Abstract
Purpose
We systematically reviewed pharmacoepidemiologic and comparative effectiveness studies that use probabilistic bias analysis to quantify the effects of systematic error including confounding, misclassification, and selection bias on study results.
Methods
We found articles published between 2010 and October 2015 through a citation search using Web of Science and Google Scholar and a keyword search using PubMed and Scopus. Eligibility of studies was assessed by one reviewer. Three reviewers independently abstracted data from eligible studies.
Results
Fifteen studies used probabilistic bias analysis and were eligible for data abstraction – nine simulated an unmeasured confounder and six simulated misclassification. The majority of studies simulating an unmeasured confounder did not specify the range of plausible estimates for the bias parameters. Studies simulating misclassification were in general clearer when reporting the plausible distribution of bias parameters. Regardless of the bias simulated, the probability distributions assigned to bias parameters, number of simulated iterations, sensitivity analyses, and diagnostics were not discussed in the majority of studies.
Conclusion
Despite the prevalence and concern of bias in pharmacoepidemiologic and comparative effectiveness studies, probabilistic bias analysis to quantitatively model the effect of bias was not widely used. The quality of reporting and use of this technique varied and was often unclear. Further discussion and dissemination of the technique are warranted.
Keywords: probabilistic bias analysis, quantitative bias analysis, pharmacoepidemiology, comparative effectiveness
Introduction
Stakeholders of pharmacoepidemiologic and comparative effectiveness research – including patients, clinicians, regulators, and policymakers – need to know the validity and degree of uncertainty of epidemiologic study findings to make important health-related decisions.1–3 This includes an assessment of both random and systematic error (bias; e.g. confounding, misclassification, selection bias). Although researchers routinely estimate random error using confidence intervals or p-values,4–6 few attempt to quantitatively evaluate the effects of bias despite concern that systematic error may be more impactful to study results in observational studies than random error;7,8 this is particularly important when making causal claims and “arguably essential” when making policy recommendations.9
Quantitative bias analyses are sensitivity analyses that can assess the magnitude, direction, and uncertainty of bias through the simulation of bias parameters – the values required to estimate the effects of bias in an analysis.1–3,7,9–15 Probabilistic bias analysis offers several advantages because it balances incorporating uncertainty about bias parameters while requiring less expertise to implement than alternative strategies (e.g. Bayesian approaches);16 further, it can address common concerns of unmeasured confounding, misclassification, and selection bias when using claims data (see eTable 1).17 Probabilistic bias analysis is an extension of simple bias analysis applied by using Monte-Carlo techniques to repeatedly sample from investigator-assigned bias parameter distributions.1–3,7,18 For each iteration, sampled bias parameter values are applied to simple bias analysis formulas to generate a single bias-adjusted estimate (Table 1); this process is repeated multiple times to generate a frequency distribution of bias-adjusted effect estimates (a simulation interval; most commonly summarized as a 95% simulation interval containing bias-adjusted estimates between the 2.5th and 97.5th percentiles).
Table 1.
General steps to implementing probabilistic bias analysis to simulate an unmeasured confounder, misclassification, or selection bias (modified from Lash, Fox, and Fink, 2009)2*
| Steps | Description |
|---|---|
| 1. Identify source of bias to be addressed in probabilistic bias analysis |
Prioritize modeling biases that may have the greatest impact on the conventional results based on a thorough review of the design, analysis, and limitations.9 Single or multiple biases can be readily incorporated into the bias analysis.2,7 |
| 2. Determine the bias model and whether it will be applied to crude estimates, adjusted estimates, or directly to individual records |
Probabilistic bias analysis can be applied to crude effect estimates or stratified data. To apply probabilistic analysis to adjusted effect estimates (beyond stratification and pooling), investigators can simulate bias at the record-level or (in some cases) apply bias models directly to adjusted effect estimates. Applying probabilistic bias analysis to individual records can be time intensive but maintains the relationships between measured variables and allows for modeling.2,16 Unmeasured Confounder: can be applied to crude estimates,2,7,11 adjusted estimates,16 and to individual records.18 Misclassification: can be applied to crude estimates and individual records.2,3,7,11 It can only be applied to adjusted estimates under the assumption of nondifferential misclassification with perfect specificity and imperfect sensitivity.16 Selection bias – unequal selection probabilities: can be applied to crude summary estimates,2,7,11 adjusted summary estimates,2,7 or applied to individual records. Selection bias – loss to follow-up: can be applied to crude estimates of individual records.2,18 |
