Abstract
Peptidyl Met residues are readily oxidized by reactive oxygen species to form Met sulfoxide. The enzyme peptide Met sulfoxide reductase (PMSR) catalyzes the reduction of Met sulfoxides back to Met. In doing so, PMSR is proposed to act as a last-chance antioxidant, repairing proteins damaged from oxidative stress. To assess the role of this enzyme in plants, we generated multiple transgenic lines with altered expression levels of the plastid form of PMSR (PMSR4). In transgenic plants, PMSR4 expression ranged from 95% to 40% (antisense) and more than 600% (overexpressing lines) of wild-type plants. Under optimal growing conditions, there is no effect of the transgene on the phenotype of the plants. When exposed to different oxidative stress conditions—methyl viologen, ozone, and high light—differences were observed in the rate of photosynthesis, the maximum quantum yield (Fv/Fm ratio), and the Met sulfoxide content of the isolated chloroplast. Plants that overexpressed PMSR4 were more resistant to oxidative damage localized in the chloroplast, and plants that underexpressed PMSR4 were more susceptible. The Met sulfoxide levels in proteins of the soluble fraction of chloroplasts were increased by methyl viologen and ozone, but not by high-light treatment. Under stress conditions, the overexpression of PMSR4 lowered the sulfoxide content and underexpression resulted in an overall increase in content.
The generation of reactive oxygen species (ROS), especially under conditions of metabolic stress, is an unavoidable side effect of life in an oxygen atmosphere. These species include singlet oxygen, hydrogen peroxide, superoxide, and hydroxyl radical. To protect against ROS, aerobic organisms have evolved both enzymatic and nonenzymatic scavenging systems (Scandalios, 1993; Smirnoff and Wheeler, 2000). The enzymatic systems include superoxide dismutases (Scandalios, 1993), catalases, and peroxidases (Defelipe et al., 1988). The nonenzymatic systems include antioxidants such as glutathione, vitamin E, carotene, and vitamin C. ROS that escape the scavenging system can cause oxidative damage to virtually all biomolecules (Klatt and Lamas, 2000). Once damaged, certain macromolecules, such as DNA, may be repaired by the cell. Research within the past decade has shown that proteins can also be repaired.
Amino acids vary in susceptibility to oxidative damage, with Met residues the most vulnerable followed by Cys and Tyr (Levine et al., 1996). ROS readily oxidize Met residues by two electrons to form the sulfoxide (MetSO), which can be in the R or S configuration. The sulfoxide, in turn, can be reduced back to Met by the peptide Met sulfoxide reductase (PMSR). This enzyme, named MsrA in Escherichia coli (Moskovitz et al., 1995), utilizes thioredoxin to reduce only the S stereoisomer back to Met. This enzyme is active with either the free amino acid sulfoxide or the peptidyl MetSO. For the R stereoisomer of the sulfoxide, a gene encoding a PMSR was only recently discovered in E. coli (Grimaud et al., 2001) and has since been found in other cells (Lowther et al., 2002; Olry et al., 2002). This enzyme, named MsrB in E. coli, is homologous to a protein from Neisseria gonorrhoeae that can catalyze the reduction of the R isomer of MetSO (Lowther et al., 2002; Olry et al., 2002).
Plant PMSR activity in turnip and bean leaves was first reported in 1966 by Doney and Thompson (1966). The first isolation of a plant gene for PMSR was from Brassica napus (Sadanandom et al., 1996). Subsequently, it has been shown that Arabidopsis (Arabidopsis thaliana) has several copies of PMSR (Sadanandom et al., 2000). One of the gene products is targeted to the chloroplast, whereas the other three are believed to be cytosolic. A fifth gene with a high degree of homology to PMSR does not contain the conserved residues believed to be essential for activity and, according to the gene annotation in The Arabidopsis Information Resource (TAIR; At2g18030), is targeted to a secretory pathway. Results thus far have not clarified the role of the various PMSRs in Arabidopsis. Sadanandom et al. (2000) examined the expression of the plastid form and one cytosolic form and found the plastid form to be expressed in green tissues, whereas the cytosolic form was expressed in all tissues examined. No change in expression of either form was observed when exposed to high or low temperature, wounding, jasmonic acid treatment, or the virulent pathogen Pseudomonas syringae. Induction of the cytosolic form was only observed 35 d postinoculation with the cauliflower mosaic virus.
To examine the role of PMSR in plants, we have transformed Arabidopsis plants to alter expression levels of the plastid form of PMSR (PMSR4). Multiple lines were generated expressing different levels of PMSR4 ranging from 95% to 40% (antisense) and more than 600% (overexpressing lines) of wild-type plants. Our results show that PMSR4 plays a role in protection from ROS in the chloroplast.
RESULTS
Oxidative Stress and Wild-Type PMSR4 Activity
Wild-type plants were exposed to three agents that have been shown to cause oxidative stress: ozone, methyl viologen, and high-light intensity. The PMSR4 activity was measured in chloroplasts isolated from foliage after these treatments and compared to nontreated controls. Because the PMSR activity was measured in isolated chloroplasts, other nonchloroplastic isozymes do not contribute to the total measured enzyme activity. Figure 1 shows PMSR4 expression as measured by western blot (Fig. 1A) and by enzyme activity (Fig. 1B) after the oxidative stress treatments. The intensity of the immunoblots was measured and revealed that PMSR content was 3.5 times higher after high-light treatment and 4.5 times higher after methyl viologen treatment. In contrast, PMSR4 content was only 1.6 times higher in plants exposed to ozone. The pattern in PMSR4 enzyme activity was similar to that obtained from the western blots.
Figure 1.
Expression of PMSR4 in wild-type plants in response to oxidative stress. A, Western blot using anti-PMSR4 antibody. B, Enzyme activity of the soluble fraction of three independent preparations of isolated chloroplasts from 32 plants as described in “Materials and Methods.” Control (CON), Control plants without treatment. High light (HL), Plants exposed to 600 μmol photons m−2 s−1 at 8°C for 48 h. Paraquat (PQ), Plants sprayed with 10 μm methyl viologen (paraquat). Ozone (O3), Plants exposed to 0.46 μL L−1 O3 for 6 h during 4 d. PMSR4 activity results show the mean and sd (values above bars). Means with the same letter above the values are not statistically different (P ≤ 0.05). Purity of the chloroplast preparation was assessed by AAT activity gels. C, AAT activity gel. Total crude extracts (T) exhibit two activity bands corresponding to cytosol (cytosolic) and chloroplast (chloroplastic) isozymes. Chloroplast preparations (C) show only one band. D, Loading control for western blot shown in A. The nitrocellulose membrane was stained with Red Punceau.
