonspecific | 10,000:1 |
| Proteinase A | Nonspecific | 100:1 |
| Subtilisin | Nonspecific | 200,000:1 |
| Pepsin | Aromatic, acidic, hydrophobic | 10:1 |
Different proteases can be, and should be, used in limited proteolysis experiments. Listed above are examples of proteases and the protein:protease ratios that were used in limited proteolysis experiments at pH 6.8 on the glycogenolytic enzyme phosphorylase kinase (76). While these ratios will likely differ for other proteins, these are reasonable starting points for optimization. Other proteases that are commonly used include Proteinase K, elastase, and Asp-N.
Another important experimental variable to optimize is the quenching step, because different proteases may be typically inhibited differently. The ideal quenching step, however, is one that can be used for all proteases in the study. If more than one quenching method is used, it should be shown that neither the results nor the protein are affected. Finally, and as discussed further in 2.3.3, quenching must be both rapid and complete.
2.3.2 Materials
2.3.3 Quenching of proteolysis
Not just for reproducibility, but also to avoid too much proteolysis, it is important to ensure that quenching conditions are indeed stopping the hydrolysis of the parent protein. Ideally, the activity of the proteases should be stopped instantly and completely. The criterion of instantly rules out many of the specific inhibitors for proteases, especially irreversible, active-site-directed, chemical inhibitors, as they may act relatively slowly. Quenching by changing conditions, such as pH, can be useful; however, if the quenching pH must be altered prior to analysis, the possibility for renewed proteolysis must be considered. Often quenching is achieved by adding trifluoroacetic acid or acetonitrile, although protein precipitation may occur. Denaturants can also be used to quench; however, some proteases still show residual activity in the presence of denaturants, plus the denatured protein that is being studied will likely be an even better substrate for proteolysis than its native counterpart. When analysis is performed by MALDI, proteolysis has sometimes been quenched by addition of the matrix solution or by pipetting an aliquot of the hydrolysis mixture directly onto the plate (66,73). The bottom line is that whatever quenching condition one chooses to employ, it is imperative to experimentally test it to establish with certainty that quenching does, in fact, occur.
2.3.4 Mass Spectrometry
MALDI and ESI MS are both capable of analyzing limited proteolysis data. MALDI-MS is tolerant of buffers and does not require desalting the samples, both desirable features. ESI-MS does require desalting, but chromatographic separation of complex mixtures allows for sequencing of more peptides, particularly desirable in complex mixtures.
2.3.5 Peptide identification
Given that limited proteolysis is typically used on purified, known proteins, the use of standard protocols, which employ probabilities and false discovery rates, is not essential. Peptide identification in limited proteolysis is similar to that used in HDX-MS (1.6.1) and general peptide identification is discussed in more detail in Chapter 8. Typically, a region will be targeted, rather than a specific residue, and if different proteases with different specificities are used, it is likely there will be overlapping peptides covering the same region. This indicates consistency of the data and the flexibility and exposure of that region. Proteolysis will likely result in subdigestions, i.e., after a region has been initially cleaved, the protease may continue to act on that peptide, resulting in multiple smaller peptides from that same region. These subdigestions can be ignored in favor of the longer peptides that cover the same region. In fact, by considering subdigestions cautiously, one can avoid over-interpreting the putative importance of specific residues within the larger region that encompasses them.
2.3.6 General protocol
Proteolysis – Incubate protein with protease at the optimized ratio determined previously (2.3.1) under conditions (buffer, pH, and temperature) best suited for protein stability.
Quenching – After incubation for appropriate time(s), remove aliquot and quench reaction (2.3.3).
MS – Prepare samples following protocol established for the MS to be used. Be aware of maintaining quenched conditions, so as not to resume proteolysis. Keep all peptides for analysis. See 2.6 for discussion on peptide release.
2.5 Representative results and data presentation
Organization and presentation of data are largely dependent on the main point of the experiment, the type of experiments performed, and the protein(s) involved. Table 2 and Figure 6 show several possible ways to present results.
Table 2.
Representative dataa
| Trypsin | Pepsin | Arg C | |||
|---|---|---|---|---|---|
|
| |||||
| WT | Mutant | WT | Mutant | WT | Mutant |
| 1–20 | 1–23 | ||||
| 83–90 | 83–90 | 83–89 | 83–89 | 83–90 | 83–90 |
| 150–161 | 150–161 | 150–158 | 150–158 | 150–161 | 150–161 |
When comparing mutants, activated proteins, or complex formation, it is necessary that all conformers or states are included in the table in a format that makes comparison easy. Because using multiple proteases is advisable, shown here is a table in which the different protease results are compared side by side for the different conformers of protein. The titles (wild type and mutant) can be exchanged for non-activated vs. activated protein forms, complexed protein vs. free, etc.
Figure 6. Representative results.
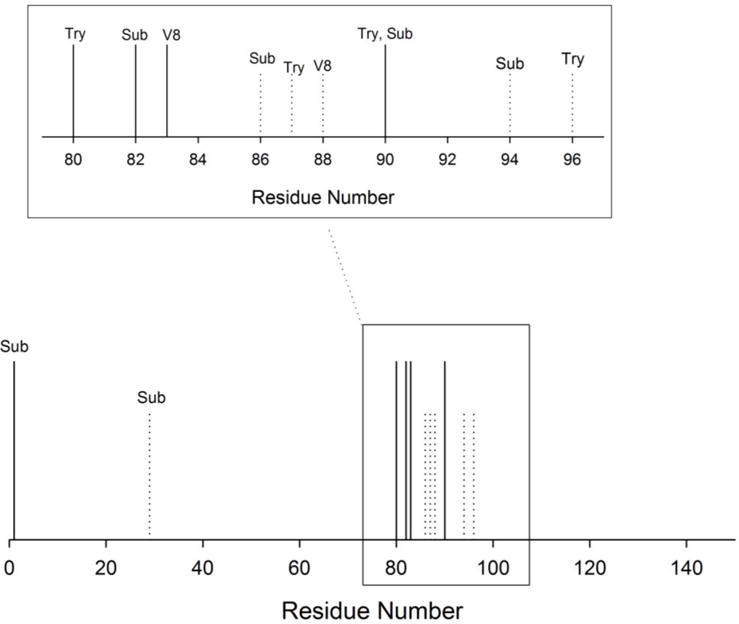
When mapping exposed loops and regions of disorder, it is helpful to visually demonstrate results in a straightforward way representing the protein structure and sites of cleavage. Demonstrated here is a way to conveniently show regions that are targeted by proteases; when using multiple proteases, this figure also demonstrates clearly that different proteases are targeting the same region, further implying flexibility and exposure. Depending on the size of the protein, the line representing residues could be substituted for the actual sequence. Alternatively, if the protein is too large, the representative residue lines used in this figure may more clearly portray the results, and regions that are cleaved can be magnified to highlight cleavage details.
2.6 Caveats concerning limited proteolysis
A possibility that is not often considered is whether all peptides formed during limited proteolysis are actually released from the parent protein following quenching. This is not an important concern when product analysis is carried out by MALDI, as all peptides should be observed, regardless; however, the binding of proteolyzed peptides may be a concern with other analytical methods, as some peptides could be missing in the final product analysis. The noncovalent binding of otherwise free peptides by a proteolyzed parent has been observed with the protein phosphorylase kinase, a 1.3 MDa complex of multiple subunits. Following selective chymotryptic hydrolysis of its largest subunit (to the extent that no remaining trace of it was observed on SDS-PAGE), there were only small changes in the structure of the proteolyzed parent as observed by electron microscopy (77), despite the fact that the degraded subunit accounts for 43% of that parent complex’s total mass. Consequently, evaluating a variety of conditions for the quenching of proteolysis, or between proteolysis and the removal of remaining parent protein prior to analysis, could prove advantageous in assuring maximum release of generated peptides. Note also that if the parent protein is precipitated prior to analysis, peptides derived from it could be trapped within the precipitant.
A caveat that was discussed in 2.3.5 is the production of smaller peptides from the subdigestion of initially released larger parent peptides, which may potentially produce peptides too small to detect. If a proteolysis time-course is run, these subdigestion peptides are likely to be observed later than their parent peptides. A time course can also show the later secondary appearance of less readily cleaved peptides from different regions of the protein. A caveat concerning interpretation of the appearance of these unique secondary peptides is that, instead of representing regions less readily cleaved, they could also represent a new conformation of the protein induced by an initial proteolysis event. A new proteolytically induced conformation introduces special concern for proteins whose function is controlled by so-called intrasteric regulation (78), i.e. a region of primary structure in the protein that is autoregulatory through its interaction with other regions of the protein, generally the active site (79). For many of these proteins, the autoregulation can be overcome by limited proteolysis, resulting in a new conformation with a different activity. Thus, an important control to include in limited proteolysis experiments is the determination of functional changes following proteolysis. This concern also suggests that keeping the extent of proteolysis relatively limited is prudent.
2.7 Side chain modification as a complementary technique to partial proteolysis
Historically the goal of this method for structural analyses was to identify a protein’s relatively reactive nucleophilic amino acid side chains, which by definition are accessible to the electrophilic reagent used to covalently modify them. Thus, the residues modified are likely to be on the surface of the protein and could be within, or adjacent to, the exposed loops implicated by partial proteolysis. Identification of modified residues can, therefore, corroborate results from partial proteolysis. Over the years, more complex methods of side chain modification having a considerably wider range of amino acid targets, such as oxidation by hydroxyl radicals (80–82), have been developed, but the underlying idea of preferentially modifying surface residues remains the same. An increase in the variety of side chains that can be modified does, however, add greatly to the power of the technique, making it complementary to HDX. Unlike HDX, however, the covalent modifications are irreversible, potentially facilitating analysis.
This general method of side chain modification could reasonably be called chemical or protein “footprinting”; however, beginning with DNA analysis, the term footprinting has connoted protection from cleavage as being directly involved in the identification of regions of interest, e.g., protection from cleavage by DNA-binding proteins. Similarly, the term “protein footprinting” has also been used to denote cleavage of a protein, direct or indirect, at the specific residues modified by a chemical reagent (83,84). This same term has also been used, however, to describe the analyses through side chain modification of nearly every characteristic of proteins, from dynamics to structure to binding, but with cleavage occurring only secondarily in the generation of peptides for MS analyses. Consequently, to avoid potential confusion in terminology, we call this approach side chain modification, rather than protein footprinting.