| 3. Identify bias parameters |
Unmeasured confounder: Requires (when simulating binary confounder) estimates of the prevalence of the confounder in exposed groups and the strength between the confounder and outcome.† Misclassification: Requires estimates of classification probabilities (sensitivity/specificity and/or positive predictive value/negative predictive value). Addressing outcome misclassification in case-control studies also requires estimates of the sampling fractions for cases and controls.28 Selection bias – unequal selection probabilities: Requires estimates of each exposure-disease selection probability (assuming binary exposure and outcome) or the selection bias odds ratio.2,11 Selection bias – loss to follow-up: Requires estimates of outcome risk by exposure status and person-time to event.2,18 |
| 4. Assign probability distributions to bias parameters |
Use internal validation data, external validation data, and expert judgement to define a range and central tendency of plausible values; choose a distribution shape that reflects the uncertainty about the bias parameters (see eFigure 1 for more detail).2,9 |
| 5. Sample from bias parameter distributions using Monte Carlo techniques and apply values to bias formulas to generate a single bias-adjusted estimate |
Single bias parameters values are sampled from assigned probability distributions and applied to the bias model for a single bias-adjusted estimate. Investigators can create correlated bias parameter distributions.2,7,9 Misclassification: Under the scenario of differential misclassification, classification probabilities are hypothesized to be different between groups but are likely correlated; creating correlated bias parameter distributions is recommended.2,3,7,9 |
| 6. Save the single bias-adjusted estimate and repeat step 5 |
Repeatedly sample from bias parameters to generate a frequency distribution of bias adjusted estimates (a simulation interval). |
| 7. Apply sensitivity analyses and diagnostics to bias model |
Vary the location and spread of bias parameter distributions and applying different types of probability distributions to bias parameters.2,9 When using correlated bias parameter distributions, compare initial results to a bias model with independent bias parameter distributions.9 Diagnostics include generating histograms of bias parameters to ensure that the sampled distributions correspond to assigned distributions; the shape of the generated simulation interval should also be examined for implausible results.2,9 Misclassification: Some combinations of classification probabilities will generate implausible, negative cell-counts in contingency tables. These iterations are normally discarded. The frequency of discarded iterations should be reported.2,3,9 |
| 8. Summarize bias analysis results | Report median bias-adjusted estimate and simulation interval to summarize results of probabilistic bias analysis (also consider graphical presentations).2,9 Best practices include providing all methods, results, sensitivity analyses and diagnostics, and programming code used to implement the analysis (using online appendices).2,9 |
| 9. Interpret bias analysis results | Restate the assumptions of the bias analysis and focus on whether the modeled bias plausibly explains the conventional estimate.2,9 Do not interpret the simulation interval as having frequentist properties (e.g., null-hypothesis tests) or fully Bayesian properties (e.g., credibility interval).2,9 The simulation interval can be used for insight into the direction and magnitude of bias as well as providing a better estimate of total error (systematic and random error) about an effect estimate.2,9 |
Modified from the “General steps to probabilistic bias analysis” provided in Lash et al, 2009, p. 132.2
Several factors have increased the ease of implementing probabilistic bias analysis including the availability of resources detailing best practices on implementation and reporting,2,9 online appendices, and high-speed computing.19 However, no study has documented how probabilistic bias analyses have been conducted and reported in pharmacoepidemiologic and comparative effectiveness studies. To address this, we aimed to systematically review pharmacoepidemiologic and comparative effectiveness studies that used probabilistic bias analysis.
METHODS
Study selection
We implemented two separate search strategies: 1) MEDLINE (PubMed) and EMBASE (Scopus) from January 2010 to October 2015 using the keywords “bias analysis” OR “uncertainty analysis” OR “Monte-Carlo sensitivity analysis” OR “Monte Carlo sensitivity analysis” OR “MCSA” or “probabilistic sensitivity analysis”; and 2) Web of Science to perform citations searches of papers that referenced seminal papers.3,14,18,20 Two key works were not indexed in Web of Science,2,21 so we also used Google Scholar.