Purity of Chloroplast Preparations
Because our studies of transgenic plants involve characterization of the chloroplast, we needed to initially determine the integrity of our chloroplast preparations. The NADP-dependent glyceraldehyde-3-phosphate dehydrogenase (G3PDH) activity is a marker enzyme for the chloroplast, whereas phosphoenolpyruvate carboxylase (PEPC) is a marker enzyme for the cytosol. Transgenic and wild-type plants were grown under optimal growth conditions and the activity of these enzymes in the crude (chloroplast-containing) extracts and chloroplast preparations of the different lines was determined (Table I). After chloroplast isolation, the activity of the marker enzyme G3PDH was 2 to 3 times higher than in the crude extract, showing that the procedure enriched for the targeted organelle. When assaying the isolated chloroplast for the cytosolic marker PEPC, a maximum of 5% of the activity was recovered in the plastidial preparation. This shows the high purity of the isolated chloroplasts.
Table I.
Activities of marker enzymes in cells and chloroplasts of wild-type and transgenic plants underexpressing (lines 801, 1002, and 1003) and overexpressing (lines 802, 202, and 1001) PMSR4
| Line | Subcellular Fraction | G3PDHa | Percent Activity in Chloroplast | PEPCb | Percent Activity in Chloroplast |
|---|---|---|---|---|---|
| μmol min−1 mg−1 | μmol min−1 mg−1 | ||||
| Wild-type | Crude extract | 1.47 | 255 ± 34 | 3.34 | 2.0 ± 1.9 |
| Chloroplast | 2.23 | 0.07 | |||
| 801 | Crude extract | 1.63 | 185 ± 18 | 1.98 | 3.5 ± 2.8 |
| Chloroplast | 2.72 | 0.07 | |||
| 1002 | Crude extract | 1.05 | 281 ± 1.9 | 1.72 | 3.3 ± 2.8 |
| Chloroplast | 2.92 | 0.06 | |||
| 1003 | Crude extract | 1.80 | 264 ± 17 | 1.71 | 3.2 ± 2.2 |
| Chloroplast | 3.21 | 0.08 | |||
| 802 | Crude extract | 1.77 | 264 ± 28 | 2.04 | 3.8 ± 2.6 |
| Chloroplast | 3.29 | 0.08 | |||
| 202 | Crude extract | 1.70 | 201 ± 3 | 1.70 | 2.7 ± 1.4 |
| Chloroplast | 3.47 | 0.07 | |||
| 1001 | Crude extract | 1.32 | 192 ± 50 | 1.32 | 3.6 ± 2.3 |
| Chloroplast | 1.85 | 0.06 |
Data are presented as μmol min−1 mg−1 chlorophyll for a representative experiment. The percentage of activity in the chloroplast fraction corresponds to the mean ± sd for three independent chloroplast preparations of 32 plants grown under optimal growing conditions as described in “Materials and Methods.”
Chloroplast marker, NADP-dependent G3PDH.
Cytosolic marker, PEPC.
The purity of the chloroplast was confirmed by Asp aminotransferase (AAT) activity gels. This method separates the cytosolic and the chloroplastic isoforms of the enzyme (Schultz and Coruzzi, 1995) and detects their activity in the gels. Both isoforms were detected in the crude extract, but only one band corresponding to the chloroplastic form of AAT is present in the isolated chloroplasts (Figs. 1C, 3B, and 4B). Since the results obtained with the AAT activity gels were consistent with those of the marker enzymes, we decided to use AAT gel activity assay to routinely monitor the purity of the chloroplast preparations. Furthermore, PEPC activity was determined in several more chloroplast isolations and in no case was it detected more than 5% of cytosolic contamination (data not shown).
Figure 3.
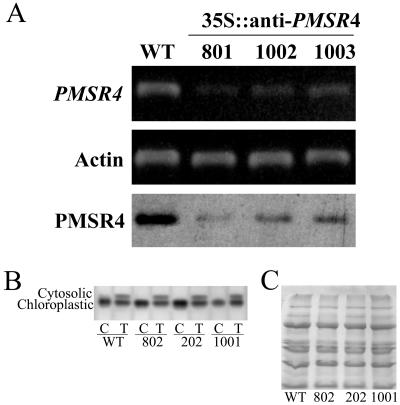
Characterization of PMSR4 expression in wild-type and transgenic Arabidopsis plants underexpressing PMSR4. RNA was isolated from 10-d-old wild-type (WT) and underexpressing (35S∷anti-PMSR4) plants and subjected to RT-PCR analysis. Actin was used as an internal control. The PMSR4 protein content of the plants was determined by western blot (labeled PMSR4 in the figure). A, Gene-specific primers as described in “Materials and Methods” were used to determine the level of the endogenous mRNA (labeled PMSR4). The western blot is in the lowest row. Isolated chloroplasts from 4-week-old plants were used for the protein expression analysis (PMSR4). For the westerns blots, 20 μg of protein were separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and reacted with anti-PMSR antibodies. B and C, AAT activity gel and western-blot loading control, respectively, similar to those shown in Figure 1.
Figure 4.
PMSR activity of isolated chloroplasts from transgenic Arabidopsis plants overexpressing (A) or underexpressing (B) PMSR4. The control values are of plants transformed with the empty vector. Results shown are mean and sd of three independent chloroplast preparations. Chloroplasts were isolated from 32 4-week-old plants grown under optimal conditions (see “Materials and Methods”). Enzyme activity was measured as described in the legend of Figure 1. Means with the same letter above the values are not statistically different (P ≤ 0.05).
Characterization of Transgenic Plants
To study the role of PMSR4 in the response of Arabidopsis plants to oxidative stress conditions, we obtained transgenic plants with altered expression of PMSR4. A full-length PMSR4 cDNA was placed in the sense (overexpression) and in the antisense (underexpression) direction, behind the cauliflower mosaic virus 35S promoter (35S∷PMSR4 and 35S∷anti-PMSR4, respectively) and transformed into Arabidopsis. The presence of the construct was confirmed by PCR analysis in several lines, and the independence of the lines was assessed by Southern-blot analysis. Three of the lines of each construct were further characterized.