There are few variables in carrying out side chain modification experiments: choice of modifying reagent and of modifying conditions (time, pH and concentration of modifier with respect to protein). The conditions used will affect the rate, and perhaps the extent, of modification, and deciding on which conditions to use is an empirical process. One wants to obtain a reasonable amount of modification in a reasonable amount of time; however, what represents a reasonable amount of modification is not necessarily obvious. Certainly, enough product should be formed to be able to argue that it truly represents the conformation of a large population of the native protein, as opposed to the conformations of minor components produced by denaturation, oxidation, post-translational modification, or minor proteolysis during protein purification. On the other hand, one doesn’t want so much modification that the conformation of the protein could be altered by the modifications or conditions employed to modify. Consequently, a control that should be run, but often isn’t, is to characterize the properties of the protein following modification. Full retention of biological function is a reassuring control, as is relatively unchanged higher order structure assessed biophysically. Many studies do not address the extent of modification, nor its reproducibility. The latter should assure that similar results are obtained with multiple independent protein preparations. If one is comparing two conformations, e.g. apo-protein vs. ligand-bound, misleading information is less likely if modifications of both are kept in the linear phase.
Part 3: Crosslinking
3.1 Introduction
Chemical crosslinking refers to the covalent coupling of separate functional moieties. In the context of proteins, this technique has been used for over 50 years to analyze the structure, function and interactions of these biomolecules by identifying protein crosslinking sites that are formed by small multi-functional reagents, termed crosslinkers. The coupling of protein crosslinking with modern MS techniques (CXMS) has led to resurgence in the field, with new instruments and crosslinking technologies being developed to facilitate identification of conjugates (crosslinked proteins and/or peptides) from ever smaller amounts (nmole to pmole) of sample. CXMS is a bottom-up approach, in that the protein is first crosslinked and then digested specifically with proteases to generate peptides for detection by MS. A limiting factor in the analysis of proteins by CXMS is the extensive array of products (including many side products) that are possible from such digests. These product arrays are too complex to be annotated manually and require the use of search engines that have been developed specifically to segregate the more numerous side products from conjugates of interest. Our intent in this chapter is to expose novice users to: (A) CXMS approaches that minimize the generation of side products and maximize structurally useful conjugates, (B) available conventional, mass and affinity tag crosslinking reagents, and (C) search engine technologies for identifying conjugates.
3.1.1 Advantages and applications
Crosslinking provides low to medium structural information for proteins that are not amenable to high resolution techniques such as NMR and X-ray crystallography. It is a versatile technique that, in its simplest form, has been used to determine nearest neighbors and the minimal subunit stoichiometry in multi-oligomeric complexes (85). In combination with Western blotting, immuno-precipitation, various protein labeling methods, top-down MS and CXMS approaches, it has been used successfully to capture protein-protein interactions (PPI) in transient and stable complexes (86), providing maximum distance information for these targets in both in vitro and in vivo applications (reviewed in (87,88)). Recent advances in the detection of peptides from complex mixtures by modern MS and supporting search engine technologies have provided a robust platform for the development of CXMS and its primary use in the identification of crosslinked peptides from digests of crosslinked proteins. CXMS provides a range of structural information, and the resolution of this information is dependent on how specifically a crosslinking (CX) site can be localized in the context of a protein target, with the identification of crosslinked amino acid side chains providing the highest structural resolution. CX sites may be used to determine the proximity of domains and amino acid side chains in protein monomers or complexes, to identify potential intramolecular or intermolecular protein binding sites, and to provide structural constraints for theoretical protein models (89–91). Many search algorithms and specialized reagents have been developed to enrich and enhance the detection of conjugates and more numerous side products in digests of crosslinked proteins (90,92–94), making this approach readily accessible to any researcher with access to MS and proteomics facilities.
3.1.2 Chemistry of crosslinking
3.1.2.1 Crosslinking reagents
The range of structural information gained from CXMS is inherently dependent upon the type of cross-linking reagent (CXR) used. The largest and most commonly used classes of CXRs are bifunctional molecules containing two reactive groups that are connected by an intervening spacer group. Bifunctional CXRs are further divided into two subgroups, based on whether they contain identical (homobifunctional) or different (heterobifunctional) reactive groups. Many different reactive groups with varying chemistries have been incorporated into CXRs (Table 3); however, there are five functional groups that are commonly used, because they are chemically compatible with targeting protein side chains in aqueous solutions at near physiological pH (85). N-hydroxysuccinimide (NHS) and imidoester groups react preferentially with the N-termini of proteins, as well as the pyrrole and ε-amines of proline and lysine, respectively. Sulfo-derivatives of the NHS group are also available to increase the solubility of CXRs with large hydrophobic spacer groups. Maleimide and alkyl halide groups are targeted primarily by the free thiols of cysteine. As opposed to the functional groups above, aryl azides are promiscuous, and upon exposure to UV, insert non-selectively as nitrenes at active hydrogen-carbon bonds or undergo ring expansion to form dehydroazepines (87), which react both with nucleophiles and active hydrogen-containing species.
Table 3.
Selected reactive groups of typical crosslinking reagents
| Reactive group chemical structure | Group name | Amino acid preferentially targeted |
|---|---|---|

|
N-Hydroxysuccinimide ester (NHS) |
Lysine |
|
Maleimide | Cysteine |
|
|
Alkylhalide | Cysteine |

|
Imidoester | Lysine |

|
Phenylazide | Non-specific |
|
|
Carbodiimideb | Aspartic and Glutamic acid |
R denotes spacer and second reactive group, except for the carbodiimide.
Zero-length crosslinking reagent that activates carboxyl groups for subsequent attack by proximal amines.
Spacers or linkers are chemical moieties that covalently couple the reactive functional groups of a crosslinker. Besides determining the distance between the reactive groups, spacers also influence the geometry of crosslinking and the solubility of the CXR. For example, CXRs with long alkyl spacers are generally hydrophobic and can include a broad range of crosslinking distances between potential nucleophiles, due to the flexibility of the linker. Spacers also contain functional groups that allow for their cleavage by specific reagents, such as periodate or DTT, which cleave intervening glycol or disulfide groups, respectively. Crosslinkers that contain these groups are members of a subclass of bifunctional reagents, termed cleavable CXRs. In addition to chemical cleavage sites, CXRs with spacers containing MS-cleavable functional groups have been developed to facilitate bond breaking by collision-induced dissociation (CID) and/or electron transfer dissociation (ETD) in mass spectrometers. Such reagents are used as reporter groups to aid in the identification of crosslinked peptides from complex mixtures (95,96). Spacers comprising affinity tags such as biotin and Click chemistry labels are employed to enrich low abundant conjugates (97,98), and even more complex forms that contain both affinity and mass tags have been synthesized to simultaneously enhance enrichment and identification of crosslinked peptides (99). CXRs containing functional spacers are often identified as trifunctional or multifunctional reagents; however, the term trifunctional also refers to CXRs that contain three reactive groups that emanate from a central spacer group or atom, each of which is capable of reacting with three distinct sites on protein targets.
Zero-length CXRs refer to molecules that directly couple amino acid side chains without an intervening spacer. These reagents generally modify and activate functional groups on specific side chains for subsequent attack by an adjacent protein nucleophile, such as the ε-amine of lysine. For example, N-substituted carbodiimides react with the carboxylates of Asp and Glu residues to form acylisourea intermediates that facilitate the formation of isoamide bonds with proximal lysine residues (Table 3). Free thiols may also target these reactive intermediates to form thioester linkages; however, these conjugates are relatively unstable by comparison with the corresponding amide linkage. For complete and thorough reviews of crosslinking reagents see the works of Wong and Hermanson (87,88).
3.1.2.2 Proteins as reactants
Proteins as reactants add to the complexity of products generated in crosslinking reactions, because they are polyvalent structures, containing multiple reactive amino acid side chains with varying relativities that are dependent upon their microenvironments in the protein complex. The microenvironment reflects for a given amino acid, its location and the dynamics of the region encompassing that location in the tertiary structure of the protein, its solvent accessibility, and interactions with and chemical composition of its nearest neighbors. On the basis of hydrophobicity, amino acids may be divided into two major classes, nonpolar and polar. Polar residues can be separated into those containing side chains with nonionizable (asparagine, glutamine, serine and threonine) and ionizable (histidine, lysine, arginine, tyrosine, cysteine, aspartate and glutamate) functional groups. With the occasional exception of tryptophan, the latter group is primarily targeted by CXRs.
3.1.2.3 Products of crosslinking
As previously mentioned, the crosslinking and subsequent digestion of a protein and/or protein complexes generates a vast array of products that must be accounted for to detect crosslinked peptides. Figure 7 shows examples of the types of products that are typically observed when two proteins are treated with a bifunctional crosslinker. In addition to crosslinking between (intermolecular) the two proteins and within (intramolecular) a single protein, more numerous mono-modifications (dead-end) occur as well. Moreover, crosslinking is a continuous process, and if not carefully controlled, results in the formation of multiple protein conjugates, progressing from crosslinked dimers to large polymeric arrays. Subsequent digestion of the crosslinked proteins by proteases for analysis by MS significantly increases the number of possible products, particularly if the CXR targets side chains that are also substrates of the protease, which results in incomplete digestion of the crosslinked protein targets (100). Estimates suggest that the number of potential peptides products from such digests increases exponentially with the size of the protein (101), necessitating the use of bioinformatics approaches to annotate all possible products.
Figure 7.
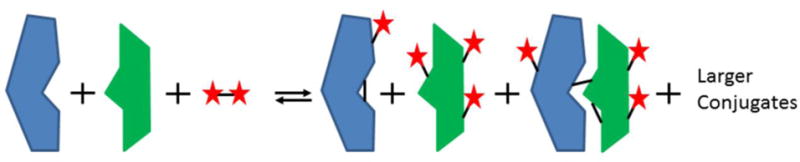
Products of protein crosslinking
3.1.2.4 Data analysis
For two reasons, analysis of CXMS data is not trivial and requires dedicated software tools. The first is that the number of candidates that must be considered is enormous in comparison to regular proteome-wide peptide analysis. The second is that the abundance of crosslinked proteins is much lower than that of non-modified proteins, and the data analysis algorithm must be sufficiently sensitive to identify small signal peaks amongst dominating, neighboring peaks.
A number of software tools have been developed in the past decade for CXMS data analysis. In the following sections, we will explain the basic data analysis principles, look into the computational algorithms behind these tools, examine their pros and cons, and finally provide our perspectives on future development of data analysis algorithms and software tools for CXMS analysis.
3.2 Methodology
Crosslinking is a specialized form of general protein chemical modification, both of which are empirical processes. It is simply impossible to predict under which conditions and with which CXR a given protein will undergo crosslinking. Variables such as time, reaction component concentrations, pH and CXRs must be screened to maximize the specificity and selectivity of crosslinking. Specificity refers to the preferential, stable modification of a protein side chain functional group by a specific class of CXR reactive group. Specificity, on the other hand, denotes the potential for detecting observed protein interactions by crosslinking. Both of these parameters are inter-related and the extent to which one is controlled significantly affects the other, and ultimately, successful crosslinking of a protein to obtain maximum yields of a desired conjugate. Crosslinking is the first step in the CXMS pathway to identifying CX sites in any protein or complex of interest. Optimization of subsequent proteolysis and detection steps is also critical and the corresponding protocols, instrumentation and software will be discussed in the following sections.
