Abstract
We have investigated the subcellular localization, the domain topology, and the amino acid residues that are critical for the function of the presumptive Arabidopsis thaliana auxin influx carrier AUX1. Biochemical fractionation experiments and confocal studies using an N-terminal yellow fluorescent protein (YFP) fusion observed that AUX1 colocalized with plasma membrane (PM) markers. Because of its PM localization, we were able to take advantage of the steep pH gradient that exists across the plant cell PM to investigate AUX1 topology using YFP as a pH-sensitive probe. The YFP-coding sequence was inserted in selected AUX1 hydrophilic loops to orient surface domains on either apoplastic or cytoplasmic faces of the PM based on the absence or presence of YFP fluorescence, respectively. We were able to demonstrate in conjunction with helix prediction programs that AUX1 represents a polytopic membrane protein composed of 11 transmembrane spanning domains. In parallel, a large aux1 allelic series containing null, partial-loss-of-function, and conditional mutations was characterized to identify the functionally important domains and amino acid residues within the AUX1 polypeptide. Whereas almost all partial-loss-of-function and null alleles cluster in the core permease region, the sole conditional allele aux1-7 modifies the function of the external C-terminal domain.
INTRODUCTION
Auxin represents a key regulator of plant cellular and developmental processes (Leyser, 2002). As a true hormone, auxin behaves as a non-cell autonomous signal (Reed et al., 1998; Casimiro et al., 2001; Bhalerao et al., 2002). Auxins characteristically move between plant cells in a polar fashion. Classic studies originally observed that the radiolabeled auxin indole-3-acetic acid (IAA) moved in a basipetal (apex to base) direction in shoot tissues (reviewed in Goldsmith, 1977). Recent molecular studies suggest that auxins also move in an acropetal direction via epidermal cells on the flanks of the shoot apical meristem and newly formed leaf primordia (Benkova et al., 2003; Reinhardt et al., 2003). In roots, distinct acropetal (base to tip) and basipetal (tip to base) auxin transport streams also exist (Rashotte et al., 2000). Acropetal auxin transport occurs in central root tissues, whereas basipetal auxin transport takes place via lateral root cap and epidermal tissues (Rashotte et al., 2000; Swarup et al., 2001). Evidence also exists for a faster phloem-based auxin redistribution from auxin source tissues such as young leaves (Ljung et al., 2001; Marchant et al., 2002) to the root (Swarup et al., 2001). Intuitively, the speed and distance often required to move auxin from shoot to root apices, particularly in larger plants, could only realistically be achieved via the vascular system (Baker, 2000). Nevertheless, the profound impact of auxin transport inhibitors and mutations on plant morphology underlines that polar auxin transport plays a key role over short distances during auxin-regulated developmental processes such as embryo patterning (Friml et al., 2003).
At the cellular level, IAA is transported through a combination of membrane diffusion and carrier-mediated mechanisms (Delbarre et al., 1996). Plants employ specialized influx and efflux carriers to transport IAA from cell to cell (reviewed in Palme and Gälweiler, 1999; Parry et al., 2001a). Molecular genetic studies in Arabidopsis thaliana have resulted in the identification of the presumptive IAA influx and efflux carrier component AUX1 (Bennett et al., 1996) and PIN1 (Galweiler et al., 1998), respectively. Localization studies have revealed that the polarity of auxin transport is underpinned by asymmetric localization of auxin influx and efflux carriers; for example, the asymmetric localization of putative IAA carriers AUX1 and PIN1 to the basal and apical plasma membranes (PM) of root protophloem cells facilitates the polar movement of IAA in an acropetal direction (Swarup et al., 2001).
Recent studies have revealed that AUX1 activity is required for both polar- and phloem-based IAA transport pathways in Arabidopsis root and shoot tissues. Root gravitropic curvature requires AUX1 (and PIN2) to facilitate basipetal auxin transport via lateral root cap and epidermal tissues (Müller et al., 1998; Swarup et al., 2001), whereas the positioning of new leaf primordia is dependent on AUX1 (and PIN1)-mediated acropetal auxin transport via L1 cells on the flanks of the shoot apical meristem (Reinhardt et al., 2003). AUX1-dependent phloem transport is important for mobilizing IAA from newly formed leaf primordia (Marchant et al., 2002), which represent a major site of auxin biosynthesis (Ljung et al., 2001), to developing lateral root primordia in the root (Bhalerao et al., 2002).
AUX1 belongs to the amino acid/auxin permease (AAAP) family of proton-driven transporters (Bennett et al., 1996; Young et al., 1999). However, transport assays on aux1 roots did not observe a reduction in Trp uptake compared with the wild type. Instead, the mutation disrupted 2,4-D (Marchant et al., 1999) and IAA (but not 1-NAA) accumulation (Rahman et al., 2001), thereby mimicking the described substrate specificity of the auxin influx carrier (Delbarre et al., 1996). Auxin reporter-based assays and direct IAA transport measurements have detected reduced basipetal auxin transport in aux1 root apical tissues (Rashotte et al., 2000, 2003; Swarup et al., 2001). Basipetal auxin transport has been shown to be important for root gravitropism (Rashotte et al., 2000), which is likely to provide the basis for the mutant's root agravitropic phenotype (Swarup et al., 2001). The aux1 agravitropic phenotype can be phenocopied in wild-type seedlings using the auxin influx carrier inhibitor, 1-NOA (Parry et al., 2001b; Ottenschläger et al., 2003). The aux1 and 1-NOA induced agravitropic phenotypes can be rescued using the membrane permeable auxin, 1-NAA, whereas auxins that require influx carrier activity such as 2,4-D are unable to rescue aux1 root gravitropism (Yamamoto and Yamamoto, 1998; Marchant et al., 1999; Parry et al., 2001b).
Although a large body of evidence supports an auxin permease function, little is known about the functionally important residues and domains that are critical for AUX1 activity. The molecular and phenotypic characterization of an aux1 allelic series offers a powerful experimental approach to identify important residues and domains. Such an approach is facilitated by the ease of screens required to isolate auxin transport mutants in Arabidopsis (Estelle and Somerville, 1987; Luschnig et al., 1998). A large number of aux1 alleles are already reported in the literature (Maher and Martindale, 1980; Estelle and Somerville, 1987; Okada and Shimura, 1990; Roman et al., 1995). Although the majority of alleles behave as null mutations, several have been reported to have partial loss of function or conditional phenotypes (Maher and Martindale, 1980; Rahman et al., 2001). However, the molecular basis of only a few of these mutations has been determined to date (Bennett et al., 1996; Marchant and Bennett, 1998). This study describes the lesions in 17 new or existing aux1 alleles to investigate the molecular basis of their mutant phenotypes. Missense mutations that result from the substitution of a single, functionally important amino acid are among the most useful genetic resources with which to conduct a protein structure-function study. We have assembled a large aux1 allelic series for this purpose, including 13 newly isolated and two previously identified missense mutations, enabling us to identify key amino acid residues and functional domains in AUX1. The current study also provides important new evidence that AUX1 functions as an auxin influx carrier. Consistent with its proposed transport activity, we demonstrate that AUX1 is PM-localized, its topology is conserved with the amino acid permease NAT2/AAP1, and partial-loss-of-function and null aux1 alleles cluster in the central permease region.
RESULTS
AUX1 Encodes a Plasma Membrane Protein
As a component of the auxin uptake carrier, AUX1 is predicted to function at the plant cell plasma membrane (PM). We employed two distinct approaches—biochemical fractionation and confocal microscopy—to confirm AUX1 PM localization.
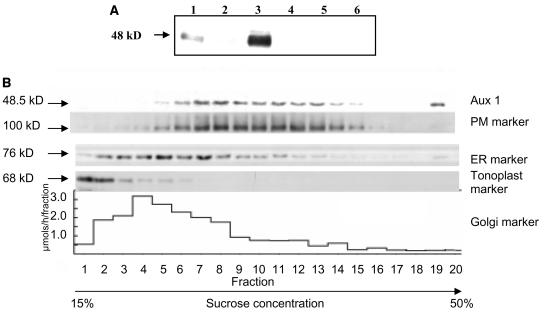
Membranes were prepared using Arabidopsis root cultures (Sanderfoot et al., 2001), because AUX1 localization revealed a pattern identical to that described for young seedling root cells by Swarup et al. (2001) (data not shown). Total, soluble, and microsomal fractions were prepared from Arabidopsis root cultures, protein-gel-blotted, and then probed using an anti-AUX1 peptide antiserum (see Figure 1A). The 19-amino-acid N-terminal sequence of AUX1 was selected as the peptide antigen for antibody production because it represents the most divergent segment of protein coding sequence among the four members of the Arabidopsis AUX/LAX family (Parry et al., 2001a). To confirm that the anti-AUX1 antiserum did not cross-react with other members of the AUX1 protein family, samples were prepared from the wild type (Ws) and the T-DNA–tagged aux1-100 null mutant (Bennett et al., 1996). A cross-reacting band was detectable in the wild-type samples but not aux1-100 (Figure 1A) confirming the specificity of the antiserum for AUX1. In the wild-type extract, AUX1 was detected in the total protein sample (lane 1), was absent from the soluble fraction (lane 2), and was enriched in the microsomal fraction (lane 3), consistent with the predicted membrane localization.
Figure 1.
AUX1 Comigrates with Fractions Enriched for Plasma Membranes.