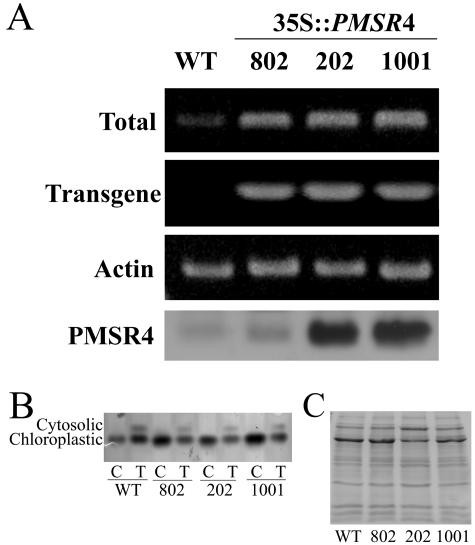
The expression level of PMSR4 in the transgenic plants was analyzed by semiquantitative reverse transcription (RT)-PCR and by western blot. For overexpressing plants with the construct in the sense orientation (Fig. 2), RT-PCR showed a large increase in the PMSR message. Using specific primers (see “Materials and Methods”) that amplified only the mRNA transcribed from the transgene, we confirmed that the increase in the PMSR4 mRNA levels was due exclusively to the effect of the transformation. Furthermore, analysis of the plants showed that the transformation did not induce gene silencing by cosuppression. All of the lines showed overexpression of PMSR4 mRNA (Fig. 2A). Similar analysis of the transgenic antisense lines showed a considerable reduction in the PMSR4 mRNA as seen by RT-PCR analysis (Fig. 3A). The reduction in the mRNA levels was in accord with the decrease observed in the protein levels (Fig. 3A).
Figure 2.
Characterization of PMSR4 expression in wild-type and transgenic Arabidopsis plants overexpressing PMSR4. RNA was isolated from 10-d-old wild-type (WT) and overexpressing (35S∷PMSR4) plants and subjected to RT-PCR analysis. Actin was used as an internal control. The PMSR4 protein content of the plants was determined by western blot (labeled PMSR4 in the figure). A, Gene-specific primers as described in “Materials and Methods” were used to determine the combined level of the endogenous mRNA and the transgenic mRNA (labeled Total) or the level of transgenic mRNA (labeled Transgene in the figure). The western blot is in the lowest row. Isolated chloroplasts from 4-week-old plants were used for the protein expression analysis (PMSR4). For the westerns blots, 7.5 μg of protein were separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and reacted with anti-PMSR antibodies. B and C, AAT activity gel and western-blot loading control, similar to those shown in Figure 1.
The peptidyl PMSR activity in the chloroplast extracts from these transgenic lines was measured using 9-fluorenylmethyloxycarbonyl (Fmoc)-MetSO as the peptide mimic substrate (Fig. 4). The overexpressing lines (Fig. 4A) expressed higher PMSR4-specific activity than the untransformed wild-type plants. The PMSR4-specific activity in the overexpressing line 1001 was nearly 6 times higher than the activity of the untransformed wild-type plants. The enzyme activity of line 202 was almost 3 times that of the wild type. Consistent with the results of the western blot shown in Figure 2, the activity of line 802 was not much higher than that of the wild type, but the difference was statistically significant (P ≤ 0.05).
In contrast, the PMSR4-specific activities of the underexpressing lines (Fig. 4B) were significantly lower than that of the untransformed wild type, ranging from 0.15 nmol min−1 mg−1 protein in line 1003 to 0.27 nmol min−1 mg−1 protein in line 801.
MetSO content in the soluble fraction of the chloroplast preparations was also measured (Table II). Under optimal growing conditions, there were no statistical differences (P ≤ 0.05) in the percentage of MetSO among the different transgenic lines and the control (wild-type) plants. MetSO content of the chloroplast proteins was between 27% and 33% of the total Met pool.
Table II.
Percent basal MetSO content of isolated chloroplasts from wild-type and transgenic plants underexpressing (35S∷anti-PMSR4) or overexpressing (35S∷PMSR4) PMSR4
| 35S∷PMSR4
|
35S∷anti-PMSR4
|
||||||
|---|---|---|---|---|---|---|---|
| Wild Type | 802 | 202 | 1001 | 801 | 1002 | 1003 | |
| Percent MetSO | 28 ± 5 | 31 ± 13 | 28 ± 11 | 27 ± 10 | 28 ± 5 | 33 ± 10 | 27 ± 12 |
Chloroplasts were isolated from 4-week-old plants grown under optimal growing conditions. The results represent the percentage of MetSO in the total Met pool of the soluble proteins of the chloroplasts and correspond to the mean ± sd of three independent chloroplast preparations of 32 plants each.
PMSR4 Overexpression and Oxidative Stress
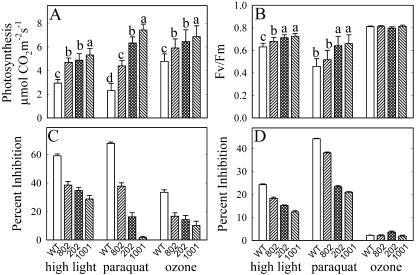
The wild-type and transgenic plants underexpressing and overexpressing PMSR4 were subjected to the oxidative stress treatments of high-light/low-temperature methyl viologen and ozone. Photosynthetic rates and chlorophyll a fluorescence were measured and the percentage reduction was calculated (see “Materials and Methods”). Under optimal growing conditions, no phenotypic variations between the transgenic overexpressing lines and the untransformed wild-type plants were observed (data not shown). The photosynthetic activity ranged from 7.1 to 7.6 μmol CO2 m−2 s−1 and the maximum quantum yield (Fv/Fm ratio) was around 0.83. However, oxidative stress treatments induced a reduction in the photosynthetic rate (Fig. 5A) and in the Fv/Fm ratio (Fig. 5B) in wild-type and overexpressing plants. The overexpression of PMSR4 reduced the severity of the stress-induced impact on the photosynthetic rate and the Fv/Fm ratio. The treatment that resulted in the largest difference in response between the wild-type and the overexpressing lines was methyl viologen (i.e. overexpression of the chloroplast PMSR4 afforded the most protection from methyl viologen stress). Whereas the photosynthetic rate and maximum quantum yield were inhibited 67% and 27%, respectively, in wild-type plants, these functions were inhibited only 38% and 19% in the overexpressing line 802; 16% and 8% in the overexpressing line 202, and 0.5% and 11% in the overexpressing line 1001.
Figure 5.
Effect of different oxidative stress treatments on the photosynthetic rate (A) and chlorophyll a fluorescence (B) of Arabidopsis plants overexpressing PMSR4. Four-week-old plants were subjected to high light (600 μmol photons m−2 s−1 at 8°C for 48 h), 20 μm methyl viologen (paraquat), or 0.46 μL L−1 O3 for 6 h for 4 d. Photosynthesis was measured in leaves 4 and 5 from the apex from at least 12 plants/treatment. Chlorophyll a fluorescence (Fv/Fm) was measured in the same leaves. The percentage of reduction of photosynthesis (C) and of Fv/Fm (D) was calculated (see “Materials and Methods”) to show the differences in the basal levels (before treatment) of each set of plants. Each experiment was repeated three times with similar results. In each bar, sd is shown as an error bar. For each treatment, means with the same letter (above the bar) were not statistically different (P ≤ 0.05). Comparisons among means are shown only for the original values (not for the calculated percent of inhibition) and are not present in case there are no statistical differences among the lines (Fv/Fm ozone treatment).