3.2.1 pH
Most CXRs contain electrophilic reactive groups that are targeted by protein nucleophiles in reactions. These reactions generally involve either displacement of a leaving group or direct addition to a double bond with adjacent electron withdrawing groups on the CXR to form a covalent bond between it and the attacking amino acid side-chain. In terms of Lewis acid-base theory, the reactivity of an amino acid side chain is directly related to the nucleophilicity or electron-pair donating capability of its side chain functional group, which in turn can be expressed in terms of the ratio of concentrations of its electron donor/base (A−) and electron acceptor/acid (HA) forms in solution. This ratio can be estimated theoretically using the Henderson-Hasselback equation, which implies mathematically that for a nucleophile to exist equally in its conjugate base and acid forms, the pH value must equal its pKa.
For one and two unit increases in pH, the percentage of the basic form increases correspondingly from 50 to 95 and 99 percent, respectively. Thus at alkaline pH values, the nucleophilicity for basic R-S− and R-NH2 protein nucleophiles is greater than their corresponding acid forms (R-SH and R-NH3+) at low pH values.
The relative order of nucleophilicity for protein functional groups involved in crosslinking reactions is: R-S− > R-NH2 > R-COO− ≅ R-O−. With the exception of zero-length crosslinkers, most conventional, commercially available CXRs are designed to react preferentially with thiolate or amine-containing protein nucleophiles. Examining the range of theoretical pKa’s for cysteine (8.8–9.1) and lysine (9.3–9.5) (87), one might assume that they would be poor nucleophiles at neutral pH; however, in the microenvironments of proteins, these side chains are often reactive and covalently modified by CXRs. Thus optimization of pH is critical in controlling the outcome of crosslinking. For example, crosslinking at high pH values might seem prudent to increase the reactivity of amino acid side chains; however, it also significantly diminishes the selectivity of a CXR for its intended target and may diminish the specificity of crosslinking by increasing unwanted side reactions and the formation of large conjugates, rendering the results uninterpretable. Moreover, hydrolysis of many CXR reactive groups increases significantly and competes with crosslinking at high pH values, generating excessive dead-end modifications. A general rule of thumb is that pH and all other variables in the crosslinking reaction should be adjusted through screening to maximize the formation of detectable desirable low mass conjugates.
3.2.2 Uses of conventional and mass/affinity tag CXRs
In the following section, conventional CXRs are defined as those not containing mass tag/reporter and/or affinity tags. Because crosslinking is an empirical process, a CXR is generally chosen for a protein target from screens of reagents with multiple chemistries under multiple conditions. That having been said, there are many commercially available CXRs with properties that are advantageous for specific types of analyses. Zero to short (2–3 Å) length CXRs are preferable for detecting protein interactions, in that their product conjugates are more likely to represent an actual interaction within or between protein targets, i.e. the specificity of crosslinking is maximized. For such analyses, conventional or specialized mass/affinity tag-containing reagents with large crosslinking spans (> 2–3 Å) should be avoided. In low resolution crosslinking experiments in which the identification of one or more binding partners (low resolution experiments) is sufficient, longer span CXRs with affinity or mass tags are more advantageous, simply because they generally increase the likelihood of isolating and/or detecting crosslinked products. Hydrophobic, water insoluble CXRs are typically used for screening protein interactions that are stabilized by hydrophobic binding surfaces, whereas hydrophilic, water soluble, reagents are often employed for labeling charged, solvent accessible residues on the surfaces of proteins. Homobifunctional CXRs are used primarily in one-step crosslinking reactions, in which all components are present in the reaction. Heterobifunctional reagents containing two chemically distinct functional groups are often exploited for use in two-step crosslinking experiments. In such experiments, a protein target is first modified under conditions that favor the reactivity of one functional group, followed by purification of the labeled complex to remove non-covalently bound reagent and to facilitate its exchange into conditions that favor reaction of the second CXR functional group. For example, CXRs containing both photo- and thermo-reactive functional groups (Table 1) are typically used in these reactions, with the protein first labeled with the thermo-reactive group and then purified in the dark, following which the modified complex is exposed to UV radiation to activate and promote crosslinking by the photoactive group.
Some specialized CXRs contain functional groups that enhance their purification (affinity tags), using affinity purification media designed to capture the tag and/or mass tags that generate unique isotopic signatures that aid in the detection and identification of crosslinks and dead-end side modifications. For the most part these reagents use the same chemistries as conventional crosslinkers, most of which incorporate NHS groups to target lysine ε-amines. Many strategies have been introduced to follow sequentially labeled precursor ions (ionized intact crosslinked peptide) and their collision products with mass/affinity tag combination CXRs created to reduce the complexity of the product pool and to facilitate cleavage of both peptide arms of the crosslink. Several notable examples include CLIP (98), which utilizes a bis-NHS CXR, with a terminal Click alkyne tag for enrichment (using biotin azide) and an NO2 reporter group that both enhances water solubility and acts as a neutral loss reporter during MS-induced fragmentation. Several groups have developed CXRs that fragment during MS/MS to release small molecules that provide mass signatures for crosslinked peptides (95), termed protein interaction reporters (102). Using a different ligation approach, Trnka and Burlingame synthesized a novel CXR, diformyl ethynlbenzene (DEB), which forms Schiff bases with lysine ε-amines that are subsequently reduced to secondary amines with cyanoborohydride (91). The authors demonstrate that reduction to the amine, rather than the common acetylation product formed by NHS groups, provides two additional protonation centers with incorporation of the intervening rigid ring spacer, decreasing the m/z ratio of the conjugate for more optimal fragmentation by high resolution ETD and electron capture dissociation (ECD) fragmentation methods, which provide more complete fragmentation along the peptide backbone. Moreover, the reagent contains a clickable tag for purification and fragments during MS/MS to produce diagnostic reporter groups. Digestion of DEB crosslinked proteins also generates high charge state gas phase precursor ions (4–6+), which allows for their exclusion from native and dead-end modified peptide ions using charge dependent precursor selection (91). More recently Ihling et al. have developed a CXR with a spacer that contains an N-oxy-tetramethylpiperidine linked to benzene (TEMPO), which contains a CID-labile NO-C bond (103). This reagent facilitates free radical initiated peptide sequencing (FRIPS), generating open shell radicals that provide signatures for determining the sequence and location of the CX site on crosslinked peptides by successive MS2 and MS3 analyses. More solutions for reducing the complexity of crosslinking products are likely as the list of these reagents that exploit high resolution tandem MS continues to grow.
3.2.3 Equipment
The initial stages in the analysis of proteins by crosslinking requires very basic equipment, commonly found in most biochemical laboratories. These include various SDS-PAGE apparatus (large and mini gel versions) to analyze protein crosslinking products, circulating water baths and incubators to control for temperature and light boxes for viewing stained gels. In-gel proteolysis techniques require a laminar flow hood, bench-top centrifuge and vacuum centrifuge. After optimizing the yield and proteolysis of a desired conjugate, MS technologies are employed to analyze the digest. There are many different configurations for mass spectrometers; however, high resolution instruments with fast duty cycles almost always produce the best data for analysis by search engines. This is because considerable accuracy is required to limit the mass degeneracy associated with the many mass combinations possible from digests of crosslinked proteins (100), and because faster duty cycles (percentage of a time window required to make a measurement) increase the potential for analyzing low intensity ions typically associated with crosslinked peptides during a given run. Orbitrap MS instruments best fulfill these requirements (104). In addition to the parameters listed above, Orbitraps come in different tandem MS configurations, with the most advance being capable of carrying out CID, ETD and higher-energy C-trap dissociation (HCD) fragmentation of precursor ions. See Chapter 5 for more detailed descriptions of mass spectrometers.
3.2.4 Data analysis using search engines
The goal of data analysis is to identify crosslinked peptides. Crosslinked peptides include inter-crosslink peptides, intra-crosslink peptides, and dead-end peptides. Identification of intra-crosslink peptides and dead-end peptides may be achieved by using software tools that were designed originally to identify regular (i.e. non-crosslinked) peptides from shotgun proteomics experiments; however, their identification is extremely difficult. This is because inter-crosslinked peptides include two peptides and the search algorithm must search each experimental spectrum (i.e. query spectrum) against all of the possible pairs of peptides. Figure 8 illustrates a general data analysis procedure that comprises several steps that are explained in detail below.
Figure 8.
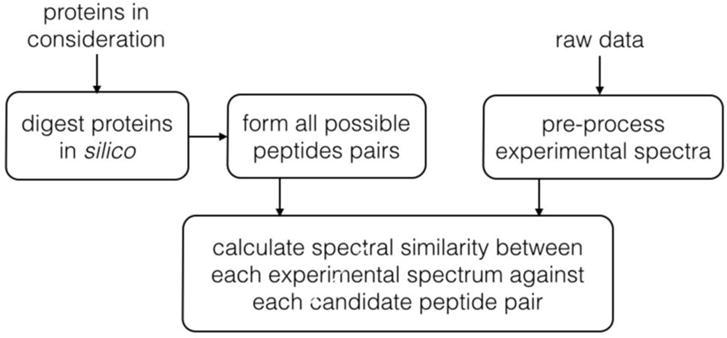
Data analysis theoretical and experimental flow of information
In the first step, sample proteins are digested in silico to generate all of the possible peptides using a digestion rule, which uses the known chemistry of the protease selected to determine where cleavage should take place along the amide backbone. For example, if trypsin is selected, then the algorithm generates all possible peptides arising from cleavage C-terminal to lysine and arginine, except when these residues are located N-terminal in the primary sequence to proline. Experimentally it is not uncommon, however, for trypsin to miss one or more of these cleavage sites. Moreover, peptides that are too short or extremely hydrophilic are often lost in wash steps prior to injection in the mass spectrometer, further depleting the expected data set. Likewise, large peptides with masses greater than 4000 Da. are not efficiently cleaved and transmitted. Therefore algorithms must be flexible enough to accommodate these and other results that deviate from ideal theoretical conditions. To accommodate these possibilities, search engines typically incorporate two user input parameters that may be adjusted to narrow the range of peptide chain length, depending upon the capabilities of the instrument being used, and the number of missed cleavage sites (NMC).
In the second step, the peptides generated in the first step are combined in pairs, and their masses are calculated and annotated in extremely large data bases, based on sequence and potential chemical modifications. Possible modifications are defined from rule sets that take into account the possible chemistries and residues that are potentially targeted by any given CXR. Depending on the flexibility of the search engine, a user may manually limit the number of potential crosslinks and dead-end products that applied to any peptides during the run. More powerful programs may also calculate for crosslinking among three or more peptides using the parameters described above.