Eligible articles had to 1) be a pharmacoepidemiology or comparative effectiveness study with a pharmaceutical, biologic, medical device, or medical procedure as exposure and a health outcome; 2) be an observational study; and 3) apply probabilistic bias analysis to estimate the effects of systematic error including selection bias, unmeasured confounding, and/or exposure or outcome misclassification. We excluded 1) methodological studies or studies using simulated data only; 2) randomized control trials, mediation analyses, systematic reviews, meta-analyses, opinions, and non-research letters; 3) articles not published in English; and 4) duplicate publications.
We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines.22 Article titles and abstracts were screened by one reviewer (JNH). Articles which met our eligibility criteria were included in our full text review. Three reviewers (JNH, KLL, SC) independently performed data abstraction on all eligible articles. Discrepancies were resolved through group discussion.
Data abstraction
We used two sources on implementing and reporting bias analysis to guide our data abstraction process.2,9 Table 1 provides a general overview.1–3,9,14,18,20 We abstracted information on the specific bias addressed in the analysis, the bias model used, sources used for simulating the bias parameters (e.g., internal validation studies, external validation studies, expert judgement), believed location (central tendency and spread) and type of bias parameters distributions assigned (e.g. uniform, triangular, normal; see eFigure 1), the number of simulated iterations, and sensitivity analyses or diagnostics.
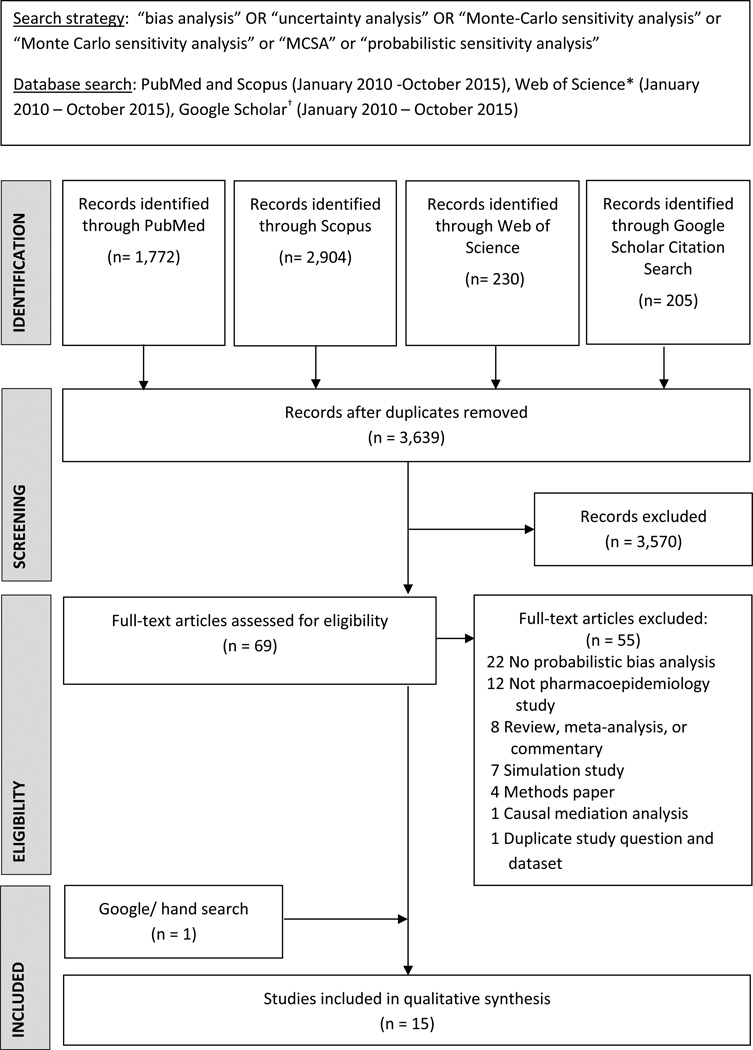
Figure 1.