PMSR4 overexpression also enhances the resistance of the plants against the oxidative stress imposed by the treatment of high-light intensity under low temperature. The inhibition caused by high-light treatment in wild-type plants was 59% in photosynthetic rate and 24% in Fv/Fm ratio. In plants overexpressing PMSR4, lines 802, 202, and 1001, the photosynthetic rate was inhibited to a lesser extent at 38%, 34%, and 28%, respectively. The reduction of Fv/Fm ratio was 18% (line 802), 15% (line 202), and 12% (line 1001).
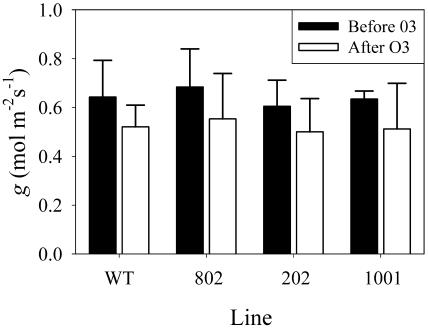
Treatment with ozone resulted in statistically significant differences in the photosynthetic rate between the wild-type and the transgenic lines, but not in the Fv/Fm ratio (Fig. 5B) or in the stomatal conductance (Fig. 6). The inhibition of photosynthesis in the wild type was 33%, and in the overexpressing plants, it was 16%, 14%, and 10% in lines 802, 202, and 1001, respectively.
Figure 6.
Effect of ozone on the stomatal conductance (g) of plants overexpressing PMSR4. Four-week-old plants were subjected to 0.46 μL L−1 O3 for 6 h for 4 d. Stomatal conductance was measured before (black bars) and after (white bars) ozone treatment in leaves 4 and 5 from the apex from at least 12 plants per treatment. Wild type (WT) corresponds to untransformed plants. Lines 802, 202, and 1001 are overexpressing lines. Each experiment was repeated three times with similar results. sd is presented as an error bar.
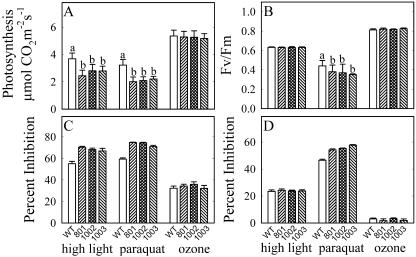
PMSR4 Underexpression and Oxidative Stress
Underexpression of PMSR4 resulted in greater sensitivity to high light and to methyl viologen, but not to ozone (Fig. 7). In plants exposed to high light under low temperature, photosynthesis and Fv/Fm ratio were reduced 55% and 23%, respectively, in the wild type, whereas the reduction in the transgenic lines was 70% and 24% (line 801), 68% and 23% (line 1002), and 66% and 23% (line 1003). The reduction in photosynthesis and Fv/Fm ratio in the plants treated with methyl viologen was 59% and 46% (wild type), 74% and 54% (line 801), 74% and 55% (line 1002), 71% and 57% (line 1003).
Figure 7.
Effect of different oxidative stress treatments on the photosynthetic rate (A) and the chlorophyll a fluorescence (B) of plants underexpressing PMSR4. Four-week-old plants were exposed to 600 μmol photons m−2 s−1 at 8°C for 48 h (high light), 20 μm methyl viologen (paraquat), or 0.46 μL L−1 O3 for 6 h during 4 d. Photosynthesis was measured in leaves 4 and 5 from the apex of at least 12 plants/treatment. Fv/Fm was determined in the same leaves. The percentage of reduction of photosynthesis (C) and of Fv/Fm (D) in response to the treatments was calculated (see “Materials and Methods”) to show the differences in the basal levels (before treatment) of each set of plants. Each experiment was repeated three times with similar results. sd is presented as an error bar. For each treatment, means with the same letter (above the bar) were not statistically different (P ≤ 0.05). Comparisons among means are shown only for the original values (not for the calculated percent of inhibition) and are not present in case there are no statistical differences among the lines.
MetSO Content of Chloroplasts
The MetSO content of the soluble fraction of chloroplasts was also determined in wild-type and transgenic lines after treatment with oxidative stress agents. The MetSO contents of the overexpressing lines 202 and 1001 after high-light treatment were slightly, but significantly, lower than that of wild type (Fig. 8A). The percent MetSO was 30% in wild-type plants and 25% in line 1001. No statistical differences were observed between the wild-type and the other overexpressing line 802. Treatment of the plants with methyl viologen resulted in a large increase in the MetSO content of all plants; however, in the overexpressing plants, the MetSO content was lower (50%) than in the wild-type plants (58%). A similar MetSO pattern was observed when plants were treated with ozone. Whereas the wild-type plants contained 56% MetSO, the overexpressing lines contained statistically significant lower amounts of MetSO (approximately 35% for all lines).
Figure 8.
MetSO content of soluble proteins from isolated chloroplasts of plants after exposure to oxidative stress. A, Plants overexpressing PMSR4. B, Plants underexpressing PMSR4. MetSO content is expressed as the percent of MetSO in relation to the total Met pool (Met + MetSO). For details on the treatments, see “Materials and Methods.” sd is shown as an error bar and represents the average of three independent chloroplast preparations. In each section, means with the same letter above the bars are not statistically different (P ≤ 0.05).
In plants underexpressing PMSR4 (Fig. 8B), the high-light treatment resulted in formation of approximately 30% MetSO in all the lines. There were no significant differences among them. In plants underexpressing PMSR4, methyl viologen treatment resulted in MetSO content significantly increasing to a much higher value (as much as 80% in line 1003) relative to wild type. A similar MetSO profile was also observed when plants were treated with ozone. Plants underexpressing PMSR4 contained as much as 85% MetSO content after ozone treatment, a value much higher than that observed in wild-type plants.