(A) Cellular extracts from the wild type (ecotype Ws) (lanes 1 to 3), and the aux1-100 (lanes 4 to 6) were membrane-fractionated. Ten micrograms of total (lanes 1 and 4), soluble (lanes 2 and 5), and microsomal proteins (lanes 3 and 6) were separated by SDS-PAGE, blotted, then probed with the anti-AUX1 peptide antiserum. Protein size is indicated on the left.
(B) Comparison of the distribution of AUX1 with membrane markers in Arabidopsis microsomes separated on a continuous sucrose gradient. Equal volumes (10 μL) of each membrane fraction were electrophoresced by 7.5% SDS-PAGE, blotted, and then probed with the anti-AUX1 peptide antiserum. The peaks in the distribution of plasma membrane, Golgi, ER, and tonoplast determined by immunological and biochemical markers are indicated. Protein sizes are indicated on the left.
Further resolution of AUX1 subcellular localization was obtained using sucrose density-based organelle fractionation (Figure 1B). Microsomes isolated from the wild type (Landsberg erecta) were subjected to subfractionation on a continuous sucrose gradient (15 to 50%). A selection of biochemical and immunological organelle markers were used (see Methods) to identify tonoplast, Golgi, PM, and ER membrane fractions (Figure 1B). The antipeptide antiserum detected the majority of AUX1 in sucrose gradient fractions comigrating with the plasma membrane H+-ATPase marker (Figure 1B). A small proportion of AUX1 was also detected in the highest density part of the gradient which also contains traces of the PM and ER markers (Figure 1B); the signal may represent a small proportion of AUX1 that resides in another compartment such as the ER, or it may be from larger aggregated membrane fragments which can tend to accumulate here during gradient fractionation. However as seen in Figure 1 AUX1 separates predominantly with the PM-enriched fractions in sucrose gradient centrifugation.
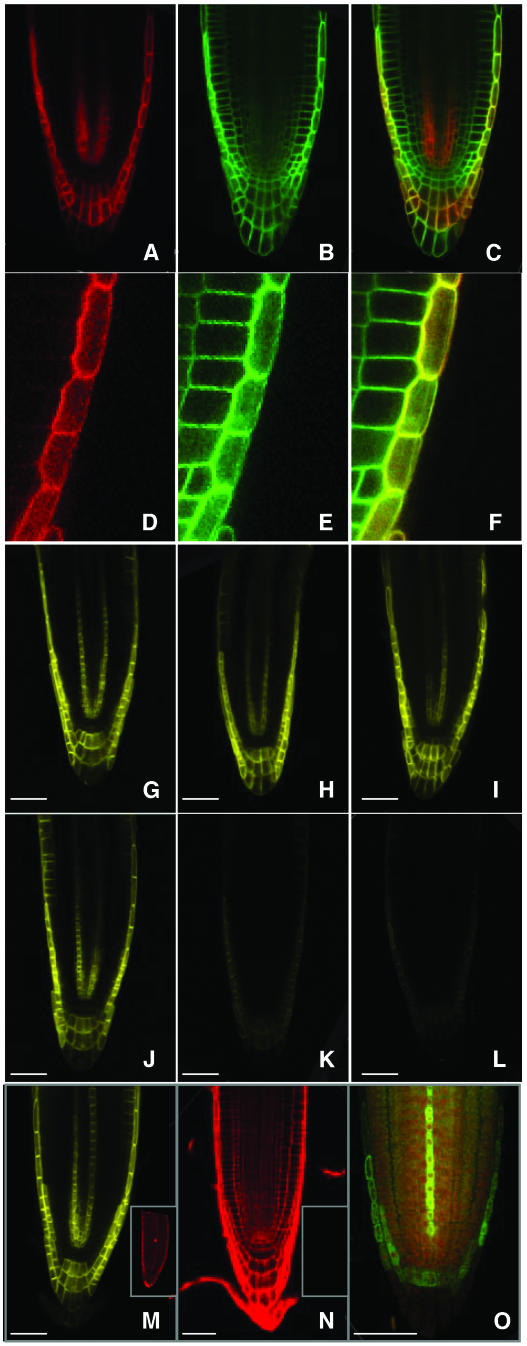
A confocal microscopy-based approach independently verified our subcellular fractionation results. A transgenic Arabidopsis line expressing an N-terminal translational fusion between the AUX1 and yellow fluorescent proteins (termed N-YFP-AUX1) under the control of the native AUX1 promoter (see Methods) was crossed with the 35S promoter-driven PM marker line EGFP-LTI6a (Grebe et al., 2003). The spectral separation capabilities of confocal microscopy enabled us to individually image the YFP-AUX1 (Figures 2A and 2D) or EGFP-LTI6a (Figures 2B and 2E) fusion proteins in seedling roots expressing both transgenes. The N-YFP-AUX1 protein appeared to localize to the PM of root cells (Figures 2A and 2D), in a manner identical to the localization of the EGFP-LTI6a PM marker (Figures 2B and 2E). Overlaying these images revealed that N-YFP-AUX1 and EGFP-LTI6a were colocalized (Figures 2C and 2F).
Figure 2.
AUX1-YFP Fusion Proteins Are Plasma-Membrane–Localized.
(A) Confocal image of a transgenic Arabidopsis root expressing the N-YFP-AUX1 fusion (red).
(B) Confocal image of a transgenic Arabidopsis root expressing the PM marker, EGFP-LTI6a (green).
(C) Superimposed image from (A) and (B) demonstrating colocalization at the PM (yellow).
(D) A transgenic Arabidopsis lateral root cap cell expressing N-YFP-AUX1 (red).
(E) The same transgenic Arabidopsis lateral root cap cells expressing the PM marker, EGFP-LTI6a (green).
(F) Superimposed image from (D) and (E) illustrating colocalization of N-YFP-AUX1 and EGFP-LTI6a fluorescence in the lateral root cap cell PM (yellow).
(G) to (I) Confocal images of transgenic Arabidopsis root expressing N-YFP-AUX1 (yellow) at 0 (G), 30 (H), and 60 (I) min after treatment with MS media (buffered to pH 5).
(J) to (L) Confocal images of transgenic Arabidopsis root expressing N-YFP-AUX1 (yellow) at 0 (J), 30 (K), and 60 (L) min after treatment with MS media containing 20 mM propionic acid (buffered to pH 5).
(M) Confocal image of a transgenic Arabidopsis aux1 root apex expressing AUX1-YFP116 (yellow). Inset image of same root counterstained with propidium iodide (red).
(N) Confocal image of a transgenic Arabidopsis aux1 root apex expressing AUX1-YFP165, counterstained with propidium iodide (red). Inset of same root imaged using the YFP channel, but failing to fluoresce yellow.
(O) Immunolocalization of the AUX1-YFP165 fusion protein (green) in transgenic Arabidopsis aux1 root using anti-GFP antibodies. Background stained with propidium iodide (red).
YFP has a pKa value above 7.1, with fluorescence emission abrogated below pH 6.0 (Llopis et al., 1998), meaning that the fluorescing N-YFP-AUX1 fusion protein is unlikely to be exposed to the pH conditions (4.8 to 5.3) measured in the apoplastic space of root cells (Kosegarten et al., 1999). Indeed, transgenic root cells continued to emit YFP fluorescence even when the external media was buffered to pH 5.0 (Figures 2G to 2I). The YFP domain of the N-terminal AUX1 fusion is not therefore oriented on the apoplastic face of the PM of root cells. If oriented in the cytoplasm, we would expect N-YFP-AUX1 fluorescence to be sensitive to cytoplasmic acidification. Treatment of Arabidopsis seedling roots with the weak membrane-permeable acid, 20 mM propionic acid (buffered to pH 5), has been reported to cause rapid cytoplasmic acidification, dropping from pH 7.2 to 6.4 within 60 min (Moseyko and Feldman, 2001). When we exposed transgenic root cells to identical experimental conditions, N-YFP-AUX1 fluorescence was significantly reduced within 30 min (Figure 2K). We conclude from our confocal results that the N-terminal YFP domain of the AUX1 fusion must be oriented cytoplasmically. When this information is combined with our subcellular fractionation results (Figure 1B), we can conclude that the YFP domain of the AUX1 fusion must be oriented on the cytoplasmic face of the PM of root cells.
Mapping AUX1 Topology
As a member of the auxin-amino acid permease family (Young et al., 1999), AUX1 is predicted to function as an integral membrane protein whose transmembrane (TM) domains repeatedly span the PM (Bennett et al., 1996). Eleven TM helix prediction programs have been employed to model AUX1 topology (see Methods) using the ARAMEMNON database (Schwacke et al., 2003). Their consensus predicts that the AUX1 protein contains 10 TM domains and that its N- and C-terminal domains reside outside the cell (Figure 3A). However, the latter prediction is inconsistent with the ability of the N-YFP-AUX1 fusion protein to fluoresce in the PM of transgenic root cells (Figure 2) and the absence of either a cleavable or noncleavable signal sequence that would be required by the AUX1 N-terminus to traverse the ER membrane. Given the discrepancy between the predicted and experimental data, we decided to directly test AUX1 topology in planta by creating a series of additional YFP translational fusions in the first and second hydrophilic loops predicted within the AUX1 protein sequence by the ARAMEMNON database (Figure 3A). We would then assess whether the particular AUX1 hydrophilic domain that YFP was inserted into was oriented internally (cytoplasmic) or externally on the basis of the presence or absence of fluorescence, respectively.