Experimental spectra are pre-processed in the third step to account for variations in noise in tandem MS signals and to normalize low and high abundance peaks, both of which are generally important in conjugate identification. Pre-processing of an experimental spectrum separates signal peaks from noise peaks, removes the latter, and normalizes the resulting signal peaks so that low and high intensity peaks are scaled differently. Normalization permits amplification of low intensity peaks, which are often characteristic of crosslinked peptides and thus allows them to be weighted to a greater extent in subsequent scoring rounds. Programs that are designed to carry out this procedure are generally capable of detecting more conjugates than those that simply analyze a given number of the most intense peaks in the spectrum.
Usually, the terminal step in processing is to score the spectral similarity between processed experimental spectra from step three and theoretically generated spectra for all potential candidates generated in step two. Existing programs calculate spectral similarity in different ways, either by cross-correlation or simply summarizing the number of matches detected between peaks from experimental MS/MS and theoretical fragmentation spectra. Candidates are first generated, and these consist of all of the crosslinks with calculated masses that fall within a defined range bracketing the precursor mass measured in the experimental MS/MS spectrum. For each of the candidate crosslinks, a theoretical fragmentation spectrum is generated. As opposed to the general processing of non-modified peptides, only b- and y-ions are generally considered for crosslinks. This is because each crosslink contains two or more peptides and their theoretical fragmentation spectra become very complicated if other ion types, such as a-ions and those arising from loss of H2O and NH3 are also considered. Existing software tools are summarized in Table 4.
Table 4.
Search Engines
| Name | Language | Publication year | Platform | Reference |
|---|---|---|---|---|
| PeptideMap | 1997 | (105) | ||
| ASAP and MS2Asign | 2000 | (106) | ||
| GPMAW | 2001 | (107) | ||
| X-Link | 2002 | (108) | ||
| Popitam | 2003 | (109) | ||
| MS2PRO | 2003 | (110) | ||
| Links and MS2Link | 2004 | (111) | ||
| CLPM | 2005 | (112) | ||
| XLINK | 2006 | (113) | ||
| VIRTUAL-MSLAB | 2006 | (114) | ||
| SearchXLinks | 2006 | (115,116) | ||
| Pro-Crosslink | 2006 | (117) | ||
| X!Link | 2007 | (118,119) | ||
| X-Links | 2007 | (120) | ||
| CrossSearch | 2008 | (100) | ||
| MS-3D | 2008 | (121) | ||
| xComb | 2010 | (122) | ||
| xQuest | 2010 | (123,124) | ||
| Mass-Matrix | 2010 | (125) | ||
| CRUX | 2010 | (126) | ||
| MS-Bridge | 2010 | (127) | ||
| Xlink-Identifier | C++ | 2011 | Web | (128) |
| CrossWork | 2011 | (129) | ||
| StavroX | 2012 | (130) | ||
| pLink | 2012 | (131) | ||
| SQID-XLinK | C | 2012 | Windows | (132) |
| Hekate | Perl and SQL | 2013 | Debian Linux | (133) |
| XLPM | Perl and MySQL | 2014 | Web | (134) |
| MXDB | 2014 | (135) | ||
| AnchorMS | Python with PHP front-end | 2014 | Web | (136) |
| SIM-XL | C# | 2015 | Windows | (137) |
3.3 General protocols
Because crosslinking is an entirely empirical process, the following sections will focus primarily on developing screens, rather than explicit protocols, to determine the best conditions and reagents for optimizing the yield and digestion of a desired conjugate from protein targets. Because CXMS is a bottom-up process, we will assume in these screens that protein reactants are purified to near homogeneity.
3.3.1 Crosslinking screens
Ideally in any reagent screen, it is advisable to first analyze the target protein under conditions that allow its native state or that support a known function and/or interaction with a specific partner. The concentration of protein should be sufficient for visualization using general gel staining procedures. Under such conditions, either the time or concentration can be varied for the CXR. In one-step crosslinking screens, the concentration of CXR is generally varied in molar excess from 10 to 500 over the protein target, initially for a fixed time of 15 minutes. Conversely, greater than 500 molar excesses of CXR are incubated with the protein for short time periods, ranging between 1 and 10 minutes. Using small gel formats with 15 to 20 wells, 3–4 reagents may be assessed per gel, and as many as 16 reagents may be tested in one day. If any conjugates are observed, then reaction conditions may be varied to optimize formation of the desired conjugate. During the screening process, care must be taken to assure that accessory components (e.g., buffers, salts, stabilizing reagents) are compatible with the crosslinker being used. For example, amine containing buffers such as TRIS should be avoided when using NHS-substituted CXRs or any other functional group that targets amines. To avoid large quantities of side-product formation, excessive amounts of crosslinker should be avoided, and only the amount required to generate a sufficient yield of the desired conjugate should be used. Additionally, extremely high pH values should be avoided, because most conventional CXR reactive groups are susceptible to hydrolysis and are either rapidly deactivated or preferentially mono-modify the protein target to form dead-end side products.
Screens using heterobifunctional CXRs to form conjugates in two-step crosslinking protocols are more complicated than one-step screens, because of the intermediate purification step required between successive modification steps with each of the two functional groups of the CXR (see 3.2.2). A rapid assessment of conditions required for two-step crosslinking can be achieved by using small one-mL spin columns loaded with desalting gel media to partially purify the complex after the first modification step and to exchange it into reaction media that are compatible with the second modification step. For example when screening conditions for optimizing crosslinking with a heterobifunctional CXR containing photo-reactive azido and NHS functional groups, time- and concentration-dependent modification of the protein target with the NHS group is carried out in the first step, as described above under one-step crosslinking. To avoid activating the azido group, reactions should be carried out using Eppendorf tubes that are not transparent to light and the total volume for each condition should not exceed 100 μl. Reactions are simply quenched by removing excess reagent with the desalting spin column using a benchtop centrifuge. The desalting gel should be equilibrated in a buffer solution that is compatible with the second photolysis step. Multiple samples may be loaded onto crystallization trays that contain shallow wells, and exposed simultaneously to UV light using a simple hand-held lamp that is placed over the tray for 2–3 minutes. The reactions are then quenched using SDS-buffer and loaded onto gels to analyze product formation. Although the spin columns do not remove all excess CXR and do not permit a complete exchange of conditions, they provide an efficient method for narrowing the conditions required for optimal crosslinking of the target.
3.3.2 Digestion of conjugates for MS analyses
In-gel or hetero-phase and solution digestions are the two most common approaches for hydrolyzing crosslinked proteins, and MS facilities generally provide basic protocols to follow for sample submission or provide services to perform these procedures. However, the preparation of protein samples, and specifically crosslinked proteins, for MS analyses is a critical and often overlooked component of CXMS. The ultimate goal of this process is to maximize the coverage of the crosslinked protein, which requires optimal cleavage and recovery of the peptide components of the conjugate. Both in-gel and solution methods require similar components, which include a targeting protease or chemical to catalyze hydrolysis at specific sites along the amide chain, a denaturant to unfold the protein to enhance maximal cleavage along the backbone, a reductant (typically either DTT or 2-mercaptoethanol) to reduce cystine disulfides and an alkylating reagent (iodoacetic acid or iodoacetamide) to modify free thiols generated by reduction. The latter two steps are carried out to prevent refolding. Proteins have unique properties and are targeted to different extents by specific proteases, with the covalent attachments introduced by crosslinking further complicating such interactions. As with crosslinking, proteolysis is also an empirical process and must be optimized by varying solution conditions and the general components discussed above (138). Typically, the reaction steps are carried out in the following order: denaturation, reduction, alkylation and proteolysis. Historically, denaturants such as urea, guanidinium hydrochloride and SDS were used and subsequently diluted after reduction and alkylation steps to concentrations tolerated by the protease; however, they interfere and are poorly tolerated by MS. To address this problem, more MS-friendly denaturants such as Rapigest™ (Waters, Milford MA) (139), sodium deoxycholate (SDC) (139) or sodium 3-[(2-methyl-2-undecyl-1,3-dioxolan-4-yl)methoxyl]-1-propanesulfonate (ALS) (140) have been developed. Alternatively, spin concentrators and various filters have been developed to facilitate exchange of secondary chemicals and denaturants without significant loss of the conjugate prior to proteolysis (139,141). Additional methods for improving protein denaturation, including thermal (IR and microwave radiation), ultrasonic and solvent-based techniques, are summarized in an excellent review by Hustoft et al. (142). After denaturation, engineered forms of trypsin are generally used to carry out proteolysis, because they specifically cleave amide backbones after lysine and arginine, function well in low concentrations of multiple denaturants and are relatively resistant to autolysis. A recent report suggests that tandem application of trypsin and Lys-C (lysine–specific protease) promotes more efficient cleavage of protein substrates than trypsin alone (138). Despite all the possible choices in such reactions, some of the following parameters are good starting points for in solution digestions. First, the conjugate may be reduced with DTT (10 M excess) and denatured concomitantly in either 0.1% ALS or SDC at elevated temperatures (~ 50–85 °C) for 1 hr. This is followed by alkylation with iodoacetic acid (40 M excess) in the dark for 30 minutes at 30 °C. After alkylation, DTT is added in excess of iodoacetic acid to prevent alkylation of trypsin. The denatured protein may then be exchanged into 25 mM ammonium bicarbonate using a 3000 MW cutoff spin concentrator (EMD Millipore) and digested overnight at 30 °C using a 25-fold excess (w/w) of sequencing grade trypsin (Promega). Peptides may be recovered by several rounds of centrifugation and washes with 10% acetonitrile in 25 mM ammonium bicarbonate. Peptides are then concentrated to remove acetonitrile or lyophilized in a vacuum concentrator.
In-gel digestion uses the resolving power of SDS-PAGE to isolate the desired conjugate from complex mixtures of crosslinking products, significantly reducing the number of products to be analyzed. On the other hand, it also can hinder peptide recovery, depending to a great extent on the type of extraction procedure used. Several aspects of this technique are unique compared to in-solution methods, based on the polyacrylamide matrix, which limits diffusion of the reactants and protease necessary for generating peptides (143). Thus, the ratios of protease to substrate are generally much higher than those typically used in solution. Additionally, the gel sections containing the conjugate must be treated with solvents (typically 50% acetonitrile in 25 mM ammonium bicarbonate) to remove SDS and other gel solution components that inhibit the activity of the protease. Another important consideration is that there are many handling steps that can potentially introduce contaminants, particularly keratins. Thus all reagents must be prepared carefully and any instruments used must be cleaned scrupulously before carrying out the procedure. Gloves and sterile sleeve protectors should be worn at all times. Specific details for gel-phase proteolysis conditions are outlined in a published protocol (143) and can be accessed online at the UCSF mass spectrometry website.