DISCUSSION
Plant Oxidative Stress and PMSR
Due to the ease of Met oxidation to yield MetSO and the ability of PMSR to repair the MetSO, Met has been proposed to act as a last-chance antioxidant for proteins (Levine et al., 1999). Whereas most organisms have one gene encoding the PMSR that reduces the S enantiomer of MetSO (Hoshi and Heinemann, 2001), plant genomes encode multiple copies possibly due to different subcellular localization (Perl-Treves and Perl, 2002). This study used transgenic plants with altered PMSR4 expression levels to assess its role in oxidative stress. The chloroplast should be highly sensitive to alterations in PMSR expression because photosynthesis and photorespiration are responsible for the production of large amounts of ROS (Foyer, 2002). While in vitro studies have documented the role of PMSR4 in repair of peptidyl MetSO (Gustavsson et al., 1999, 2002), the results from in vivo studies have not been as clear (Ferguson and Burke, 1994; Sadanandom et al., 1996). Only recently, Bechtold et al. (2004) found a correlation between decreased PMSR2 levels and increased sensitivity to oxidative stress in Arabidopsis.
Expression of PMSR4
Our results show that wild-type plants increase PMSR4 expression in response to oxidative stress (Fig. 1). This expression is dependent upon the type of oxidative stress and where it is localized. Oxidative stress, localized in the chloroplast and induced by high light and methyl viologen, caused an increase in PMSR4 expression. In contrast, ozone, which probably reacts before reaching the chloroplast, had little, if any, effect on PMSR4 expression. Similar results were found by Sadanandom et al. (2000) working with other nonchloroplast-localized stress conditions. These results suggest that the signal-inducing PMSR4 expression originates in the chloroplast. Evidence of such compartment-specific signaling has also been observed for cytosol-expressed PMSR2 from Arabidopsis (Bechtold et al., 2004). The authors found that a null mutant of this enzyme did not exhibit increased sensitivity to a 10-fold excess of light, methyl viologen, drought, or infection with cauliflower mosaic virus or P. syringae. The mutant was more susceptible only to oxidative stress in the dark phase of short-day conditions. These data are consistent with PMSR2 being responsive to oxidative stress localized only in the mitochondria, but not from other cell compartments.
High-Light Stress
High-light stress caused a decrease in both photosynthetic rate and in chlorophyll a fluorescence (Table III). However, no change was observed in the MetSO content of the soluble proteins. These results are actually consistent with the known mechanism of high-light damage. Upon high-light stress, there is a decrease in quantum yield and photosynthetic productivity referred to as photoinhibition. This has been suggested to be caused by ROS oxidation of PSII proteins (Aro et al., 1993). Although our measurement of MetSO content was only of the soluble proteins and Met oxidation of membrane proteins of PSII would not be detected, our results are consistent with PMSR4 being active with membrane-associated proteins. This would explain the large differences in Fv/Fm ratio and photosynthetic rate between the wild-type and the overexpressing lines.
Table III.
Summary of plant response to oxidative stress
| Treatment
|
Measurement
|
Response
|
||
|---|---|---|---|---|
| Wild Type | Overexpressing | Underexpressing | ||
| High | Photosynthesis | ↓↓ | Protect | Inhibit |
| Light | Fv/Fm | ↓↓↓ | Protect | No change |
| MetSO | No change | ↓ | No change | |
| Paraquat | Photosynthesis | ↓↓↓ | Protect | Inhibit |
| Fv/Fm | ↓↓ | Protect | Inhibit | |
| MetSO | ↑ | ↓ | ↑ | |
| Ozone | Photosynthesis | ↓↓ | Protect | No change |
| Fv/Fm | No change | No change | No change | |
| MetSO | ↑ | ↓ | ↑ | |
The number of arrows reflects the magnitude of the increase or decrease.
Consistent with the findings from the overexpressing lines, with plants underexpressing PMSR4, the reduction of the photosynthetic rate is not accompanied by an increase in MetSO content. This again suggests that when plants are exposed to high light, PMSR4 plays a key role in repairing membrane proteins in the chloroplast. Aro et al. (1993) found that the degradation and replacement of the D1 protein (and at lower level D2 protein) in PSII play a protective role against photodamage under both stress and nonstress conditions. Both of these are membrane proteins that have high turnover rates (replaced quickly in the cell) and Met residues may play a key role in this turnover. Both proteins have several Met residues, which, upon oxidation, can induce structural changes in the protein triggering the signaling for proteolysis (Aro et al., 1993; Satoh, 1998). Such oxidation of D1 and D2 Met residues after high-light stress has been observed with purified proteins from pea plants (Sharma et al., 1997b). If oxidation of these Met residues facilitates their proteolysis, then PMSR4 may play a key role in regulating D1 and D2 turnover and, in turn, in the integrity of PSII.
Ozone Stress
Due to its reactivity, ozone is proposed not to reach the chloroplast, reacting in the apoplast and the cytosol of plant cells (Langebartels et al., 2002). However, Torsethaugen et al. (1997) demonstrated that ozone treatment results in a dose-dependent reduction of photosynthesis. Similarly, our results show that, in all the lines analyzed, ozone caused a reduction in photosynthesis but not in Fv/Fm ratio. In plants underexpressing PMSR4, despite higher levels of MetSO in the soluble proteins of chloroplasts, no significant difference was observed in photosynthesis when compared to wild-type plants. This indicates that the response of plants to ozone is not impacted by the basal level of PMSR4 expression. One plausible explanation is that Met oxidation in the chloroplast-soluble proteins reached a level beyond which it is not possible to obtain further reduction in the photosynthetic rate. Yet, when PMSR4 is overexpressed, MetSO content is lowered in the soluble proteins, resulting in protection of photosynthesis. In the latter case, PMSR4-dependent repair lowered the MetSO content below the threshold value limiting photosynthesis.
The finding that ozone caused a decrease in photosynthesis but not in Fv/Fm ratio indicates that ozone has little impact on the proteins of PSII. In contrast, the changes in MetSO content indicate that ozone treatment results in oxidation of the soluble proteins of the chloroplast. Because Rubisco is the major protein of the chloroplast, it is a likely target for Met oxidation and repair in vivo. Previous research has attributed the decrease in photosynthesis after ozone treatment to be due to a decrease in Rubisco content (Pell et al., 1992; Landry and Pell, 1993). This decrease in Rubisco has been attributed to oxidation, inactivation (Roshchina and Roshchina, 2003), and proteolysis (Pell et al., 1992; Landry and Pell, 1993). Our results here are consistent with these findings (Table III). The repair of proteins by PMSR4 overexpression in the chloroplast would then lower the MetSO content and, in turn, result in higher photosynthetic rates.
Another possible mechanism for protection is the ability of PMSR4 overexpression to protect guard cells in the stomata. Ozone treatment has been shown to cause a guard cell-dependent decrease in CO2 exchange and thus in photosynthetic rates mainly due to changes in stomatal conductance (Torsethaugen et al., 1999). However, our results (Fig. 6) show that stomatal conductance of plants overexpressing PMSR4 is not statistically different from wild-type plants. This shows that PMSR4 protection of photosynthesis after ozone treatment is not linked to changes in the aperture and closure of stomata.
