Abstract
To investigate the nuclear-controlled mechanisms of [4Fe-4S] cluster assembly in chloroplasts, we selected Arabidopsis thaliana mutants with a decreased content of photosystem I (PSI) containing three [4Fe-4S] clusters. One identified gene, ACCUMULATION OF PHOTOSYSTEM ONE1 (APO1), belongs to a previously unknown gene family with four defined groups (APO1 to APO4) only found in nuclear genomes of vascular plants. All homologs contain two related motifs of ∼100 amino acid residues that could potentially provide ligands for [4Fe-4S] clusters. APO1 is essentially required for photoautotrophic growth, and levels of PSI core subunits are below the limit of detection in the apo1 mutant. Unlike other Arabidopsis PSI mutants, apo1 fails to accumulate significant amounts of the outer antenna subunits of PSI and PSII and to form grana stacks. In particular, APO1 is essentially required for stable accumulation of other plastid-encoded and nuclear-encoded [4Fe-4S] cluster complexes within the chloroplast, whereas [2Fe-2S] cluster–containing complexes appear to be unaffected. In vivo labeling experiments and analyses of polysome association suggest that translational elongation of the PSI transcripts psaA and psaB is specifically arrested in the mutant. Taken together, our findings suggest that APO1 is involved in the stable assembly of several [4Fe-4S] cluster–containing complexes of chloroplasts and interferes with translational events probably in association with plastid nucleoids.
INTRODUCTION
Photosystem I (PSI) mediates light-driven electron transfer from plastocyanin to ferredoxin and is involved in light energy conversion by balancing linear and cyclic electron transport (Chitnis, 2001; Munekage et al., 2002). The two proteins PsaA and PsaB form the heterodimer of the PSI reaction center that binds pairs of the primary electron donor and acceptor chlorophylls (P700 and A0, respectively), the phylloquinone A1, and the [4Fe-4S] cluster Fx. Two additional [4Fe-4S] clusters, which are bound to the plastid-encoded PsaC, transfer the electrons from Fx to ferredoxin. The remaining chlorophyll molecules bound to the core complex and to the outer antenna proteins LhcA1 to LhcA4 are involved in light absorbing and energy transfer (Golbeck, 2003). The three-dimensional structures of the cyanobacterial and the plant PSI have been resolved at 2.5 and 4.4 Å resolution, respectively (Jordan et al., 2001; Ben-Shem et al., 2003). In higher plants the core complex comprises 15 subunits that are assembled together with the cofactors in a complex and concerted manner. The functions of several subunits have been identified (Scheller et al., 2001).
It is widely accepted that posttranscriptional processes represent important regulatory steps in the accumulation of thylakoid membrane complexes (Leon et al., 1998; Barkan and Goldschmidt-Clermont, 2000). Translation, cotranslational insertion of proteins into photosynthetic membranes, and cofactor assembly are proposed to be coordinately regulated and probably enable fine tuning of photosystem adjustment as a response to tissue-specific, developmental, and environmental signals (Stern et al., 1997; Goldschmidt-Clermont, 1998; Merchant and Dreyfuss, 1998). Inhibitory mechanisms in darkness and/or activation of translation in light couple plastid protein synthesis to light supply (Bruick and Mayfield, 1999).
Supply of chlorophyll and [4Fe-4S] clusters is essential for the stable accumulation of reaction center proteins in higher plants (Eichacker et al., 1990; Mullet et al., 1990; Kim et al., 1994; Lezhneva et al., 2004). However, in cyanobacteria the presence of the three [4Fe-4S] clusters is not required for the biosynthesis of trimeric P700-A1 cores (Shen et al., 2002). Little is known about the biogenesis of Fe-S clusters in the chloroplast as compared with that in eubacteria and mitochondria (reviewed in Beinert et al., 2000; Lill and Kispal, 2000; Frazzon and Dean, 2003). Conserved chloroplast NIFS (nitrogen fixation subunit S) and NFU (NIFU-like protein) proteins are involved in the activation of sulfur from l-Cys and in the assembly of [2Fe-2S] clusters, respectively (Pilon-Smits et al., 2002; Léon et al., 2003; Touraine et al., 2004; Yabe et al., 2004). Members of the universally conserved FSC ([4Fe-4S] cluster; HCF101) protein family appear to be specifically required for the assembly of [4Fe-4S] clusters not only in the chloroplast but presumably also in other cellular compartments like mitochondria, the cytoplasm, and the nucleus (Lezhneva et al., 2004).
According to the evolutionary origin from free living cyanobacteria, plastids have retained the basic eubacterially derived ribosomal machinery, and many genes that have been transferred from the endosymbiont to the nucleus encode basic ribosomal components (Sugiura et al., 1998). Plastid mRNA binding proteins have been identified, but their precise roles in translational processes often remain to be elucidated (Fedoroff, 2002).
Genetic studies mainly performed in Synechocystis, Chlamydomonas reinhardtii, maize (Zea mays), and Arabidopsis thaliana provided evidence for the nuclear control of plastid translation (McCormac and Barkan, 1999; Rattanachaikunsopon et al., 1999; Zerges, 2000; Cohen et al., 2001; Dauvillée et al., 2003). The function and composition of trans-acting regulatory factors involved in translation changed during genome–plastome coevolution. This came along with alterations of their target sites often found to be located in the 5′ untranslated region (Bruick and Mayfield, 1999). Combined genetic and transplastomic approaches uncovered that chloroplast translation in Chlamydomonas is coupled with the assembly of thylakoid membrane complexes, the so-called CES (controlled by epistasy of synthesis) process (Stampacchia et al., 1997; Choquet and Wollman, 2002; Wostrikoff et al., 2004). However, the sublocalization of translational events within the chloroplast as well as the nuclear-controlled mechanisms of cofactor incorporation during translation or the assembly process of the photosynthetic complexes remain to be elucidated.
We aimed to genetically dissect the specific nuclear control of PSI biogenesis and identified a nuclear-encoded factor, ACCUMULATION OF PHOTOSYSTEM ONE1 (APO1), that is required for accumulation of PSI and other plastid-encoded and nuclear-encoded [4Fe-4S] cluster proteins of the chloroplast. Members of the APO gene family are only found in vascular plants so far and are predicted to be localized in chloroplasts and mitochondria. All homologs contain a highly conserved region spanning ∼100 amino acid residues (APO motif 1) that is repeated at the C terminus (APO motif 2). The two motifs are separated by a nonconserved spacer of variable length. Each of the groups, APO1 to APO4, displays distinct and conserved features suggesting that they are all important for the plant cell and possess related functions in different cellular compartments.
RESULTS
Selection, Phenotype, and Growth of apo1
In a systematic screen for nuclear-encoded factors responsible for acclimation and maintenance of PSI biogenesis, we identified Arabidopsis mutants incapable of photoautotrophic growth (Meurer et al., 1996a). We selected 87 mutants with visible phenotypes that exhibited an albinotic or pale phenotype and seedling lethality. Mutant plants that were grown heterotrophically often survived, indicating photosynthetic deficiencies as primary defects. All analyses were performed with plants grown in vitro. On sugar-supplemented medium, the otherwise albinotic apo1 mutant showed a very pale and high chlorophyll fluorescence (hcf) phenotype, indicating substantial loss of chlorophyll and that absorbed light energy cannot be used efficiently, which is therefore dissipated as increased red chlorophyll fluorescence (Figure 1A). The T-DNA–induced apo1 mutation of Arabidopsis was selected because it caused a complete loss of PSI activity (see below). Growth at 10 μmol photons m−2 s−1 was slower, but leaf morphology and development were unaltered in apo1 in comparison to wild-type plants (Figure 1A). Under moderate light intensities of 50 μmol photons m−2 s−1, mutant plants bleached rapidly, indicating increased light sensitivity.
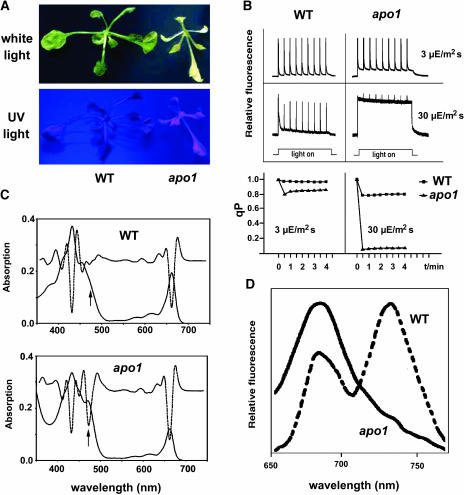
Figure 1.
Phenotype, Chlorophyll Fluorescence, and Absorbance Spectra of apo1 Mutant Plants.
(A) Three-week-old mutant plants growing on sucrose medium at 10 μmol photons m−2 s−1 are pale and show a hcf phenotype under UV light. WT, Wild type.
(B) Chlorophyll fluorescence induction and qP at low (3 μmol photons m−2 s−1) and moderate (30 μmol photons m−2 s−1) light intensities. Leaves were exposed to a series of superimposed 800-ms saturating light flashes.