Figure 3.
Topological Predictions of AUX1 Secondary Structure.
(A) Schematic diagram illustrating AUX1 secondary structure and topology modified from the output from the ARAMEMNON DB (see http://aramemnon.botanik.uni-koeln.de) using 11 protein modeling algorithms (denoted down left-hand side). Boxes denote TM domains; lines indicate hydrophilic regions; whereas the positions where YFP was inserted to test AUX1 topology are highlighted with black arrowheads. The term out refers to the predicted topology of the C-terminal domain on the outer face of the PM.
(B) Schematic diagram illustrating AUX1 secondary structure and protein topology based on experimental evidence. White circles represent hydrophilic regions. Black circles represent PRED-TMR predictions for the TM regions. Amino acid substitutions in the 15 aux1 missense alleles are demarcated in a white circle with a bold outline; predicted sites for N-glycosylation with black arrowheads; and the positions where YFP was inserted to test AUX1 topology are highlighted with small gray arrows.
To create these AUX1-YFP fusions, an in-frame KpnI restriction enzyme site was engineered into the AUX1 gene by PCR mutagenesis after codons 116 and 165, then the full length YFP coding sequence (except start and stop codons) was cloned at the engineered KpnI sites of the AUX1 gene to create the AUX1-YFP116 and AUX1-YFP165 transgenes, respectively (see Methods). Transgenic aux1 roots expressing each AUX1pro:AUX1-YFP transgene was imaged using confocal microscopy (Figure 2). The AUX1-YFP116 protein exhibited strong protein fluorescence, indicative that its domain was orientated in the cytoplasm (Figure 2M). By contrast, transgenic root cells expressing the AUX1-YFP165 fusion protein did not fluoresce (Figure 2N), consistent with its domain being extracytoplasmic. Immunolocalization of the AUX1-YFP165 fusion was necessary to demonstrate that the loss of fluorescence was because of its protein topology, rather than destabilization. Anti-GFP antibodies revealed that the AUX1-YFP165 protein was expressed and targeted identically to both N-YFP-AUX1 and AUX1-YFP116 (Figure 2O). Hence, loss of AUX1-YFP165 fluorescence was consistent with its hydrophilic loop projecting into the low pH environment of the apoplastic space.
Our experimental results demonstrate that the first hydrophobic region which the N-YFP and YFP116 fusions flank must be composed of not one, but two TM domains for both fusion proteins to orient YFP on the cytoplasmic PM face (Figure 3A). This explanation is further strengthened by the observed extracytoplasmic orientation of the AUX1-YFP165 fusion. We therefore conclude from our YFP-based topological studies that AUX1 contains not 10, but 11 TMs (Figure 3B), consistent with results obtained for another member of the auxin-amino acid permease family, NAT2/AAP1, using an epitope-tagged recombinant NAT2/AAP1 protein expressed in animal tissue culture cells (Chang and Bush, 1997).
Mapping Functionally Important Amino Acid Residues and Domains in AUX1
Little is known about the functionally important residues and domains that are critical for AUX1 activity. Molecular and phenotypic characterization of a series of allelic mutations provides a very effective experimental approach to investigate such functionally important features (Tornero et al., 2002). We have assembled a large aux1 allelic series, including 13 newly isolated (this study) and two previously identified missense mutations (Maher and Martindale, 1980; Estelle and Somerville, 1987). New aux1 alleles were isolated from two EMS-mutagenized populations (totaling 25,000 M1 plants) that were screened for seedling roots exhibiting resistance to a concentration of the synthetic auxin 2,4-D that inhibits wild-type root elongation (Marchant and Bennett, 1998). Forty-one of the seedlings recovered were later shown to be allelic to aux1 (A. Marchant and M. J. Bennett, unpublished results). The 41 aux1 alleles were isolated from 31 of the 50 M2 pools screened, and were therefore likely to represent a large number of independent mutations within the AUX1 gene. To determine the molecular basis of each mutation, RT-PCR was performed to amplify the mutated aux1 cDNA template which was then sequenced. Each mutation was confirmed by sequencing the corresponding region of genomic DNA. The molecular lesions for 17 new and existing aux1 alleles were determined (see Supplemental Table 1 online). With the exception of T-DNA–tagged aux1-100 and the aux1-21 alleles in which a single base deletion resulted in a frame shift, all other mutations arose from single base substitutions within the transcribed AUX1 sequence. Of the 22 aux1 alleles described in this study, 15 represent independent novel missense alleles (see Supplemental Table 1 online).
The frequency of the 15 missense alleles along the length of the amino acid sequence was plotted to identify functionally important regions in AUX1. The 485 amino acids were first divided into five windows of 100 amino acids, then a q-value was calculated for each window to assess the statistical probability of the observed distribution being significantly different from a Poisson prediction of random distribution. Windows with high q-values are those in which significantly more or fewer missense mutations are observed than would be expected by random distribution. Eight of the 15 missense alleles occurred in the 201 to 300 window (Table 1), which exhibits a q-value implying a >99% probability that the larger number of mutations occurring in this window is not by chance (data not shown). The q-value is still >99% when taking into account the GC content of this window (data not shown), given the preference of EMS for causing G-to-A mutations (Sega, 1984). This region therefore appears to be particularly important for AUX1 function.
Table 1.
Summary of Missense Substitutions in the aux1 Allelic Series and Their Properties
| Mutation Name | Location/Domaina | WT AAb | Locus (AA No.)c | aux1 AAd | Signaturee | Conservationf | Penetranceg | PHCDh |
|---|---|---|---|---|---|---|---|---|
| aux1-120 | Tm2 | Ser | 57 | Phe | NDi | 100% | Total | 155 |
| aux1-112 | Tm4 | Gly | 178 | Asp | Potential N-myristoylation site | 100% identical | Total | 94 |
| aux1-111 | Tm5 | Ser | 215 | Phe | ND | 100% | Total | 155 |
| aux1-104 | Tm6 | Gly | 238 | Glu | ND | 80% | Total | 98 |
| aux1-106 | Tm6 | Gly | 247 | Glu | ND | 80% | Total | 98 |
| aux1-113 | Tm6 | Ala | 250 | Thr | ND | 80% | Partial | 58 |
| aux1-114 | Tm6/Tm7 loop | Met | 259 | Ile | ND | 80% | Partial | 10 |
| aux1-116 | Tm6/Tm7 loop | Pro | 262 | Ser | ND | 80% | Partial | 74 |
| aux1-2 | Tm7 | Ala | 272 | Val | ND | 100% | Partial | 64 |
| aux1-117 | Tm7/Tm8 loop | Gly | 292 | Glu | Highly conserved motif in AAAP genes | 80% | Total | 98 |
| aux1-102 | Tm7/Tm8 loop | Met | 305 | Arg | ND | 100% | Total | 91 |
| aux1-103 | Tm8 | Thr | 324 | Met | ND | 80% | Total | 81 |
| aux1-118 | Tm10 | Ser | 386 | Asn | ND | 80% | Total | 46 |
| aux1-105 | Tm11 | Gly | 439 | Arg | ND | 80% | Total | 125 |
| aux1-7 | C-term tail | Gly | 459 | Asp | ND | 80% | Total | 94 |
Tm, transmembrane domain.
WT AA, wild-type amino acid residue.
Locus AA, number of amino acid residue in the AUX1 coding sequence.
aux1 AA, amino acid substitution in the aux1 allele.
Signature, AAAP (amino acid-auxin permease) signature.
Conservation, conservation over AUX1 and four paralogues (see Fig. 5).
Penetrance, severity of genetic defect in the aux1 allele.
PHCD, physicochemical distance.
ND, no data.
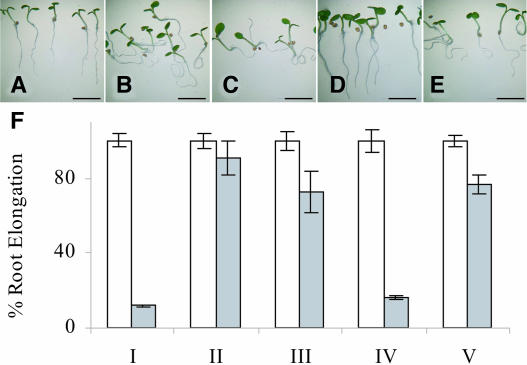
The number of non-sense mutations versus other classes of mutations was double the theoretically expected value (15% versus 7.23% when expressed as a percentage of the total number of loss-of-function mutations), suggesting that mutations which do not cause a loss of function must occur, but were not recovered in our 2,4-D screen. This could imply that fewer loss-of-function mutations occur in some regions of the AUX1 sequence than others. Only one missense mutation was recovered in the N-terminal third of the AUX1 primary sequence (Figure 3B), suggesting that this region may tolerate amino acid substitutions more than the 201 to 300 window. To probe the functional importance of the N-terminal third of AUX1, we examined whether the N-YFP-AUX1, AUX1-YFP116, and AUX1-YFP165 fusion proteins retained biological activity. Each of the AUX1pro:AUX1-YFP transgenes described in Figure 2 had originally been transformed into the null aux1-22 mutant background, allowing us to assess whether any of the AUX1-YFP fusions retained biological activity on the basis of complementation of the aux1 root defect. Root gravitropic assays revealed that the AUX1pro:AUX1-YFP116 transgene was able to rescue the aux1 agravitropic phenotype (Figure 4D) and restore a wild-type level of 2,4-D sensitivity (Figure 4F). Neither the N-YFP-AUX1 (Figure 4C) nor AUX1-YFP165 (Figure 4E) fusion proteins rescued aux1-22 gravitropism, despite a slight reversal of the aux1-22 2,4-D insensitive root phenotype (Figure 4F). The reduced activity of either fusion is unlikely to be attributable to the YFP insertion causing AUX1 misfolding, because we would expect the proteins to be degraded, as observed for a C-terminal AUX1-YFP fusion (data not shown) and several missense alleles (see Supplemental Figure 1 online). Neither can the reduction in N-YFP-AUX1 or AUX1-YFP165 activity be explained by loss of protein stability or targeting, because both fusion proteins were clearly detectable and exhibited a pattern of localization identical to the functional AUX1-YFP116 fusion (Figure 2M). Instead, the N-terminal and loop 3 regions appear to be important for AUX1 function once the protein is localized at the root cell PM, by possibly impairing functionally important protein-protein interactions. By contrast, YFP inserted into the cytoplasmic loop 2 does not disrupt AUX1 protein function. Our AUX1-YFP116 results suggest that selected regions within the N-terminal third of the AUX1 primary sequence can indeed tolerate modifications without loss of protein function.