Flow diagram of studies considered for inclusion
*Web of Science used to index any paper citing one of 4 seminal papers in probabilistic bias analysis.3,14,18,20
†Google Scholar used to index any paper citing a seminal book and paper that were not available in Web of Science database.2,21
We abstracted information on the bias analysis results including the median bias-adjusted effect estimate and simulation interval and compared these results to the conventional analysis by calculating the percent change in effect estimates and interval widths. When many bias analyses were conducted, we abstracted information on the least and most extreme bias scenarios; if bias analysis results were visually displayed (without numeric data), we abstracted results discussed in the text. We abstracted how investigators interpreted bias analysis results in comparison to conventional estimates and whether frequentist or Bayesian properties were assigned to simulation intervals; simulation intervals do not have frequentist or purely Bayesian interpretations and should not be used for null-hypothesis testing or interpreted as Bayesian credible intervals.2,7,9,21
We summarize issues in data abstraction for simulating an unmeasured confounder, misclassification, and selection bias below. See eTable 2 for bias analysis formulas.
Unmeasured confounding
Bias parameters needed to simulate an unmeasured confounder (assuming binary exposure, confounder, and outcome) include confounder prevalence in the exposed (p1) and unexposed (p0) and strength of the association between confounder and outcome (RRCD);2,11,23,24 alternative (but equivalent) formulations includes only parameterizing the relative risk due to confounding (RRC) – the ratio of the crude effect estimate and the standardized risk ratio (standardized to the exposed group).2,7,10,25 Finding literature to parameterize RRC alone can be difficult; investigators may assign RRC distributions that result in a narrower simulation interval due to incorporating less uncertainty in the range of values assigned to a single bias parameter distribution than would be incorporated when simulating multiple bias parameters (p1,p0, and RRCD).2 Extensions of basic formulas to continuous or multinomial confounders and unmeasured confounding in the presence of effect measure modification are available.2,26
When applying probabilistic bias analysis to adjusted effect estimates (beyond stratification and pooling), approaches have been developed for directly adjusting the summary estimate or through simulating confounding at the record-level.16,18,27 Data may be presented as: 1) a single discrete bias scenario; 2), multiple discrete scenarios showing the sensitivity of bias analysis results to varying bias parameter distributions; and 3) continuous changes in bias-adjusted estimates over a continuous interval of bias parameters.
We abstracted information on the specific unmeasured confounder being simulated; details on the approach implemented including bias parameters simulated and whether the method was applied to the effect estimate or individual records; and presentation of the results.
Misclassification
Bias parameters needed for binary exposure/outcome/confounder misclassification include classification probabilities (e.g., sensitivity/specificity or positive/negative predictive values (PPV/NPV)).2,3,7,11,18 Probabilistic bias analysis can be applied to summary effect estimates to adjust for outcome misclassification with perfect specificity and imperfect sensitivity;7,16 otherwise, record-level simulation is required.3 Record-level simulation involves sampling sensitivity/specificity or PPV/NPV values from the assigned bias parameter distributions to determine if individual records should be reclassified based on the assumed classification probabilities; for each simulated iteration, individuals are potentially reclassified based on Bernoulli trials;3 after reclassifying individual observations, standard statistical models are applied to the bias-adjusted dataset to generate a single bias-adjusted effect estimate. The process is repeated to generate a simulation interval. Investigators simulating nondifferential misclassification assume sensitivity/specificity are the same between groups; simulating differential misclassification assumes that sensitivity/specificity are different between groups, though introducing correlation between sensitivity/specificity between groups is recommended.2,3,7
We abstracted information on the type of misclassification simulated; probability distribution assigned; diagnostics including counting the number of negative cell counts; and whether investigators adjusting for outcome misclassification in case-control studies appropriately accounted for differing sampling fractions for cases and controls.7,28
Selection bias
Investigators can apply probabilistic bias analysis to selection bias when participation is related to exposure and outcome (differential selection) or loss to follow-up.2,7,11 Bias parameters needed to simulate differential selection into the study (assuming binary exposure and outcome) include selection proportions for four combinations of exposure and outcome.2,7,11 Alternatively, a single distribution can be assigned to the selection bias odds ratio, though the simulation interval may be narrower than when simulating unique selections proportion because it may incorporate less uncertainty than simulating multiple bias parameters.2 Investigators can also assess the impact of loss to follow-up by simulating bias parameters for outcome risk by exposure status and person-time until the event by applying crude or record-level simulation.2,18
RESULTS
Figure 1 shows that 69 articles were eligible for full text review. Of these, 55 were excluded: 22 studies did not conduct a probabilistic bias analysis; 12 were not pharmacoepidemiologic/comparative effectiveness studies; 8 were reviews, metaanalyses, or commentaries; 7 studies used simulated data only; 4 were methods papers, 1 applied probabilistic bias analysis to a causal mediation analysis, and 1 was a duplicate publication (we kept the earlier of the two publications).27, 29 Fifteen were eligible- 14 identified through our keyword and citation search29–42 and 1 through Google/ hand searching.43
Nine studies simulated an unmeasured confounder29–37 and 6 simulated misclassification (eTable 3).38–43 No studies applied probabilistic bias analysis to selection bias or multiple biases. Six studies simulating confounding were conducted by the same team,29, 32-36 as were 2 studies simulating misclassification.39, 40
All studies addressing an unmeasured confounder29–37 described a specific binary30–37 or multinomial confounder(s)29 to be simulated (Table 2). Sources used to assign bias parameter values varied; six studies31–35, 37 used prior literature to estimate bias parameters including observational studies,31–33,35, 37 trials,31,33,37 government reports,37 online databases,37 and systematic reviews/meta-analyses.34, 35 Bias parameter estimates were sometimes based on small studies with imprecise estimates.32, 33 One study clearly reported the probability distributions assigned (including type and range of values) for all bias parameters.31 Two reported the number of simulated iterations applied - each used ≥5,000 iterations.29, 31
Table 2.