(C) Absorbance spectra of mutant and wild-type leaves grown at 10 μmol photons m−2 s−1. The increased band at 473 nm (see arrows) in the second order derivative spectra (dotted line) is indicative of an increased carotenoid accumulation in the mutant relative to chlorophyll levels.
(D) Low-temperature (77K) fluorescence emission spectra. The PSII band at 688 nm in the apo1 mutant was normalized to the PSI band at 735 nm in the wild type.
Fluorometric and Spectroscopic Characteristics of the Photosynthetic Apparatus and Pigment Content in apo1 Revealed Specific Deficiencies of PSI
The light-induced in vivo chlorophyll fluorescence measurements can serve as a sensitive tool to estimate the excitation state of PSII reaction centers. The ratio of variable fluorescence to maximum fluorescence (Fv/Fm) reflects the potential capacity of the photochemical reaction of PSII (Krause and Weis, 1991). The fluorescence is quenched by the use of light energy (photochemical quenching [qP]) measured at intensities of 3 and 30 μmol photons m−2 s−1. Fv/Fm reached levels of 0.62 ± 0.06 in apo1 as compared with 0.81 ± 0.02 in the wild type, indicating partial loss of PSII capacity or a stimulated emission of PSI antenna pigments that contribute to a higher minimum fluorescence with no effect on Fv. At the lower light regime, qP was reduced to 0.84 in the mutant as compared with 0.98 in the wild type (Figure 1B). At still moderate light intensities of 30 μmol photons m−2 s−1, dramatic deficiencies were observed in qP processes in apo1. Whereas wild-type plants exhibited a qP of 0.81 under steady state conditions, apo1 failed to quench most of the variable fluorescence (qP < 0.06; Figure 1B). Such a behavior is typical for mutants affected in electron transport processes independent of PSII (Meurer et al., 1996a).
This inference was confirmed by measuring the light-induced redox kinetics of the PSI pigment P700. Under normal conditions, P700 is completely reduced in the dark and oxidized to its maximum level in selective PSI light, whereas under steady state conditions in heterochromatic light, the P700 redox state is adjusted somewhere in between (Klughammer and Schreiber, 1994). Although the dual wavelength pulse amplitude–modulated system was set at maximal sensitivity, light-induced P700 redox changes were below the limit of detection in apo1, indicating complete loss of PSI function (data not shown).
In plants grown heterotrophically, levels of chlorophylls were ∼10-fold reduced in apo1 as compared with wild-type seedlings (1.84 ± 0.09 mg/g fresh weight in the wild type and 0.17 ± 0.03 mg/g fresh weight in mutants). The ratio of chlorophyll a to chlorophyll b was altered from 2.88 ± 0.04 in the wild type to 2.64 ± 0.12 in the mutant. The photosynthetic pigment absorbance spectra demonstrated that the typical maxima at 430 and 670 nm for chlorophyll a, at 450 nm for chlorophyll b, and at 473 nm for carotenoids were present in the mutant (Figure 1C). The second derivative of the absorbance spectra showed a much broader carotenoid band at 473 nm in apo1, which is indicative of a relatively high carotenoid content and of an increased sensitivity to light (Figure 1C; Havaux and Niyogi, 1999).
Low-Temperature Fluorescence Emission Analysis Demonstrates Lack of the Entire PSI Complex in apo1
Fluorometric studies at 77K showed that the PSII-specific band at 685 nm was present in the mutant and not shifted to shorter wavelengths. This indicates that PSII reaction centers are present at normal levels relative to the outer light-harvesting complex II (LHCII) and that excitons can be efficiently transferred from LHCII to the inner PSII antenna (Krause and Weis, 1991). The usually most-abundant PSI-specific absorbance band at 735 nm was reduced to a hardly detectable shoulder in the spectra of apo1 (Figure 1D). Therefore, it appears that PSII complexes are functionally assembled, whereas the whole PSI complex, including the outer antenna, severely and specifically fails to accumulate in apo1.
Intrinsic and Peripheral PSI Subunits Are Severely Reduced in apo1
Stationary protein levels were normalized by equal loading of mutant and wild-type membrane and soluble proteins of the chloroplast. Immunological analysis of apo1 revealed that nuclear-encoded and plastid-encoded proteins of PSII (subunits PsbB, PsbC, PsbD, and PsbO1/2) and cytochrome b6f complex (subunits PetA and PetB) accumulated to significant levels between 50 and 100% (Figure 2A). Amounts of the ATP synthase (subunits AtpA and AtpG) and the soluble PetH and PetE proteins encoding ferredoxin-NADP+ reductase and plastocyanin, respectively, were unaltered in apo1. By contrast, only traces, if any, of the nuclear-encoded and plastid-encoded intrinsic PSI proteins PsaA/B, PsaC, PsaD, and PsaF could be detected in apo1 (Figure 2A).
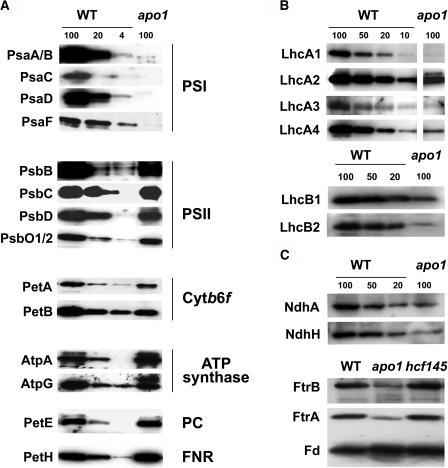
Figure 2.
Accumulation of Plastid Proteins.
Loading 100 corresponds to 8 μg of membrane or soluble proteins of the wild type. The quantity of apo1 thylakoid membranes (100%) was adjusted to wild-type levels of the ATP synthase. For quantification, dilution series of the wild type were used. Three-week-old wild-type and apo1 leaves were used for analysis.
(A) Immunoblot analysis of thylakoid membrane and soluble plastid proteins. Designations of proteins and of the corresponding complexes are labeled at left and right, respectively.
(B) Immunoblot analysis of the outer PSI and PSII antenna proteins in apo1 and wild type. Lhc, Light-harvesting complex.
(C) Immunoblot analysis of Fe-S cluster–containing plastid proteins. The plastid-encoded thylakoid membrane and the nuclear-encoded soluble protein complexes NDH and FTR, respectively, contain [4Fe-4S] clusters. Ferredoxin (FD) binds [2Fe-2S] clusters.
Levels of the peripheral antenna proteins of PSI were strongly reduced in apo1, amounting to ∼10% for LhcA1 and LhcA3 and ∼20% or less for LhcA2 and LhcA4 as compared with those of the wild type (Figure 2B), confirming the conclusions deduced from 77K fluorescence data. The inability of apo1 to accumulate significant amounts of the four antenna proteins of PSI contrasts findings on other known Arabidopsis PSI mutants because they are primarily and specifically affected in the accumulation of all core subunits but not to this extent of the outer antenna (Lezhneva and Meurer, 2004; Lezhneva et al., 2004). Remarkably, the outer PSII antenna proteins LhcB1 and LhcB2 were also significantly reduced to <20% of the wild type (Figure 2B).
Accumulation of Fe-S Cluster–Containing Proteins in apo1
Recent findings show that Fe-S cluster assembly is important for stable accumulation of PSI and other proteins or complexes within the chloroplast (Lezhneva et al., 2004; Touraine et al. 2004; Yabe et al., 2004). To investigate whether APO1 has additional targets that participate in chloroplast biogenesis, we studied the accumulation of several plastid Fe-S cluster–containing complexes and proteins (Figure 2C). Ferredoxin, which contains a [2Fe-2S] cluster, is not affected in size or in abundance in apo1. By contrast, the mutants have significantly reduced levels of [4Fe-4S] cluster–containing complexes like the ferredoxin-thioredoxin reductase (FTR) and NAD(P)H-dependent dehydrogenase (NDH). The deficiency in accumulation of [4Fe-4S] clusters does not appear to be characteristic for mutants lacking PSI because hcf145, which is also missing the PSI core complex, shows normal levels of other chloroplast [4Fe-4S] cluster proteins (Figure 2C; Lezhneva and Meurer, 2004; Lezhneva et al., 2004).
Accumulation and Integrity of PSI Transcripts Is Unaffected in apo1
The abundance and sizes of nuclear (psaD, psaE, psaF, psaG, and psaH) and all plastid transcripts encoding PSI proteins (the trimeric psaA-psaB-rps14, psaC, psaI, and psaJ) were identical in mutant and wild-type plants as determined by RNA gel blot analysis and by expression profiling using macroarrays that were equipped with probes of all plastid genes (Figure 3A; data not shown). Transcript levels of the nuclear cab and rbcS genes, which are well known to be significantly reduced when the functional state of the chloroplast is arrested at an early stage (Surpin et al., 2002), were unaltered as well in apo1 (Figure 3A).
