Abstract
Over the past 20 years, protein engineering has been extensively used to improve and modify the fundamental properties of fluorescent proteins (FPs) with the goal of adapting them for a fantastic range of applications. FPs have been modified by a combination of rational design, structure-based mutagenesis, and countless cycles of directed evolution (gene diversification followed by selection of clones with desired properties) that have collectively pushed the properties to photophysical and biochemical extremes. In this review, we attempt to provide both a summary of the progress that has been made during the past two decades, and a broad overview of the current state of FP development and applications in mammalian systems.
Prototypical FPs
The initial demonstration in 1994 that the Aequorea victoria jellyfish (class Hydrozoa) green fluorescent protein (avGFP; Figure 1A) could function as a genetically encodeable fluorescent tag [1] was followed by rapid-fire protein engineering efforts to fine tune the biochemical and fluorescent properties and expand the color palette to encompass blue, cyan, and yellow variants [2,3]. Further genomic exploration of marine organisms soon led to the discovery of additional prototypical fluorescent proteins (FPs) (i.e., homologs of avGFP) from class Anthozoa [4], including the Discosoma striata mushroom anemone, which gave rise to DsRed and subsequent mFruit progeny (Figure 1B) [5,6], and the sea anemone Entacmaea quadricolor, which yielded eqFP, TagRFP, mKate, and mRuby derivatives, among others [7–10]. FPs from these species extended the color palette into the orange, red, and far red. Accompanying this evolution of prototypical FPs, new classes of non-prototypical FPs based on the binding of flavin mononucleotide (FMN) [11], phycocyanobilin [12], biliverdin [13], or bilirubin [14], have emerged in recent years. Figure 2A shows the trajectory of FP development, with emphasis on the wild-type roots of modern variants.
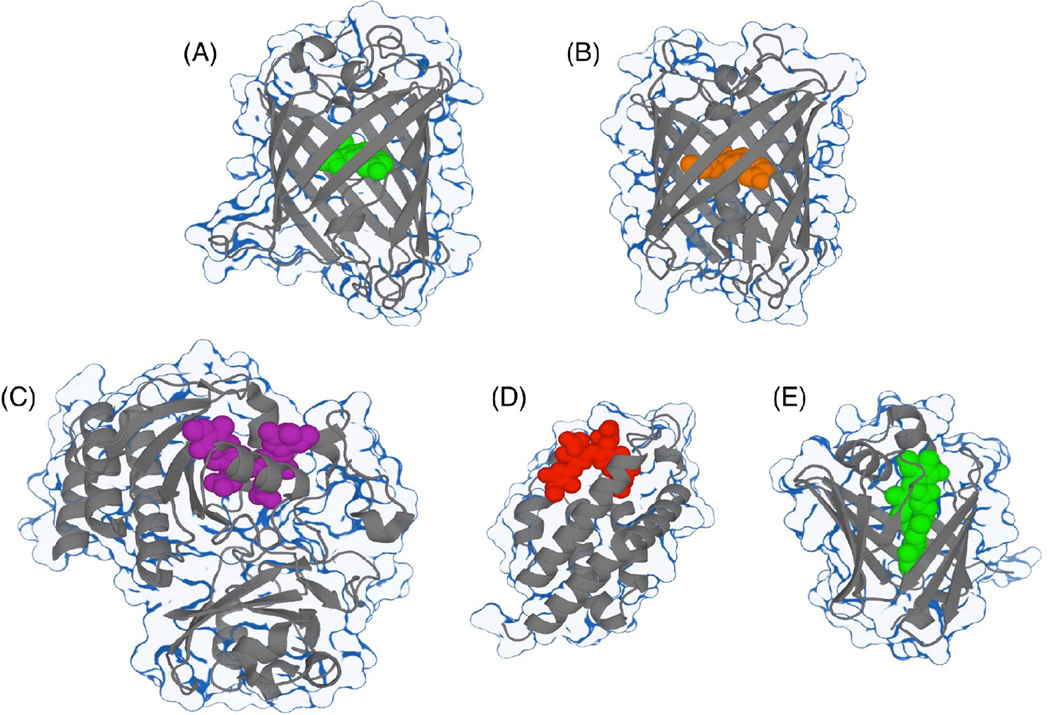
Figure 1. Structural representations of FPs.
(A) Aequorea GFP with the chromophore represented in green (PDB ID 1EMA) [168]. (B) The monomeric DsRed-derived RFP known as mCherry with the chromophore represented in orange (PDB ID 2H5Q) [169]. (C) IFP2.0 derived from a bacteriophytochrome with bound biliverdin in purple (PDB ID 4CQH) [33]. (D) A representation of a cyanobacterial allophycocyanin α-subunit that is homologous to smURFP, with bound biliverdin shown in red (PDB 4PO5) [170]. (E) Structure of UnaG with bound bilirubin shown in green (PDB ID 4I3B) [14].
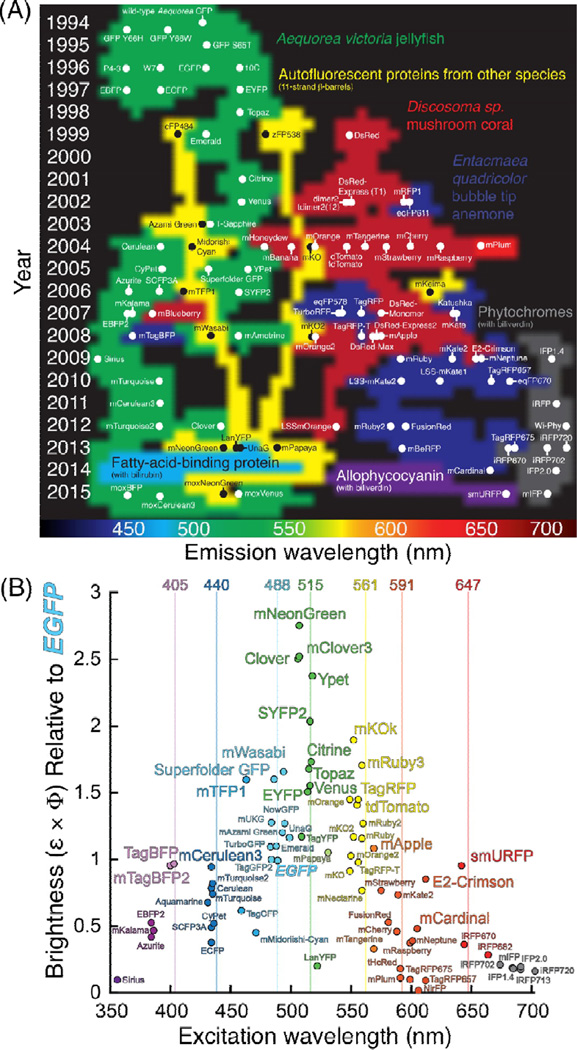
Figure 2. An overview of genetically encoded fluorophores introduced since the advent of wild-type avGFP in 1994.
(A) This chart includes FPs that are 11-stranded (3-barrel homologs of avGFP, as well as proteins that are fluorescent when bound to a biliverdin or bilirubin chromophore. Placement along the vertical axis represents the year in which the FP was first introduced, while placement along the horizontal axis represents the peak emission wavelength. (B) Intrinsic fluorescent brightness (ε × Φ) of selected FPs are plotted relative to EGFP as a function of peak excitation wavelength. Vertical lines at 405 nm, 440 nm, 488 nm, 515 nm, 561 nm, 591 nm, and 647 nm correspond to commonly available laser lines on fluorescence microscopes.
One of the primary goals of FP engineering has been to develop brighter FPs. The brighter an FP, the lower the intracellular concentration that can be reliably imaged with sufficient fluorescent contrast relative to the autofluorescent background. A fluorophore’s intrinsic fluorescent brightness (see Glossary) is proportional to the product of the molecular extinction coefficient (ε) and the fluorescence quantum yield (Φ). However, practically speaking, the apparent brightness in cells is highly dependent on additional environmental factors. Therefore, many mutational strategies have targeted improving gene expression by optimizing codon usage, facilitating protein folding, promoting the extent and rate of chromophore maturation, and increasing protein stability by reducing the influence of environmental perturbations such as pH and O2. Because most FPs are naturally dimeric or tetrameric, substantial engineering has also been aimed at engineering monomeric variants [15]. Figure 2B provides a summary of FP brightness with respect to their wavelength of maximum excitation, and with reference to commonly available laser lines.
Despite 20 years of FP engineering, there remains substantial room for improvement in FPs. For instance, two new GFPs, mClover3 [16] and mNeonGreen [17], have recently emerged as the new benchmark for brightness (approximately a 2.5-fold increase compared to mEGFP). mClover3 was engineered from avGFP by combining key insights accumulated from years of evolution, including identification of positions that influenced the H-bonding network surrounding the chromophore, stacking residues that shift the energy of emission, and residues that influence photostability and folding [16,18].. In contrast, mNeonGreen emerged from an underexplored lineage and was created by incorporating 21 mutations into the yellow FP derived from Branchiostoma lanceolatum, LanYFP [17], suggesting that there remains untapped potential in additional marine species. These examples reveal that there may be considerable room for improvement for prototypical FPs using approaches such as meta-analysis of protein engineering efforts and identifying new lineages of β-barrel FPs. Yet another approach is to discover or engineer non-prototypical classes of FPs such as FMN-binding proteins that have the advantage (relative to avGFP homologs) of developing fluorescence in low oxygen environments [11,19,20].
Although FPs are undoubtedly powerful tools for a variety of cell biological applications, as researchers have pushed the boundaries to longer-term imaging and more specialized sub-cellular compartments, limitations of the existing toolset have been uncovered. For example, long-term expression of red FPs often leads to accumulation of puncta that colocalize with lysosomes, perhaps due to resistance of highly stable RFPs to lysosomal proteases [21–24]. Many FPs characterized as monomeric have been shown to interact when expressed as a fusion to membrane proteins [25,26]. Finally, inadvertent glycosylation sites and cysteine residues can lead to formation of intermolecular disulfide bonds, leading to higher order oligomers and interfering with chromophore formation potentially causing mislocalization particularly in the crowded and oxidizing environment of the secretory pathway [24]. Some recent engineering efforts have attempted to alleviate these limitations. For example, FusionRed was designed from mKate with mutations that reduce toxicity, improve performance in fusions, and enhance monomericity [23], and cysteine-less blue, cyan, green, and yellow FPs (oxFPs) have been developed for use in oxidizing compartments [24].