3.3.3 Data input
As discussed in 3.2.3, the use of search engines generally requires little from the user. Most are designed with interfaces that allow the user to upload the sequence(s) and reagents being tested. Additionally, some parameters such as the number of allowed side products and crosslinks per conjugate may also be available. Typically it’s wise to limit these parameters in the first rounds of an analysis, first to minimize computing space and time, and second to avoid extensive data output. Some programs ask the user to specify the reactive groups of the CXR and the mass of the intervening spacer (after modification), as well as the mass of dead-end products. Users with limited knowledge of cross-linking or chemistry should avoid the latter programs.
3.4 Caveats
Perhaps the greatest mistake made by even experienced users of the crosslinking technique is to over interpret results. First, there is a tendency in the literature for users to define a detected CX site on a protein as a binding site, no matter the span of the CRX. The specificity and therefore the probability that crosslinking represents an actual binding event is greatest when zero-length chemically coupled residues on opposing binding partners are identified. CX sites that are detected using CXR reagents with crosslinking spans greater than 2–3 Å should be discussed in terms of the proximity of the linked residues, defined by the range of distances that the spacer can occupy in solution (144). Another common misconception is that the absence of crosslinking indicates absence of interaction (145). In this case there are many more reasons why crosslinking does not occur, based on incompatibility of the CXR with the chemistry, geometry and/or solvent accessibility of the protein-protein interaction surface(s) being studied. There are many examples for which CX sites are purportedly identified by simply matching the experimental mass of a precursor ion with the theoretical mass of a crosslinked peptide. With large proteins it has been demonstrated that a large number of dead-end and crosslinked peptides may account for a single precursor ion within the resolving limits (~ 3 ppm) of a high resolution FT instrument (100). Even when the masses of a precursor and its corresponding fragments are well matched to those for a theoretically generated candidate, there is a reasonable potential for misidentification, based on the limited resolving capabilities (> 200 ppm) of typical collision cells, i.e. significant error in the identification fragment ions generated by tandem MS. High resolution measurements of fragment ions are now possible in new Orbitrap MS instruments using HCD, significantly increasing the potential for boosting confidence levels in matching assignments. Finally, corroborating evidence from alternative methods is always desirable for any interaction that is detected or suggested by crosslinking.
3.5 Representative results and complementary techniques
Because of its versatility, CXMS has been used in combination with many complementary techniques developed to detect protein-protein interactions, to determine the structure of proteins and their complexes and to theoretically model non-homologous proteins. Several examples of these combined approaches will be discussed in terms of how each complements the other. With the development of MS instruments that are capable of transmitting large macromolecular complexes (146), top-down MS has become a well-established method for analyzing the interaction of proteins and/or subunits in large protein complexes that are not amenable to NMR and X-ray crystallographic methods (147), providing a potent alternative and complementary approach to crosslinking (94). The basic approach relies on the transmission of a partially hydrated protein complex in near-native conditions, in which the topological arrangement and interactions of its protein components are probed either by CID after injection (148) or the introduction of sub-stoichiometric amounts of small molecules that destabilize the complex prior to injection (149). Maps defining the interactions of integral subunits in the complex are constructed based on the composition and number of subcomplexes detected (150). Top-down MS also is capable of detecting differences in the stability of a complex in different conformational states (151). For example Lane et al. showed that the native, non-activated form of the (αβγδ)4 phosphorylase kinase complex (PhK) is more stable than its active phosphorylated form by demonstrating that the percentage of intact phosphorylated complex decreased with respect to that of the native under identical conditions (151). In that study, phosphorylation of the complex also perturbed interactions of the subunits in the complex, resulting in preferential interactions among the regulatory β subunits, also detected by crosslinking in a previous study (152). In a parallel study, these investigators combined CXMS, immuno EM, cryoEM, modeling and biochemical data to determine the location of the regulatory β subunits in the PhK complex (89). The topology and location of the subunits in the connecting bridge region of the bilobal complex was determined using top-down MS, and CXMS was used to constrain an atomic model of the β subunit (generated by I-TASSER (153)) to facilitate its docking in the bridges of the cryoEM 3D structure. Aebersold and coworkers have also used CXMS to provide distance constraints in combination with tandem affinity purification to model a protein phosphatase 2A network of interactions (154). CXMS has become the method of choice for constraining theoretical models (155), and is widely used in integrative structural modeling (ISM), an approach in which theoretical models of variable resolution are scored, based on their agreement with constraints provided by different forms of experimental data, commonly referred to as input data (156). ISM approaches using CXMS have been used to model complex macromolecular assemblies, including the yeast eIF3:eIF5 complex (90) and the photoreceptor phosphodiesterase hetero-oligomer (157).
References
- 1.Baldwin RL. Early days of protein hydrogen exchange: 1954–1972. Proteins. 2011;79:2021–2026. doi: 10.1002/prot.23039. [DOI] [PubMed] [Google Scholar]
- 2.Hvidt A, Linderstrom-Lang K. Exchange of hydrogen atoms in insulin with deuterium atoms in aqueous solutions. Biochimica et biophysica acta. 1954;14:574–575. doi: 10.1016/0006-3002(54)90241-3. [DOI] [PubMed] [Google Scholar]
- 3.Schellman JA, Schellman CG. Kaj Ulrik Linderstrom-Lang (1896–1959) Protein Sci. 1997;6:1092–1100. doi: 10.1002/pro.5560060516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheinblatt M. Determination of an acidity scale for peptide hydrogens from nuclear magnetic resonance kinetic studies. Journal of the American Chemical Society. 1970;92:2505–2509. doi: 10.1021/ja00711a048. [DOI] [PubMed] [Google Scholar]
- 5.Molday RS, Englander SW, Kallen RG. Primary structure effects on peptide group hydrogen exchange. Biochemistry. 1972;11:150–158. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- 6.Rosa JJ, Richards FM. An experimental procedure for increasing the structural resolution of chemical hydrogen-exchange measurements on proteins: application to ribonuclease S peptide. J Mol Biol. 1979;133:399–416. doi: 10.1016/0022-2836(79)90400-5. [DOI] [PubMed] [Google Scholar]
- 7.Wagner G, Wuthrich K. Amide protein exchange and surface conformation of the basic pancreatic trypsin inhibitor in solution. Studies with two-dimensional nuclear magnetic resonance. Journal of molecular biology. 1982;160:343–361. doi: 10.1016/0022-2836(82)90180-2. [DOI] [PubMed] [Google Scholar]
- 8.Katta V, Chait BT. Conformational changes in proteins probed by hydrogen-exchange electrospray-ionization mass spectrometry. Rapid communications in mass spectrometry: RCM. 1991;5:214–217. doi: 10.1002/rcm.1290050415. [DOI] [PubMed] [Google Scholar]
- 9.Pascal BD, Willis S, Lauer JL, Landgraf RR, West GM, Marciano D, Novick S, Goswami D, Chalmers MJ, Griffin PR. HDX workbench: software for the analysis of H/D exchange MS data. Journal of the American Society for Mass Spectrometry. 2012;23:1512–1521. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villar MT, Miller DE, Fenton AW, Artigues A. SAIDE: A Semi-Automated Interface for Hydrogen/Deuterium Exchange Mass Spectrometry. Proteomica. 2010;6:63–69. [PMC free article] [PubMed] [Google Scholar]
- 11.Englander JJ, Rogero JR, Englander SW. Protein hydrogen exchange studied by the fragment separation method. Analytical biochemistry. 1985;147:234–244. doi: 10.1016/0003-2697(85)90033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass spectrometry reviews. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith DL, Deng Y, Zhang Z. Probing the non-covalent structure of proteins by amide hydrogen exchange and mass spectrometry. Journal of mass spectrometry: JMS. 1997;32:135–146. doi: 10.1002/(SICI)1096-9888(199702)32:2<135::AID-JMS486>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 15.Hamuro Y, Coales SJ, Southern MR, Nemeth-Cawley JF, Stranz DD, Griffin PR. Rapid analysis of protein structure and dynamics by hydrogen/deuterium exchange mass spectrometry. Journal of biomolecular techniques: JBT. 2003;14:171–182. [PMC free article] [PubMed] [Google Scholar]
- 16.Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. Journal of the American Society for Mass Spectrometry. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Busenlehner LS, Armstrong RN. Insights into enzyme structure and dynamics elucidated by amide H/D exchange mass spectrometry. Archives of biochemistry and biophysics. 2005;433:34–46. doi: 10.1016/j.abb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Analytical chemistry. 2006;78:1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- 19.Chalmers MJ, Busby SA, Pascal BD, Southern MR, Griffin PR. A two-stage differential hydrogen deuterium exchange method for the rapid characterization of protein/ligand interactions. Journal of biomolecular techniques: JBT. 2007;18:194–204. [PMC free article] [PubMed] [Google Scholar]
- 20.Hoofnagle AN, Resing KA, Ahn NG. Practical methods for deuterium exchange/mass spectrometry. Methods in molecular biology. 2004;250:283–298. doi: 10.1385/1-59259-671-1:283. [DOI] [PubMed] [Google Scholar]
- 21.Englander SW, Downer NW, Teitelbaum H. Hydrogen exchange. Annual review of biochemistry. 1972;41:903–924. doi: 10.1146/annurev.bi.41.070172.004351. [DOI] [PubMed] [Google Scholar]