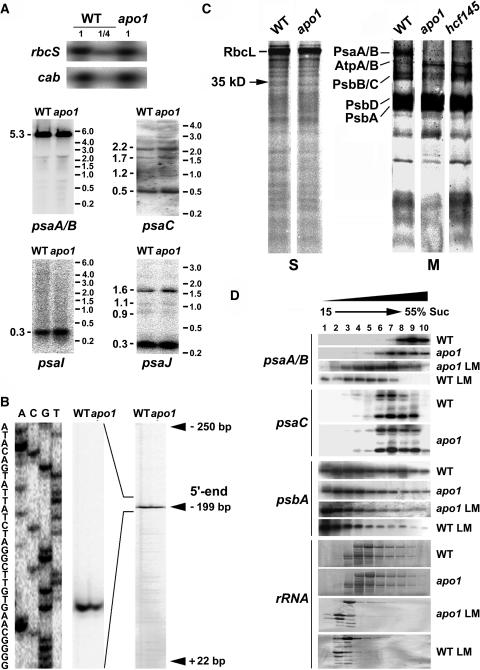
Figure 3.
Quantities and Integrity of psaA-psaB Transcripts and Protein Labeling Studies.
(A) RNA gel blot analysis of the plastid PSI genes and the nuclear cab and rbcS genes. Eight and two (1/4) μg of total RNA from three-week-old mutant and wild-type leaves were analyzed using gene-specific probes. Sizes of the standard (right) and the bands (left) are indicated in kilobases.
(B) Primer extension analysis of mutant and wild-type mRNA shows that the transcript 5′ termini at position −199 nt relative to the start codon of the psaA message are intact in apo1.
(C) In vivo labeling of plastid soluble (S) and membrane (M) proteins separated by SDS-PAGE in apo1, hcf145, and the wild type (WT). Wild-type and mutant proteins with equivalent amounts of radioactivity (100.000 cpm) were loaded.
(D) Polysome sedimentation in 15% to 55% sucrose (Suc) gradients by ultracentrifugation and subsequent RNA gel blot analysis of fractionated samples. Probes used are indicated at left. The filter used for the psaC probe was rehybridized with the psaA-psaB probe. The filter of the lincomycin-treated (LM) material was used for psaA-psaB and rehybridized with the psbA probe. Lincomycin treatment of wild-type and mutant plants was performed 4 h before polysome preparation. rRNAs have been detected by staining the blots.
RNA gel blot analysis demonstrates that the psaA-psaB genes yield just a single prominent, tricistronic RNA of 5.3 kb in Arabidopsis. Primer extension studies uncovered that the 5′ transcript terminus of the psaA-psaB-rps14 tricistron is unaltered in apo1 when compared with the wild type (Figure 3B). Therefore, we conclude that the apo1 mutant exhibits normal promoter usage, integrity, and abundance of psaA-psaB-rps14 transcripts.
Radiolabeling of the Plastid PsaA and PsaB Proteins Is Impaired in apo1
PSI protein abundance could be limited by translational disturbances in the mutant. Therefore, pulse labeling of membrane-bound and soluble proteins of the chloroplast was performed with [35S]Met in the presence of the cytoplasmic translation inhibitor cycloheximide. With the exception of one plastid-encoded protein of 35 kD, biosynthesis of other detectable soluble proteins in the stroma was unchanged in apo1 (Figure 3C). Labeling of ATP synthase subunits α and β (AtpA and AtpB) was slightly increased in the mutant relative to the wild-type control based on equal loading of radioactive label. Incorporation of radioactivity into the PSII proteins PsbA (D1) and PsbD (D2) was comparable to that in wild type, but labeling of the chlorophyll binding proteins PsbB and PsbC was reduced. These data demonstrate the pleiotropy of the phenotype.
As a control, the labeling pattern of apo1 was compared with that of another Arabidopsis PSI mutant, hcf145, accumulating only ∼10% of the tricistronic psaA-psaB-rps14 mRNA (Lezhneva and Meurer, 2004). The hcf145 mutant specifically exhibits decreased levels of PsaA and PsaB radiolabeling (Figure 3C) and, similar to apo1, increased incorporation of label into AtpA and AtpB. A failure to radiolabel the PSI core proteins PsaA and PsaB in apo1 could reflect that the rates of translation of the two PSI reaction center subunits are repressed in apo1. However, it also remains possible that the synthesis of PsaA and PsaB is unchanged in apo1, but the proteins or the nascent chains are rapidly degraded during the time course of the experiment.
Polysome Association of psaA and psaB Transcripts Is Decreased in apo1
If translation of psaA and psaB mRNAs is perturbed in apo1, changes in polysome association for the psaA and psaB transcripts could be expected. Polysomal loading was analyzed by sucrose density gradient centrifugation. The sizes and distribution of polysomes loaded with psaC and psbA transcripts did not differ in the mutant when compared with the wild type (Figure 3D). However, polysomes associated with tricistronic psaA-psaB-rps14 transcripts were shifted to decreased sucrose concentrations, suggesting a failure in the functional loading of psaA-psaB-rps14 transcripts with ribosomes in the mutant.
To test whether initiation or elongation of translation is affected in apo1, the translation inhibitor lincomycin that prevents reinitiation and forces previously initiated ribosomes to run off was applied to wild-type and mutant plants 4 h before polysome isolation. As expected, the inhibitor caused banding of psaA-psaB-rps14 mRNAs at much lower sucrose concentrations in wild type, whereas in apo1 lincomycin induced a less prominent liberation of psaA-psaB transcripts from ribosomes, indicating that translational elongation is retarded in the mutant (Figure 3D). Lincomycin treatment of the mutant was as effective as in the wild type because psbA-containing and rRNA-containing polysomes were shifted toward the top of the gradient to the same extent in both cases. Therefore, it appears that APO1 is not needed for initiation but rather specifically for elongation of psaA and psaB translation, cofactor assembly during translation, or targeting of the nascent chains to their right place. However, the changes in the size of polysomes could also reflect an assembly mediated regulation of translation (Wostrikoff et al., 2004).
Ultrastructure of apo1 Chloroplasts
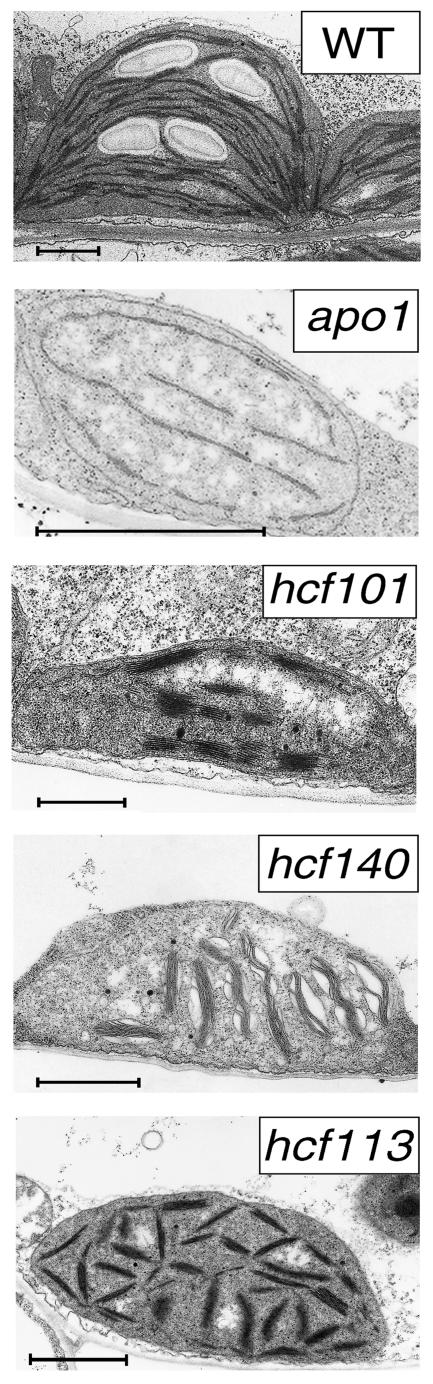
The ultrastructure of the apo1 was compared with that of three other PSI-specific mutants, hcf101, hcf140, and hcf113, and the wild type (Figure 4). Wild-type chloroplasts showed a well-developed membrane system consisting of interconnected grana and a random distribution of starch grains. The apo1 plastids were ∼3 to 4 times smaller than those of the wild type and appeared to be swollen. The mutant forms rudimentary thylakoids consisting only of stroma lamellae and fails to accumulate grana stacks. This indicates that membrane development is arrested at a very early morphological stage and distinguishes apo1 from other PSI mutants, which form larger (hcf101), swollen (hcf140), or unoriented (hcf113) grana stacks but only fragmentary stroma lamellae (Figure 4). The small size of the chloroplasts in apo1 also explains the low level of chlorophyll based on the fresh weight.
Figure 4.
Chloroplast Ultrastructure of the Wild Type and the PSI Mutants apo1, hcf101, hcf140, and hcf113.
Bars = 1 μm.