Infrared FPs and Indicators: Advantages and Caveats
Long-wavelength light between 650 and 900 nm penetrates the furthest through animal tissue because the combined effects of tissue absorbance (i.e., from hemoglobin, water, and lipids) and light scattering, are at a minimum [27,28]. Accordingly, infrared FPs (IFPs) are preferred for use as protein tags and genetically encoded indicators for in vivo imaging applications [13]. As avGFP homologs with excitation in the near infrared range have been neither discovered in nature or engineered in the lab, researchers have turned to bacterial phytochromes (BphPs) as templates for engineering IFPs (Figure 1C). BphPs belong to the phytochrome red/far-red photoreceptor superfamily [29,30] and typically exhibit maximum absorbance at approximately 650 – 700 nm, but are natively not fluorescent. In nature, the chromophore of BphPs is a covalently bound biliverdin (BV), a linear tetrapyrrole that is produced by heme oxygenase 1 (HO-1) as the chief catabolic metabolite of heme. This strategy builds upon the pioneering work of Lagarias and coworkers who developed red and near-infrared fluorescent ‘Phytofluors’ from phytochrome proteins [12,31]. In contrast to BV-binding BphPs, the earlier Phytofluor proteins required linear tetrapyrrole molecules, such as phycoerythrobilin and phycocyanobilin, that are not produced by mammalian cells.
The first example of utilizing BphPs in mammalian imaging was IFP1.4, which was engineered from a truncated DrBphP from Deinococcus radiodurans [13]. The utility of IFP1.4 was demonstrated by fluorescence imaging of the liver in intact mice. Since thousands of bacteriophytochrome-like sequences have been reported, this work opened a new door in engineering long-wavelength FPs. Indeed, soon after, iRFP was engineered from RpBphP2 from Rhodopseudomonas palustris and used for imaging of mouse liver with improved brightness [32]. iRFP has similar molecular brightness compared to IFP1.4, but it is significantly brighter in cells and thus it appears to utilize endogenous BV better than IFP1.4. IFP2.0, the improved version of IFP1.4, was demonstrated to have similar brightness compared to iRFP when imaging brain tumors in intact mice [33]. Yet other IFPs include: Wi-Phy, derived from truncated DrBphP [34]; iRFP720, a redshifted iRFP mutant; iRFP670 and iRFP702, both derived from RpBphP6 [35]; mIFP, engineered from BrBphP (see below) [36]; and iBlueberry: a rationally designed mIFP variant with a ~40 nm blue shift in both excitation and emission [37].
To optimally serve as a protein fusion tag, an IFP should be monomeric so as not to perturb the stoichiometry of the protein of interest, which may affect protein function and/or trafficking. Based on sequence and structural analysis of the dimer interface, a bacteriophytochrome from Bradyrhizobium that is monomeric in its truncated form was identified from a sequence database, and was engineered into a naturally monomeric IFP (mIFP) [36]. mIFP can be used to label various proteins in live cells, Drosophila, and zebrafish, and, unlike dimeric iRFP, can be used to efficiently label the fine structure of dendrites in Drosophila. However, while mIFP is very useful for tagging and imaging of proteins, it is about 5 times less photostable than iRFP. Generally speaking, this is not a limiting factor for whole animal imaging since the low excitation light intensities typically used [37] permit acquisition of tens of thousands of images within the photobleaching half-time of mIFP. However, the relatively low photostability can limit the duration of time-lapse imaging experiments with mIFP-tagged proteins in live cells. Accordingly, in addition to improving the intrinsic brightness, a future direction for mIFP development should focus on increasing the photostability of monomeric IFPs or engineering a monomeric iRFP for protein labeling.
One caveat of BphP-derived IFPs is that the fluorescence depends on BV concentration, which varies in different cells and organisms. In Drosophila and zebrafish, BV concentration is limited [33,36]. In mammals, neurons appear to have low concentration of BV. Usage of BphP-derived IFPs is thus limited in these organisms and cell types. However, this may provide an opportunity for inducible fluorescence by delivering exogenous BV, which might open new applications such as pulse-chase labeling. Other than exogenous addition of BV, one way of overcoming this limitation is co-expression of HO-1, which converts heme into BV. For example, HO-1 co-expression improves IFP2.0 fluorescence in Drosophila, mice brain tumors [33], and zebrafish [36]. HO-1 has been reported to have anti-inflammatory and anti-oxidative functions via its products, and its expression is highly induced in response to pathophysiological stress [38]. Although HO-1 overexpression may introduce biological perturbation, such toxicity has not been observed in Drosophila and zebrafish. Nevertheless, an appropriate control is recommended when BphP-derived IFPs are used in cells and animals even without overexpression of HO-1. This is because expression of BV-binding IFPs may perturb the endogenous pool of BV and BV-derived bilirubin that is lipophilic and has antioxidant and cytoprotective roles in protecting lipids from oxidation, complementing the water soluble glutathione that protects water soluble proteins [39].
Since the BphP-derived IFPs have a completely different protein structure (Figure 1C) compared to the 11-stranded β-barrel coelenterate FPs (Figure 1A,B), IFPs provide new opportunities for engineering genetically encoded indicators. An infrared fluorogenic executioner caspase reporter (iCasper) was recently developed, based on the unique interactions between mIFP and its chromophore [40] (Figure 3). iCasper was used in imaging embryonic development in Drosophila and revealed interesting spatiotemporal coordination between cell apoptosis and embryonic morphogenesis, as well as dynamics of apoptosis during tumorigenesis in the brain of Drosophila [40]. The iCasper technology should be easily adaptable for various protease activities, enabling the dissection of signaling pathways that regulate protease activity, high-throughput screening of protease inhibitors for drug development, and in vivo biological studies. The development of the iCasper technology demonstrates that BphP-derived IFPs provide a new and promising scaffold for designing fluorogenic indicators (e.g., of kinase activity and membrane potential) that will be ideal for visualizing spatiotemporal dynamics of cell signaling in vivo.
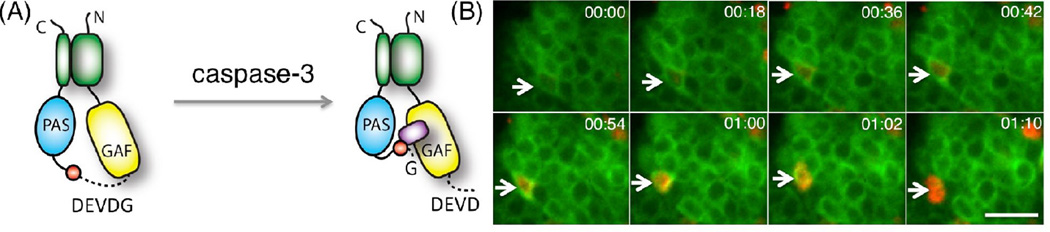
Figure 3. Design of an IFP protease indicator.
(A) Cartoon showing the cleavage and activation of iCasper. iCasper enables imaging of caspase-3 activation during apoptosis, one type of programed cell death. (B) iCasper enables the visualization of the spatiotemporal dynamics of apoptosis in vivo. Time-lapse images of neurons in the ventral nerve cord in Drosophila reveal caspase activation and apoptotic cell shape change during CNS development. The neuron undergoing apoptosis is indicated with the arrow. The number at the top right corner of each panel refers to time (h:min). Note that the neurons also express membrane targeted GFP, which is much brighter than the split GFP fluorescence from iCasper. Scale bar, 10 µm.
A new class of far-red FPs based on the allophycocyanin oc-subunit from a cyanobacterial phycobiliprotein
In an effort to further expand the color palette of FPs and improve upon the relatively low quantum yields and poor stability [13,32] of BphP-derived IFPs, a new class of FP has recently been developed from an allophycocyanin α-subunit (APCα) from a cyanobacterium, Trichodesmium erythraeum [41]. Native APC is a highly fluorescent hexamer that requires an auxiliary lyase to incorporate phycocyanobilin (PCB). To develop a useful FP from native APC, the protein was engineered to be self-sufficient (i.e., no lyase required) to covalently incorporate a BV rather than PCB chromophore. The protein was then subjected to twelve rounds of directed evolution for bright fluorescence and low cytotoxicity to Escherichia coli. This effort ultimately led a bright APCα-derived FP that was designated small Ultra-Red FP (smURFP) [41] because its light blue color when viewed under white light is reminiscent of the ‘Smurf’ cartoon characters (Figure 4A). SmURFP is a homodimer of 15 kDa subunits (Figure 1D) and has excitation and emission maxima at 642 nm and 670 nm, respectively. SmURFP has an exceptionally large extinction coefficient (ε = 180,000 M−1cm−1) and a modest quantum yield (Φ = 0.18) resulting in an intrinsic fluorescent brightness that is similar to EGFP. Accordingly, smURFP is the brightest far-red (conventionally defined as FPs with emission maxima between 633 and 670 nm) or near-infrared (emission maximum greater than 670 nm) FP yet reported (Figure 2B). Due to the limiting concentration of BV in mammalian cells [42], treatment of transfected cells with 1–5 µM BV dimethyl ester (BVMe2) is typically necessary to obtain the brightest fluorescence. Yet other favorable properties of smURFP include extremely good photostability and good expression in neurons (Figure 4B) with no apparent formation of fluorescent puncta.
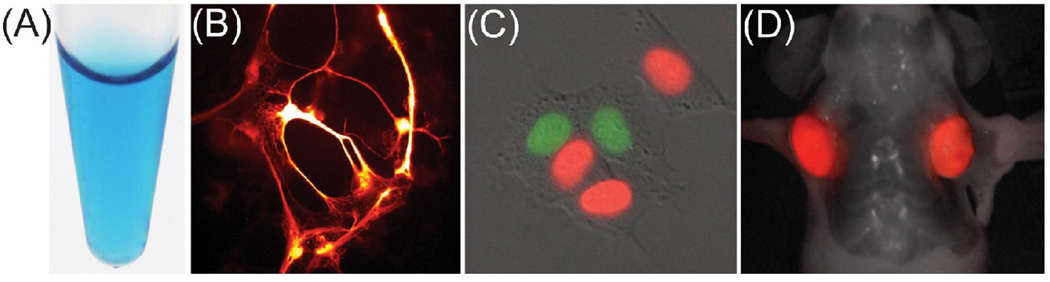
Figure 4. smURFP: a far-red, biliverdin-binding fluorescent protein derived from the allophycocyanin α-subunit from the cyanobacteria Trichodesmium erythraeum.
(A) Purified smURFP (left) shows color reminiscent of the cartoon character’s (Smurf) skin. (B) SmURFP expressed in primary rat neurons. (C) Far-red/near-infrared FUCCI expressed in HEK293A cells. Red-tinted fluorescence corresponds to smURFP, which illuminates the G0/G1 phases, while green-tinted fluorescence corresponds to IFP2.0, which illuminates the S/G2/M phases. (D) HT1080 tumor xenografts stably expressing smURFP. Fluorescence is visible without injection of exogenous BV.
Using smURFP and IFP2.0, a far-red/near-infrared Fluorescent Ubiquitination-based Cell Cycle Indicator (FUCCI) [43] was created (Figure 4C). This new indicator could be combined with the previously reported visible FP-based FUCCI for imaging cell cycle progression in two distinct cell types [41]. SmURFP expressed in HT1080 tumor rodent xenografts show fluorescence even without addition of exogenous BV (Figure 4D), though fluorescence intensity was less than mCherry or mCardinal under similar conditions.