- 22.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weis DD, Wales TE, Engen JR, Hotchko M, Ten Eyck LF. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. Journal of the American Society for Mass Spectrometry. 2006;17:1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Ferraro DM, Lazo N, Robertson AD. EX1 hydrogen exchange and protein folding. Biochemistry. 2004;43:587–594. doi: 10.1021/bi035943y. [DOI] [PubMed] [Google Scholar]
- 25.Krishna MM, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods. 2004;34:51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Miller DE, Prasannan CB, Villar MT, Fenton AW, Artigues A. HDXFinder: Automated Analysis and Data Reporting of Deuterium/Hydrogen Exchange Mass Spectrometry. J Am Soc Mass Spectrom. 2012;23:425–429. doi: 10.1007/s13361-011-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pascal BD, Chalmers MJ, Busby SA, Griffin PR. HD desktop: an integrated platform for the analysis and visualization of H/D exchange data. J Am Soc Mass Spectrom. 2009;20:601–610. doi: 10.1016/j.jasms.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. Journal of the American Society for Mass Spectrometry. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 29.Lou X, Kirchner M, Renard BY, Kothe U, Boppel S, Graf C, Lee CT, Steen JA, Steen H, Mayer MP, Hamprecht FA. Deuteration distribution estimation with improved sequence coverage for HX/MS experiments. Bioinformatics. 2010;26:1535–1541. doi: 10.1093/bioinformatics/btq165. [DOI] [PubMed] [Google Scholar]
- 30.Lindner R, Lou X, Reinstein J, Shoeman RL, Hamprecht FA, Winkler A. Hexicon 2: automated processing of hydrogen-deuterium exchange mass spectrometry data with improved deuteration distribution estimation. Journal of the American Society for Mass Spectrometry. 2014;25:1018–1028. doi: 10.1007/s13361-014-0850-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Marshall AG. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J Am Soc Mass Spectrom. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 32.Connelly GP, Bai Y, Jeng MF, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 33.Liu YH, Konermann L. Enzyme conformational dynamics during catalysis and in the ‘resting state’ monitored by hydrogen/deuterium exchange mass spectrometry. FEBS letters. 2006;580:5137–5142. doi: 10.1016/j.febslet.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 34.Hu W, Walters BT, Kan ZY, Mayne L, Rosen LE, Marqusee S, Englander SW. Stepwise protein folding at near amino acid resolution by hydrogen exchange and mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7684–7689. doi: 10.1073/pnas.1305887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wintrode PL, Rojsajjakul T, Vadrevu R, Matthews CR, Smith DL. An obligatory intermediate controls the folding of the alpha-subunit of tryptophan synthase, a TIM barrel protein. Journal of molecular biology. 2005;347:911–919. doi: 10.1016/j.jmb.2005.01.064. [DOI] [PubMed] [Google Scholar]
- 36.Yang H, Smith DL. Kinetics of cytochrome c folding examined by hydrogen exchange and mass spectrometry. Biochemistry. 1997;36:14992–14999. doi: 10.1021/bi9717183. [DOI] [PubMed] [Google Scholar]
- 37.Busby SA, Chalmers MJ, Griffin PR. Improving digestion efficiency under H/D exchange conditions with activated pepsinogen coupled cloumns. Int J Mass Spectrom. 2007;259:130–139. [Google Scholar]
- 38.Ahn J, Jung MC, Wyndham K, Yu YQ, Engen JR. Pepsin immobilized on high-strength hybrid particles for continuous flow online digestion at 10,000 psi. Analytical chemistry. 2012;84:7256–7262. doi: 10.1021/ac301749h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y, Kaveti S, Engen JR. Extensive deuterium back-exchange in certain immobilized pepsin columns used for H/D exchange mass spectrometry. Analytical chemistry. 2006;78:1719–1723. doi: 10.1021/ac0518497. [DOI] [PubMed] [Google Scholar]
- 40.Tsybin YO, Haselmann KF, Emmett MR, Hendrickson CL, Marshall AG. Charge location directs electron capture dissociation of peptide dications. Journal of the American Society for Mass Spectrometry. 2006;17:1704–1711. doi: 10.1016/j.jasms.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 41.Demmers JA, Rijkers DT, Haverkamp J, Killian JA, Heck AJ. Factors affecting gas-phase deuterium scrambling in peptide ions and their implications for protein structure determination. Journal of the American Chemical Society. 2002;124:11191–11198. doi: 10.1021/ja0125927. [DOI] [PubMed] [Google Scholar]
- 42.Ferguson PL, Konermann L. Nonuniform isotope patterns produced by collision-induced dissociation of homogeneously labeled ubiquitin: implications for spatially resolved hydrogen/deuterium exchange ESI-MS studies. Analytical chemistry. 2008;80:4078–4086. doi: 10.1021/ac8001963. [DOI] [PubMed] [Google Scholar]
- 43.Ferguson PL, Pan J, Wilson DJ, Dempsey B, Lajoie G, Shilton B, Konermann L. Hydrogen/deuterium scrambling during quadrupole time-of-flight MS/MS analysis of a zinc-binding protein domain. Analytical chemistry. 2007;79:153–160. doi: 10.1021/ac061261f. [DOI] [PubMed] [Google Scholar]
- 44.Jorgensen TJ, Bache N, Roepstorff P, Gardsvoll H, Ploug M. Collisional activation by MALDI tandem time-of-flight mass spectrometry induces intramolecular migration of amide hydrogens in protonated peptides. Molecular & cellular proteomics: MCP. 2005;4:1910–1919. doi: 10.1074/mcp.M500163-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Jorgensen TJ, Gardsvoll H, Ploug M, Roepstorff P. Intramolecular migration of amide hydrogens in protonated peptides upon collisional activation. Journal of the American Chemical Society. 2005;127:2785–2793. doi: 10.1021/ja043789c. [DOI] [PubMed] [Google Scholar]
- 46.Kim MY, Maier CS, Reed DJ, Deinzer ML. Site-specific amide hydrogen/deuterium exchange in E. coli thioredoxins measured by electrospray ionization mass spectrometry. Journal of the American Chemical Society. 2001;123:9860–9866. doi: 10.1021/ja010901n. [DOI] [PubMed] [Google Scholar]
- 47.Xu G, Takamoto K, Chance MR. Radiolytic modification of basic amino acid residues in peptides: probes for examining protein-protein interactions. Analytical chemistry. 2003;75:6995–7007. doi: 10.1021/ac035104h. [DOI] [PubMed] [Google Scholar]
- 48.Sharp JS, Becker JM, Hettich RL. Protein surface mapping by chemical oxidation: structural analysis by mass spectrometry. Analytical biochemistry. 2003;313:216–225. doi: 10.1016/s0003-2697(02)00612-7. [DOI] [PubMed] [Google Scholar]
- 49.Hambly DM, Gross ML. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. Journal of the American Society for Mass Spectrometry. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 50.Takamoto K, Chance MR. Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annual review of biophysics and biomolecular structure. 2006;35:251–276. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 51.Konermann L, Stocks BB, Pan Y, Tong X. Mass spectrometry combined with oxidative labeling for exploring protein structure and folding. Mass spectrometry reviews. 2010;29:651–667. doi: 10.1002/mas.20256. [DOI] [PubMed] [Google Scholar]
- 52.Linderstrom-Land K, Ottesen M. A new protein from ovalbumin. Nature. 1947;159:807. doi: 10.1038/159807a0. [DOI] [PubMed] [Google Scholar]
- 53.Neurath H. Limited proteolysis, protein folding and physiological regulation. In: Jaenicke R, editor. Protein Folding. Elsevier/North-Holland Biomedical Press, University of Regensburg; Regensburg, West Germany: 1979. [Google Scholar]
- 54.Bloxham DP, Ericsson LH, Titani K, Walsh KA, Neurath H. Limited proteolysis of pig heart citrate synthase by subtilisin, chymotrypsin, and trypsin. Biochemistry (Mosc) 1980;19:3979–3985. doi: 10.1021/bi00558a014. [DOI] [PubMed] [Google Scholar]
- 55.Fontana A, de Laureto PP, Spolaore B, Frare E. Identifying disordered regions in proteins by limited proteolysis. Methods Mol Biol. 2012;896:297–318. doi: 10.1007/978-1-4614-3704-8_20. [DOI] [PubMed] [Google Scholar]
- 56.Fontana A, Fassina G, Vita C, Dalzoppo D, Zamai M, Zambonin M. Correlation between sites of limited proteolysis and segmental mobility in thermolysin. Biochemistry (Mosc) 1986;25:1847–1851. doi: 10.1021/bi00356a001. [DOI] [PubMed] [Google Scholar]
- 57.Bantscheff M, Weiss V, Glocker MO. Identification of linker regions and domain borders of the transcription activator protein NtrC from Escherichia coli by limited proteolysis, in-gel digestion, and mass spectrometry. Biochemistry (Mosc) 1999;38:11012–11020. doi: 10.1021/bi990781k. [DOI] [PubMed] [Google Scholar]
- 58.Hubbard SJ. The structural aspects of limited proteolysis of native proteins. Biochim Biophys Acta. 1998;1382:191–206. doi: 10.1016/s0167-4838(97)00175-1. [DOI] [PubMed] [Google Scholar]
- 59.Hubbard SJ, Eisenmenger F, Thornton JM. Modeling studies of the change in conformation required for cleavage of limited proteolytic sites. Protein Sci. 1994;3:757–768. doi: 10.1002/pro.5560030505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 61.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 62.Suh MJ, Pourshahian S, Limbach PA. Developing limited proteolysis and mass spectrometry for the characterization of ribosome topography. J Am Soc Mass Spectrom. 2007;18:1304–1317. doi: 10.1016/j.jasms.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng Y, De Franceschi G, Kahraman A, Soste M, Melnik A, Boersema PJ, de Laureto PP, Nikolaev Y, Oliveira AP, Picotti P. Global analysis of protein structural changes in complex proteomes. Nat Biotechnol. 2014;32:1036–1044. doi: 10.1038/nbt.2999. [DOI] [PubMed] [Google Scholar]
- 64.Orru S, Dal Piaz F, Casbarra A, Biasiol G, De Francesco R, Steinkuhler C, Pucci P. Conformational changes in the NS3 protease from hepatitis C virus strain Bk monitored by limited proteolysis and mass spectrometry. Protein Sci. 1999;8:1445–1454. doi: 10.1110/ps.8.7.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zappacosta F, Pessi A, Bianchi E, Venturini S, Sollazzo M, Tramontano A, Marino G, Pucci P. Probing the tertiary structure of proteins by limited proteolysis and mass spectrometry: the case of Minibody. Protein Sci. 1996;5:802–813. doi: 10.1002/pro.5560050502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bothner B, Dong XF, Bibbs L, Johnson JE, Siuzdak G. Evidence of viral capsid dynamics using limited proteolysis and mass spectrometry. J Biol Chem. 1998;273:673–676. doi: 10.1074/jbc.273.2.673. [DOI] [PubMed] [Google Scholar]
- 67.Fontana A, Zambonin M, Polverino de Laureto P, De Filippis V, Clementi A, Scaramella E. Probing the conformational state of apomyoglobin by limited proteolysis. J Mol Biol. 1997;266:223–230. doi: 10.1006/jmbi.1996.0787. [DOI] [PubMed] [Google Scholar]