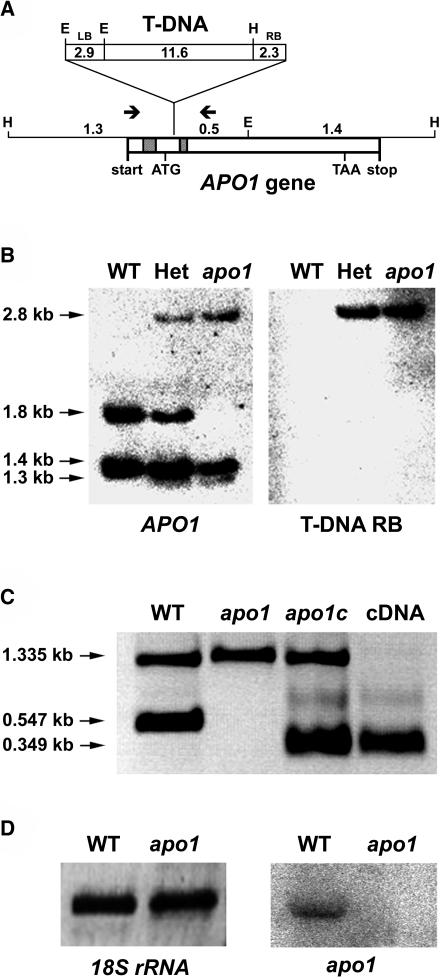
Molecular Mapping and T-DNA Tagging of apo1
The apo1 mutation segregated in a Mendelian manner and was mapped between the two microsatellite markers nga128 and chr1-110 on the lower part of chromosome 1 at position 23.395 ± 0.8 Mb. Neither known genes encoding plastid ribosomal proteins nor PSI genes map in this region (Legen et al., 2001), suggesting that APO1 encodes a novel protein.
The kanamycin resistance conferred by the introduced T-DNA cosegregated with the mutant phenotype in 384 analyzed segregants, indicating that APO1 is tagged by the insertion. The localization of APO1 to the lower part of chromosome 1 was confirmed by isolation and sequencing of the T-DNA left-border and right-border flanking regions by inverse PCR (Figure 5A). Using APO1 and T-DNA right-border probes for DNA gel blot analyses confirmed a single T-DNA insertion that cosegregates strictly with the apo1 mutation (Figure 5B). The location of the insertion site close to the 5′ end of APO1 at position 24.153 Mb of chromosome 1 (position +103 relative to the APO1 start codon) is consistent with the mapping data. The full-length cDNA of APO1 was isolated using a rapid amplification of cDNA ends (RACE) approach (Frohman et al., 1988). Interestingly, the APO1 start codon is localized in the second exon (Figure 5A). Expression of the full-length APO1 cDNA under the control of the 35S RNA Cauliflower mosaic virus promoter functionally complemented the mutant phenotype, resulting in the development of fully green seedlings which grew photoautotrophically and showed all physiological chlorophyll fluorescence and P700 redox characteristics which are typical for the wild type (Figure 5C; data not shown).
Figure 5.
Inactivation of APO1 by T-DNA Insertion and Complementation of apo1.
(A) Schematic view of the T-DNA insertion in APO1. The two introns are indicated by shaded boxes. HindIII (H) and EcoRI (E) restriction sites and the T-DNA left (LB) and right (RB) borders are shown. Sizes are given in kilobases. The transcription start and stop are indicated. The arrows show the positions of primers used in (C). The position of the translational start (ATG) and stop (TAA) codons are indicated.
(B) Genomic DNA gel blot analysis of EcoRI and HindIII double-digested mutant and wild-type (WT) DNA results in polymorphisms that can be deduced from (A). The used probes of the T-DNA right border (RB) and the APO1 gene recognize one and the same 2.8-kb fragment. Het, Heterozygous plants for apo1.
(C) PCR analysis of wild-type, mutant, complemented mutant lines (apo1c), and the apo1 cDNA. Control primers, which amplify another chromosomal region of 1.335 kb, were used in the same reaction with the APO1 gene-specific primers. APO1 exon-specific primers apo1-f and apo1-r2 did not amplify a 0.547-kb product of genomic DNA in the apo1 mutant and complemented lines, but a 0.349-kb fragment originating from the expressed cDNA.
(D) RNA gel blot analysis of mutant and wild type (WT) was performed with a probe specific for apo1. Equal loading (8 μg) is shown by hybridization with a probe specific for 18S rDNA.
Expression of apo1 Is Stimulated in Illuminated Leaves
APO1 transcripts of 1.65 kb could be detected in the wild type but were absent in apo1, consistent with the T-DNA insertional inactivation of the APO1 gene (Figure 5D). Because transcript abundance of APO1 was very low even in the wild type, the precise expression levels of APO1 were estimated by real-time RT-PCR using 18S rRNA for normalization. Twelve-day-old dark-grown wild-type seedlings expressed 23% ± 5% of the light control. Expression in roots, stems, siliques, and flowers was 28% ± 3%, 63% ± 6%, 23% ± 4%, and 69% ± 6%, respectively, relative to that found in leaves. Although substantial transcript levels were already present in dark and in roots, expression of APO1 appears to be stimulated during photomorphogenesis. This implies important roles of APO1 in the dark and also in nonphotosynthetic tissues.
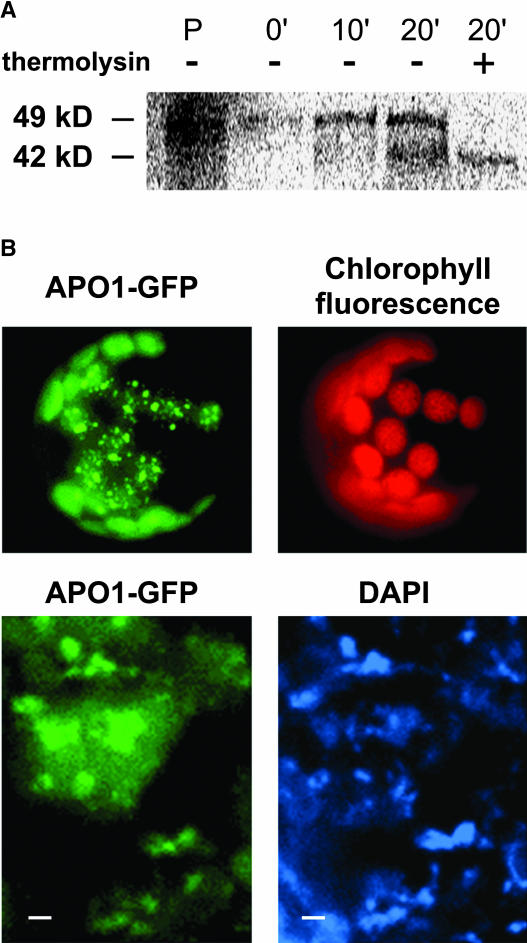
Localization of APO1 to Plastid Nucleoids
In vitro synthesized APO1 precursor proteins of 49 kD were imported into chloroplasts and processed to a mature size of 42 kD (Figure 6A), demonstrating the presence of a cleavable transit peptide even if no chloroplast target sequence was identified with any of the prediction programs available in public databases.
Figure 6.
Sublocalization of APO1 within the Chloroplast.
(A) Import of radiolabeled APO1 proteins into isolated chloroplasts and subsequent detection of gel-separated proteins by phosphor imaging. The precursor of 49 kD (P) was imported and proteolytically processed to 42 kD at the indicated periods of chloroplast incubation. After import chloroplasts were either treated (+) or not treated (−) with thermolysin to digest nonimported proteins.
(B) The APO1 protein was fused to GFP (APO1-GFP) and transiently expressed in tobacco protoplasts. GFP fluorescence was exclusively found in spots inside the chloroplast as revealed by chlorophyll fluorescence (top). Transformed protoplasts were incubated with DAPI and visualized at higher magnification (bottom). Bars = 1 μm.
To sublocalize APO1 within the chloroplast, the APO1 cDNA was translationally fused to the green fluorescence protein (GFP) and transiently expressed in isolated protoplasts. The green fluorescence was found exclusively in chloroplasts consistent with the findings of the in organello experiments (Figure 6B). Although there is a diffuse GFP signal apparently throughout the stroma, much of the APO1-GFP fusion resided in distinct spots within the organelle. Comparing the fluorescence induced by GFP with that induced by 4′,6-diamidino-2-phenylindole (DAPI)–stained cells indicates that the fluorescent labels of both samples overlapped. Therefore, APO1 appears to be associated with plastid nucleoids (Figure 6B). The relative intensity of the APO1-GFP and DAPI fluorescence varied in the individual spots, indicating that APO1 has a higher affinity to a distinct fraction of nucleoids. It is important to note that APO1-GFP association with nucleoids does not seem to reflect an unspecific aggregation of overexpressed protein because the compartmentalization was also apparent in chloroplasts with barely detectable APO1-GFP fluorescence and not observed with other highly expressed chloroplast-targeted GFP fusions showing a uniform distribution (Meurer et al., 1998a).