A Bilirubin-Inducible FP from the Vertebrate Subphylum
Recent years have seen an increasing recognition of the prevalence of biological fluorescence in vertebrate animals. One such vertebrate is the Japanese eel, Anguilla japonica [44], which exhibits green fluorescence from its skeletal muscle. The gene responsible for this green fluorescence was cloned [14] and found to encode for a 16.9 kD polypeptide in the fatty-acid binding protein (FABP) family [45–47]. The protein, which was named UnaG (Figure 1E), had no intrinsic fluorescence but showed bright green emission in mammalian cells and biological mixtures even under anaerobic conditions. Extensive screening revealed the chromophore to be bilirubin, a lipophilic bilin that is the reduction product of BV, is a clinical diagnostic for liver function, and is responsible for the diseases jaundice and kernicterus [48–50]. UnaG binds to bilirubin non-covalently with high-affinity (Kd = 98 pM) and high-specificity. These favorable properties enabled UnaG to be used to quantify serum levels of bilirubin from human subjects [14].
As an alternative to avGFP for live cell fluorescence imaging, UnaG has the inherent advantages of a smaller gene size (which may facilitate packaging in viral vectors with limited DNA capacity), and the ability to develop fluorescence in anaerobic conditions when bilirubin is supplied from serum. Accordingly, UnaG fills an important niche in the toolbox of genetically encoded fluorophores and is likely to serve as the prototype for a growing class of fluorescence-based indicators [51–53].
Harnessing FP Photochromism for Superresolution Microscopy or Enhancing Contrast
While most FP engineering efforts have focused on making improved tools for traditional fluorescence microscopy, there has been a parallel effort to develop FPs that switch between molecular states (photochromism). FP photochromism was first demonstrated in 1997 by Moerner, Tsien and colleagues who established that single molecules of yellow FPs exhibited intermittent fluorescence emission (i.e., blinking), and that molecules trapped in a long-lived dark “off” state could be converted back into the “on” state by illumination with high energy (405 nm) light [54]. Subsequent spectroscopic studies have largely reinforced the model of dark state conversion proposed by Moerner and colleagues, and have revealed multiple mechanisms for excited state and dark state transitions, including conformational dynamics of the chromophore such as cis-trans isomerization [55,56], excited state proton transfer and excited state solvation dynamics [57–59], and triplet state conversion [60]. However, it was not until 2002 when mutagenesis was first used to accentuate this photochromism, leading to the first designed photoactivatable avGFP (PA-GFP) [61]. The notion that proteins could be engineered for improved photochromism helped pave the way for the rapid expansion of super-resolution microscopy platforms that exploit transitions between molecular states to localize molecules with high precision.
Since the original development of microscopies that rely on switching of molecules between different molecular states to improve spatial resolution, such as Stimulated Emission Depletion (STED) [62,63], Photo-Activated Localization Microscopy (PALM) [64] and Stochastic Optical Reconstruction Microscopy (STORM) [65], there has been a rapid proliferation of techniques that exploit complex photochromism of FPs to improve spatial resolution and/or enhance fluorescence contrast (Table 1). Photochromism in FPs can be reversible or irreversible, inducible or spontaneous, and can occur on a wide range of time scales from ~1 µs recovery from triplet states [60], to 10–1000 ms recovery from transient dark states that can be either photoprotective or photoreactive [66], or even much slower recovery from kinetically trapped dark states from which irradiation can induce transition back to bright state [54]. Not surprisingly, different modalities exploit different photophysical properties, and have different requirements for optimal performance as outlined in Table 1. While a number of FPs have been explicitly engineered based on irreversible photochromism for PALM/STORM requirements [67–70], only a few FPs have been screened for microscopies that exploit reversible photochromism [71,72]. Sophisticated screens could be developed to explore the parameter space [73,74], perhaps leading to greater insight into the mechanism(s) of these processes and probes optimized for specific imaging modalities.
Table 1.
Fluorescence imaging modalities that exploit FP photochromism
| Microscopy | Photochromism | Commonly used FPs |
Desired properties for optimal performance |
Reference |
|---|---|---|---|---|
| PALM/STORM | Photo-activation, photo- conversion, reversible photoswitching |
mEos3.2, mMaple 2 or 3, PA-mCherry |
High number of photons emitted/switching cycle; ratio of rate constant for on & off switching |
[67–69] |
| RESOLFT | Reversible photo-switching |
rsEGFP2 | Rapid switching kinetics between on/off states; high contrast ratio between on/off states; number of switching cycles prior to photobleaching |
[71] |
| SOFI, pcSOFI | Spontaneous or light-induced dark state conversion |
Dronpa, rsTagRFP, Skylan-S |
Fluorescence fluctuations that are slow compared to timescale of acquisition time (10s of ms); photostable enough to allow visualization of fluctuations over duration of experiment |
[72,161– 163] |
| SAFIRe | Dark state lifetime |
Long lived dark state with red- shifted absorption |
[164,165] | |
| OLID, OPIOM | Photoswitching kinetics |
Photoswitching dynamics matched to modulation of excitation light |
[166,167] |
Resolving Biochemical Activities in Superresolution
It has become increasingly clear that biochemical activities within the cell are often spatially compartmentalized into regions, with sizes as small as tens of nanometers, known as micro- or nano-domains [75]. While a large number of FP-based indicators have been developed to track these biochemical events in living cells using standard fluorescence microscopy [76,77], there are now a growing number of examples in which fluorescent indicators have been paired with superresolution imaging methods to produce cellular maps of biochemical activities in high resolution.
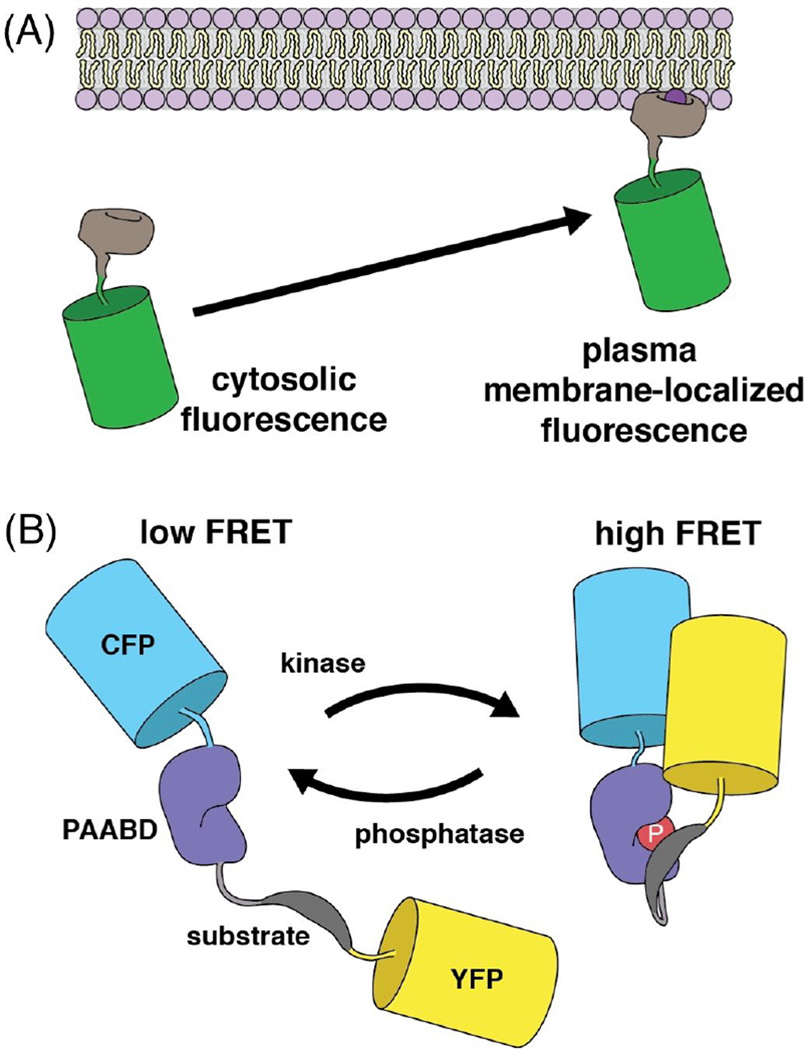
A typical fluorescent indicator consists of a “sensing element” to detect the target biochemical activity and a “reporting element” to translate the biochemical event into a change in the fluorescent readout [78]. Depending on the reporting element, the indicators can be categorized into different classes, such as those based on translocation of fluorescence (Figure 5A), change in Förster resonance energy transfer (FRET) efficiency (Figure 5B), and change in the fluorescence of a single fluorescent protein (Figure 6A).
Figure 5. Schematic representations of indicators for imaging of intracellular signaling activity.
(A) Translocation-based indicator design. (B) Intramolecular FRET-based design of a kinase activity indicator.
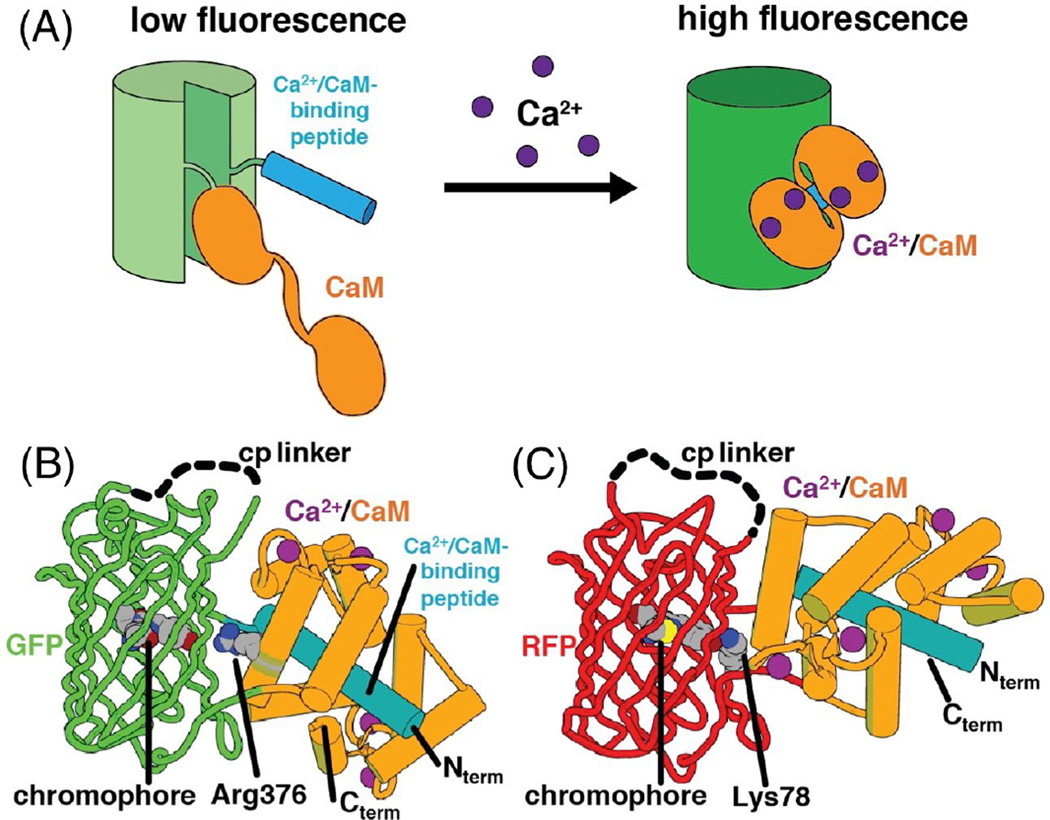
Figure 6. Single FP-based GECIs.