Figure 4.
The AUX1-YFP116 Fusion Retains Biological Activity.
Root gravitropic phenotype of 5-d-old (A) wild type; (B) aux1-22; (C) aux1-22 N-YFP-AUX1; (D) aux1-22 AUX1-YFP116; and (E) aux1-22 AUX1-YFP165 seedlings.
(F) 2,4-D sensitive root elongation phenotype of 5-d-old (i) wild type; (ii) aux1-22; (iii) aux1-22 N-YFP-AUX1; (iv) aux1-22 AUX1-YFP116; and (v) aux1-22 AUX1-YFP165 seedlings. White bars denote root growth on MS alone, whereas gray bars denote root growth on MS plus 10−7 M 2,4-D.
A search of the entire AUX1 primary amino acid sequence (see Methods) revealed it to be consistent with earlier assignments to the subfamily of membrane-spanning permeases with specificity for amino acids or related compounds. Additionally, consensus sites were identified for posttranslational modifications including N-myristoylation (14 sites), N-glycosylation (four sites), and protein kinase C and Casein kinase II phosphorylation sites (four and two sites, respectively). Of the 15 missense mutations, 13 occur within both the core permease region of the protein and nine of these map to individual predicted transmembrane regions (see Figure 3B). A further mutation (aux1-112) maps to a potential N-myristoylation site, located close to the end of predicted TM4 (Table 1).
A Clustal primary sequence alignment between AUX1, LAX1, LAX2, LAX3, and AAP1 revealed that several of the aux1 missense alleles result from amino acid substitutions at highly conserved residues (Figure 5). For example, the Gly residue substituted in the aux1-117 allele was highlighted by Young et al. (1999) as part of one of the most highly conserved motifs (GYAAFG) shared among the AAAP family of proteins. Nevertheless, the importance of the majority of the residues identified in the aux1 allelic series could not have been inferred on the basis of their sequence conservation alone, highlighting the effectiveness of our forward genetic approach compared with site-directed mutagenesis to identify functionally important amino acids.
Figure 5.
Alignment of the Amino Acid Permease AAP1 and the Arabidopsis AUX1 Gene Family.
The AAP1, AUX1, LAX1, LAX2, and LAX3 protein sequences were aligned using Clustal. Identical amino acids are boxed; residues substituted in an aux1 missense allele and conserved between aligned sequences are shaded.
The aux1 Allelic Series Contains Null, Partial-Loss-of-Function, and Conditional Alleles
The root gravitropic response of each missense mutant was assayed to discriminate between null and partial-loss-of-function aux1 alleles. Vertically grown seedlings were given a 90° gravity stimulus for 24 h before measuring the angle of root curvature (Parry et al., 2001a). All wild-type (Ws) roots (n = 50) responded to the 90° gravity stimulus by turning between 80° and 120° within 24 h (Figure 6). Eighteen of the 22 aux1 alleles exhibited a fully agravitropic root growth behavior (Figure 6; see Supplemental Table 1 online) identical to the non-sense allele wav5-33 (Okada and Shimura, 1990; Marchant and Bennett 1998). Roots from the missense alleles aux1-2, aux1-113, aux1-114, and aux1-116 (Figure 6) exhibited a nonrandom pattern of root growth after the gravity stimulus, suggesting that they may retain partial AUX1 function. Elongation assays observed that the four aux1 alleles exhibited a sensitivity to auxin which was intermediate to null aux1 and wild-type roots (see Supplemental Figure 2 online), implying that aux1-2, aux1-113, aux1-114, and aux1-116 represented partial-loss-of-function alleles.
Figure 6.
The aux1 Allelic Series Contains Partial-Loss-of-Function, Null, and Conditional Mutations.
Three-day-old vertically grown seedlings were given a 90° gravity stimulus for 24 h before measuring the angle of root curvature (Parry et al., 2001b). The angles of root curvature were grouped in nine classes of 40° (0 to 40, 41 to 80, 81 to 120, 121 to 160, 161 to 200, 201 to 240, 241 to 280, 281 to 320, and 321 to 360°) and the gravitropic response of the roots was expressed as percentage in a wheel diagram.
It was necessary to establish the basis for the partial-loss-of-function phenotypes of the aux1-2, aux1-114, aux1-113, and aux1-116 alleles compared with the null phenotype of other aux1 mutants (Figure 6; see Supplemental Table 1 online). To assess whether this simply reflected the differential stability of partial-loss-of-function versus null mutant proteins, root microsomes from all loss of function aux1 missense alleles described were characterized by protein gel blot immunodetection using an anti-AUX1 anti-peptide antiserum (see Methods). AUX1 was detected in all null and partial-loss-of-function missense alleles, with the exception of aux1-103 and aux1-112 (see Supplemental Figure 1 online). Instead, the phenotypic severity predicted for each aux1 missense mutant appeared to more closely correlate with the chemical relatedness of the amino acid side chain that substituted the original wild-type residue (termed the physiochemical distance; PHCD) (Table 1; Grantham, 1974). Missense substitutions in null aux1 alleles had PHCD values above 91 (91 to 155), whereas partial-loss-of-function aux1 alleles has PHCD values below 74 (10 to 74). The single exception to this trend is the Ser-to-Asn substitution at amino acid 386 in the aux1-118 allele which has a low PHCD value of 46, but causes a null phenotype.
Rahman et al. (2001) reported that the aux1-7 allele was conditional; the root defects could be rescued by a gamma-pyronyl-triterpenoid saponin, chromosaponin I (CSI). We tested whether any other aux1 alleles were conditional by examining if CSI could restore their root gravitropic phenotypes of a selection of aux1 missense mutants (Figure 7). The effects of 60-μM CSI on the root gravitropic response of the aux1-2, 1-7, 1-114 (Figure 7B) and aux1-102, 1-104, 1-105, 1-111, 1-113, 1-116, 1-117, and 1-120 alleles (data not shown) was examined after a 90° gravity stimulus (Rahman et al., 2001). With the exception of aux1-7, 60-μM CSI failed to restore the gravitropic phenotype of the remaining aux1 missense alleles (Figure 7B). Conversely, CSI treatment completely blocked the partial gravitropic response of aux1-2 and aux1-114 alleles, and also impaired the wild-type controls response (Figure 7B).
Figure 7.
The Root Agravitropic Phenotype of Conditional aux1-7 Mutants Can Be Rescued by CSI.
Root gravitropic response of Col, aux1-2, aux1-7, and aux1-114 alleles and transgenic lines HAaux1-7-1 and HAaux1-7-2 (expressing the HA-AUX1-7 protein in an aux1-22 background) in the absence (A) or presence (B) of 60 μM CSI after a 90° gravity stimulus (see above).
The aux1-7 Missense Substitution Disrupts AUX1 Activity
The allele-specific rescue of aux1-7 gravitropism by CSI suggests that the saponin does not bypass the requirement for a functioning influx carrier by nonspecifically permeabilizing plant cell to auxin. Instead, our results suggest that CSI rescues aux1-7 gravitropism by either interacting with (1) the mutant C-terminal domain to restore AUX1-7 function or (2) via an another mutation in the aux1-7 background. To discriminate between these possibilities, we examined whether CSI was able to rescue gravitropism of a null aux1-22 line expressing a HA-AUX1-7 transgene. We reengineered the HA-AUX1 transgene (Swarup et al., 2001) to include the aux1-7 mutation (Table 1), then transformed the HA-aux1-7 transgene into a null aux1-22 background. Unlike the wild-type HA-AUX1 transgene (Swarup et al., 2001), HA-aux1-7 transgene lines (termed HAaux1-7-1 and HAaux1-7-2) did not rescue the agravitropic phenotype of the aux1-22 mutant allele (Figure 7A). However, root gravitropism could be restored when either HA-aux1-7-1 or HAaux1-7-2 seedlings were grown in the presence of CSI (Figure 7B). Hence, the allele-specific rescue of aux1-7 gravitropism by CSI must be attributable to a direct interaction between the saponin and mutant C-terminal domain.