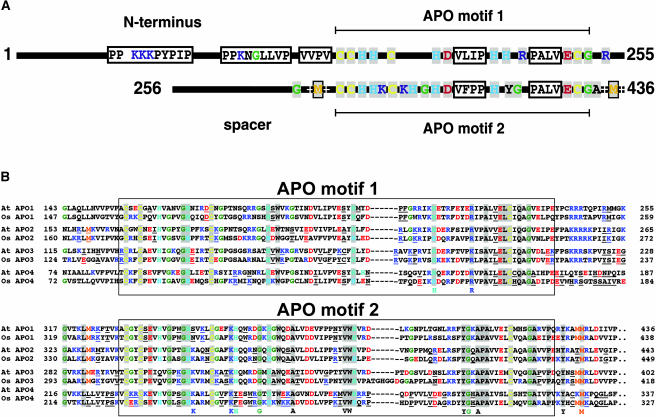
APO1 Belongs to a Novel Gene Family in Vascular Plants with Four Defined Groups Containing a Conserved Repeat Motif
The APO1 gene encodes a previously unknown protein and shows similarities to genes only found in vascular plants (Figure 7; data not shown). The absence of similarities in the sequenced genomes of eubacteria, archaebacteria, or Chlamydomonas indicates that the APO gene family evolved after the divergence of the green lineage or has been lost in related lineages. Using public databases (http://bioinf.cs.ucl.ac.uk/psipred/; http://us.expasy.org), all attempts to identify sequence motifs or domains within APO1 that are indicative of a function failed. Because the instability index of APO1 is computed to be 44.97, the protein is classified to be unstable (Guruprasad et al., 1990). According to structural predictions, APO1 consists of extensive random coiled (58%) and, to a lesser extent, of alpha helical (28%) and extended strand (14%) structures (theoretical pI 9.27).
Figure 7.
Sequence Analysis of the APO Gene Family in Arabidopsis and Rice.
(A) Scheme of the primary APO1 structure with the conserved and repeated APO motifs 1 and 2. Conserved amino acid residues in group 1 and differences between motifs 1 and 2 present in this group are highlighted.
(B) Sequence alignment of motifs 1 and 2 in APO1 to APO4 in Arabidopsis (At) and rice (Os). Amino acid residues that are conserved in all members are on gray background. Amino acid residues that are specific for each defined group in Arabidopsis and rice and different in the others are underlined. Conserved differences between the motifs 1 and 2 in all four groups are listed below the alignment.
The APO1 protein contains 436 amino acid residues and has a deduced molecular mass of 49.6 kD consistent with the size of in vitro translated APO1 proteins (Figures 6A and 7A). Three additional homologous genes of unknown function, APO2 to APO4, are present in the Arabidopsis genome. Interestingly, each of the four APO proteins possesses an orthologous counterpart in the rice genome (Figure 7B). Therefore, the APO gene family consists of four distinguishable groups that are present in both monocotyledonae and dicotyledonae. APO1 to APO4 show much fewer similarities in the N terminus than in the remaining part of the proteins, indicating different localizations and/or functions. APO2 is also predicted to be localized in the chloroplast. In rice (Oryza sativa), APO3 is computed to be present in the chloroplast as well, whereas the Arabidopsis protein is predicted to be localized in mitochondria. APO4 is computed to represent a mitochondrial protein in both organisms.
All members of the previously unknown APO gene family contain a 100 amino acid residue–spanning region (APO motif 1) with conserved Cys, His, Gly, and acidic and basic amino acids (Figure 7). The highly conserved APO motif 1 is duplicated at the C terminus (designated APO motif 2). These two motifs are always separated from each other by a less-conserved spacer that is also variable in length but in groups 1 to 3 contains one conserved Met embedded within positively charged amino acid residues directly upstream of APO motif 2 (Figure 7). A similar sequence is also present downstream of APO motif 2 close to the C terminus in all groups. APO motif 1 is followed by a short stretch containing positive charges in groups 1 to 3. The highly conserved signature of both motifs in APO1, which fits to all members of the APO gene family in vascular plants, can be defined as C-x2-C-x3-(H,Q)-x4-GH-x4-C-x11-H-x-W-x6-D-x8-H-x(20-26)-PA-x2-E(L,I)C-x3-G. The conserved Cys in both motifs could provide ligands for tetranuclear Fe-S centers (Sticht and Rösch, 1998). Several conserved differences between the two motifs could indicate different functions (Figure 7B). For example, both motifs, APO 1 and 2, contain a conserved H in another position. Two conserved R residues are present in APO motif 1 and two K residues in motif 2. In addition, APO motif 2 contains conserved G, VW, YG, and A residues, which are not present in APO motif 1 in any of the groups (Figure 7).
DISCUSSION
Role of APO1 in psaA-psaB Translation, PSI Accumulation, and Grana Stacking
Regulation of PSI accumulation is important for establishing photosynthetic efficiency and the stoichiometric adjustment, for instance, if PSI-to-PSII ratios have to be balanced in response to exogenous or endogenous signals (Depège et al., 2003). Modulations of psaA and psaB gene expression at the level of transcription (Allen and Pfannschmidt, 2000), transcript stability (Barkan and Goldschmidt-Clermont, 2000), translation (Zerges, 2000), and assembly of PSI (Schwabe and Kruip, 2000) represent key steps for such regulatory processes. APO1 is indispensable and plays a crucial role in the accumulation of the entire PSI complex and probably in photosystem adjustment in vivo.
In higher plants the psaA and psaB genes are cotranscribed with rps14 encoding the S14 protein of the 30S plastid ribosomes. We assume that translation of rps14, which is processed from the primary transcript (Lezhneva and Meurer, 2004), occurs independently and is functional in apo1 because the general translational machinery is intact in the mutant.
The findings that ribosomal loading of psaA-psaB transcripts and, therefore, initiation of translation takes place in apo1 and that liberation of psaA-psaB transcripts from ribosomes is retarded after lincomycin treatment in the mutant suggest that APO1 operates essentially at the level of translation elongation (Figure 3D). For example, when aconitase in prokaryotes and the cytosole of eukaryotes loses its [4Fe-4S] cluster under iron starvation or oxidative stress, the interconverted iron regulatory protein binds specific iron regulatory elements on mRNAs either to stabilize the transcripts or to block translation. How this specificity is achieved remains unknown (Kiley and Beinert, 2003) but shows that reversible [4Fe-4S] cluster binding can be used to regulate translation.
Synthesis or supply of chlorophyll and [4Fe-4S] clusters appears to play a substantial role in stabilization of PsaA and PsaB in chloroplasts (Mullet et al., 1990; Kim et al., 1994; Lezhneva et al., 2004). Because APO1 possesses Cys that could bind [4Fe-4S] clusters, it is reasonable to propose a possible role of APO1 in [4Fe-4S] cofactor assembly or cotranslational incorporation into PsaA and/or PsaB proteins. The function of APO1 could also be required for either still unknown steps in other pathways, like chlorophyll biosynthesis or incorporation of chlorophyll into PSI and antenna proteins. This would explain the deficiency of LHC proteins and the loss of grana stacking in the apo1 mutant.
Roles of APO1 in Accumulation of Other Chloroplast [4Fe-4S] Cluster–Containing Complexes
APO1 could have additional targets that interfere with chloroplast and/or thylakoid membrane development because several differences between apo1 and other mutants specifically deficient in PSI became apparent. (1) The apo1 mutant shows an albinotic phenotype when grown photoautotrophically. (2) The thylakoid membrane system is poorly developed and, surprisingly, lacks grana stacks but contains extended stroma lamellae. (3) The size of the chloroplast is up to 4 times smaller than in the wild type or in other PSI mutants (Figure 4). (4) The impairment of the accumulation not only of the entire PSI complex but also of the outer PSI and PSII light-harvesting antennae as well as the absence of the corresponding fluorescence band at 77K are unique to apo1 when compared with other PSI mutants (Figures 1 and 2; Lezhneva et al., 2004). (5) Members of the APO gene family are predicted to be localized in other cellular compartments like mitochondria. (6) Not only levels of PSI but also those of two other [4Fe-4S] cluster–containing protein complexes, FTR and NDH, are reduced in apo1. This suggests a specific role of APO1 in the accumulation of [4Fe-4S] cluster–containing complexes and could explain the pleiotropic phenotype.
Recently, a nuclear gene, Copper Response Defect1 (CRD1), which is responsible for PSI accumulation under copper starvation, was described from Chlamydomonas (Moseley et al., 2000, 2002). The CRD1 gene shows similarities to genes in photosynthetic organisms and has been suggested to be required for the synthesis of protochlorophyllide (Tottey et al., 2003). The failure of apo1 to accumulate the core as well as the outer antenna of PSI is in common with the crd1 mutant in Chlamydomonas under copper starvation. However, the Arabidopsis crd antisense lines are mostly affected in the levels of chlorophyll binding proteins but not in those of the photosynthetic core complexes (Tottey et al., 2003). Therefore, the function of APO1 may be substantially different from that of CRD1.