(A) Schematic representation of mechanism of response of a GCaMP-type Ca2+ indicator. (B) Structure of GCaMP6m (PDB ID 3WLD) [171]. Arg376 modulates the fluorescent response upon binding to Ca2+ by electrostatic stabilization of the anionic state of the chromophore. (C) Structure of R-GECO1 (PDB ID 4I2Y) [106]. Lys78 is proposed to act similarly to Arg376 of GCaMP6m for modulation of the Ca2+-dependent fluorescent response.
Our understanding of phosphoinositide (PI) dynamics has greatly benefited from a series of indicators designed to track the changes in various PI species in individual cells [79]. These probes, which generally consist of a standard FP (the reporting element) fused to a protein domain that specifically binds a PI (the sensing element), typically rely upon translocation of the probe to the specific membrane compartment to report on the generation of the PI species under study. Given the controversial model of lipid rafts and interest in probing lipid nano-domains in the plasma membrane [80], it is tempting to use these PI indicators to map PI distribution in the live-cell membrane in high resolution, but this task is not trivial. One of the complicating factors is that direct observation of lipid clusters at the nanometer length scale is hampered by the diffraction of light. To overcome this limitation, researchers have combined FP-based phosphoinositide indicators and superresolution imaging to quantify PIs in the plasma membrane and visualize their distributions. In a recent study [81], PALM was used to visualize and quantify phosphatidylinositol 3-phosphate (PI3P), a lipid involved in endocytosis and membrane transport. A photoswitchable FP, mEos2 [82], was fused to a PI3P-binding domain (a double FYVE domain of EEA1) to construct a superresolution-compatible PI3P indicator. Although colocalization experiments using different markers suggest mEos2 did not alter the localization pattern of the FYVE domain in this case despite its propensity to dimerize [69], the non-dimerizing variant mEos3.2 should provide a better alternative to eliminate any potential dimerization artifacts. In a separate study, van den Bogaart et al. used a phosphatidylinositol 4,5-bisphosphate (PIP2) indicator (the Pleckstrin Homology domain of protein lipase C delta fused to Citrine) in combination with STED microscopy to map PIP2 clusters on the plasma membrane of a cell [83].
Perhaps the most widely used class of indicators are those developed for tracking protein-protein interactions. While many of these indicators are based on FRET, protein fragment complementation [84] is another popular approach. Protein fragment complementation requires two non-fluorescent FP fragments that do not self-associate on their own, but that can recombine to form a functional FP if brought into close proximity with the help of an interacting pair of proteins. This property, which is the basis for Bimolecular Fluorescence Complementation (BiFC) and several related protein fragment complementation assays (PCAs), has been exploited to detect protein-protein interactions in living cells and even whole animals [85], although the effectively irreversible nature of BiFC has limited its application in tracking dynamic interactions. In order to generate high-resolution maps of specific protein-protein interactions, specific pairs of fragments have been identified for photoactivatable or photoswitchable proteins and used for imaging protein-protein interactions in superresolution [86–89]. For example, a BiFC-PALM approach was recently developed using PAmCherry1 [88]. Nickerson et al. studied interactions between the small GTPase Ras and its downstream effector Raf using this approach and observed nanoscale clustering and diffusion of individual KRas G12D/CRaf RBD (Ras-binding domain) complexes on the cell membrane [88]. The Zhang laboratory has recently described a strategy in which protein-protein interactions induce the reconstitution of fluorescent proteins capable of exhibiting single-molecule fluctuations suitable for Stochastic Optical Fluctuation Imaging (SOFI). Subsequently, spatial maps of these interactions can be resolved in super-resolution in living cells based on statistical analysis of the fluorescence fluctuations. This strategy, termed reconstituted fluorescence-based SOFI (refSOFI), was used to investigate the interaction between the endoplasmic reticulum (ER) Ca2+ sensor STIM1 and the pore-forming channel subunit ORAI1 at the ER-plasma membrane junction [87]. Limitations of these BiFC-based approaches include irreversibility and long maturation time ranging from 30 min to hours. It can be anticipated that new indicators with better temporal resolution and reversibility will continue to emerge to help reveal a more detailed map of biochemical activities in cells.
Genetically Encoded Calcium Ion Indicators
Paralleling the development of improved FPs and indicators of biochemical activities have been concerted efforts to develop improved genetically encoded calcium ion (Ca2+) indicators (GECIs). The first examples of single FP-based Ca2+ indicators (Figure 6A) were reported in 1999 [90], just two years after the debut of the FRET-based “cameleon”-type indicators [91]. Cameleon-type indicators produce a ratiometric fluorescent signal as a result of Ca2+-dependent changes in the efficiency of FRET from a blue-shifted donor to a red-shifted acceptor. In contrast, single FP-based indicators produce Ca2+-dependent fluorescent changes (typically, but not always, an intensiometric fluorescent signal) as a result of modulation of the chromophore’s proteinaceous environment. The first generation single FP-based indicators (“camgaroo”-type) were created through the clever insertion of calmodulin at position 145 of EYFP [90]. In camgaroo, the conformational change associated with binding of Ca2+ to calmodulin modulated the chromophore environment such that the pKa shifted to a lower value and the fluorescence intensity increased 7-fold. The second generation of single FP-based Ca2+ indicators, specifically the Pericam construct from Miyawaki [92] and the G-CaMP construct from Nakai [93], exploited a circularly permuted FP topology that had been first predicted by Tsien and coworkers [90]. These second generation indicators were composed of an FP that was circularly permuted such that the new termini were in close proximity to the chromophore. Fused to the termini were calmodulin and a calmodulin-binding peptide that underwent a Ca2+-dependent interaction and caused the requisite modulation of the chromophore environment.
Early efforts to further improve single FP-based Ca2+ indicators, specifically the G-CaMP design [93], were hampered by the lack of a protein structural model, or even a solid hypothesis about the mechanism by which the indicator functioned [94]. Two independent reports of the X-ray crystal structure of a second generation variant [95,96] served as a catalyst for future structure guided engineering efforts, soon facilitating the development of GCaMP3 [97]. GCaMP3 is generally considered to be the ‘breakthrough’ version that was practically useful for routine neuronal activity imaging in a variety of contexts, including transgenic mice [98]. A series of additional improved versions have since been reported [99–102]. The current state-of-the-art variants are the GCaMP6 (Figure 6B) series produced by the Genetically-Encoded Neuronal Indicator and Effector (GENIE) project at the Howard Hughes Medical Institute (HHMI) Janelia Research Campus [103]. Transgenic mice expressing fast and slow variants of GCaMP6 under a neuron-specific Thy-1 promoter are now available [104].
In addition to serving as the catalyst for rapid improvements in the GCaMP series, the X-ray crystal structures of GCaMP2 [95,96] also helped accelerate the development of variants with altered fluorescent hues. In 2011 the Campbell lab reported a series of new indicators, including blue, blue-green emission ratiometric, and red fluorescent variants, that they designated the GECO-series [105]. Parallel efforts from the GENIE project also led to a series of color variants that included blue, cyan, yellow, and red Ca2+ indicators [106]. The most promising new indicators to arise from these efforts were the two red fluorescent variants: the Campbell lab’s mApple-derived R-GECO1 (Figure 6C), and the GENIE project’s mRuby-derived RCaMP. Generally speaking, red fluorescent indicators are preferable to green ones due to the lower autofluorescence, lower phototoxicity, and greater tissue penetration associated with longer wavelength excitation. As with the GCaMP series, the red indicators have continued to be improved [107,108], with the latest variants (i.e., the R-GECO1-derived R-CaMP2 [109] and the GENIE project’s jRCaMP1a,b and jRGECO1a variants [110]) offering performance that approaches that of GCaMP6.
The toolbox of single FP-based indicators continues to expand in a number of directions, with other engineering efforts focused on making improved yellow indicators [111], a long Stokes-shift indicator [112], variants with lower Kd values that are suited for imaging Ca2+ in the endoplasmic reticulum [113,114], and photoconvertible variants that can be selectively “highlighted” by spatially defined conversion to a spectrally distinct fluorescence hue [115,116]. Interestingly, it has recently been reported that GCaMP6 itself can be “highlighted” by photoconversion to a red state that retains Ca2+ responsiveness [117]. An exciting twist on the utility of photoconvertible Ca2+ indicators is the recently reported CaMPARI variant [118]. CaMPARI is a GCaMP-type indicator that exhibits Ca2+-dependent photoconversion to a red state, and thus acts as an integrator of neuronal activity during a period of illumination.
FP-based Indicators of Transmembrane Voltage
While GECIs are the current workhorse of neuronal activity imaging, neuroscientists have long recognized the need for effective genetically encoded voltage indicators (GEVIs). Neurons process and transmit signals (an action potential) via changes in their transmembrane voltage. A negative-inside transmembrane voltage is established across the membrane by ion pumps and transporters, and can be modulated with millisecond kinetics by the action of neurotransmitter- and voltage-gated ion channels [119]. If voltage changes could be visualized in real time with a fluorescent indicator, then neuroscientists would be able to study questions such as how inputs at different synapses are integrated to induce action potential firing, and how neurons in a circuit fire in a coordinated manner. One major challenge for optical imaging of voltage changes is that a single action potential is complete within a few milliseconds and so image acquisition rates in the range of 100 to 1,000 Hz are essential [119]. The correspondingly short image acquisition times and high intensity excitation light sources necessitate that practically useful voltage indicators be particularly bright and photostable. Further complicating the use of voltage indicators is the fact that an indicator of membrane potential must be confined to the two-dimensional surface of the plasma membrane, rather than the three-dimensional volume of the cytoplasm. In contrast, elevated cytoplasmic Ca2+ levels persist for hundreds of milliseconds following an action potential [120], which enables relatively facile detection of these changes with much higher signal-to-noise than is typically achievable for voltage indicators.