Summary of probabilistic bias analysis when simulating unmeasured confounding, sorted by study design.
| Study | Unmeasured confounder simulated |
Source of bias parameter estimates |
Type of probability distribution assigned to bias parameters* |
Bias parameters for which assigned range of values were reported† |
Simulated iterations |
|---|---|---|---|---|---|
|
Cohort studies | |||||
| Albert et al. (2012)30 | Use of endocrine therapy (Binary) |
Not reported | Not reported | Prevalence of confounder in exposed and unexposed |
Not reported |
| Bannister-Tyrell et al. (2014)31 | Severe labor pain intensity (Binary) |
Expert judgement (prevalence); prior studies (association) |
Trapezoidal | Prevalence of confounder in exposed and unexposed; association between confounder and outcome |
10,000 |
| Corrao et al. (2011)32 | Depression (Binary) |
Prior studies | Not reported | Not reported | Not reported |
| Corrao et al. (2014)33 | Obesity (Binary) |
Prior studies | Not reported | Not reported | Not reported |
| Corrao et al. (2014)34 | Severity of hypercholesterolemia (Binary) |
Prior study | Not reported | Not reported | Not reported |
|
Case-control studies | |||||
| Corrao et al. (2011)29 | Severity of hypertension; CDS; BMI (Multinomial) |
External dataset (prevalence); expert judgement |
Normal | Association between confounder and outcome |
5,000 |
| Corrao et al. (2013)35 | Severe hypercholest- erolemia (Binary) |
Prior studies | Not reported | Not reported | Not reported |
| Ghirardi et al. (2014)36 | Severity of osteoporosis (Binary) |
Not reported | Not reported | Not reported | Not reported |
| Schmidt et al. (2010)37 | Smoking (Binary) |
Prior studies | Not reported | Association between confounder and outcome |
Not reported |
See eFigure 1 in the online supplementary appendix for further description of probability distribution types.
Bias parameters for unmeasured confounding include confounder prevalence in exposed and unexposed and strength of association between the confounder and outcome. We report whether investigators discussed the range of plausible values assigned to bias parameters
Three approaches to simulating an unmeasured confounder were used: 2 studies simulated a single bias scenario,31, 37 5 studies presented multiple discrete bias scenarios with varying strength/prevalence of the unmeasured confounder,29, 30, 32, 35, 36 and 2 studies simulated a range of plausible bias-adjusted effect estimates over a predefined interval of the exposure-confounder odds ratio (the strength and overall prevalence of the confounder were held constant).33, 34 Most studies simulated an unmeasured confounder by applying bias-adjustment formulas to the summary estimate,29, 31-37 though one study performed record-level adjustment.30
For misclassification (Table 3), 3 studies simulated outcome misclassification (2 cohort,39, 40 1 case-control41) and 3 studies simulated exposure misclassification (2 cohort,38, 43 1 case-control42). Sources used to estimate the distribution of plausible bias parameters varied. Two studies simulating outcome misclassification used an internal validation study to estimate plausible distributions of sensitivity/specificity estimates of their outcome definitions.39, 40 One study simulating outcome misclassification used an external study with a similar patient population for sensitivity/specificity estimates.41 One study simulating exposure misclassification used validation data from three external studies with different patient populations to define bias parameter distributions.43 One study used a combination of internal and external validation data to simulate misclassification of a time-varying exposure (simulating PPV, NPV and time-to-initiation).38 One study provided no details on the bias analysis.42 Types and ranges of values assigned to probability distributions were discussed in 4 studies38, 39, 41, 43 and simulated iterations reported in three - each used ≥1,000 iterations.38, 39, 41
Table 3.