- 68.Villa JA, Cabezas M, de la Cruz F, Moncalian G. Use of limited proteolysis and mutagenesis to identify folding domains and sequence motifs critical for wax ester synthase/acyl coenzyme A:diacylglycerol acyltransferase activity. Appl Environ Microbiol. 2014;80:1132–1141. doi: 10.1128/AEM.03433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Graves DJ, Hayakawa T, Horvitz RA, Beckman E, Krebs EG. Studies on the subunit structure of trypsin-activated phosphorylase kinase. Biochemistry (Mosc) 1973;12:580–585. doi: 10.1021/bi00728a003. [DOI] [PubMed] [Google Scholar]
- 70.Potter RL, Taylor SS. The structural domains of cAMP-dependent protein kinase I. Characterization of two sites of proteolytic cleavage and homologies to cAMP-dependent protein kinase II. J Biol Chem. 1980;255:9706–9712. [PubMed] [Google Scholar]
- 71.Fontana A, de Laureto PP, Spolaore B, Frare E, Picotti P, Zambonin M. Probing protein structure by limited proteolysis. Acta Biochim Pol. 2004;51:299–321. [PubMed] [Google Scholar]
- 72.Scaloni A, Miraglia N, Orru S, Amodeo P, Motta A, Marino G, Pucci P. Topology of the calmodulin-melittin complex. J Mol Biol. 1998;277:945–958. doi: 10.1006/jmbi.1998.1629. [DOI] [PubMed] [Google Scholar]
- 73.Cohen SL, Ferre-D’Amare AR, Burley SK, Chait BT. Probing the solution structure of the DNA-binding protein Max by a combination of proteolysis and mass spectrometry. Protein Sci. 1995;4:1088–1099. doi: 10.1002/pro.5560040607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Monti M, Pucci P. Mass Spectrometry of Protein Interactions. John Wiley & Sons, Inc; 2006. Limited Proteolysis Mass Spectrometry of Protein Complexes; pp. 63–82. [Google Scholar]
- 75.Suckau D, Kohl J, Karwath G, Schneider K, Casaretto M, Bitter-Suermann D, Przybylski M. Molecular epitope identification by limited proteolysis of an immobilized antigen-antibody complex and mass spectrometric peptide mapping. Proc Natl Acad Sci U S A. 1990;87:9848–9852. doi: 10.1073/pnas.87.24.9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trempe MR, Carlson GM. Phosphorylase kinase conformers. Detection by proteases. J Biol Chem. 1987;262:4333–4340. [PubMed] [Google Scholar]
- 77.Trempe MR, Carlson GM, Hainfeld JF, Furcinitti PS, Wall JS. Analyses of phosphorylase kinase by transmission and scanning transmission electron microscopy. J Biol Chem. 1986;261:2882–2889. [PubMed] [Google Scholar]
- 78.Kemp BE, Pearson RB. Intrasteric regulation of protein kinases and phosphatases. Biochim Biophys Acta. 1991;1094:67–76. doi: 10.1016/0167-4889(91)90027-u. [DOI] [PubMed] [Google Scholar]
- 79.Kobe B, Kemp BE. Active site-directed protein regulation. Nature. 1999;402:373–376. doi: 10.1038/46478. [DOI] [PubMed] [Google Scholar]
- 80.Xu G, Chance MR. Radiolytic modification and reactivity of amino acid residues serving as structural probes for protein footprinting. Anal Chem. 2005;77:4549–4555. doi: 10.1021/ac050299+. [DOI] [PubMed] [Google Scholar]
- 81.Kiselar JG, Chance MR. Future directions of structural mass spectrometry using hydroxyl radical footprinting. J Mass Spectrom. 2010;45:1373–1382. doi: 10.1002/jms.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang H, Gau BC, Jones LM, Vidavsky I, Gross ML. Fast photochemical oxidation of proteins for comparing structures of protein-ligand complexes: the calmodulin-peptide model system. Anal Chem. 2011;83:311–318. doi: 10.1021/ac102426d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hanai R, Wang JC. Protein footprinting by the combined use of reversible and irreversible lysine modifications. Proc Natl Acad Sci U S A. 1994;91:11904–11908. doi: 10.1073/pnas.91.25.11904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tu BP, Wang JC. Protein footprinting at cysteines: probing ATP-modulated contacts in cysteine-substitution mutants of yeast DNA topoisomerase II. Proc Natl Acad Sci U S A. 1999;96:4862–4867. doi: 10.1073/pnas.96.9.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nadeau OW, Carlson GM. Protein Interactions Captured by Chemical Cross-linking. In: Golemis E, Adams PD, editors. Protein-protein interactions: A Molecular Cloning Manual. 2nd. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2005. pp. 105–127. [Google Scholar]
- 86.Nadeau OW. Protein Interaction Analysis: Chemical Cross-linking. In: Ganten D, Ruckpaul K, editors. Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine. Springer; Berlin: 2006. [Google Scholar]
- 87.Hermanson GT. Bioconjugate techniques. 2nd. Elsevier Academic Press; Amsterdam; Boston: 2008. [Google Scholar]
- 88.Wong SS. Chemistry of Protein Conjugation and Cross-linking. CRC Press; Boca Raton, FL: 1993. [Google Scholar]
- 89.Nadeau OW, Lane LA, Xu D, Sage J, Priddy TS, Artigues A, Villar MT, Yang Q, Robinson CV, Zhang Y, Carlson GM. Structure and location of the regulatory beta subunits in the (alphabetagammadelta)4 phosphorylase kinase complex. J Biol Chem. 2012;287:36651–36661. doi: 10.1074/jbc.M112.412874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Politis A, Schmidt C, Tjioe E, Sandercock AM, Lasker K, Gordiyenko Y, Russel D, Sali A, Robinson CV. Topological models of heteromeric protein assemblies from mass spectrometry: application to the yeast eIF3:eIF5 complex. Chem Biol. 2015;22:117–128. doi: 10.1016/j.chembiol.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Trnka MJ, Burlingame AL. Topographic studies of the GroEL-GroES chaperonin complex by chemical cross-linking using diformyl ethynylbenzene: the power of high resolution electron transfer dissociation for determination of both peptide sequences and their attachment sites. Molecular & cellular proteomics: MCP. 2010;9:2306–2317. doi: 10.1074/mcp.M110.003764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Paramelle D, Miralles G, Subra G, Martinez J. Chemical cross-linkers for protein structure studies by mass spectrometry. Proteomics. 2013;13:438–456. doi: 10.1002/pmic.201200305. [DOI] [PubMed] [Google Scholar]
- 93.Singh P, Panchaud A, Goodlett DR. Chemical cross-linking and mass spectrometry as a low-resolution protein structure determination technique. Anal Chem. 2010;82:2636–2642. doi: 10.1021/ac1000724. [DOI] [PubMed] [Google Scholar]
- 94.Stengel F, Aebersold R, Robinson CV. Joining forces: integrating proteomics and cross-linking with the mass spectrometry of intact complexes. Molecular & cellular proteomics: MCP. 2012;11:R111 014027. doi: 10.1074/mcp.R111.014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chowdhury SM, Munske GR, Tang X, Bruce JE. Collisionally activated dissociation and electron capture dissociation of several mass spectrometry-identifiable chemical cross-linkers. Anal Chem. 2006;78:8183–8193. doi: 10.1021/ac060789h. [DOI] [PubMed] [Google Scholar]
- 96.Tang X, Munske GR, Siems WF, Bruce JE. Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal Chem. 2005;77:311–318. doi: 10.1021/ac0488762. [DOI] [PubMed] [Google Scholar]
- 97.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 98.Chowdhury SM, Du X, Tolic N, Wu S, Moore RJ, Mayer MU, Smith RD, Adkins JN. Identification of cross-linked peptides after click-based enrichment using sequential collision-induced dissociation and electron transfer dissociation tandem mass spectrometry. Anal Chem. 2009;81:5524–5532. doi: 10.1021/ac900853k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vellucci D, Kao A, Kaake RM, Rychnovsky SD, Huang L. Selective enrichment and identification of azide-tagged cross-linked peptides using chemical ligation and mass spectrometry. J Am Soc Mass Spectrom. 2010;21:1432–1445. doi: 10.1016/j.jasms.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nadeau OW, Wyckoff GJ, Paschall JE, Artigues A, Sage J, Villar MT, Carlson GM. CrossSearch, a user-friendly search engine for detecting chemically cross-linked peptides in conjugated proteins. Molecular & cellular proteomics: MCP. 2008;7:739–749. doi: 10.1074/mcp.M800020-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J. Structural Analysis of Multiprotein Complexes by Cross-linking, Mass Spectrometry, and Database Searching. Molecular & cellular proteomics: MCP. 2007;6:2200–2211. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- 102.Hoopmann MR, Weisbrod CR, Bruce JE. Improved strategies for rapid identification of chemically cross-linked peptides using protein interaction reporter technology. Journal of proteome research. 2010;9:6323–6333. doi: 10.1021/pr100572u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ihling C, Falvo F, Kratochvil I, Sinz A, Schafer M. Dissociation behavior of a bifunctional tempoactive ester reagent for peptide structure analysis by free radical initiated peptide sequencing (FRIPS) mass spectrometry. J Mass Spectrom. 2015;50:396–406. doi: 10.1002/jms.3543. [DOI] [PubMed] [Google Scholar]
- 104.Jedrychowski MP, Huttlin EL, Haas W, Sowa ME, Rad R, Gygi SP. Evaluation of HCD- and CID-type fragmentation within their respective detection platforms for murine phosphoproteomics. Molecular & cellular proteomics: MCP. 2011;10:M111 009910. doi: 10.1074/mcp.M111.009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fenyo D. A software tool for the analysis of mass spectrometric disulfide mapping experiments. Comput Appl Biosci. 1997;13:617–618. doi: 10.1093/bioinformatics/13.6.617. [DOI] [PubMed] [Google Scholar]
- 106.Young MM, Tang N, Hempel JC, Oshiro CM, Taylor EW, Kuntz ID, Gibson BW, Dollinger G. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc Natl Acad Sci U S A. 2000;97:5802–5806. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peri S, Steen H, Pandey A. GPMAW–a software tool for analyzing proteins and peptides. Trends Biochem Sci. 2001;26:687–689. doi: 10.1016/s0968-0004(01)01954-5. [DOI] [PubMed] [Google Scholar]
- 108.Taverner T, Hall NE, O’Hair RA, Simpson RJ. Characterization of an antagonist interleukin-6 dimer by stable isotope labeling, cross-linking, and mass spectrometry. J Biol Chem. 2002;277:46487–46492. doi: 10.1074/jbc.M207370200. [DOI] [PubMed] [Google Scholar]
- 109.Hernandez P, Gras R, Frey J, Appel RD. Popitam: towards new heuristic strategies to improve protein identification from tandem mass spectrometry data. Proteomics. 2003;3:870–878. doi: 10.1002/pmic.200300402. [DOI] [PubMed] [Google Scholar]
- 110.Kruppa GH, Schoeniger J, Young MM. A top down approach to protein structural studies using chemical cross-linking and Fourier transform mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:155–162. doi: 10.1002/rcm.885. [DOI] [PubMed] [Google Scholar]
- 111.Kellersberger KA, Yu E, Kruppa GH, Young MM, Fabris D. Top-down characterization of nucleic acids modified by structural probes using high-resolution tandem mass spectrometry and automated data interpretation. Analytical chemistry. 2004;76:2438–2445. doi: 10.1021/ac0355045. [DOI] [PubMed] [Google Scholar]