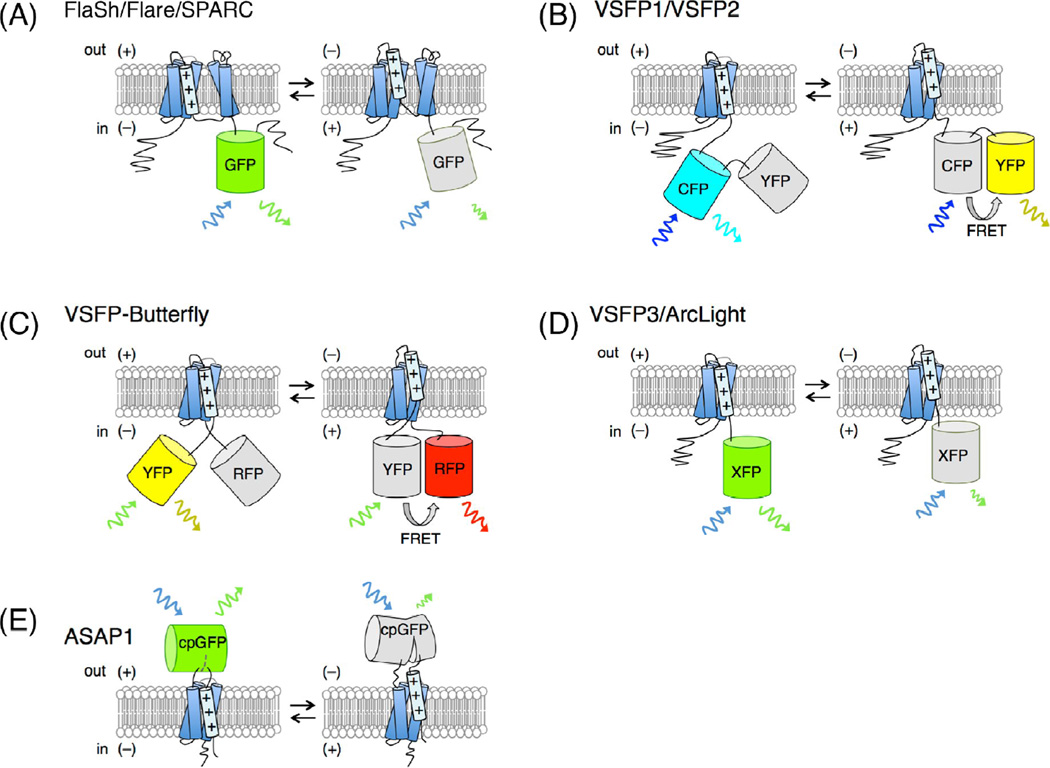
The field of GEVI development is as old as the field of GECI development, but has matured at a slower rate due to the more challenging nature of GEVI design (Figure 7) and optimization. The first GEVI, FlaSh, consisting of a GFP domain inserted in an intracellular segment of a voltage-gated potassium channel, was published in 1997 [121], the same year as the first GECI, Cameleon [91]. FlaSh exhibits a 5% decrease in GFP fluorescence upon membrane depolarization in the physiological range of −70 to +30 mV, but with rather slow kinetics. Subsequently, SPARC, a GFP insertion in an intracellular loop of a voltage-gated sodium channel, was found to show very small (<0.5%) changes in fluorescence upon depolarization but with very fast kinetics (0.8 ms activation time-constant) [122]. In both FlaSh and SPARC, the mechanism of voltage sensing by the GFP is unknown. In addition, a GEVI named VSFP1 was created by fusion of CFP and YFP in tandem at the C terminus of an isolated voltage-sensing domain (VSD) from a voltage-gated potassium channel [123]. VSFP1 shows a 2% increase in YFP fluorescence upon depolarization with very fast kinetics (0.7 ms activation time-constant), presumably because changes in the distribution of FP orientations upon voltage-induced VSD movements induce an increase in FRET. However, poor membrane expression of FlaSh, a FlaSh derivative named Flare, and VSFP1 in mammalian neurons prevented their actual use in neuroscience applications [124].
Figure 7. Designs of FP-based GEVIs.
(A) In FlaSh/Flare/SPARC, GFP is inserted in an intracellular segment of a homotetrameric potassium channel (FlaSh/Flare) or of a pseudotetrameric sodium channel (SPARC). Only one repeat of the 6-helix transmembrane motif is shown for clarity. (B) In VSFP1/2, a FRET pair is fused to the intracellular segment following a 4-helix VSD. (C) VSFP-Butterfly is composed of a FRET pair of FPs (YFP and RFP) fused to the termini of the VSD. (D) In VSFP3/ArcLight, a single FP is fused following the VSD. (E) In ASAP1, a circularly permuted GFP (cpGFP) is inserted into an extracellular loop of the VSD.
The discovery of voltage-sensing phosphatases in 2005 [125] provided a VSD that was well expressed at the membrane and also generated larger changes in the fluorescence of attached FP domains, serving as the basis for a series of increasingly effective voltage indicators of different architectures [126]. Fusion of this new VSD to CFP and YFP at the C terminus (VSFP2), or to one FP at the N terminus and another at the C terminus (VSFP-Butterfly), gave ~10% fluorescence changes with fast (1 – 3 ms) kinetics [127,128]. In addition, fusions of single FP domains at the C terminus (VSFP3s) showed small (1.6 – 3.5%) and fast (1.8 – 3.8 ms) fluorescence decreases upon depolarization [129]. ArcLight, a fusion of VSD and a pH-sensitive FP with an Ala-to-Asp mutation on the β-barrel surface, shows a larger (~35%) but slower (~10 ms) fluorescence decrease [130]. ASAP1, an insertion of a circularly permuted GFP in an extracellular loop of the VSD, shows both large (~25%) and fast (2 ms) fluorescence decreases upon depolarization [131]. Finally, Abdelfattah et al. recently reported FlicR1, a bright and fast red fluorescent voltage indicator based on fusion of a circularly permuted RFP to the C terminus of a VSD, that exhibits fluorescence increases of ~3% for a single action potential [132].
Yet another GEVI design is based on the genetic fusion of an FP to an opsin protein. Here, the FP serves as a FRET donor to the opsin [133]. Depolarization increases opsin absorption at orange wavelengths, increasing FRET from the FP and decreasing FP brightness [134,135]. Among these “FRET-opsin” GEVIs, the brightest and fastest are the Ace-mNeon series [136].
The utilization of FP-based GEVIs is still in its infancy. VSFP-Butterfly was used to visualize rapid synchronized voltage responses in neuronal populations in the mouse sensory cortex [128]. ArcLight detected odorant-induced depolarizations and hyperpolarizing responses (which are undetectable with calcium sensors) in living flies [137]. ASAP2f was used to deduce differences in voltage-calcium coupling in the fly visual system [138]. Ace-mNeon has been used to visualize subcellular voltage changes in mouse cortex and to measure voltage propagation rates in fly olfactory neurons [136]. A remaining challenge is to use FP-based GEVIs for temporally precise monitoring of voltage transients in individual neurons in the mammalian brain, where two-photon excitation would be helpful for background suppression and fast-scanning approaches will be required for temporal resolution.
Optogenetics with FPs
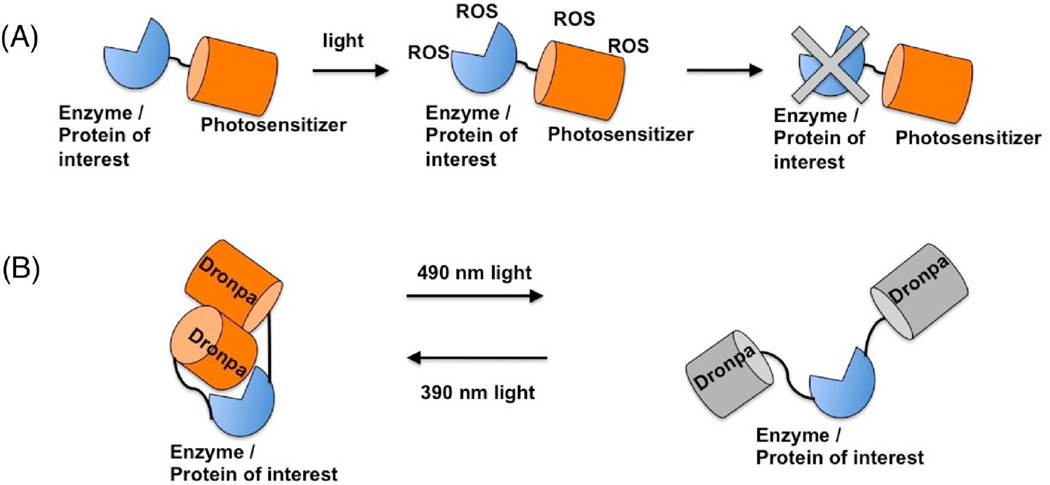
An optogenetic actuator is a genetically encoded protein that undergoes an illumination-dependent change in function that induces, disrupts, or otherwise changes, a cellular function. While channelrhodopsin-2, the blue-light activated cation channel, is often considered the archetypical optogenetic actuator [139], FPs were actually some of the earliest optogenetic actuators ever developed. The earliest examples of FP-based optogenetics were based on the principles of chromophore (which could also be a fluorophore) assisted light inactivation (CALI) (Figure 8A). The mechanism of CALI involves the photochemical generation of reactive oxygen species (ROS) by the triplet state of the excited chromophore [140,141]. ROS generated during illumination can diffuse over short distances and oxidize specific amino acid residues of nearby proteins, leading to cross-linking and photo-induced cleavage [142]. Due to the reactive nature and short lifetime of the ROS, CALI is typically specific to the protein or protein complex to which the chromophore is tethered. This photo-destructive approach can be used to irreversibly knock-out protein functions and disrupt cellular pathways. This CALI effect was observed with EGFP fusion proteins and was used to inhibit galactosidase [143], focal adhesion kinase [144], MyoII regulatory light chain [145], and Connexin 43 [146]. EGFP is a relatively poor mediator of CALI due to the protected environment of the chromophore embedded within the β-barrel, and the fact that EGFP was evolved to have high fluorescence quantum yield and a limited triplet state [143]. Efforts to increase the ROS generation by FP under illumination has produced KillerRed [147,148], a dimeric red FP that is capable of generating superoxide radical anion during green light illumination [149,150]. As the dimeric nature of KillerRed complicates its use in CALI experiments as a fusion protein, Takemoto et al. developed a monomerized version of KillerRed, named SuperNova, which can be used for CALI and is less likely to perturb the localization of a fusion protein [151].
Figure 8. Two current approaches to control protein and/or cellular functions with FPs.