Summary of probabilistic bias analysis when simulating misclassification, sorted by study design
| Study | Type of misclassification simulated |
Source of bias parameter estimates |
Type of probability distribution assigned to bias parameters* |
Bias parameters for which assigned range of values were reported† |
Simulated iterations |
|---|---|---|---|---|---|
|
Cohort studies | |||||
| Ahrens et al. (2012)38 | Differential exposure misclassification (binary) |
Internal validation data and external validation study |
Beta | PPV and time of initiation‡ |
100,000 |
| Brunet et al. (2015)43 | Nondifferential exposure misclassification (binary) |
External validation studies |
Trapezoidal | Sensitivity and specificity |
Not reported |
| Huybrechts et al. (2014)39 | Nondifferential outcome misclassification (binary) |
Internal validation study |
Triangular | Sensitivity and specificity |
1,000 |
| Palmsten et al. (2013)40 | Nondifferential outcome misclassification (binary) |
Internal validation study |
Not reported | Not reported | Not reported |
|
Case-control studies | |||||
| Barron et al. (2013)41 | Nondifferential Outcome misclassification; differential outcome misclassification (binary) § |
External validation study; expert judgement |
Trapezoidal | Sensitivity and specificity |
5,000 |
| 5,000 | |||||
| Mahmud et al. (2011)42 | Nondifferential exposure misclassification |
Not reported | Not reported | Not reported | Not reported |
NPV, negative predictive value; PPV, positive predictive value
See eFigure 1 in the online supplementary appendix for further description of probability distribution types.
Bias parameters for misclassification include classification probabilities (sensitivity/specificity and/or PPV/NPV). We report whether investigators discussed the range of plausible values assigned to bias parameters
Simulated a time-varying exposure required estimating time-to-initiation, which investigators simulated by fitting probability distributions to the range of times observed in their cohort. Only sensitivity and specificity point estimates were provided for the calculation of NPV.
Simulated both nondifferential and differential outcome misclassification
All studies reported adjusting for misclassification at the record-level; 38–43 5 used the ‘sensmac’ SAS macro.39–43 Two studies reported sensitivity analyses and diagnostics.38,41 One case-control study modeled both nondifferential and differential outcome misclassification and was the only study to report introducing correlation between bias parameter distributions.41 They also shifted bias parameter distributions upwards to limit the number of simulations producing negative cell counts.;41 this study did not report accounting for differing sampling fractions for cases and controls in the bias adjustment formulas.28 The cohort study adjusting for misclassification of a time-varying exposure used different probability distributions (beta or empirical distribution) as a sensitivity analysis and provided histograms of the simulation interval in comparison to conventional results.38
Probabilistic bias analysis changed the conventional effect estimate by ≥10% in 8 studies30–33,36,37,40,41 and by ≥30% in 4 studies (eTables 4–5).30,31,40,41 In the 13 studies that reported simulation intervals (incorporating both systematic and random error),29– 36,38–41,43 the bias analysis interval was generally larger relative to the conventional confidence interval reported (range: −30.6–1090.0%).
Seven of 9 studies simulating an unmeasured confounder used the simulation interval to conduct null-hypothesis testing;29–34,36 6 of these studies referred to the simulation interval as a confidence interval.29,31–34,36 Two studies interpreted the probabilistic bias analysis by comparing the median bias-adjusted estimates to the conventional effect estimates.35,37
One study applying probabilistic bias analysis to misclassification reported using the simulation interval to conduct null-hypothesis testing,41 and one referred to the simulation interval as a confidence interval.40 Interpretations of bias analysis varied: 2 described differences in effect estimates and interval widths,38,41 2 described differences in effect estimates only,39,40 and 2 stated that there were no changes in results42,43 – 1 of which only provided qualitative information on the bias analysis.42
DISCUSSION
Given the pervasiveness and concern about bias in observational studies, we found few studies that quantitatively examined the effects of bias through the application of probabilistic bias analysis between January 2010 and October 2015. Dissemination of the technique was limited with many of the included papers being published by the same teams of researchers or including a primary proponent of bias analysis as a coauthor (Timothy Lash).37,38 In general, we found that there was large variation in how probabilistic bias analysis was used and reported and that in many cases, key information needed to understand and critique the bias analysis was missing.