Methyl Viologen Stress
Upon illumination, methyl viologen is reduced by PSI. Reduced methyl viologen, in turn, reduces O2 yielding superoxide. The resulting superoxide, in the presence of iron or copper, can lead to generation of the hydroxyl radical. Thus, methyl viologen has the capacity to oxidize all soluble and membrane-associated proteins of the chloroplast. Indeed, among the different oxidative stress treatments, methyl viologen caused the strongest inhibition of photosynthesis and of maximum quantum yield. Methyl viologen also caused the largest increase in MetSO content of chloroplast-soluble proteins.
Our results indicate that PMSR4 plays a significant role in plant response to methyl viologen. Underexpression caused a greater decrease in the Fv/Fm ratio, a greater increase in MetSO formation in the chloroplast, and a greater decrease in assimilation capacity. In contrast, plants overexpressing PMSR4 were more resistant to methyl viologen as demonstrated by lower MetSO formation, higher Fv/Fm ratio, and higher photosynthetic rates (line 1001, Fig. 5C).
Summary of PMSR Function
The apparently wide range of functions of PMSR4 is still not clearly resolved and the natural substrates for the enzyme are yet to be identified. For example, Gustavsson et al. (1999) have shown that the chloroplast chaperone-like protein HSP21 has a Met-rich domain. Upon oxidation, the chaperone-like activity is lost; however, when the oxidized HSP21 is treated with recombinant PMSR4, the activity of HSP21 is recovered (Gustavsson et al., 2002). PMSR4 may also have a role in the regulation of chloroplast proteins. For example, the β-subunit of cytochrome b559 and the psbI gene product of PSII have four and two oxidizable residues, respectively. However, upon light treatment, only one specific Met residue in each protein is oxidized (Sharma et al., 1997a). The presence of these specific residues that are highly susceptible to oxidation is not well understood, but given the presence of PMSR4 in the chloroplast and its induction after high-light stress, the oxidation of these Met residues and their subsequent reduction by PMSR4 might play a regulatory role of PSII.
The lack of phenotypic variation between underexpressing and wild-type plants under nonstress conditions suggests that PMSR4 does not play a crucial role in ROS metabolism under low-stress conditions. However, PMSR4 appears to play a key role in plant response under conditions of oxidative stress. Our findings also indicate that, under basal nonoxidative stress conditions, plant chloroplasts contain a high level of MetSO and appear to be tolerant of these high levels. The average value of 27% to 33% MetSO in the soluble fraction of the chloroplasts of different lines under optimal growing conditions is considerably higher than the protein MetSO content reported for yeast (Saccharomyces cerevisiae), bacteria (Moskovitz et al., 1997), and mammals (Fliss et al., 1983). Also, MetSO content in the chloroplast is higher than the MetSO content found in the whole-cell proteins of cotton, pea, wheat, and potato (Ferguson and Burke, 1994). This is consistent with the highly oxidative environment of the chloroplast and is consistent with the proposal that Met serves as a last-chance antioxidant.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) ecotype Columbia seeds were planted in a commercial soil mix (Redi-earth Plug and Seedling mix; Scotts-Sierra, Marysville, OH) and grown in lighted growth chambers, with approximately 120 μmol photon m−2 s−1 on a 23°C/21°C, 12-h day/night cycle. Plants were fertilized weekly with 20:20:20 fertilizer (Peters Professional; Scotts-Sierra).
Construction of Transgenic Arabidopsis Plants
The full-length cDNA encoding PMSR4 from Arabidopsis (expressed sequence tag 226P20T7; GenBank accession no. X97326) was obtained from the Arabidopsis Biological Resource Center (Columbus, OH). The cDNA was cloned in the sense (overexpressing) and the antisense (underexpressing) direction into the pBI121 vector after removal of the β-glucuronidase gene from the vector using BamHI and SstI. The vector utilizes the 35S promoter. The orientation of the cDNA in each case was confirmed by restriction analysis. Fifteen plants were vacuum transformed with either the sense or the antisense construct and also with an empty vector as a control. Seeds from each plant were harvested separately. Seeds from the transformed plants were screened for resistance on 0.5× Murashige and Skoog plates supplemented with 50 μg mL−1 kanamycin. Resistant seedlings were transplanted and allowed to self. Seeds were harvested and screened again for kanamycin resistance. Resistant seedlings were transplanted and seed collected from individual T2 plants was again screened for kanamycin resistance to identify plants homozygous for kanamycin resistance. Homozygous T3 kanamycin-resistant plants were used in all the experiments.
Gene Expression Analysis
RNA was extracted from 10-d-old plants grown in solid medium (0.5× Murashige and Skoog, 1% Suc, 0.7% bacto-agar). Leaf tissue was ground in liquid nitrogen and total RNA was extracted from 100 mg of tissue (RNeasy; Qiagen, Chatsworth, CA). RNA (1 μg) was treated with DNAse I and reverse transcribed to synthesize the first-strand cDNA using the Retroscript System (Ambion, Austin, TX) in a two-step reaction with previous denaturing of the RNA at 80°C and cDNA synthesis at 44°C using oligo(dT). The synthesized cDNA (2 μL) was used for PCR amplification of the genes of interest using gene-specific primers as in Zhong et al. (2003). For the overexpressing lines, the total amount of PMSR4 mRNA (the product of the natural endogenous PMSR4 plus the product of the introduced PMSR4 under the 35S promoter) was determined with the PMSR4-intL primer (5′-ATGAACAACCTTTTCAACAGACTCGG-3′) and the PMSR4-intR primer (5′-TTAGCCATAGCATCGGATTGGATC-3′), which were specific for the coding region of PMSR4. The amount of transcript product of the PMSR4 transgene under the 35S promoter control was determined using the same left primer as before (PMSR4-intL) and the Nost-R primer (5′-TCGCAAGACCGGCAACAGGATTC-3′) derived from the sequence of the Nos terminator in pBI121. For the antisense lines, the PMSR4 mRNA level was determined using the PMSR4-intR and the PMSR4-intL primers. The mRNA levels where normalized using the PCR-amplified cDNA of a constitutively active actin cDNA (Jambunatan et al., 2001).
Oxidative Stress Treatments
Methyl viologen was applied to mature rosettes of 4-week-old plants by spraying with 10 μm methyl viologen with 0.05% Tween 20. Approximately 3 mL of methyl viologen per plant were applied 3 h before the beginning of the light period. The plants were then returned to the growth chamber.