(A) FPs capable of generating ROS during illumination (photosensitizer), such as EGFP, miniSOG, KillerRed and SuperNova, can be used to disrupt the function of protein(s) of interest tethered to the photosensitizer. This is the principle of CALI. (B) Light controlled, reversible oligomerization of Dronpa can sterically block the catalytic or functional site on fusion protein. This oligomerization process is controlled with two different wavelengths of light.
MiniSOG is an engineered variant of a flavin mononucleotide (FMN)-binding Light, Oxygen, Voltage (LOV) domain from Arabidopsis thaliana that generates ROS during blue light illumination [152]. CALI with miniSOG was used to inhibit synaptic proteins [153,154], disrupt the CaMKII analogue UNC-43 in C. elegans [155], and in experiments to ablate cells [156]. SOPP, a newer variant engineered from miniSOG, is reported to have ~ 8-fold improved singlet oxygen quantum yield, but is yet to be tested in living cells [157]. A newly discovered LOV domain from Pseudomonas putida (Pp2FbFP L30M) is reported to have singlet oxygen quantum yield 3-fold higher than miniSOG [158], which may be a suitable template for future development of flavin-binding FPs as optogenetic tools to alter cellular function.
Other than utilizing FPs for CALI and cell ablation experiments, the utilization of FPs to control the activities of intracellular enzymes and proteins has been demonstrated [159]. For example, an engineered variant of the photoswitchable Dronpa FP enables switching between a tetrameric and monomeric state by illumination with different wavelengths of light (Figure 8B). By using light to control the oligomerization state of this Dronpa mutant, it is possible to block the active site of attached enzymes and proteases, leading to the optogenetic control of protein functions. Currently, its use may be limited due to the concentration-dependent nature of the association; however, the light induced oligomerization and dissociation of Dronpa can be further exploited to develop novel methods of optogenetic control of intracellular pathways.
With the rapid adoption of optogenetic approaches and recent advances towards achieving all optical observation and manipulation of biological pathways [160], the potential utilization of FPs for simultaneous observation and manipulation may further revolutionize the way intracellular events are studied in the coming decades.
CONCLUDING REMARKS
The authors of this review are enthusiastic developers and users of FP technology, and one of us (RYT) was among the very first researchers to recognize the utility of engineered FPs as tools for fluorescence imaging. In the mid-1990s it was surprising and gratifying that color variants of avGFP could be created. We have been astounded by the myriad applications that have been enabled by FP technology over the last two decades. We have no reason to fear that FPs have yet given up all their secrets, and it is a safe bet that the coming years will see yet further advances. While some of the general directions for these advances are apparent (e.g., FPs from new organisms, near-infrared fluorescence, new classes of indicators, and new interactions with photons), history has taught us to expect the unexpected.
OUTSTANDING QUESTIONS BOX.
What are the molecular features that control photophysical properties such as photobleaching and photochromism and how can we engineer FPs in which we can precisely control these properties?
Can we engineer BphP-derived IFPs to have brightness in cells that is similar or greater than GFP? How far can IFPs be redshifted? Can we develop IFP-based indicators for Ca2+, membrane potential, and other biochemical activities?
Can monomeric and spectrally unique smURFP variants, that retain high brightness and stability, be engineered? Can photostability be further enhanced?
How might a transgenic animal be treated or genetically modified to increase the amount of available BV? Can an optimal BV analog be created to penetrate both the cellular membrane and/or the blood brain barrier while remaining in the blood for long periods of time?
Are there naturally occurring FPs, homologous to avGFP or otherwise, with properties that have not yet been observed in nature or in the lab?
What are the limits in terms of brightness, fluorescent response, and kinetics for single FP-based GECIs and GEVIs? Can the lessons learned during the optimization of GCaMP be applied to the GEVIs and other indicators?
Can new designs of FP-based biosensors increase the range of biochemical events that are amenable to fluorescence monitoring and visualization?
Can we develop FPs variants to both observe and manipulate cellular functions simultaneously in the cell using fully optical approaches?
TRENDS BOX.
Monomeric red and far-red fluorescent proteins (FPs), and indicators, now perform nearly as well as the best green FPs (and indicators).
Reversible and irreversible photochromism in FPs can be exploited to increase optical resolution and improve contrast compared to traditional fluorescence microscopy.
Infrared FPs (IFPs) are becoming ever more useful as labels for various proteins that allow correct localization and whole animal imaging. IFPs can serve as an additional fluorescent “color” for simultaneous imaging with visible FP-labeled proteins.
Bacterial phytochrome (BphP)-based IFPs provide a new scaffold for engineering fluorogenic indicators, which are ideal to visualize spatiotemporal dynamics of cell signaling in vivo.
Small Ultra-Red FP (SmURFP) is the brightest far-red non-prototypical FP (comparable to EGFP) and is extremely photostable. SmURFP may prove particularly useful as a photostable FP for superresolution imaging and as a FRET acceptor for biosensing applications.
The engineering of new fluorescent indicators that combine features of prototypical FP-based indicators with photochromic proteins can reveal the cellular maps of biochemical activities in superresolution.
FPs can be used as optogenetic actuators to manipulate cellular and protein functions through chromophore assisted light inactivation or light-controlled protein oligomerization.
GLOSSARY
- Intensiometric fluorescent signal
a change in fluorescence intensity (i.e., either an increase or decrease) at a single wavelength
- Intrinsic fluorescent brightness
the product of the molecular extinction coefficient (ε) and the fluorescence quantum yield (Φ)
- Optogenetic actuator
A genetically encoded protein that undergoes an illumination-dependent changes in function that, induces, disrupts, or otherwise changes, a cellular function
- Photo-Activated Localization Microscopy (PALM) and Stochastic Optical Reconstruction Microscopy (STORM)
Superresolution imaging modality in which an image is constructed from a multitude of single fluorophore localizations, where each localization is determined to a resolution higher than the diffraction limit
- Ratiometric fluorescent signal
a change in the ratio of fluorescence intensity at one wavelength relative to the fluorescent intensity at a second wavelength
- Stimulated Emission Depletion (STED)
A superresolution imaging modality in which a first laser beam is used to excite fluorescence and a second donut-shaped laser beam is used to confine the size of the excitation spot to smaller than the diffraction limit
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Chalfie M, et al. Green Fluorescent Protein as a Marker for Gene-Expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 2.Cubitt AB, et al. Understanding, improving and using green fluorescent proteins. Trends Biochem. Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 3.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 4.Matz MV, et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 5.Campbell RE, et al. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 7.Wiedenmann J, et al. A far-red fluorescent protein with fast maturation and reduced oligomerization tendency from Entacmaea quadricolor (Anthozoa, Actinaria) Proc. Natl. Acad. Sci. U. S. A. 2002;99:11646–11651. doi: 10.1073/pnas.182157199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merzlyak EM, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 9.Shcherbo D, et al. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 10.Kredel S, et al. mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS ONE. 2009;4:e4391. doi: 10.1371/journal.pone.0004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drepper T, et al. Reporter proteins for in vivo fluorescence without oxygen. Nat Biotechnol. 2007;25:443–445. doi: 10.1038/nbt1293. [DOI] [PubMed] [Google Scholar]
- 12.Fischer AJ, Lagarias JC. Harnessing phytochrome’s glowing potential. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17334–17339. doi: 10.1073/pnas.0407645101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu X, et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumagai A, et al. A bilirubin-inducible fluorescent protein from eel muscle. Cell. 2013;153:1602–1611. doi: 10.1016/j.cell.2013.05.038. [DOI] [PubMed] [Google Scholar]
- 15.Campbell RE, et al. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bajar BT, et al. Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci Rep. 2016;6:20889. doi: 10.1038/srep20889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaner NC, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam AJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman S, et al. The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc Natl Acad Sci U S A. 2008;105:20038–20043. doi: 10.1073/pnas.0807551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee A, et al. Engineering and characterization of new LOV-based fluorescent proteins from Chlamydomonas reinhardtii and Vaucheria frigida. ACS Synth Biol. 2015;4:371–377. doi: 10.1021/sb500237x. [DOI] [PubMed] [Google Scholar]
- 21.Kimura S, et al. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 22.Katayama H, et al. GFP-like proteins stably accumulate in lysosomes. Cell Struct. Funct. 2008;33:1–12. doi: 10.1247/csf.07011. [DOI] [PubMed] [Google Scholar]
- 23.Shemiakina II, et al. A monomeric red fluorescent protein with low cytotoxicity. Nat. Commun. 2012;3:1204. doi: 10.1038/ncomms2208. [DOI] [PubMed] [Google Scholar]
- 24.Costantini LM, et al. A palette of fluorescent proteins optimized for diverse cellular environments. Nat. Commun. 2015;6:7670. doi: 10.1038/ncomms8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costantini LM, et al. Assessing the tendency of fluorescent proteins to oligomerize under physiologic conditions. Traffic. 2012;13:643–649. doi: 10.1111/j.1600-0854.2012.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cranfill PJ, et al. Quantitative assessment of fluorescent proteins. Nat. Methods. 2016;13:557–562. doi: 10.1038/nmeth.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weissleder R, Ntziachristos V. Shedding light onto live molecular targets. Nat. Med. 2003;9:123–128. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- 28.Jöbsis FF. Noninvasive, infrared monitoring of cerebral and myocardial oxygen sufficiency and circulatory parameters. Science. 1977;198:1264–1267. doi: 10.1126/science.929199. [DOI] [PubMed] [Google Scholar]
- 29.Karniol B, et al. Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem. J. 2005;392:103–116. doi: 10.1042/BJ20050826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auldridge ME, Forest KT. Bacterial phytochromes: more than meets the light. Crit Rev Biochem Mol Biol. 2011;46:67–88. doi: 10.3109/10409238.2010.546389. [DOI] [PubMed] [Google Scholar]
- 31.Murphy JT, Lagarias JC. The phytofluors: a new class of fluorescent protein probes. Curr. Biol. 1997;7:870–876. doi: 10.1016/s0960-9822(06)00375-7. [DOI] [PubMed] [Google Scholar]
- 32.Filonov GS, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu D, et al. An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat. Commun. 2014;5:3626. doi: 10.1038/ncomms4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auldridge ME, et al. Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J. Biol. Chem. 2012;287:7000–7009. doi: 10.1074/jbc.M111.295121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods. 2013;10:751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu D, et al. A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat. Methods. 2015;12:763–765. doi: 10.1038/nmeth.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu D, et al. Rational design of a monomeric and photostable far-red fluorescent protein for fluorescence imaging in vivo. Protein Sci. 2016;25:308–315. doi: 10.1002/pro.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu ML, et al. Heme oxygenase-1 in inflammation and cardiovascular disease. Am. J. Cardiovasc. Dis. 2011;1:150–158. [PMC free article] [PubMed] [Google Scholar]