The Chlamydomonas mutants tab1 and tab2, which are primarily affected in initiation of psaB translation, are also unable to translate the psaA mRNA as a result of a secondary effect of the mutation, designated the CES process (Stampacchia et al., 1997; Dauvillée et al., 2003; Wostrikoff et al., 2004). Provided that the CES process, which operates for plastid-encoded proteins, also occurs in higher plants, a failure to synthesize PsaA could be caused by the lack of psaB translation in apo1. Nonetheless, because levels of the nuclear-encoded FTR complex also are significantly reduced in apo1, it seems unlikely that APO1 is primarily involved in the CES process itself.
Comparison of the apo1 and hcf101 Mutant Phenotypes
Although HCF101 has been proposed to be involved in [4Fe-4S] cluster biogenesis, unlike apo1, the hcf101 mutant is able (1) to accumulate substantial amounts of the outer antenna proteins of PSI and PSII, (2) to form grana stacks, (3) to incorporate radiolabel into PsaA/B proteins in vivo, and (4) to functionally load psaA-psaB transcripts with ribosomes (Lezhneva et al., 2004). Ribosomal loading of psaC, however, is unaltered in apo1 as well as in hcf101 mutants, although the PsaC protein contains two [4Fe-4S] clusters (Figure 3D; Lezhneva et al., 2004). Because of the stronger and pleiotropic phenotype of apo1, the function of the two proteins seems to be required for different but related pathways.
For example, it is reasonable to assume that APO1 is essential for [4Fe-4S] (FX) cofactor incorporation into PsaA and/or PsaB proteins during translation and, therefore, stabilization of the nascent peptide chains, whereas HCF101 could to be required for the incorporation of the [4Fe-4S] clusters FA and FB into the PsaC protein. Alternatively, APO1 could be involved in early stages of [4Fe-4S] cluster incorporation, and HCF101 could function in subsequent processes. On the other hand, the two factors could be required for diverse stages of the [4Fe-4S] cluster metabolism, like biogenesis, insertion, or stability. Similarly, it also remained uncertain at which step rubredoxin functions in [4Fe-4S] cluster metabolism during PSI assembly in cyanobacteria (Shen et al., 2002). In this context, it should be mentioned that HCF101 and APO1 are also involved in the stable assembly of the nuclear-encoded, soluble FTR complex. It also remains to be shown at which stage during the assembly of the FTR the two factors are required.
Association of Chloroplast Gene Expression to Plastid Nucleoids
Although it has been reported that initiation of both transcription and translation occurs in membrane-associated nucleoids of Escherichia coli (Simon and Nisman, 1977), little is known about the nucleoid organization, the interacting proteins, and processes leading to phase separation between nucleoids and the stroma or membranes in the chloroplast system (Kobayashi et al., 2002; Sato et al., 2003). In bacteria the specific association of ribosomes with nucleoids is a dynamic process involving active synthesis of RNA (Mascarenhas et al., 2001). The situation in the chloroplast may resemble the mitochondrial/eubacterial system, in which it has recently been shown that efficient expression of genes involves a complex series of interactions that localize transcriptionally active complexes in the nucleoids to the inner membrane surface to coordinate translational and transcriptional events (Rodeheffer and Shadel, 2003).
There is increasing evidence that chloroplast gene expression occurs at the inner envelope membrane, and it has been shown that nucleoids are associated with the inner envelope membrane in differentiating chloroplasts (Sato et al., 1999; Zerges, 2000). These findings are relevant to the possible roles of APO1. The localization of APO1 close to the nucleoids suggests that translation of plastid-encoded genes and early PSI biogenesis are associated with the DNA-containing subplastidial compartment. It is feasible that initiation and the first steps of translation elongation take place in transcriptionally active nucleoids, but late translational events occur at different places, for example, in the stroma or in association with membranes. APO1 seems to be preferentially associated with transcriptionally and translationally active nucleoids, reflecting its higher affinity to individual nucleoids inside the chloroplast (Figure 6B).
The Novel APO Repeat Gene Family Contains Four Distinct Groups in Vascular Plants
Remarkably, the APO1 start codon is localized in the second exon, indicating that the first exon is untranslated. APO1 is a member of a novel gene family with unknown function encoding proteins consisting of 327 to 449 amino acid residues and exclusively found in vascular plants (Figure 7B). The size variation is mainly as a result of different lengths of the N-terminal part in the individual groups APO1 to APO4 (Figure 7A). The APO gene family is characterized by a duplicated region of ∼100 amino acid residues (APO motifs 1 and 2) that is highly conserved among all members. These characteristics are indicative of a symmetric structure of APO proteins and probably of a symmetric ligand. On the other hand, APO1 could bind two similar ligands. Interestingly, four genes are present in Arabidopsis, and the corresponding orthologs were also found in rice (Figure 7B). The orthologs are similar in size and share the same sequence characteristics that are specific for each of the four APO groups. APO2 and APO3 in rice are computed to be localized in the chloroplast as well. Therefore, the function of APO1 could be partially redundant. The conservation of these four gene copies in vascular plants indicates that they might have specific functions not only in the chloroplast but also in other cellular compartments.
METHODS
Strains, Medium, and Culture Conditions
The apo1 mutant, accession Wassilewskija, was selected from a T-DNA insertion collection (Feldmann, 1991) obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH). Wild-type and mutant strains were grown under greenhouse conditions or on sucrose-supplemented medium as described (Meurer et al., 1996a). In all analyses, phenotypically wild-type plants of heterozygous progenies grown under the same conditions were compared with apo1. Selection of mutant plants was facilitated by a chlorophyll fluorescence imaging system (FluorCam690M; Photon System Instruments, Brno, Czech Republic).
For translation inhibition, hypocotyls of 3-week-old wild-type and mutant plants were clipped in a solution containing nutrients and 400 mg/L lincomycin.
Chlorophyll Fluorescence Induction, P700 Redox Kinetics, Emission Spectra at 77K, and Absorbance Spectra of Pigments
Chlorophyll fluorescence measurements at room temperature were performed using a pulse amplitude–modulated fluorometer equipped with a data acquisition system PDA100 to record fast changes (PAM 101; Walz, Effeltrich, Germany). Consecutive saturating light pulses of 800 ms were applied by halogen lamps. The connected emitter–detector subunit allowed us to measure the light-induced P700 redox kinetics by absorbance changes at 830 nm.
Chlorophyll fluorescence emission spectra at 77K of grinded leaves were recorded as described (Meurer et al., 1996b).
For pigment isolation, 150 mg of lyophilized leaves were ground in 1 mL of acetone containing 25 mg of Na2CO3. After centrifugation, the pigment absorbance spectra were registered in the wavelength region from 350 to 750 nm. Chlorophyll amounts were calculated according to Arnon (1949).
Map-Based Cloning, T-DNA Tagging, and Complementation of apo1
For genetic crosses, male plants of the accession Landsberg erecta were used to generate the F2 mapping population. Single mutant seedlings were chosen for mapping the APO1 gene using the microsattelite marker nga128 (http://www.arabidopsis.org/) and the marker chr1-110 (5′-CAC ACA TAT TAA CGA GTG GAT TGA CG-3′ and 5′-GGA CTC AAA TAT GTG ACA AGA GTA AGA CTC-3′) on chromosome 1 at positions 20.234 and 25.364 Mb, respectively. T-DNA flanking sequences were isolated by inverse PCR. SalI-digested genomic DNA was religated and subsequently subjected to PCR using the primers 5′-CAC CTG TCC TAC GAG TTG C-3′ and 5′-GCA TAG ATG CAC TCG AAA TCA GCC-3′. PCR products were cloned and sequenced. The 5′ end of the APO1 cDNA was determined by RACE experiments with the 5′ RACE primer SMART (BD Biosciences Clontech, Palo Alto, CA) and the gene-specific primer apo1-r2 (5′-ACT TGT CTG CTC TCT ATG CTT CTG ATT-3′) located within the coding region. The full-length cDNA of APO1 was amplified by PCR using the 5′ end primer apo1-f (5′-CAC GGT CTG AGC TGA TTG CGT GTT CTC-3′) and the TAA stop codon–containing primer apo1-r (5′-CCA AGG ACT TAT GCG ACC ATG TCG GCT TCC-3′). The product obtained was cloned into the SmaI site of the binary plant transformation vector pS001 (Meurer et al., 1998a). Heterozygous offsprings were transformed via Agrobacterium tumefaciens using the floral dip method (Clough and Bent, 1998).
Expression Studies of Nuclear and Plastid Genes
RNA gel blot analysis and the hybridization probes were described by Meurer et al. (1996a). Additional plastid probes were generated by amplifying specific regions of the Arabidopsis thaliana plastome (Sato et al., 1999). Poly(A+) mRNA was isolated using Oligo(dT) beads according to the manufacturer's instructions (Dynal, Oslo, Norway) and used for RACE experiments. Total RNA was isolated and subjected to RNA gel blot analysis as described (Meurer et al., 1996b). Quantitative two-step RT-PCR for apo1 and wild-type mRNA was performed using the LightCycler system (Roche Molecular Biochemicals, Mannheim, Germany) applying the SYBR Green protocol (Wittwer et al., 1997). Real-time PCR was performed with APO1 exon-specific primers (5′-GCT TCT GGT TTC TCC TGC TTG TAG AGG TG-3′ and 5′-ACT TGT CTG CTC TCT ATG CTT CTG ATT-3′) and, as an internal control, with 18S rDNA primers (5′-GCT CAA AGC AAG CCT ACG CTC TGG-3′ and 5′-GGA CGG TAT CTG ATC GTC TTC GAG CC-3′). Serially diluted samples of the APO1 cDNA were used for the calibration curve.