Ozone was applied to 4-week-old plants at 0.46 μL L−1 for 6 h during 4 d. Ozone was generated by passing oxygen through an ozonator (OREC V1-0, Ozone Research and Equipment, Phoenix), and O3 concentrations in the growth chamber were monitored continuously with a UV photometric O3 analyzer (model 49; Thermo Environmental Instruments, Franklin, MA).
To determine the effect of high light, 4-week-old plants were exposed to 600 μmol photon m−2 s−1 light intensity at 8°C for 48 h.
Photosynthetic rates, stomatal conductance, and the maximal quantum yield of PSII photochemistry (Fv/Fm ratio) were measured 24 h before and after treatments. Photosynthetic rates and stomatal conductance were measured with a LI-COR 6400 gas exchange system fitted with an Arabidopsis chamber (LI-COR, Lincoln, NE). The gas exchange cuvette was maintained at 23°C block temperature, 400 μL L−1 CO2, and 600 ± 30 μmol photon m−2 s−1 light intensity. Chlorophyll a fluorescence was measured using a pulse amplitude-modulated fluorometer PAM-2000 equipped with an Arabidopsis chamber and clip (Walz, Effeltrich, Germany). Photosynthesis, stomatal conductance, and chlorophyll a fluorescence were determined in leaves 4 and 5 from the apex of at least 8 (photosynthesis) or 12 (fluorescence) plants. Photosynthesis and stomatal conductance were measured 4 h after the beginning of the light period. Fv/Fm ratio was determined in dark-adapted leaves before the beginning of the light period. The percentage of reduction in photosynthesis and in Fv/Fm ratio was calculated as the reduction in the variable after the treatment in relation to the value measured before the treatment. After measuring photosynthesis, plants were placed in the dark for 6 h and then leaves were harvested for chloroplast isolation.
Chloroplast Preparation
Chloroplasts were isolated from fully expanded leaves of 4-week-old plants (32/preparation). The purity of the preparations was determined by Asp aminotransferase activity gels according to the organelle preparation protocol (Weigel and Glazebrook, 2002). The chloroplast marker enzyme G3PDH was also measured (Stitt et al., 1989). Cytosolic contamination of the chloroplast was determined by the cyotosolic marker enzyme PEPC (Stitt et al., 1989).
Western-Blot Analysis
Isolated chloroplasts were resuspended in 50 mm Tris-Cl, pH 7.5, 0.1% Triton X-100, 10% glycerol, 5 mm β-mercaptoethanol, 10 mm MgCl2, 50 mm KCl, and sonicated on ice for 2 min. To remove the chloroplast membranes, the sample was centrifuged at 4°C for 15 min. Protein content was determined by the Bio-Rad protein assay (Hercules, CA) following the manufacturer's instructions. SDS-PAGE was performed using the Bio-Rad Mini-Protean system and the separated proteins were transferred to nitrocellulose using a Bio-Rad Transfer Blot Cell. PMSR4 was visualized using antibodies raised against the recombinant PMSR4 at 1:1,000 concentration and detected using the Supersignal West Chemiluminescent System (Pierce, Rockford, IL). Immunoblots were digitalized and the intensity of the bands measured with Scion Image freeware (Scion; http://www.scioncorp.com).
Measurement of MetSO Content
Lysed chloroplasts (100 μL) were centrifuged at 16,000g at 4°C and the supernatant proteins were isolated according to the method of Ferguson and Burke (1994). The MetSO content was measured as per Levine et al. (1996). The protein extracts were incubated with cyanogen bromide (CNBr) along with nontreated controls. CNBr does not react with MetSO, whereas it oxidizes Met to homo-Ser. Subsequent acid hydrolysis with dithiothreitol reduces MetSO back to Met. Thus, in samples treated with CNBr, the Met content is equal to the MetSO content. Analysis of the untreated samples (no CNBr) yields the total Met content. Thus, the percent MetSO content can be determined by comparison between CNBr-treated samples and nontreated samples.
PMSR Activity Assay
PMSR activity was measured as per Ferguson and Burke (1994) with some modifications. MetSO (Sigma, St. Louis) was derivatized with Fmoc according to the procedure of Ferguson and Burke (1992). Reaction mixture contained 3 mm Fmoc-MetSO, 150 mm dithiothreitol, 50 mm Tris-Cl, pH 8.0, in a total volume of 120 μL. Activity was measured in the soluble fraction of chloroplasts. The reaction was carried out at room temperature and quenched with 1 volume of cold acetone. Protein was precipitated at −20°C for 30 min followed by centrifugation at 16,000g. The supernatant was passed through a 0.2-μm filter. Fmoc-MetSO and the Fmoc-Met formed were quantified by HPLC using a Supelcosil LC-18 column (Supelco, Bellefonte, PA). The mobile phase consisted of 50 mm acetate buffer, pH 3.9 (solvent A), and acetonitrile (solvent B). Solvent B was increased from 38% to 90% in 20 min, and then further increased to 96% for 3 min and then returned to initial. The amount of Met and MetSO in each reaction was determined by peak area.
Statistics
Data were subjected to ANOVA using statistical software (SAS System for Windows version 8.2; SAS Institute, Cary, NC). Means were compared by Duncan's test at P ≤ 5%.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requester.
This work was supported in part by the Instituto Colombiano Para el Desarrollo de la Ciencia y la Tecnología “Francisco José de Caldas”–COLCIENCIAS and the Universidad Nacional de Colombia (doctoral fellowship to H.M.R).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.046656.