- 39.Sedlak TW, et al. Bilirubin and glutathione have complementary antioxidant and cytoprotective roles. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5171–5176. doi: 10.1073/pnas.0813132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.To TL, et al. Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proc. Natl. Acad. Sci. U. S. A. 2015;112:3338–3343. doi: 10.1073/pnas.1502857112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez EA, et al. A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein. Nat. Methods. 2016 doi: 10.1038/nmeth.3935. Published online 01 August 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marx V. Probes: seeing in the near infrared. Nat. Methods. 2014;11:717–720. doi: 10.1038/nmeth.3001. [DOI] [PubMed] [Google Scholar]
- 43.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 44.Hayashi S, Toda Y. A novel fluorescent protein purified from eel muscle. Fish. Sci. 2009;75:1461–1469. [Google Scholar]
- 45.Storch J, Thumser AE. The fatty acid transport function of fatty acid-binding proteins. Biochim. Biophys. Acta. 2000;1486:28–44. doi: 10.1016/s1388-1981(00)00046-9. [DOI] [PubMed] [Google Scholar]
- 46.Schaap FG, et al. Evolution of the family of intracellular lipid binding proteins in vertebrates. Mol. Cell. Biochem. 2002;239:69–77. [PubMed] [Google Scholar]
- 47.Zimmerman AW, Veerkamp JH. New insights into the structure and function of fatty acid-binding proteins. Cell. Mol. Life Sci. 2002;59:1096–1116. doi: 10.1007/s00018-002-8490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapitulnik J. Bilirubin: an endogenous product of heme degradation with both cytotoxic and cytoprotective properties. Mol. Pharmacol. 2004;66:773–779. doi: 10.1124/mol.104.002832. [DOI] [PubMed] [Google Scholar]
- 49.Notter MF, Kendig JW. Differential sensitivity of neural cells to bilirubin toxicity. Exp. Neurol. 1986;94:670–682. doi: 10.1016/0014-4886(86)90246-3. [DOI] [PubMed] [Google Scholar]
- 50.Wennberg RP, et al. Toward understanding kernicterus: a challenge to improve the management of jaundiced newborns. Pediatrics. 2006;117:474–485. doi: 10.1542/peds.2005-0395. [DOI] [PubMed] [Google Scholar]
- 51.Erapaneedi R, et al. A novel family of fluorescent hypoxia sensors reveal strong heterogeneity in tumor hypoxia at the cellular level. EMBO J. 2016;35:102–113. doi: 10.15252/embj.201592775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.To TL, et al. Structure-guided design of a reversible fluorogenic reporter of protein-protein interactions. Protein Sci. 2016;25:748–753. doi: 10.1002/pro.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Navarro R, et al. A Novel Destabilizing Domain Based on a Small-Molecule Dependent Fluorophore. ACS Chem Biol. 2016;11:2101–2104. doi: 10.1021/acschembio.6b00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dickson RM, et al. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 55.Habuchi S, et al. Evidence for the isomerization and decarboxylation in the photoconversion of the red fluorescent protein DsRed. J. Am. Chem. Soc. 2005;127:8977–8984. doi: 10.1021/ja047023o. [DOI] [PubMed] [Google Scholar]
- 56.Andresen M, et al. Structure and mechanism of the reversible photoswitch of a fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13070–13074. doi: 10.1073/pnas.0502772102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi X, et al. Ultrafast Excited-State Dynamics in the Green Fluorescent Protein Variant S65T/H148D. 2. Unusual Photophysical Properties. Biochemistry. 2007;46:12014–12025. doi: 10.1021/bi700904a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haupts U, et al. Dynamics of fluorescence fluctuations in green fluorescent protein observed by fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13573–13578. doi: 10.1073/pnas.95.23.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Konold P, et al. Hydrogen bond flexibility correlates with Stokes shift in mPlum variants. J. Phys. Chem. B. 2014;118:2940–2948. doi: 10.1021/jp412371y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Donnert G, et al. Major signal increase in fluorescence microscopy through dark-state relaxation. Nat. Methods. 2007;4:81–86. doi: 10.1038/nmeth986. [DOI] [PubMed] [Google Scholar]
- 61.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 62.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 63.Willig KI, et al. Nanoscale resolution in GFP-based microscopy. Nat. Methods. 2006;3:721–723. doi: 10.1038/nmeth922. [DOI] [PubMed] [Google Scholar]
- 64.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 65.Rust MJ, et al. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dean KM, et al. Analysis of red-fluorescent proteins provides insight into dark-state conversion and photodegradation. Biophys. J. 2011;101:961–969. doi: 10.1016/j.bpj.2011.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subach FV, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang M, et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat. Methods. 2012;9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- 69.Wang S, et al. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc. Natl. Acad. Sci. U. S. A. 2014;111:8452–8457. doi: 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nienhaus K, Nienhaus GU. Fluorescent proteins for live-cell imaging with super-resolution. Chem. Soc. Rev. 2014;43:1088–1106. doi: 10.1039/c3cs60171d. [DOI] [PubMed] [Google Scholar]
- 71.Grotjohann T, et al. rsEGFP2 enables fast RESOLFT nanoscopy of living cells. Elife. 2012;1:e00248. doi: 10.7554/eLife.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang X, et al. Development of a reversibly switchable fluorescent protein for super-resolution optical fluctuation imaging (SOFI) ACS Nano. 2015;9:2659–2667. doi: 10.1021/nn5064387. [DOI] [PubMed] [Google Scholar]
- 73.Subach FV, et al. Directed molecular evolution to design advanced red fluorescent proteins. Nat. Methods. 2011;8:1019–1026. doi: 10.1038/nmeth.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dean KM, et al. High-speed multiparameter photophysical analyses of fluorophore libraries. Anal. Chem. 2015;87:5026–5030. doi: 10.1021/acs.analchem.5b00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cambi A, Lidke DS. Nanoscale membrane organization: where biochemistry meets advanced microscopy. ACS Chem. Biol. 2012;7:139–149. doi: 10.1021/cb200326g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miyawaki A, Niino Y. Molecular spies for bioimaging--fluorescent protein-based probes. Mol. Cell. 2015;58:632–643. doi: 10.1016/j.molcel.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 77.Newman RH, et al. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem. Rev. 2011;111:3614–3666. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sample V, et al. Genetically encoded molecular probes to visualize and perturb signaling dynamics in living biological systems. J. Cell Sci. 2014;127:1151–1160. doi: 10.1242/jcs.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Halet G. Imaging phosphoinositide dynamics using GFP-tagged protein domains. Biol. Cell. 2005;97:501–518. doi: 10.1042/BC20040080. [DOI] [PubMed] [Google Scholar]
- 80.Owen DM, et al. The lipid raft hypothesis revisited--new insights on raft composition and function from super-resolution fluorescence microscopy. Bioessays. 2012;34:739–747. doi: 10.1002/bies.201200044. [DOI] [PubMed] [Google Scholar]
- 81.Puchner EM, et al. Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory. Proc. Natl. Acad. Sci. U. S. A. 2013;110:16015–16020. doi: 10.1073/pnas.1309676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McKinney SA, et al. A bright and photostable photoconvertible fluorescent protein. Nat. Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van den Bogaart G, et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479:552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miller KE, et al. Bimolecular Fluorescence Complementation (BiFC) Analysis: Advances and Recent Applications for Genome-Wide Interaction Studies. J. Mol. Biol. 2015;427:2039–2055. doi: 10.1016/j.jmb.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Han Y, et al. In vivo imaging of protein-protein and RNA-protein interactions using novel far-red fluorescence complementation systems. Nucleic Acids Res. 2014;42:e103. doi: 10.1093/nar/gku408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu Z, et al. Super-resolution imaging and tracking of protein-protein interactions in sub-diffraction cellular space. Nat. Commun. 2014;5:4443. doi: 10.1038/ncomms5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hertel F, et al. RefSOFI for Mapping Nanoscale Organization of Protein-Protein Interactions in Living Cells. Cell Rep. 2016;14:390–400. doi: 10.1016/j.celrep.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nickerson A, et al. Photoactivated localization microscopy with bimolecular fluorescence complementation (BiFC-PALM) for nanoscale imaging of protein-protein interactions in cells. PLoS ONE. 2014;9:e100589. doi: 10.1371/journal.pone.0100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xia P, et al. Superresolution imaging reveals structural features of EB1 in microtubule plus-end tracking. Mol. Biol. Cell. 2014;25:4166–4173. doi: 10.1091/mbc.E14-06-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baird GS, et al. Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 92.Nagai T, et al. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc. Natl. Acad. Sci. U. S. A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nakai J, et al. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat. Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 94.Tallini YN, et al. Imaging cellular signals in the heart in vivo: Cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4753–4758. doi: 10.1073/pnas.0509378103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Q, et al. Structural basis for calcium sensing by GCaMP2. Structure. 2008;16:1817–1827. doi: 10.1016/j.str.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Akerboom J, et al. Crystal structures of the GCaMP calcium sensor reveal the mechanism of fluorescence signal change and aid rational design. J. Biol. Chem. 2009;284:6455–6464. doi: 10.1074/jbc.M807657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zariwala HA, et al. A Cre-dependent GCaMP3 reporter mouse for neuronal imaging in vivo. J. Neurosci. 2012;32:3131–3141. doi: 10.1523/JNEUROSCI.4469-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Muto A, et al. Genetic visualization with an improved GCaMP calcium indicator reveals spatiotemporal activation of the spinal motor neurons in zebrafish. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5425–5430. doi: 10.1073/pnas.1000887108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Akerboom J, et al. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. J. Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun XR, et al. Fast GCaMPs for improved tracking of neuronal activity. Nat. Commun. 2013;4:2170. doi: 10.1038/ncomms3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Muto A, et al. Real-time visualization of neuronal activity during perception. Curr. Biol. 2013;23:307–311. doi: 10.1016/j.cub.2012.12.040. [DOI] [PubMed] [Google Scholar]
- 103.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dana H, et al. Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo. PLoS ONE. 2014;9:e108697. doi: 10.1371/journal.pone.0108697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao Y, et al. An expanded palette of genetically encoded Ca2+ indicators. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Akerboom J, et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ohkura M, et al. An improved genetically encoded red fluorescent Ca2+ indicator for detecting optically evoked action potentials. PLoS ONE. 2012;7:e39933. doi: 10.1371/journal.pone.0039933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu J, et al. Improved orange and red Ca2+ indicators and photophysical considerations for optogenetic applications. ACS Chem. Neurosci. 2013;4:963–972. doi: 10.1021/cn400012b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Inoue M, et al. Rational design of a high-affinity, fast, red calcium indicator R-CaMP2. Nat. Methods. 2015;12:64–70. doi: 10.1038/nmeth.3185. [DOI] [PubMed] [Google Scholar]