Polysome Analysis
Isolation of polysomes from leaves of 3-week-old plants was performed essentially as described in Barkan (1998). Polysome aliquots were layered onto 15% to 55% sucrose gradients and centrifuged for 65 min at 272.000 g and 4°C in a SW60 Ti rotor (Beckman, Munich, Germany). Fractions of 0.4 mL were collected, and the RNA obtained was subjected to RNA gel blot analysis.
Chloroplast Import Experiments
The APO1 cDNA was amplified with primers 5′-GTA ATA CGA CTC ACT ATA GGG CTT ATA TAG TCA ACA-3′ and 5′-CCA AGG ACT TAT GCG ACC ATG TCG GCT TCC-3′ that generate a T7 promoter upstream of the coding region, allowing in vitro transcription. Isolated chloroplasts were incubated for 0, 5, 10, and 20 min with labeled in vitro translation products of apo1 generated in cell-free wheat germ extracts in the presence of [35S]Met (Roche Molecular Biochemicals). Chloroplasts were purified, washed twice, and treated with thermolysin when indicated, and isolated proteins were subjected to SDS-PAGE (Lezhneva et al., 2004). Gels were dried and exposed to phosphor imaging plates (BASIII Fuji Bio Imaging plates and BAS2000 software package [Tokyo, Japan] and the AIDA software package version 3.25 beta; Raytest, Straubenhardt, Germany).
GFP Fusion
PCR products of the APO1 cDNA were generated using primers 5′-ACT TAT ATA GTC GAC ATG CTT CTG GTT TCT-3′ and 5′-GAT ATC CAC GTC GAC CGA TCA CTC TCT TCC-3′ and cloned into the SalI site of the GFP expression vector pOL-LT (Mollier et al., 2002), producing a translational fusion of the APO1 protein containing APO motif 1 with GFP. GFP was transiently expressed in tobacco (Nicotiana tabacum) protoplasts using the polyethylene glycol protocol (Lyznik et al., 1991). Fluorescence was visualized using a fluorescence microscope equipped with a digital camera (Axioplan; Zeiss, Jena, Germany).
Primer Extension
Primer extension reactions were performed with 50 μg of DNase-treated RNA using the Superscript II reverse transcriptase (Invitrogen, Karlsruhe, Germany) and the fluorochrome-labeled primer 5′-GTG AGC ATC AGC ATG TAG GTT CCA GAT CC-3′ annealing to the psaA 5′ coding region. Sequencing and product analysis was performed using the LI-COR 4200IR2 two-laser system (MWG Biotech, Ebersberg, Germany).
Immunological Analysis
Membrane proteins of 3-week-old plants were isolated substantially as described (Meurer et al., 1996a). The supernatant of the first centrifugation, which contained the soluble proteins, was precipitated with 15% trichloroacetic acid. The sediment was washed twice with 80% acetone and resuspended in 100 mM Na2CO3, 10% sucrose, and 50 mM dithioerythritol. Proteins separated by SDS-PAGE were transferred to polyvinylidene difluoride membranes (Amersham Buchler, Braunschweig, Germany), incubated with specific antibodies (Meurer et al., 1996a), and visualized by the enhanced chemiluminescence technique (Amersham Buchler).
Radioactive Labeling of Plastid Membrane Proteins
In vivo labeling of leaf proteins was performed as described (Meurer et al., 1996b), with the exception that the hypocotyls of plants were first cut in the antibiotic-containing solution and subsequently immersed into the appropriate medium supplemented with [35S]Met (50 mCi; specific activity >1000 Ci/mmol) for 20 min.
Chloroplast Ultrastructure
Sample preparation for ultrastructural analysis and electron microscopy were performed as described (Meurer et al., 1998b).
The GenBank accession number of the Arabidopsis APO1 cDNA reported in this article is AY466161. Accession numbers or names for other genes mentioned in this article are At5g57930.1, At5g61930.1, and At3g21740.1 (Arabidopsis APO2 to APO4 genes, respectively), AL662950, AK104342, AP003840, and AK064525 (rice APO1 to APO4 genes, respectively).
Acknowledgments
We thank Elli Gerick for excellent technical assistance. We also thank Ahmed Nada and Lea Ester for providing import-competent chloroplasts. Itzhak Ohad is acknowledged for critical reading of the manuscript. This research was supported by the German Science Foundation (grant ME 1794/1-3 to J.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jörg Meurer (joerg.meurer@lrz.uni-muenchen.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024935.
References
- Allen, J.F., and Pfannschmidt, T. (2000). Balancing the two photosystems: Photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon, D.I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297, 38–57. [Google Scholar]
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Beinert, H., Holm, R.H., and Munck, E. (2000). Iron-sulfur clusters: Nature's modular, multipurpose structures. Science 277, 653–659. [DOI] [PubMed] [Google Scholar]
- Ben-Shem, A., Frolow, F., and Nelson, N. (2003). Crystal structure of plant photosystem I. Nature 426, 630–635. [DOI] [PubMed] [Google Scholar]
- Bruick, R.K., and Mayfield, S.P. (1999). Light-activated translation of chloroplast mRNAs. Trends Plant Sci. 4, 190–195. [DOI] [PubMed] [Google Scholar]
- Chitnis, P.R. (2001). Photosystem I: Function and physiology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 539–626. [DOI] [PubMed] [Google Scholar]
- Choquet, Y., and Wollman, F.A. (2002). Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 529, 39–42. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cohen, A., Yohn, C.B., and Mayfield, S. (2001). Translation of the chloroplast-encoded psbD is arrested post-initiation in a nuclear mutant of Chlamydomonas reinhardtii. J. Plant. Physiol. 158, 1069–1075. [Google Scholar]
- Dauvillée, D., Stampacchia, O., Girard-Bascou, J., and Rochaix, J.D. (2003). Tab2 is a novel conserved RNA binding protein required for translation of the chloroplast psaB mRNA. EMBO J. 22, 6378–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depège, N., Bellafiore, S., and Rochaix, J.D. (2003). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299, 1572–1575. [DOI] [PubMed] [Google Scholar]
- Eichacker, L.A., Soll, J., Lauterbach, P., Rüdiger, W., Klein, R.R., and Mullet, J.E. (1990). In vitro synthesis of chlorophyll a in the dark triggers accumulation of chlorophyll a apoproteins in barley etioplasts. J. Biol. Chem. 265, 13566–13571. [PubMed] [Google Scholar]
- Fedoroff, N.V. (2002). RNA-binding proteins in plants: The tip of an iceberg? Curr. Opin. Plant Biol. 5, 452–459. [DOI] [PubMed] [Google Scholar]
- Feldmann, K.A. (1991). T-DNA insertion mutagenesis in Arabidopsis: Mutational spectrum. Plant J. 1, 71–82. [Google Scholar]
- Frazzon, J., and Dean, D.R. (2003). Formation of iron-sulfur clusters in bacteria: An emerging field in bioinorganic chemistry. Curr. Opin. Chem. Biol. 7, 166–173. [DOI] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. (1988). Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbeck, J.H. (2003). The binding of cofactors to photosystem I analyzed by spectroscopic and mutagenic methods. Annu. Rev. Biophys. Biomol. Struct. 32, 237–256. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M. (1998). Coordination of nuclear and chloroplast gene expression in plant cells. Int. Rev. Cytol. 177, 115–180. [DOI] [PubMed] [Google Scholar]
- Guruprasad, K., Reddy, B.V., and Pandit, M.W. (1990). Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 4, 155–161. [DOI] [PubMed] [Google Scholar]
- Havaux, M., and Niyogi, K.K. (1999). The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc. Natl. Acad. Sci. USA 96, 8762–8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, P., Fromme, P., Witt, H.T., Klukas, O., Saenger, W., and Krauss, N. (2001). Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411, 909–917. [DOI] [PubMed] [Google Scholar]
- Kiley, P.J., and Beinert, H. (2003). The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 6, 181–185. [DOI] [PubMed] [Google Scholar]
- Kim, J., Eichacker, L.A., Rüdiger, W., and Mullet, J.E. (1994). Chlorophyll regulates accumulation of the plastid-encoded chlorophyll proteins P700 and D1 by increasing apoprotein stability. Plant Physiol. 104, 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klughammer, C., and Schreiber, U. (1994). An improved method, using saturating light pulses, for the determination of photosystem I quantum yield via P700+-absorbance changes at 830 nm. Planta 192, 261–268. [Google Scholar]
- Kobayashi, T., Takahara, M., Miyagishima, S.Y., Kuroiwa, H., Sasaki, N., Ohta, N., Matsuzaki, M., and Kuroiwa, T. (2002). Detection and localization of a chloroplast-encoded HU-like protein that organizes chloroplast nucleoids. Plant Cell 14, 1579–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause, G.H., and Weis, E. (1991). Chlorophyll fluorescence and photosynthesis: The basics. Annu. Rev. Plant. Physiol. Plant Mol. Biol. 42, 313–349. [Google Scholar]