- 112.Tang Y, Chen Y, Lichti CF, Hall RA, Raney KD, Jennings SF. CLPM: a cross-linked peptide mapping algorithm for mass spectrometric analysis. BMC Bioinformatics. 2005;6(Suppl 2):S9. doi: 10.1186/1471-2105-6-S2-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seebacher J, Mallick P, Zhang N, Eddes JS, Aebersold R, Gelb MH. Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and integrated computational data processing. J Proteome Res. 2006;5:2270–2282. doi: 10.1021/pr060154z. [DOI] [PubMed] [Google Scholar]
- 114.de Koning LJ, Kasper PT, Back JW, Nessen MA, Vanrobaeys F, Van Beeumen J, Gherardi E, de Koster CG, de Jong L. Computer-assisted mass spectrometric analysis of naturally occurring and artificially introduced cross-links in proteins and protein complexes. Febs J. 2006;273:281–291. doi: 10.1111/j.1742-4658.2005.05053.x. [DOI] [PubMed] [Google Scholar]
- 115.Schnaible V, Wefing S, Resemann A, Suckau D, Bucker A, Wolf-Kummeth S, Hoffmann D. Screening for disulfide bonds in proteins by MALDI in-source decay and LIFT-TOF/TOF-MS. Analytical chemistry. 2002;74:4980–4988. doi: 10.1021/ac025807j. [DOI] [PubMed] [Google Scholar]
- 116.Wefing S, Schnaible V, Hoffmann D. SearchXLinks. A program for the identification of disulfide bonds in proteins from mass spectra. Analytical chemistry. 2006;78:1235–1241. doi: 10.1021/ac051634x. [DOI] [PubMed] [Google Scholar]
- 117.Gao Q, Xue S, Doneanu CE, Shaffer SA, Goodlett DR, Nelson SD. Pro-CrossLink. Software tool for protein cross-linking and mass spectrometry. Analytical chemistry. 2006;78:2145–2149. doi: 10.1021/ac051339c. [DOI] [PubMed] [Google Scholar]
- 118.Lee YJ, Lackner LL, Nunnari JM, Phinney BS. Shotgun cross-linking analysis for studying quaternary and tertiary protein structures. J Proteome Res. 2007;6:3908–3917. doi: 10.1021/pr070234i. [DOI] [PubMed] [Google Scholar]
- 119.Lee YJ. Probability-based shotgun cross-linking sites analysis. J Am Soc Mass Spectrom. 2009;20:1896–1899. doi: 10.1016/j.jasms.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 120.Anderson GA, Tolic N, Tang X, Zheng C, Bruce JE. Informatics strategies for large-scale novel cross-linking analysis. J Proteome Res. 2007;6:3412–3421. doi: 10.1021/pr070035z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu ET, Hawkins A, Kuntz ID, Rahn LA, Rothfuss A, Sale K, Young MM, Yang CL, Pancerella CM, Fabris D. The collaboratory for MS3D: a new cyberinfrastructure for the structural elucidation of biological macromolecules and their assemblies using mass spectrometry-based approaches. J Proteome Res. 2008;7:4848–4857. doi: 10.1021/pr800443f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Panchaud A, Singh P, Shaffer SA, Goodlett DR. xComb: a cross-linked peptide database approach to protein-protein interaction analysis. J Proteome Res. 2010;9:2508–2515. doi: 10.1021/pr9011816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Leitner A, Walzthoeni T, Aebersold R. Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nature protocols. 2014;9:120–137. doi: 10.1038/nprot.2013.168. [DOI] [PubMed] [Google Scholar]
- 124.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xu H, Hsu PH, Zhang L, Tsai MD, Freitas MA. Database search algorithm for identification of intact cross-links in proteins and peptides using tandem mass spectrometry. J Proteome Res. 2010;9:3384–3393. doi: 10.1021/pr100369y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McIlwain S, Draghicescu P, Singh P, Goodlett DR, Noble WS. Detecting cross-linked peptides by searching against a database of cross-linked peptide pairs. J Proteome Res. 2010;9:2488–2495. doi: 10.1021/pr901163d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chu F, Baker PR, Burlingame AL, Chalkley RJ. Finding chimeras: a bioinformatics strategy for identification of cross-linked peptides. Mol Cell Proteomics. 2010;9:25–31. doi: 10.1074/mcp.M800555-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Du X, Chowdhury SM, Manes NP, Wu S, Mayer MU, Adkins JN, Anderson GA, Smith RD. Xlink-identifier: an automated data analysis platform for confident identifications of chemically cross-linked peptides using tandem mass spectrometry. J Proteome Res. 2011;10:923–931. doi: 10.1021/pr100848a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rasmussen MI, Refsgaard JC, Peng L, Houen G, Hojrup P. CrossWork: software-assisted identification of cross-linked peptides. J Proteomics. 2011;74:1871–1883. doi: 10.1016/j.jprot.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 130.Gotze M, Pettelkau J, Schaks S, Bosse K, Ihling CH, Krauth F, Fritzsche R, Kuhn U, Sinz A. StavroX–a software for analyzing crosslinked products in protein interaction studies. J Am Soc Mass Spectrom. 2012;23:76–87. doi: 10.1007/s13361-011-0261-2. [DOI] [PubMed] [Google Scholar]
- 131.Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, Luo SK, Ding YH, Wang LH, Hao Z, Xiu LY, Chen S, Ye K, He SM, Dong MQ. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 132.Li W, O’Neill HA, Wysocki VH. SQID-XLink: implementation of an intensity-incorporated algorithm for cross-linked peptide identification. Bioinformatics. 2012;28:2548–2550. doi: 10.1093/bioinformatics/bts442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Holding AN, Lamers MH, Stephens E, Skehel JM. Hekate: software suite for the mass spectrometric analysis and three-dimensional visualization of cross-linked protein samples. J Proteome Res. 2013;12:5923–5933. doi: 10.1021/pr4003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jaiswal M, Crabtree N, Bauer MA, Hall R, Raney KD, Zybailov BL. XLPM: efficient algorithm for the analysis of protein-protein contacts using chemical cross-linking mass spectrometry. BMC Bioinformatics. 2014;15(Suppl 11):S16. doi: 10.1186/1471-2105-15-S11-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang J, Anania VG, Knott J, Rush J, Lill JR, Bourne PE, Bandeira N. Combinatorial approach for large-scale identification of linked peptides from tandem mass spectrometry spectra. Mol Cell Proteomics. 2014;13:1128–1136. doi: 10.1074/mcp.M113.035758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mayne SL, Patterton HG. AnchorMS: a bioinformatics tool to derive structural information from the mass spectra of cross-linked protein complexes. Bioinformatics. 2014;30:125–126. doi: 10.1093/bioinformatics/btt617. [DOI] [PubMed] [Google Scholar]
- 137.Lima DB, de Lima TB, Balbuena TS, Neves-Ferreira AG, Barbosa VC, Gozzo FC, Carvalho PC. SIM-XL: A powerful and user-friendly tool for peptide cross-linking analysis. J Proteomics. 2015 doi: 10.1016/j.jprot.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 138.Glatter T, Ludwig C, Ahrne E, Aebersold R, Heck AJ, Schmidt A. Large-scale quantitative assessment of different in-solution protein digestion protocols reveals superior cleavage efficiency of tandem Lys-C/trypsin proteolysis over trypsin digestion. Journal of proteome research. 2012;11:5145–5156. doi: 10.1021/pr300273g. [DOI] [PubMed] [Google Scholar]
- 139.Leon IR, Schwammle V, Jensen ON, Sprenger RR. Quantitative assessment of in-solution digestion efficiency identifies optimal protocols for unbiased protein analysis. Molecular & cellular proteomics: MCP. 2013;12:2992–3005. doi: 10.1074/mcp.M112.025585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nomura E, Katsuta K, Ueda T, Toriyama M, Mori T, Inagaki N. Acid-labile surfactant improves in-sodium dodecyl sulfate polyacrylamide gel protein digestion for matrix-assisted laser desorption/ionization mass spectrometric peptide mapping. J Mass Spectrom. 2004;39:202–207. doi: 10.1002/jms.578. [DOI] [PubMed] [Google Scholar]
- 141.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nature methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 142.Hustoft HK, Reubsaet L, Greibrokk T, Lundanes E, Malerod H. Critical assessment of accelerating trypsination methods. J Pharm Biomed Anal. 2011;56:1069–1078. doi: 10.1016/j.jpba.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 143.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature protocols. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 144.Green NS, Reisler E, Houk KN. Quantitative evaluation of the lengths of homobifunctional protein cross-linking reagents used as molecular rulers. Protein Sci. 2001;10:1293–1304. doi: 10.1110/ps.51201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nadeau OW, Carlson GM. Chemical cross-linking in studying protein-protein interactions. In: Golemis E, editor. Protein-protein interactions: a molecular cloning manual. Cold Spring Harbor Laboratory Press; New York: 2002. pp. 75–91. [Google Scholar]
- 146.Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 147.Benesch JL, Robinson CV. Mass spectrometry of macromolecular assemblies: preservation and dissociation. Curr Opin Struct Biol. 2006;16:245–251. doi: 10.1016/j.sbi.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 148.Benesch JL, Ruotolo BT, Simmons DA, Robinson CV. Protein complexes in the gas phase: technology for structural genomics and proteomics. Chem Rev. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 149.Hernandez H, Robinson CV. Dynamic protein complexes: insights from mass spectrometry. J Biol Chem. 2001;276:46685–46688. doi: 10.1074/jbc.R100024200. [DOI] [PubMed] [Google Scholar]
- 150.Hernandez H, Dziembowski A, Taverner T, Seraphin B, Robinson CV. Subunit architecture of multimeric complexes isolated directly from cells. EMBO Rep. 2006;7:605–610. doi: 10.1038/sj.embor.7400702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lane LA, Nadeau OW, Carlson GM, Robinson CV. Mass spectrometry reveals differences in stability and subunit interactions between activated and nonactivated conformers of the (alphabetagammadelta)4 phosphorylase kinase complex. Molecular & cellular proteomics: MCP. 2012;11:1768–1776. doi: 10.1074/mcp.M112.021394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fitzgerald TJ, Carlson GM. Activated states of phosphorylase kinase as detected by the chemical cross-linker 1,5-difluoro-2,4-dinitrobenzene. J Biol Chem. 1984;259:3266–3274. [PubMed] [Google Scholar]
- 153.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl FU, Ban N, Malmstrom L, Aebersold R. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 155.Rappsilber J. The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J Struct Biol. 2011;173:530–540. doi: 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schneidman-Duhovny D, Pellarin R, Sali A. Uncertainty in integrative structural modeling. Curr Opin Struct Biol. 2014;28:96–104. doi: 10.1016/j.sbi.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zeng-Elmore X, Gao XZ, Pellarin R, Schneidman-Duhovny D, Zhang XJ, Kozacka KA, Tang Y, Sali A, Chalkley RJ, Cote RH, Chu F. Molecular architecture of photoreceptor phosphodiesterase elucidated by chemical cross-linking and integrative modeling. J Mol Biol. 2014;426:3713–3728. doi: 10.1016/j.jmb.2014.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]