- 110.Dana H, et al. Sensitive red protein calcium indicators for imaging neural activity. Elife. 2016;5:e12727. doi: 10.7554/eLife.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao Y, et al. Microfluidic cell sorter-aided directed evolution of a protein-based calcium ion indicator with an inverted fluorescent response. Integr. Biol. (Camb.) 2014;6:714–725. doi: 10.1039/c4ib00039k. [DOI] [PubMed] [Google Scholar]
- 112.Wu J, et al. A long Stokes shift red fluorescent Ca2+ indicator protein for two-photon and ratiometric imaging. Nat. Commun. 2014;5:5262. doi: 10.1038/ncomms6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu J, et al. Red fluorescent genetically encoded Ca2+ indicators for use in mitochondria and endoplasmic reticulum. Biochem. J. 2014;464:13–22. doi: 10.1042/BJ20140931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Suzuki J, et al. Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nat. Commun. 2014;5:4153. doi: 10.1038/ncomms5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hoi H, et al. Highlightable Ca2+ indicators for live cell imaging. J. Am. Chem. Soc. 2013;135:46–49. doi: 10.1021/ja310184a. [DOI] [PubMed] [Google Scholar]
- 116.Berlin S, et al. Photoactivatable genetically encoded calcium indicators for targeted neuronal imaging. Nat. Methods. 2015;12:852–858. doi: 10.1038/nmeth.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ai M, et al. Green-to-Red Photoconversion of GCaMP. PLoS ONE. 2015;10:e0138127. doi: 10.1371/journal.pone.0138127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fosque BF, et al. Neural circuits. Labeling of active neural circuits in vivo with designed calcium integrators. Science. 2015;347:755–760. doi: 10.1126/science.1260922. [DOI] [PubMed] [Google Scholar]
- 119.Bean BP. The action potential in mammalian central neurons. Nat. Rev. Neurosci. 2007;8:451–465. doi: 10.1038/nrn2148. [DOI] [PubMed] [Google Scholar]
- 120.Broussard GJ, et al. Monitoring activity in neural circuits with genetically encoded indicators. Front. Mol. Neurosci. 2014;7:97. doi: 10.3389/fnmol.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron. 1997;19:735–741. doi: 10.1016/s0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- 122.Ataka K, Pieribone VA. A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys. J. 2002;82:509–516. doi: 10.1016/S0006-3495(02)75415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sakai R, et al. Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur. J. Neurosci. 2001;13:2314–2318. doi: 10.1046/j.0953-816x.2001.01617.x. [DOI] [PubMed] [Google Scholar]
- 124.Baker BJ, et al. Three fluorescent protein voltage sensors exhibit low plasma membrane expression in mammalian cells. J. Neurosci. Methods. 2007;161:32–38. doi: 10.1016/j.jneumeth.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 125.Murata Y, et al. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 126.St-Pierre F, et al. Designs and sensing mechanisms of genetically encoded fluorescent voltage indicators. Curr. Opin. Chem. Biol. 2015;27:31–38. doi: 10.1016/j.cbpa.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lundby A, et al. Biophysical characterization of the fluorescent protein voltage probe VSFP2.3 based on the voltage-sensing domain of Ci-VSP. Eur. Biophys. J. 2010;39:1625–1635. doi: 10.1007/s00249-010-0620-0. [DOI] [PubMed] [Google Scholar]
- 128.Akemann W, et al. Imaging neural circuit dynamics with a voltage-sensitive fluorescent protein. J. Neurophysiol. 2012;108:2323–2337. doi: 10.1152/jn.00452.2012. [DOI] [PubMed] [Google Scholar]
- 129.Perron A, et al. Red-shifted voltage-sensitive fluorescent proteins. Chem. Biol. 2009;16:1268–1277. doi: 10.1016/j.chembiol.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jin L, et al. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 2012;75:779–785. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.St-Pierre F, et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 2014;17:884–889. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Abdelfattah AS, et al. A Bright and Fast Red Fluorescent Protein Voltage Indicator That Reports Neuronal Activity in Organotypic Brain Slices. J. Neurosci. 2016;36:2458–2472. doi: 10.1523/JNEUROSCI.3484-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bayraktar H, et al. Ultrasensitive measurements of microbial rhodopsin photocycles using photochromic FRET. Photochem. Photobiol. 2012;88:90–97. doi: 10.1111/j.1751-1097.2011.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gong Y, et al. Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors. Nat. Commun. 2014;5:3674. doi: 10.1038/ncomms4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zou P, et al. Bright and fast multicoloured voltage reporters via electrochromic FRET. Nat. Commun. 2014;5:4625. doi: 10.1038/ncomms5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gong Y, et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science. 2015;350:1361–1366. doi: 10.1126/science.aab0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cao G, et al. Genetically Targeted Optical Electrophysiology in Intact Neural Circuits. Cell. 2013;154:904–913. doi: 10.1016/j.cell.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang HH, et al. Subcellular Imaging of Voltage and Calcium Signals Reveals Neural Processing In Vivo. Cell. 2016;166:245–257. doi: 10.1016/j.cell.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Boyden ES, et al. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 140.Jay DG. Selective destruction of protein function by chromophore-assisted laser inactivation. Proc. Natl. Acad. Sci. U. S. A. 1988;85:5454–5458. doi: 10.1073/pnas.85.15.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Beck S, et al. Fluorophore-assisted light inactivation: a high-throughput tool for direct target validation of proteins. Proteomics. 2002;2:247–255. doi: 10.1002/1615-9861(200203)2:3<247::aid-prot247>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 142.McLean MA, et al. Mechanism of chromophore assisted laser inactivation employing fluorescent proteins. Anal. Chem. 2009;81:1755–1761. doi: 10.1021/ac801663y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Surrey T, et al. Chromophore-assisted light inactivation and self-organization of microtubules and motors. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4293–4298. doi: 10.1073/pnas.95.8.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rajfur Z, et al. Dissecting the link between stress fibres and focal adhesions by CALI with EGFP fusion proteins. Nat. Cell Biol. 2002;4:286–293. doi: 10.1038/ncb772. [DOI] [PubMed] [Google Scholar]
- 145.Monier B, et al. An actomyosin-based barrier inhibits cell mixing at compartmental boundaries in Drosophila embryos. Nat. Cell Biol. 2010;12:60–65. doi: 10.1038/ncb2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tanabe T, et al. Multiphoton excitation-evoked chromophore-assisted laser inactivation using green fluorescent protein. Nat. Methods. 2005;2:503–505. doi: 10.1038/nmeth770. [DOI] [PubMed] [Google Scholar]
- 147.Bulina ME, et al. A genetically encoded photosensitizer. Nat. Biotechnol. 2006;24:95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 148.Bulina ME, et al. Chromophore-assisted light inactivation (CALI) using the phototoxic fluorescent protein KillerRed. Nat. Protoc. 2006;1:947–953. doi: 10.1038/nprot.2006.89. [DOI] [PubMed] [Google Scholar]
- 149.Vegh RB, et al. Reactive oxygen species in photochemistry of the red fluorescent protein “Killer Red”. Chem. Commun. (Camb.) 2011;47:4887–4889. doi: 10.1039/c0cc05713d. [DOI] [PubMed] [Google Scholar]
- 150.Wang Y, et al. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy. 2012;8:1462–1476. doi: 10.4161/auto.21211. [DOI] [PubMed] [Google Scholar]
- 151.Takemoto K, et al. SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation. Sci. Rep. 2013;3:2629. doi: 10.1038/srep02629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shu X, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lin JY, et al. Optogenetic inhibition of synaptic release with chromophore-assisted light inactivation (CALI) Neuron. 2013;79:241–253. doi: 10.1016/j.neuron.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhou K, et al. Position of UNC-13 in the active zone regulates synaptic vesicle release probability and release kinetics. Elife. 2013;2:e01180. doi: 10.7554/eLife.01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hoerndli FJ, et al. Neuronal Activity and CaMKII Regulate Kinesin-Mediated Transport of Synaptic AMPARs. Neuron. 2015;86:457–474. doi: 10.1016/j.neuron.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Qi YB, et al. Photo-inducible cell ablation in Caenorhabditis elegans using the genetically encoded singlet oxygen generating protein miniSOG. Proc. Natl. Acad. Sci. U. S. A. 2012;109:7499–7504. doi: 10.1073/pnas.1204096109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Westberg M, et al. Rational design of an efficient, genetically encodable, protein-encased singlet oxygen photosensitizer. J. Am. Chem. Soc. 2015;137:1632–1642. doi: 10.1021/ja511940j. [DOI] [PubMed] [Google Scholar]
- 158.Torra J, et al. Singlet oxygen photosensitisation by the fluorescent protein Pp2FbFP L30M, a novel derivative of Pseudomonas putida flavin-binding Pp2FbFP. Photochem. Photobiol. Sci. 2015;14:280–287. doi: 10.1039/c4pp00338a. [DOI] [PubMed] [Google Scholar]
- 159.Zhou XX, et al. Optical control of protein activity by fluorescent protein domains. Science. 2012;338:810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hochbaum DR, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods. 2014;11:825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dertinger T, et al. Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI) Proc. Natl. Acad. Sci. U. S. A. 2009;106:22287–22292. doi: 10.1073/pnas.0907866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dedecker P, et al. Widely accessible method for superresolution fluorescence imaging of living systems. Proc. Natl. Acad. Sci. U. S. A. 2012;109:10909–10914. doi: 10.1073/pnas.1204917109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Geissbuehler S, et al. Live-cell multiplane three-dimensional super-resolution optical fluctuation imaging. Nat. Commun. 2014;5:5830. doi: 10.1038/ncomms6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Richards CI, et al. Synchronously amplified fluorescence image recovery (SAFIRe) J. Phys. Chem. B. 2010;114:660–665. doi: 10.1021/jp909167j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jablonski AE, et al. Signal Discrimination Between Fluorescent Proteins in Live Cells by Long-wavelength Optical Modulation. J. Phys. Chem. Lett. 2012;3:3585–3591. doi: 10.1021/jz3016414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Marriott G, et al. Optical lock-in detection imaging microscopy for contrast-enhanced imaging in living cells. Proc Natl Acad Sci U S A. 2008;105:17789–17794. doi: 10.1073/pnas.0808882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Querard J, et al. Photoswitching kinetics and phase-sensitive detection add discriminative dimensions for selective fluorescence imaging. Angew. Chem. Int. Ed. Engl. 2015;54:2633–2637. doi: 10.1002/anie.201408985. [DOI] [PubMed] [Google Scholar]
- 168.Ormö M, et al. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 169.Shu X, et al. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 170.Peng PP, et al. The structure of allophycocyanin B from Synechocystis PCC 6803 reveals the structural basis for the extreme redshift of the terminal emitter in phycobilisomes. Acta Crystallogr. D Biol. Crystallogr. 2014;70:2558–2569. doi: 10.1107/S1399004714015776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ding J, et al. Structural basis of the ultrasensitive calcium indicator GCaMP6. Sci. China Life Sci. 2014;57:269–274. doi: 10.1007/s11427-013-4599-5. [DOI] [PubMed] [Google Scholar]