CSI could rescue aux1-7 gravitropism by either restoring HA-AUX1-7 protein targeting and/or permease activities. We investigated whether the aux1-7 genetic lesion disrupted AUX1 protein targeting using an immunolocalization approach. Confocal microscopy revealed that HA-AUX1-7 localization in root apical cells was identical to the wild-type HA-AUX1 control (see Supplemental Figure 3 online). For example, the mutant HA-AUX1-7 and wild-type HA-AUX1 proteins were PM-localized in lateral root cap cells. Similarly, localization of the mutant HA-AUX1-7 protein in protophloem cells was identical to wild-type HA-AUX1. We conclude from the wild-type localization of the HA-AUX1-7 protein that this mutation in the C-terminal domain influences AUX1 activity, rather than protein targeting.
DISCUSSION
The transgenic and molecular genetic strategies employed in our study have generated valuable new information about the subcellular localization, domains, their topology, and the amino acid residues that are critical for AUX1 function.
AUX1 Encodes a Polytopic Plasma Membrane Protein
Four independent pieces of experimental evidence are consistent with the bulk of AUX1 residing in the PM. Firstly, AUX1 was observed to comigrate with a PM marker during subcellular fractionation on a sucrose density gradient (Figure 1B). Secondly, confocal microscopy revealed that the N-YFP-AUX1 fusion protein colocalizes with the PM marker, EGFP-LTI6a (Figure 2C). Thirdly, the AUX1 protein must span the PM based on the differential fluorescence of the N-YFP-AUX1 (Figure 2A) and AUX1-YFP116 (Figure 2M) versus AUX1-YFP165 (Figures 2N and 2O) fusion proteins; and also consistent is the sensitivity of N-YFP-AUX1 (Figures 2J to 2L) to propionate-dependent cytoplasmic acidification. When this information is combined with our subcellular fractionation results (Figure 1B), the YFP domain of the AUX1 fusion must be oriented on the cytoplasmic face of a root cell PM.
We have taken further advantage of the pH sensitivity of YFP (Llopis et al., 1998) and the steep pH gradient that exists between the apoplastic (4.8 to 5.3) and cytoplasmic PM faces of plant root cells (6.5 to 7.3) to probe AUX1 topology. We were able to determine, on the basis of the presence or absence of fusion protein fluorescence, whether each particular hydrophilic domain that YFP was inserted into is oriented on either in or outside the cytoplasm, respectively. Insertion of YFP is likely to cause minimal disruption to protein folding as a result of the close proximity of the N- and C-termini of the GFP protein, as demonstrated by the correct targeting and stability of all three AUX1-YFP fusion proteins.
The YFP-based topology studies predict that AUX1 contains 11 TM domains, with N- and C-terminal hydrophilic domains oriented on cytoplasmic and apoplastic faces of the PM, respectively. Our experimental conclusions are consistent with results obtained for another member of the auxin-amino acid permease family, NAT2/AAP1, using an epitope-tagged recombinant NAT2/AAP1 protein expressed in animal tissue culture cells (Chang and Bush, 1997). The consensus prediction from the ARAMEMNON database successfully identified 10 out of these 11 TM domains, but the consensus and most of the individual algorithms incorrectly predicted that the initial long hydrophobic region (residues 55 to 105) was composed of one, rather than two TM helices (Figure 3A). The failure to spot TM1 and TM2 is a problem inherent to many of the algorithms which employ a rule that a minimum number of amino acids must occur between two predicted TM helices. Thus a region containing 40 to 50 hydrophobic amino acids may be predicted to contain one helix rather than two because the algorithms cannot identify where the loop is. This discrepancy highlights the importance of direct experimental validation of topology models for plant polytopic PM proteins.
Several hydrophobic membrane proteins have been reported to migrate faster than predicted on SDS-PAGE gels including the Arabidopsis STP1 monosaccharide transporter (Stoltz et al., 1994) and a putative sugar beet sugar transporter (Chiou and Bush, 1996). AUX1 is no exception, exhibiting a faster mobility on denaturing SDS-PAGE than predicted, giving an estimated molecular mass of 48 kD (Figures 1A and 1B) compared with the calculated value of 54 kD (Bennett et al., 1996). The discrepancy between the predicted versus the experimentally estimated molecular mass of AUX1 is unlikely to result from either N- or C-terminal processing, because both anti-AUX1 N-terminal and anti-HA antiserum are able to detect the native N- and HA-epitope–tagged N and C termini of AUX1, respectively (Figure 1; Swarup et al., 2001). Protein gel blot immunodetection (see Supplemental Figure 1 online) revealed that the electrophoretic mobility of AUX1 had increased in four of the aux1 alleles (aux1-102, aux1-105, aux1-117, and aux1-118). The reason for their altered behavior is currently unclear. None of the substitutions coincide with known recognition motifs for posttranslational modification (Table 1). All four alleles cause an uncharged amino acid to be replaced by a charged amino acid (Table 1). However, similar substitutions in other regions of AUX1, such as aux1-104 and aux1-106 in TM6, do not cause a mobility shift.
The aux1 Allelic Series Has Defined Functionally Important Residues and Domains
The molecular characterization of our aux1 allelic series has proved a highly effective approach to identify the amino acid residues and domains that are critical for AUX1 function. Of the 15 aux1 missense mutations described in this study, 13 occur within the core permease region. Nine of these map to individual predicted transmembrane helices (see Figure 3B); all but one (aux1-2) of these missense substitutions results in a null phenotype. The remaining four of the missense mutations in the core permease region map to loops 6 and 7. Interestingly, the null alleles aux1-102 and aux1-117 are located in external loop 7, whereas the partial-loss-of-function mutations aux1-114 and aux1-116 map to internal loop 6. External loops appear less tolerant of modification as demonstrated by the retention of AUX1 biological activity when YFP was inserted into internal loop 2 (AUX1-YFP116) versus the observed loss of function for the external loop 3 AUX1-YFP fusion, AUX1-YFP165 (Figure 5). The marked difference in severity of these mutations and insertions may suggest that external loops are more important to AUX1 function. However, analysis of a much larger series of aux1 missense alleles replacing residues in external versus internal facing hydrophilic loops would be required before drawing a definitive conclusion. Indeed, it remains possible that the phenotypic differences observed between the aux1 alleles simply reflects the replacement of uncharged amino acids with the bulky, charged Arg and Glu side chains in the case of the aux1-102 and aux1-117 alleles, respectively. Similarly, internal loop 2 is predicted to be considerably larger than external loop 3 (44 versus 12 amino acids in length; Figure 3C), which may make it more tolerant of the YFP fusion.
The central region of AUX1 appears particularly important for protein function, because nine of the missense mutations cluster between TM VI and VIII (Figure 3B). Ortiz-Lopez et al. (2000) also reported that substitutions in the central region of the AAP1 primary sequence modify protein function. Substitutions at residue Ala-254 in TM VI altered Km for Ala and His, leading Ortiz-Lopez et al. (2000) to suggest that the residue may define the substrate binding site. Our aux1 allelic series has also defined a functionally important residue (Gly-238) in TM VI of aux1-104 that causes a null phenotype. The severity of the aux1-104 allele contrasts the phenotypic effects of four closely linked partial-loss-of-function alleles (aux1-2, aux1-113, aux1-114, and aux1-116) that substitute residues in a short stretch of 26 amino acids predicted to be in TM VI-loop 6-TM VII (Figure 3B). Although it remains possible that the region defined by the partial-loss-of-function alleles can tolerate amino acid substitutions, the perfectly conserved sequence alignments with other family members for the region spanning TM VI-loop 6 suggests otherwise (Figure 5). By contrast, alignments reveal that TM VII features several substitutions between family members (Figure 5). Nevertheless, the Ala-to-Val substitution at invariant residue 272 in the aux1-2 allele reduces root growth sensitivity to exogenous 2,4-D (A. Yemm, R. Swarup, G. Parry, M. Bennett, and R. Napier, unpublished data). The aux1-2 partial-loss-of-function phenotype for root gravitropism and 2,4-D-sensitive root growth may highlight the steric hindrance of natural and synthetic auxin substrates passing through the AUX1-2 permease region, respectively. Alternatively, the aux1-2 Ala-to-Val substitution may perturb AUX1 function by altering helix-helix packing as described for the phage M13 coat protein (Deber et al., 1993).
Whereas almost all partial-loss-of-function and null alleles (13/14) cluster within the core permease region (Table 1), the sole conditional allele aux1-7 resides in the C-terminal domain. One possible explanation for the conditional behavior of the aux1-7 allele (Figure 7B) is that the C-terminal domain performs a regulatory function. Alternatively, the external C terminus may be required to fold into or near the active site of the transporter, as described for the Shaker K+ channel (Sokolova et al., 2003). Interestingly, deletion of the C-terminus of the related permease, NAT2/AAP1, has been reported to modify transport activity (Chang and Bush, 1997), suggesting that the C terminus may perform a common function among AAAP members.
Plant Membrane Composition Appears to Influence Auxin Carrier Protein Function
Saponins are reported to act by permeabilizing the PM (Keukens et al., 1995). However, the allele-specific effects of CSI on aux1-7 gravitropism (Figure 7) suggests that the pea saponin does not bypass the requirement for a functioning influx carrier by nonspecifically permeabilizing plant cell membranes to IAA. Instead, the saponin may directly interact with AUX1-7 to reestablish a functional interaction between the mutant C terminus and the permease core. Alternatively, CSI may rescue AUX1-7 function indirectly by altering its immediate lipid environment. Saponins are known to interact with sterols within artificial lipid bilayers (Nishikawa et al., 1984). Many animal transport proteins have been described to function within lipid rafts enriched with cholesterol and sphingolipids (reviewed in Zajchowski and Robbins, 2002) including GLUT4, which is preferentially targeted to lipid rafts in the PM where glucose uptake takes place (Gustavsson et al., 1996). By analogy, CSI could rescue AUX1-7 activity by targeting the auxin permease to sterol-rich lipid rafts in the PM of root cells.