We identified several common deficiencies in the implementation of probabilistic bias analysis to address unmeasured confounding. First, for studies presenting multiple bias scenarios,29,30,32–36 authors did not generally provide enough guidance to suggest which scenarios were most plausible. Although these approaches show how sensitive bias analysis results were to varying bias parameters estimates, drawing conclusions on the most plausible effects of unmeasured confounding is difficult when unrealistic scenarios are simulated and emphasized within the text. Second, it was not always clear from where bias parameter estimates were derived,30,36 and in some cases small studies were used to estimate the strength and prevalence of the unmeasured confounder.32,33 This may result in inaccurately describing the range of values for the bias parameters. Third, in all but one study,31 the type and range of probability distributions assigned to bias parameters was unclear. We found this surprising given that the incorporation of uncertainty about bias parameters is one of the largest strengths of probabilistic bias analysis.2,9 More transparency is needed to understand the methods used.
Studies applying probabilistic bias analysis to adjust for misclassification were in general more transparent when reporting the type and range of values assigned to probability distributions and the sources for these estimates. However, important details of the bias analysis were not always reported and discussions of sensitivity analyses and diagnostics were only reported in two studies.38,41 Whether the lack of sensitivity analyses and diagnostics presented was due to the computational intensity and/or technical difficulties in applying probabilistic bias analysis to misclassification, or journal word limits is unknown.19
Probabilistic bias analysis has been described as semi-Bayes because it assigns prior distributions to the bias parameters but no other model parameters.2 Although probabilistic bias analysis results can closely approximate a fully Bayesian analysis in many but not all situations,2,21,44,45 simulation intervals should not be interpreted as having frequentist (e.g., to conduct null-hypothesis tests) or fully Bayesian properties (e.g., as a credibility interval). Despite this, we found the practice of using the simulation interval to conduct null-hypothesis tests to be highly prevalent,29–34,36,41 especially when simulating unmeasured confounding with many authors even referring to a simulation interval as a confidence interval.29–34,36 Prior work suggests using the simulation interval to 1) gain insight into the direction and magnitude of bias given the assumed strength and distributions of the bias parameters by comparing the median bias-adjusted estimate to the conventional point estimate and 2) compare the width of the simulation interval (incorporating random and systematic error) to the confidence interval (random error only) to have a better understanding of the uncertainty about the effect estimate.2,9
Regardless of the bias modeled, few studies reported probabilistic bias analysis as previously suggested.2,9 Descriptions of the technique must be thorough to facilitate understanding and transparency.9 At a minimum, the investigator should state the purpose of the bias analysis, the values and probability distributions assigned to bias parameters, how these values were derived, the number of simulated iterations, and describe the sensitivity analyses and diagnostics implemented to ensure that the probabilistic bias analysis is as accurate as possible.2,9 The use of online appendices can provide further information on the analysis and are available for most journals.2,9
Probabilistic bias analysis is not an alternative to good study design and conduct, and several limitations must be recognized. Probabilistic bias analysis can be difficult to implement when there is limited information available to guide assigning probability distributions to bias parameters such as when simulating selection bias.2,28 Implementing a full probabilistic bias analysis can be time-intensive and technically complex such as when addressing multiple biases.14,19 However, probabilistic bias analysis is a flexible approach that readily complements common statistical methods as a sensitivity analysis and can produce simulation intervals with more accurate coverage probabilities than confidence intervals when bias parameter distributions adequately describe the underlying (but unknowable) bias present.7,46
This systematic review has several strengths. We used a comprehensive search strategy that included both keyword searches and citation indexes of key sources of probabilistic bias analysis. We considered multiple applications of the technique to different types of systematic error. We examined both the implementation and results of the bias analysis. However, our review also has some limitations. We focused on studies published between January 2010 and October 2015 that applied probabilistic bias analysis in pharmacoepidemiology or comparative effectiveness research. We recognize that the reporting of bias analysis can be restricted by journal word limits and may not reflect what the authors originally wrote. Probabilistic bias analyses are one way to implement a quantitative bias analysis and authors could have implemented other approaches (e.g., Bayesian).15,47–50 The background knowledge and programming expertise needed to implement such approaches can be a barrier.16
In conclusion, probabilistic bias analysis is underused in pharmacoepidemiologic and comparative effectiveness studies. The application and reporting of the technique is suboptimal despite recent publications on best practices for implementing and reporting. Further discussion/dissemination of the technique may enhance reporting and implementation of probabilistic bias analysis.