References
- Aro E, Virgin I, Andersson B (1993) Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Bechtold U, Murphy D, Mullineaux PM (2004) Arabidopsis peptide methionine sulfoxide reductase2 prevents cellular oxidative damage in long nights. Plant Cell 16: 908–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defelipe MR, Lucas MM, Pozuelo JM (1988) Cytochemical study of catalase and peroxidase in the mesophyll of Lolium rigidum plants treated with isoproturon. J Plant Physiol 132: 67–73 [Google Scholar]
- Doney RC, Thompson JF (1966) The reduction of S-methyl-l-cysteine sulfoxide and l-methionine sulfoxide in turnip and bean leaves. Biochim Biophys Acta 124: 39–49 [DOI] [PubMed] [Google Scholar]
- Ferguson DL, Burke JJ (1992) A new method of measuring protein methionine-S-oxide reductase activity. Plant Physiol 100: 529–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DL, Burke JJ (1994) Methionyl sulfoxide content and protein methionine-S-oxide reductase activity in response to water deficits or high temperature. Physiol Plant 90: 253–258 [Google Scholar]
- Fliss H, Weissbach H, Brot N (1983) Oxidation of methionine residues in proteins of activated human neutrophils. Proc Natl Acad Sci USA 80: 7160–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH (2002) The contribution of photosynthetic oxygen metabolism to oxidative stress in plants. In D Inzé, M Van Montagu, eds, Oxidative Stress in Plants. Taylor & Francis, London, pp 33–68
- Grimaud G, Ezraty B, Mitchell JK, Lafitte D, Briand C, Derrick PJ, Barras F (2001) Repair of oxidized proteins: identification of a new methionine sulfoxide reductase. J Biol Chem 276: 48915–48920 [DOI] [PubMed] [Google Scholar]
- Gustavsson N, Harndahl U, Emanuelsson A, Roepstorff P, Sundby C (1999) Methionine sulfoxidation of the chloroplast small heat shock protein and conformational changes in the oilgomer. Protein Sci 8: 2506–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson N, Kokke BP, Harndahl U, Silow M, Bechtold U, Poghosyan Z, Murphy D, Boelens WC, Sundby C (2002) A peptide methionine sulfoxide reductase highly expressed in photosynthetic tissue in Arabidopsis thaliana can protect the chaperone-like activity of a chloroplast-localized small heat shock protein. Plant J 29: 545–553 [DOI] [PubMed] [Google Scholar]
- Hoshi T, Heinemann SH (2001) Regulation of cell function by methionine oxidation and reduction. J Physiol 531: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunatan N, Siani JM, McNellis TW (2001) A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13: 2225–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt P, Lamas S (2000) Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur J Biochem 267: 4928–4944 [DOI] [PubMed] [Google Scholar]
- Landry LG, Pell EJ (1993) Modification of Rubisco and altered proteolytic activity in O3-stressed hybrid poplar (Populus-maximowizii × Trichocarpa). Plant Physiol 101: 1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langebartels C, Schraudner M, Heller W, Ernst D, Sandermann H Jr (2002) Oxidative stress and defense reactions in plants exposed to air pollutants and UV.N radiation. In D Inzé, M Van Montagu, eds, Oxidative Stress in Plants. Taylor & Francis, New York, pp 105–135
- Levine RL, Berlett BS, Moskovitz J, Mosoni L, Stadtman ER (1999) Methionine residues may protect proteins from critical oxidative damage. Mech Ageing Dev 107: 323–332 [DOI] [PubMed] [Google Scholar]
- Levine RL, Mosoni L, Berlett BS, Stadtman ER (1996) Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA 93: 15036–15040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowther WT, Weissbach H, Etienne F, Brot N, Matthews BW (2002) The mirrored methionine sulfoxide reductases of Neisseria gonorrhoeae pilB. Nat Struct Biol 9: 348–352 [DOI] [PubMed] [Google Scholar]
- Moskovitz J, Berlett BS, Poston JM, Stadtman ER (1997) The yeast peptide methionine sulfoxide reductase functions as an antioxidant in vivo. Proc Natl Acad Sci USA 94: 9585–9589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Rahman MA, Strassman J, Yancey SO, Kushner SR, Brot N, Weissbach H (1995) Escherichia coli peptide methionine sulfoxide reductase gene—regulation of expression and role in protecting against oxidative damage. J Bacteriol 177: 502–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olry A, Boschi-Muller S, Marraud M, Sanglier-Cianferani S, Van Dorsselear A, Branlant G (2002) Characterization of the methionine sulfoxide reductase activities of PILB, a probable virulence factor from Neisseria meningitidis. J Biol Chem 277: 12016–12022 [DOI] [PubMed] [Google Scholar]
- Pell EJ, Eckardt N, Enyedi AJ (1992) Timing of ozone stress and resulting status of ribulose bisphosphate carboxylase oxygenase and associated net photosynthesis. New Phytol 120: 397–405 [Google Scholar]
- Perl-Treves R, Perl A (2002) Oxidative stress: an introduction. In D Inzé, M Van Montagu, eds, Oxidative Stress in Plants. Taylor & Francis, New York, pp 1–32
- Roshchina VV, Roshchina VD (2003) Ozone and the Plant Cell. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Sadanandom A, Pifanelli P, Knott T, Robinson C, Sharpe A, Lydiate D, Murphy D, Fairbairn D (1996) Identification of a peptide methionine sulfoxide reductase gene in oleosin promoter from Brassica napus. Plant J 10: 235–242 [DOI] [PubMed] [Google Scholar]
- Sadanandom A, Poghosyan Z, Fairbairn DJ, Murphy DJ (2000) Differential regulation of plastidial and cytosolic isoforms of peptide methionine sulfoxide reductase in Arabidopsis. Plant Physiol 123: 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K (1998) Creation of photo-tolerant mutants of a cyanobacterium, Synechocystis sp. PCC 6803, by in vitro random mutagenesis of the psbA gene. In K Satoh, N Murata, eds, Stress Responses of Photosynthetic Organisms. Elsevier Science, New York, pp 3–14
- Scandalios JG (1993) Oxygen stress and superoxide dismutases. Plant Physiol 101: 7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Coruzzi GM (1995) The aspartate aminotransferase gene family of Arabidopsis encodes isozymes localized to three distinct subcellular compartments. Plant J 7: 61–75 [DOI] [PubMed] [Google Scholar]
- Sharma J, Panico M, Barber J, Morris HR (1997. a) Characterization of the low molecular weight photosystem II reaction center subunits and their light-induced modifications by mass spectrometry. J Biol Chem 272: 3935–3943 [DOI] [PubMed] [Google Scholar]
- Sharma J, Panico M, Shipton CA, Nilsson F, Morris HR, Barber J (1997. b) Primary structure characterization of the photosystem II D1 and D2 subunits. J Biol Chem 272: 33158–33166 [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Wheeler GL (2000) Ascorbic acid in plants: biosynthesis and function. Crit Rev Biochem Mol Biol 35: 291–314 [DOI] [PubMed] [Google Scholar]
- Stitt M, Lilley RM, Gerhardt R, Heldt HW (1989) Metabolite levels in specific cells and subcellular compartments of plant leaves. Meth Enzymol 174: 518–552 [Google Scholar]
- Torsethaugen G, Pell EJ, Assmann SM (1999) Ozone inhibits guard cell K+ channels implicated in stomatal opening. Proc Natl Acad Sci USA 96: 13577–13582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torsethaugen G, Pitcher LH, Zilinskas BA, Pell EJ (1997) Overproduction of ascorbate peroxidase in the tobacco chloroplasts does not provide protection against ozone. Plant Physiol 114: 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Zhong R, Morrison III WH, Freshour GD, Hahn MG, Ye Z (2003) Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol 132: 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]