Plant sterol composition has been demonstrated to be important for auxin signaling by regulating carrier protein trafficking, recycling or functionality in the membrane (reviewed in Lindsey et al., 2003). Mutations in the sterol methyltransferase1 (SMT1) gene causes major changes in sterol composition, which results in selected members of the PIN family of putative auxin efflux carrier proteins being mislocalized in root cells (Willemsen et al., 2003). Grebe et al. (2003) have recently shown that sterol trafficking and recycling of the PIN2 protein share a common endocytic pathway in root epidermal cells. However, neither study observed an association between sterol composition or trafficking and AUX1 localization or recycling, respectively (Grebe et al., 2003; Willemsen et al., 2003). Hence, any sterol-dependent rescue of AUX1-7 by CSI is likely to be mediated at the level of protein activity, rather than targeting, consistent with the wild-type pattern of HA-AUX1-7 localization (see Supplemental Figure 3 online).
METHODS
Preparation of Microsomal Membranes Fractions for Protein Gel Blot Immunodetection
Microsomal membranes were prepared using Arabidopsis thaliana root cultures grown in hormone-free media containing 0.32% Gamborgs B5 salts (Sigma, Poole, UK), 0.1% Mes, pH 5.8, and 2% sucrose at 22°C with gentle shaking (20 to 30 rpm) in diffused light for 21 d. The roots were washed with chilled sterile distilled water, dried, quickly frozen in liquid nitrogen, and then stored in −70°C. Microsomal membranes were prepared from frozen root tissue by the modified methods of Kjellbom and Larsson (1984) and Zettl et al. (1994). All the manipulations were performed at 4°C. Five grams of root tissue was homogenized in 3 to 4 mL of the homogenization buffer (0.5 M sucrose, 50 mM Hepes-OH, pH 7.5, 0.5% polyvinyl polypyrrolidone, 0.1% [w/v] sodium ascorbate, 1 mM DTT, and Complete Protease Inhibitor cocktail; Roche, Basel, Switzerland). The homogenate was filtered through 100-μm mesh nylon, and the resulting filtrate centrifuged for 12 min at 2800g at 4°C. Microsomal fraction was pelleted by further centrifugation at 200,000g for 33 min at 4°C. The microsomal pellet was washed and resuspended in a minimal volume of TEDM buffer [25 mM Tricine, pH 8.0, 1 mM EDTA, 1 mM DTT, 5 mM Mg(CH3COO)2, 1 μM pepstatin, 1 μM leupeptin, 2 μg/mL aprotinin, and 1 mM 4-(2-aminoethyl)-benzensulphonylfluoride]. Ten micrograms of protein from each fraction was separated by 7.5% SDS-PAGE and electroblotted onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories, Hercules, CA). PVDF membranes were probed with a rabbit anti-AUX1-N-terminal-peptide polyclonal antibody [raised against a synthetic peptide: MSEGVEAIVANDNGTDQVNG(C)] and secondary goat anti-rabbit IgG-HRP (Sigma) antibodies, then visualized by chemiluminescence using the ECL system (Amersham-Pharmacia Biotech, Uppsala, Sweden).
Preparation and Analysis of a Sucrose Density Gradient of Root Microsomes
Arabidopsis (Landsberg erecta) root cultures were grown aseptically on a rotary shaker (80 rpm, 24°C, 150 lux) in Gamborgs B5 salts plus vitamins (Sigma), 1.0 g L−1 Mes-KOH, pH 5.8, 2% (w/v) sucrose. Microsomal fractions were prepared as described by Uemura et al. (1995), but with the low-speed fraction centrifuged at 12,000g for 10 min. Supernatants were centrifuged at 80,000g, 4°C, for 45 min, and the microsomal pellets were washed through sucrose density gradient buffer (250 mM sucrose, 10% glycerol, 2 mM BTP-Mes, pH 7.0) and pelleted for 45 min at 100,000g before suspending in fresh sucrose density gradient buffer. For microsomal fractionation, 10 mg microsomal protein was suspended on a 15 to 50% (w/w) continuous sucrose gradient containing in 2 mM Tris-Mes, pH 7.0, 2 mM DTT and centrifuged at 80,000g for 3 h. Fractions were washed through 250 mM sorbitol, 10% glycerol, 10 mM Tris-Mes, pH 7.0, and 2 mM DTT and pelleted for 45 min at 100,000g before resuspending to 150 μL in fresh wash buffer.
Membrane marker proteins were detected immunologically and biochemically in equal volumes from each fraction. For protein gel blot analysis, 10 μL of each fraction was separated by 7.5% SDS-PAGE and electroblotted onto a PVDF membrane (Bio-Rad). Membranes were probed with anti-AUX1 (see above); the PM marker, anti-PMA2 (H+-ATPase) (Morsomme et al., 1996); the ER marker, anti-BiP (from M. Chrispeels, La Jolla, CA); and the tonoplast marker, anti- H+-PPase (Rea et al., 1992). Probed PVDF membranes were stripped with 2% SDS (w/v), 100 mM 2-mercaptoethanol, and 62.5 mM Tris-HCl, pH 6.8, at 55°C for 30 min between uses. Enzyme activities of the PM marker, vanadate-sensitive H+-ATPase activity at pH 7.0 and the Golgi marker, latent UDPase activity at pH 6.5, were assayed at 37°C in 40 mM Tris-Mes containing 100 mM sodium molybdate, 2 mM MgSO4, 50 mM KCl, 2 mM ATP, and 100 mM sodium orthovanadate, or 2 mM UDP ± 0.01% Brij 58. Activities were determined from rate of phosphate release by measurement of OD720 after addition of Ohnishi reagents (Ohnishi et al., 1975). Protein contents were determined using BSA standards. Details of marker enzyme analysis and protein determinations are given in Terry and Williams (2002).
Creation of AUX1 Protein Fusions
The NHA aux1-7 transgene was created by subcloning a PCR product from aux1-7 genomic DNA into a NHA-AUX1 construct (Swarup et al., 2001). Cloning into binary vector, Agrobacterium tumefaciens transformation, and plant transformation into an aux1 mutant background were done as described by Swarup et al. (2001). Homozygous lines carrying single transgene were then used for immunolocalization of AUX1-7 protein as described by Swarup et al. (2001).
To create AUX1-YFP fusions, an in-frame KpnI restriction enzyme site was engineered into the AUX1 gene by PCR mutagenesis after codon 2 (for NYFP-AUX1), 116 (for AUX1-YFP116), and 165 (for AUX1-YFP165). Full-length YFP sequence (except start and stop codons) was PCR-amplified using primers with in-frame KpnI restriction enzyme sites at the ends and subsequently cloned at the engineered KpnI sites of the AUX1 gene to create NYFP-AUX1, AUX1-YFP116, and AUX1-YFP165, respectively. Cloning into binary vector, Agrobacterium transformation, and plant transformation into an aux1 mutant background were done as described by Swarup et al. (2001). Homozygous lines carrying single transgenes were grown as described by Swarup et al. (2001) and visualized directly under a confocal microscope (see below). AUX1-YFP165 seedling roots were counterstained with propidium iodide (5 μg/mL) for 20 to 30 min before confocal microscopy (Figure 2O). Immunolocalization of AUX1-YFP165 fusion protein was performed as described in Swarup et al. (2001) using a polyclonal rabbit anti-GFP primary antibody (Molecular Probes, Eugene, OR) and Oregon-green–coupled anti-rabbit secondary antibody (Molecular Probes).
Microscopy
Confocal microscopy was performed using a Leica SP2 laser scanning microscope equipped with argon krypton and green HeNe lasers and an AOBS scan head system (Leica Microsystems, Bannockburn, IL) capable of spectrally separating the emission from GFP and YFP (Figures 2A and 2B). The 488- and 514-nm excitation lines of an argon ion laser were used to excite GFP and YFP, respectively. Experimental manipulation of the external and cytoplasmic pH of Arabidopsis root cells (Figures 2G to 2L) was performed as described by Moseyko and Feldman (2001). Kalman averaging was performed over 16 frames for single images. Images were processed using the Leica CONFOCAL software and Adobe Photoshop (version 7.01; Adobe Systems, San Jose, CA).
AUX1 Modeling, Statistical, and Bioinformatic Analyses
The 11 algorithms employed by the ARAMEMNON database (Schwacke et al., 2003; see http://aramemnon.botanik.uni-koeln.de; Figure 3A) were used to predict the number of AUX1 trans membrane-spanning domains and the topology of their hydrophilic loops. A variety of other Web sites were also used to predict transmembrane regions and orientation from the AUX1 protein sequence, including: CONPRED II (see http://bioinfo.si.hirosaki-u.ac.jp/∼ConPred2/), DAS (see http://www.sbc.su.se/∼miklos/DAS/maindas.html), HMMTOP 2.0 (see http://www.enzim.hu/hmmtop/html/submit.html), MEMSAT 2 (see http://bioinf.cs.ucl.ac.uk/psipred/psiform.html), PHDhtm (see http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_htm.html), PRED-TMR (see http://biophysics.biol.uoa.gr/PRED-TMR/input.html), PRED-TMR 2 (see http://biophysics.biol.uoa.gr/PRED-TMR2/input.html), SOSUI (see http://sosui.proteome.bio.tuat.ac.jp/sosui_submit.html), TMAP (see http://bioweb.pasteur.fr/seqanal/interfaces/tmap.html), TMHMM 2.0 (see http://www.cbs.dtu.dk/services/TMHMM-2.0/), Tmpred (see http://www.ch.embnet.org/software/TMPRED_form.html), and TopPred II (see http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html).