- Legen, J., Misera, S., Herrmann, R.G., and Meurer, J. (2001). Map positions of 69 Arabidopsis thaliana genes of all known nuclear encoded constituent polypeptides and various regulatory factors of the photosynthetic membrane: A case study. DNA Res. 27, 53–60. [DOI] [PubMed] [Google Scholar]
- Leon, P., Arroyo, A., and Mackenzie, S. (1998). Nuclear control of plastid and mitochondrial development in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 453–480. [DOI] [PubMed] [Google Scholar]
- Léon, S., Touraine, B., Ribot, C., Briat, J.F., and Lobréaux, S. (2003). Iron-sulphur cluster assembly in plants: Distinct NFU proteins in mitochondria and plastids from Arabidopsis thaliana. Biochem. J. 371, 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezhneva, L., Amann, K., and Meurer, J. (2004). The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 37, 174–185. [DOI] [PubMed] [Google Scholar]
- Lezhneva, L., and Meurer, J. (2004). The nuclear factor HCF145 affects chloroplast psaA-psaBrps14 transcript abundance in Arabidopsis thaliana. Plant J. 38, 740–753. [DOI] [PubMed] [Google Scholar]
- Lill, R., and Kispal, G. (2000). Maturation of cellular Fe-S proteins: An essential function of mitochondria. Trends Biochem. Sci. 25, 352–356. [DOI] [PubMed] [Google Scholar]
- Lyznik, L.A., Peng, J.Y., and Hodges, T.K. (1991). Simplified procedure for transient transformation of plant protoplasts using polyethylene glycol treatment. Biotechniques 10, 294–300. [PubMed] [Google Scholar]
- Mascarenhas, J., Weber, M.H., and Graumann, P.L. (2001). Specific polar localization of ribosomes in Bacillus subtilis depends on active transcription. EMBO Rep. 2, 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac, D.J., and Barkan, A. (1999). A nuclear gene in maize required for the translation of the chloroplast atpB/E mRNA. Plant Cell 11, 1709–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, S., and Dreyfuss, B.W. (1998). Posttranslational assembly of photosynthetic metalloproteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 25–51. [DOI] [PubMed] [Google Scholar]
- Meurer, J., Berger, A., and Westhoff, P. (1996. b). A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid psbB, psbD/C, ndhH, and ndhC operons. Plant Cell 8, 1193–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer, J., Grevelding, C., Westhoff, P., and Reiss, B. (1998. a). The PAC protein affects the maturation of specific chloroplast mRNAs in Arabidopsis thaliana. Mol. Gen. Genet. 258, 342–351. [DOI] [PubMed] [Google Scholar]
- Meurer, J., Meierhoff, K., and Westhoff, P. (1996. a). Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterization by spectroscopy, immunoblotting and Northern hybridization. Planta 198, 385–396. [DOI] [PubMed] [Google Scholar]
- Meurer, J., Plücken, H., Kowallik, K.V., and Westhoff, P. (1998. b). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17, 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollier, P., Hoffmann, B., Debast, C., and Small, I. (2002). The gene encoding Arabidopsis thaliana mitochondrial ribosomal protein S13 is a recent duplication of the gene encoding plastid S13. Curr. Genet. 40, 405–409. [DOI] [PubMed] [Google Scholar]
- Moseley, J.L., Page, M.D., Alder, N.P., Eriksson, M., Quinn, J., Soto, F., Theg, S.M., Hippler, M., and Merchant, S. (2002). Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14, 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley, J.L., Quinn, J., and Merchant, S. (2000). The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19, 2139–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet, J.E., Klein, P.G., and Klein, R.R. (1990). Chlorophyll regulates accumulation of the plastid-encoded chlorophyll apoproteins CP43 and D1 by increasing apoprotein stability. Proc. Natl. Acad. Sci. USA 87, 4038–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munekage, Y., Hojo, M., Meurer, J., Endo, T., Tasaka, M., and Shikanai, T. (2002). PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110, 361–371. [DOI] [PubMed] [Google Scholar]
- Pilon-Smits, E.A., Garifullina, G.F., Abdel-Ghany, S., Kato, S., Mihara, H., Hale, K.L., Burkhead, J.L., Esaki, N., Kurihara, T., and Pilon, M. (2002). Characterization of a NifS-like chloroplast protein from Arabidopsis. Implications for its role in sulfur and selenium metabolism. Plant Physiol. 130, 1309–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattanachaikunsopon, P., Rosch, C., and Kuchka, M.R. (1999). Cloning and characterization of the nuclear AC115 gene of Chlamydomonas reinhardtii. Plant Mol. Biol. 9, 1–10. [DOI] [PubMed] [Google Scholar]
- Rodeheffer, M.S., and Shadel, G.S. (2003). Multiple interactions involving the amino-terminal domain of yeast mtRNA polymerase determine the efficiency of mitochondrial protein synthesis. J. Biol. Chem. 278, 18695–18701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, N., Terasawa, K., Miyajima, K., and Kabeya, Y. (2003). Organization, developmental dynamics, and evolution of plastid nucleoids. Int. Rev. Cytol. 232, 217–262. [DOI] [PubMed] [Google Scholar]
- Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., and Tabata, S. (1999). Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 6, 283–290. [DOI] [PubMed] [Google Scholar]
- Scheller, H.V., Jensen, P.E., Haldrup, A., Lunde, C., and Knoetzel, J. (2001). Role of subunits in eukaryotic Photosystem I. Biochim. Biophys. Acta 1507, 41–60. [DOI] [PubMed] [Google Scholar]
- Schwabe, T.M., and Kruip, J. (2000). Biogenesis and assembly of photosystem I. Indian J. Biochem. Biophys. 37, 351–359. [PubMed] [Google Scholar]
- Shen, G., Antonkine, M.L., van der Est, A., Vassiliev, I.R., Brettel, K., Bittl, R., Zech, S.G., Zhao, J., Stehlik, D., Bryant, D.A., and Golbeck, J.H. (2002). Assembly of photosystem I. II. Rubredoxin is required for the in vivo assembly of F(X) in Synechococcus sp. PCC 7002 as shown by optical and EPR spectroscopy. J. Biol. Chem. 277, 20355–20366. [DOI] [PubMed] [Google Scholar]
- Simon, M.C., and Nisman, B. (1977). Initiation of transcription and translation in E. coli nucleoids. C. R. Acad. Sci. Hebd. Seances Acad. Sci. D. 285, 435–438. [PubMed] [Google Scholar]
- Stampacchia, O., Girard-Bascou, J., Zanasco, J.L., Zerges, W., Bennoun, P., and Rochaix, J.D. (1997). A nuclear-encoded function essential for translation of the chloroplast psaB mRNA in Chlamydomonas. Plant Cell 9, 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, D.B., Higgs, D.C., and Yang, J. (1997). Transcription and translation in chloroplasts. Trends Plant Sci. 2, 308–315. [Google Scholar]
- Sticht, H., and Rösch, P. (1998). The structure of iron-sulfur proteins. Prog. Biophys. Mol. Biol. 70, 95–136. [DOI] [PubMed] [Google Scholar]
- Sugiura, M., Hirose, T., and Sugita, M. (1998). Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet. 32, 437–459. [DOI] [PubMed] [Google Scholar]
- Surpin, M., Larkin, R.M., and Chory, J. (2002). Signal transduction between the chloroplast and the nucleus. Plant Cell 14, 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottey, S., Block, M.A., Allen, M., Westergren, T., Albrieux, C., Scheller, H.V., Merchant, S., and Jensen, P.E. (2003). Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. USA 100, 16119–16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touraine, B., Boutin, J.-P., Marion-Poll, A., Briat, J.-F., Peltier, G., and Lobréaux, S. (2004). Nfu2: A scaffold protein required for [4Fe-4S] and ferredoxin iron-sulphur cluster assembly in Arabidopsis chloroplasts. Plant J. 40, 101–111. [DOI] [PubMed] [Google Scholar]
- Wittwer, C.T., Ririe, K.M., Andrew, R.V., David, D.A., Gundry, R.A., and Balis, U.J. (1997). The LightCycler: A microvolume multisample fluorimeter with rapid temperature control. Biotechniques 22, 176–181. [DOI] [PubMed] [Google Scholar]
- Wostrikoff, K., Girard-Bascou, J., Wollman, F.A., and Choquet, Y. (2004). Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J. 23, 2696–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe, T., Morimoto, K., Kikuchi, S., Nishio, K., Terashima, I., and Nakai, M. (2004). The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell 16, 993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges, W. (2000). Translation in chloroplasts. Biochimie 82, 583–601. [DOI] [PubMed] [Google Scholar]