The frequency that missense alleles occur along the length of the AUX1 amino acid sequence was calculated as described by Tornero et al. (2002) to identify functionally important regions. The 485 amino acids were first divided into five windows of 100 amino acids, then a q-value was calculated for each window to assess the statistical probability of the observed distribution being significantly different from a Poisson prediction of random distribution (see Results).
The χ2 tests were also done to see if the residues of specific amino acids are mutated to, or are from, loss-of-function (LOF) missense mutants. At a P-value of 0.05, missense LOF mutations in aux1 are significantly more likely to occur in Gs and Cs of Gly and Met than in Gs and Cs of other amino acid codons. These amino acids could, therefore, be more important to the functional sites or stability of AUX1. The As and Ts of Arg, Asp, and Glu residues were created preferentially to As and Ts of other amino acids in missense LOF mutations. These amino acid substitutions could be particularly damaging to AUX1's structure or function because they are all charged and hydrophilic.
Bioinformatic analysis of the AUX1 sequence to identify functional domains, motifs, signatures, and sites of posttranslational modification was performed using PFAM (see http://www.sanger.ac.uk/Software/Pfam/), SMART (see http://smart.ox.ac.uk/), and PROSITE (see http://ca.expasy.org/prosite/) programs. Clustal was used to generate a multiple alignment between AUX1 (At2g38120) and related sequences LAX1 (At5g01240), LAX2 (At2g21050), LAX3 (At1g77690), and AAP1 (At1g58360).
Mutagenesis, Screening, and Genetic Analysis
Altogether, 25,000 M1 seeds of Arabidopsis ecotype Ws were initially treated with 50 mM EMS for 4 h, rinsed repeatedly, then immediately sown in 50 pools of 500 plants on soil (50:50 mix of vermiculite:potting compost). Approximately 6000 M2 seeds from each pool were selected on 2,4-D selective media for root elongation (as described in Marchant and Bennett, 1998). Auxin-resistant seedlings were crossed with the null aux1-100 mutant to test for allelism and also backcrossed at least once with Col. To determine the molecular basis of each aux1 mutation, RT-PCR was performed to amplify the AUX1 cDNA template, which was then sequenced. Each mutation was confirmed by sequencing the corresponding region of genomic DNA (see Supplemental Table 1 online). Isolation of RNA, cDNA synthesis, and DNA sequencing were performed as described by Marchant and Bennett (1998).
Phenotypic Characterization of aux1 Alleles and Transgenic AUX1 Lines
Vertically grown wild-type and aux1 seedlings were given a 90° gravity stimulus for 24 h before measuring the angle of root curvature (as described in Parry et al., 2001a). Although wild-type roots turned between 80° and 120° within 24 h, aux1 null alleles exhibited a fully agravitropic root growth behavior akin to the non-sense allele wav5-33 (Marchant and Bennett, 1998), and partial-loss-of-function alleles responded more slowly to the gravity stimulus in the 24-h assay period (Figure 6). Experiments on each aux1 allele were performed at least three times. The effects of 60 μM CSI on the root gravitropic response of Col, aux1-2, aux1-7, and aux1-114 alleles and transgenic lines HA1-1 and 2-1 were examined after a 90° gravity stimulus as described by Rahman et al. (2001). Auxin root elongation assays were performed on Col, aux1-22, and AUX1-YFP expressing aux1-22 seedlings as described by Parry et al. (2001a).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers At2g38120 (AUX1), At5g01240 (LAX1), At2g21050 (LAX2), At1g77690 (LAX3), and At1g58360 (AAP1).
Supplementary Material
Acknowledgments
We thank Marcus Grebe (Umeå Plant Science Centre, Sweden) for providing seed for the EGFP-LTI6a line; Jim Craigon (Biosciences, University of Nottingham, UK) and Jeff Dangl (University of North Carolina, Chapel Hill) for helpful discussion with statistical analyses; Jiri Friml and Niko Geldner (University of Tubingen, Germany) and Klaus Palme (University of Freiburg, Germany) for advice with immunolocalization; Laurent Laplaze (Montpellier, France) for confocal microscopy advice; Burkhard Schulz (Tubingen, Germany) and all three referees for critical feedback on the manuscript; Anne Osbourn (John Innes Centre, Norwich, UK) for useful discussion about CSI action; Olivier Maudoux and Marc Boutry (Catholic University of Louvain, Belgium) for kindly providing the anti-PMA2 antiserum; and Catherine Bellini (Institut National de la Recherche Agronomique, Versailles, France) and Marc Knight (University of Oxford, UK) for allowing us to screen their Ws and RLD EMS mutagenized populations for novel aux1 alleles, respectively. We also thank Joe Ecker (SALK Institute, La Jolla, CA) for aux1-21 and aux1-22 seed; Paddy Maher and Billy Sinclair (Open University, Edinburgh, UK) for aux1-2 seed; K. Okada for the wav5-33 seed; and the Nottingham Arabidopsis Stock Centre for the aux1-7 seed. Seed for all aux1 alleles described in this study are available at the Nottingham Arabidopsis Stock Centre from the date of publication of this article. Finally, we acknowledge research funding from the European Commission framework programs LATIN (R.S.), POPWOOD (A.M.), and EDEN (A.M. and D.Z.), the Wolfson Foundation (J.K.), and Biotechnological and Biological Sciences Research Council awards (R.S., A.M., R.M., and S.M.) and Grant JREI2003 for confocal facilities (M.J.B.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Malcolm J. Bennett (malcolm.benett@nottingham.ac.uk).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024737.
References
- Baker, D. (2000). Vascular transport of auxins and cytokinins in Ricinus. Plant Growth Regul. 32, 157–160. [DOI] [PubMed] [Google Scholar]
- Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Bennett, M.J., Marchant, A., Green, H.G., May, S.T., Ward, S.P., Millner, P.A., Walker, A.R., Schultz, B., and Feldmann, K.A. (1996). Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 273, 948–950. [DOI] [PubMed] [Google Scholar]
- Bhalerao, R., Eklof, J., Ljung, K., Marchant, A., Bennett, M.J., and Sandberg, G. (2002). Shoot derived auxin promotes early lateral root emergence in Arabidopsis seedlings. Plant J. 29, 325–332. [DOI] [PubMed] [Google Scholar]
- Casimiro, I., Marchant, A., Bhalerao, R., Beeckman, T., Graham, N., Swarup, R., Dhooge, S., Inzé, D., Sandberg, G., Casero, P., and Bennett, M.J. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H.-C., and Bush, D.R. (1997). Topology of NAT2, a prototypical example of a new family of amino acid transporters. Proc. Natl. Acad. Sci. USA 272, 30552–30557. [DOI] [PubMed] [Google Scholar]
- Chiou, T.J., and Bush, D.R. (1996). Molecular cloning, immunochemical localization to the vacuole, and expression in transgenic yeast and tobacco of a putative sugar transporter from sugar beet. Plant Physiol. 110, 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deber, C.M., Khan, A.R., Li, Z., Joensson, C., Glibowicka, M., and Wang, J. (1993). Val-to-Ala mutations selectively alter helix-helix packing in the transmembrane segment of phage M13 coat protein. Proc. Natl. Acad. Sci. USA 90, 11648–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre, A., Muller, P., Imhoff, V., and Guern, J. (1996). Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198, 532–541. [DOI] [PubMed] [Google Scholar]
- Estelle, M., and Somerville, C. (1987). Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206, 200–206. [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwartz, H., Hamann, T., Offringa, R., and Jurgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153. [DOI] [PubMed] [Google Scholar]
- Galweiler, L., Guan, C., Muller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. [DOI] [PubMed] [Google Scholar]
- Goldsmith, M.H.M. (1977). The polar transport of auxin. Annu. Rev. Plant Physiol. 28, 439–478. [Google Scholar]
- Grantham, R. (1974). Amino acid difference formula to help explain protein evolution. Science 185, 862–864. [DOI] [PubMed] [Google Scholar]
- Grebe, M., Xu, J., Mobius, W., Takashi, U., Nakano, A., Geuze, H.J., Rook, M.B., and Scheres, B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13, 1378–1387. [DOI] [PubMed] [Google Scholar]
- Gustavsson, J., Parpal, S., and Stralfors, P. (1996). Insulin-stimulated glucose uptake involves the transition of glucose transporters to the caveolae-rich fraction within the plasma membrane: Implications for type II diabetes. Mol. Med. 2, 367–372. [PMC free article] [PubMed] [Google Scholar]
- Keukens, E.A.J., de Vrije, T., van den Boom, C., de Waard, P., Plasmna, H.H., Thiel, F., Chupin, V., Jongen, W.M.F., and de Kruijff, B. (1995). Molecular basis of glycoalkoloid induced membrane disruption. Biochim. Biophys. Acta 1240, 216–228. [DOI] [PubMed] [Google Scholar]
- Kjellbom, P., and Larsson, C. (1984). Preparation and polypeptide composition of chlorophyll-free plasma-membranes from leaves of light-grown spinach and barley. Physiol. Plant. 62, 501–509. [Google Scholar]
- Kosegarten, H., Grolig, F., Esch, A., Glusenkamp, K.H., and Mengel, K. (1999). Effects of NH4+, NO3− and HCO3− on apoplast pH in the outer cortex of root zones of maize as measured by the fluorescence ration of fluorescein boronic acid. Planta 209, 444–452. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M.O. (2002). Molecular genetics of auxin signalling. Annu. Rev. Plant Biol. 53, 377–398. [DOI] [PubMed] [Google Scholar]
- Lindsey, K., Pullen, M.L., and Topping, J.F. (2003). Importance of plant sterols in pattern formation and hormone signalling. Trends Plant Sci. 8, 521–525. [DOI] [PubMed] [Google Scholar]
- Ljung, K., Bhalerao, R.P., and Sandberg, G. (2001). Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 28, 465–474. [DOI] [PubMed] [Google Scholar]
- Llopis, J., McCaffrey, J.M., Miyawaki, A., Farquhar, M., and Tsien, R.Y. (1998). Measurement of cytosolic, mitochondrial and Golgi pH in single living cells with green fluorescent protein. Proc. Natl. Acad. Sci. USA 95, 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig, C., Gaxiola, R.A., Grisafi, P., and Fink, G.R. (1998). EIR1, a root specific protein involved in auxin transport, is required for auxin transport in Arabidopsis thaliana. Genes Dev. 12, 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa, M., Nojima, S., Akiyama, T., Sankawa, U., and Inoue, K. (1984). Interaction of digitonin and its analogs with membrane cholesterol. J. Biochem. (Tokyo) 96, 1231–1239. [DOI] [PubMed] [Google Scholar]
- Maher, E.P., and Martindale, S.J.B. (1980). Mutants of Arabidopsis thaliana with altered responses to auxins and gravity. Biochem. Genet. 18, 1041–1053. [DOI] [PubMed] [Google Scholar]
- Marchant, A., and Bennett, M.J. (1998). The Arabidopsis AUX1 gene: A model system to study mRNA processing in plants. Plant Mol. Biol. 36, 463–471. [DOI] [PubMed] [Google Scholar]
- Marchant, A., Kargul, J., May, S.T., Muller, P., Delbarre, A., Perrot-Rechenmann, C., and Bennett, M.J. (1999). AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 18, 2066–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant, A., Bhalerao, R., Casimiro, I., Eklof, J., Casero, P., Bennett, M.J., and Sandberg, G. (2002). Auxin promotes lateral root formation by facilitating IAA distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsomme, P., d'Exaerde, A.D., DeMeester, S., Thines, D., Goffeau, A., and Boutry, M. (1996). Single point mutations in various domains of a plant plasma membrane H+-ATPase expressed in Saccharomyces cerevisiae increase H+-pumping and permit yeast growth at low pH. EMBO J. 15, 5513–5526. [PMC free article] [PubMed] [Google Scholar]
- Müller, A., Guan, C., Gälweiler, L., Tänzler, P., Huijser, P., Marchant, A., Parry, G., Bennett, M.J., Wisman, E., and Palme, K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17, 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseyko, N., and Feldman, L.J. (2001). Expression of a pH-sensitive green fluorescent protein in Arabidopsis thaliana. Plant Cell Environ. 24, 557–563. [DOI] [PubMed] [Google Scholar]
- Ohnishi, T., Gall, R.S., and Meyer, M.L. (1975). An improved assay of inorganic phosphate in the presence of extralabile phosphate compounds: Application to the ATPase assay in the presence of phosphocreatine. Anal. Biochem. 69, 261–267. [DOI] [PubMed] [Google Scholar]
- Okada, K., and Shimura, Y. (1990). Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science 250, 274–276. [DOI] [PubMed] [Google Scholar]
- Ortiz-Lopez, A., Chang, H.C., and Bush, D.R. (2000). Amino acid transporters in plants. Biochim. Biophys. Acta Biomembr. 1465, 275–280. [DOI] [PubMed] [Google Scholar]
- Ottenschläger, I., Wolff, P., Wolverton, C., Bhalerao, R.P., Sandberg, G., Ishikawa, H., Evans, M., and Palme, K. (2003). Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 100, 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palme, K., and Gälweiler, L. (1999). PIN-pointing the molecular basis of auxin transport. Curr. Opin. Plant Biol. 2, 375–381. [DOI] [PubMed] [Google Scholar]
- Parry, G., et al. (2001. a). Quick on the uptake: Characterization of a family of plant auxin influx carriers. J. Plant Growth Regul. 20, 217–225. [Google Scholar]
- Parry, G., Delbarre, A., Marchant, A., Swarup, R., Napier, R., Perrot-Rechenmann, C., and Bennett, M.J. (2001. b). Novel auxin transport inhibitors phenocopy the auxin influx carrier mutation aux1. Plant J. 25, 399–406. [DOI] [PubMed] [Google Scholar]
- Rahman, A., Ahamed, A., Amakawa, T., Goto, N., and Tsurumi, S. (2001). Chromosaponin I specifically interacts with AUX1 protein in regulating the gravitropic response of Arabidopsis roots. Plant Physiol. 125, 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte, A.M., Brady, S.R., Reed, R.C., Ante, S.J., and Muday, G.K. (2000). Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiol. 122, 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte, A.M., Poupart, J., Waddell, C.S., and Muday, G.K. (2003). Transport of the two natural auxins, indole-3-butyric acid and indole-3-acetic acid, in Arabidopsis. Plant Physiol. 133, 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea, P.A., Britten, C.J., and Sarafian, V. (1992). Common identity of substrate-binding subunit of vacuolar H+-translocating inorganic pyrophosphatase of higher-plant cells. Plant Physiol. 100, 723–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R., Brady, S., and Muday, G. (1998). Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol. 118, 1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Pescue, E.-R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260. [DOI] [PubMed] [Google Scholar]
- Roman, G., Lubarsky, B., Kieber, J.J., Rothenberg, M., and Ecker, J.R. (1995). Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics 139, 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Kovaleva, V., Bassham, D.C., and Raikhel, N.V. (2001). Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell 12, 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke, R., Schneider, A., van der Graaff, E., Fischer, K., Catoni, E., Desimone, M., Frommer, W.B., Flugge, U.-I., and Kunze, R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sega, G.A. (1984). A review of the genetic effects of ethyl methanesulfonate. Mutat. Res. 134, 113–142. [DOI] [PubMed] [Google Scholar]
- Sokolova, O., Accardi, A., Gutierrez, D., Lau, A., Rigney, M., and Grigorieff, N. (2003). Conformational changes in the C terminus of Shaker K+ channel bound to the rat Kv beta 2-subunit. Proc. Natl. Acad. Sci. USA 100, 12607–12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltz, J., Stadler, R., Opekarova, M., and Sauer, N. (1994). Functional reconstitution of the solubilized Arabidopsis thaliana STP1 monosaccharide-H+ symporter in lipid vesicles and purification of the histidine tagged protein from transgenic Saccharomyces cerevisiae. Plant J. 6, 225–233. [DOI] [PubMed] [Google Scholar]
- Swarup, R., Friml, J., Marchant, A., Ljung, K., Sandberg, G., Palme, G., and Bennett, M.J. (2001). Localisation of the auxin permease AUX1 in the Arabidopsis root apex reveals two novel functionally distinct hormone transport pathways. Genes Dev. 15, 2648–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry, M.J., and Williams, L.E. (2002). Fractionation of plant tissue for biochemical analyses. In Molecular Plant Biology, Vol. 2: A Practical Approach, P. Gilmartin and C. Bowler, eds (Oxford: Oxford University Press), pp. 147–171.
- Tornero, P., Chao, R.A., Luthin, W.N., Goff, S.A., and Dangl, J.L. (2002). Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell 14, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura, M., Joseph, R.A., and Steponkus, R.L. (1995). Cold acclimation of Arabidopsis thaliana—effect on plasma membrane lipid composition and freeze-induced lesions. Plant Physiol. 109, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen, V., Friml, J., Grebe, M., van den Toorn, A., and Scheres, B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15, 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, M., and Yamamoto, K. (1998). Differential effects of 1-naphthaleneacetic acid, indole-3-acetic acid and 2:4-dichlorophenoxyacetic acid on the gravitropic response of roots in an auxin resistant mutant of Arabidopsis, aux1. Plant Cell Physiol. 39, 660–664. [DOI] [PubMed] [Google Scholar]
- Young, G.B., Jack, D.L., Smith, W., and Saier, M.H., Jr. (1999). The amino acid/auxin:proton symport permease family. Biochim. Biophys. Acta 1415, 306–322. [DOI] [PubMed] [Google Scholar]
- Zajchowski, L.D., and Robbins, S.M. (2002). Lipid rafts and little caves: Compartmentalized signalling in membrane microdomains. Eur. J. Biochem. 269, 737–752. [DOI] [PubMed] [Google Scholar]
- Zettl, R., Schell, J., and Palme, K. (1994). Photo-affinity labeling of Arabidopsis thaliana plasma membrane vesicles by 5-azido-[7-3H]indole-3-acetic acid: Identification of glutathione S-transferase. Proc. Natl. Acad. Sci. USA 91, 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.