Supplementary Material
Take-Home Messages.
Probabilistic bias analysis is a type of quantitative bias analysis that can be used to evaluate the effects of systematic error (e.g. confounding, misclassification, and selection bias).
Probabilistic bias analysis uses Monte Carlo sampling techniques to repeatedly sample plausible values from probability distributions assigned to bias parameters (values required to estimate the effect of bias in an analysis) and apply these values to simple bias analysis formulas to generate a frequency distribution of bias-adjusted effect estimates.
Commentaries have provided guidance on how to apply and report probabilistic bias analysis, no study has systematically investigated how probabilistic bias analysis has been used and applied in the pharmacoepidemiology and comparative effectiveness literature.
Reporting of probabilistic bias analysis in pharmacoepidemiology or comparative effectiveness studies is suboptimal. The probability distributions assigned to bias parameters, number of simulated iterations, sensitivity analyses, and diagnostics were not discussed in the majority of studies.
Further discussion and dissemination of probabilistic bias analysis are warranted given the concern of bias in observational studies and the varying quality of implementation and reporting.
Acknowledgments
Funding: This work was funded by grants to Jacob Hunnicutt (1TL1TR001454) and Dr. Lapane (1R21CA198172;1TL1TR001454; R56NR015498)
Footnotes
Prior postings and presentations: None
Conflict of interest statement: All of the authors have no conflicts of interest to report.
Ethics statement: This study did not require ethics approval as no human subjects were involved.
REFERENCES
- 1.Phillips CV. Quantifying and reporting uncertainty from systematic errors. Epidemiology. 2003;14:459–466. doi: 10.1097/01.ede.0000072106.65262.ae. [DOI] [PubMed] [Google Scholar]
- 2.Lash TL, Fox MP, Fink AK. Applying Quantitative Bias Analysis to Epidemiologic Data. New York: Springer; 2009. [Google Scholar]
- 3.Fox M, Lash T, Greenland S. A method to automate probabilistic sensitivity analyses of misclassified binary variables. Int J Epidemiol. 2005;34:1370–1376. doi: 10.1093/ije/dyi184. [DOI] [PubMed] [Google Scholar]
- 4.Poole C. Beyond the confidence interval. Am J Public Health. 1987;77:195–199. doi: 10.2105/ajph.77.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothman KJ, Greenland S, Lash TL. Precision and statistics in epidemiologic studies. In: Rothman KJ, Greenland S, Lash TL, editors. Modern Epidemiology. 3rd. Philadelphia: Lippincott Williams & Wilkens; 2008. pp. 148–167. [Google Scholar]
- 6.Poole C. P-values or narrow confidence intervals: which are more durable? Epidemiology. 2001;12:291–294. doi: 10.1097/00001648-200105000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Greenland S, Lash TL. Bias analysis. In: Rothman KJ, Greenland S, Lash TL, editors. Modern Epidemiology. 3rd. Philadelphia: Lippincott Williams & Wilkens; 2008. pp. 345–380. [Google Scholar]
- 8.Grimes D, Schulz K. Bias and causal associations in observational research. Lancet. 2002;349:248–52. doi: 10.1016/S0140-6736(02)07451-2. [DOI] [PubMed] [Google Scholar]
- 9.Lash TL, Fox MP, MacLehose RF, et al. Good practices for quantitative bias analysis. Int J Epidemiol. 2014;43:1969–1985. doi: 10.1093/ije/dyu149. [DOI] [PubMed] [Google Scholar]
- 10.Flanders W, Khoury M. Indirect assessment of confounding: graphic description and limits on effect of adjusting for covariates. Epidemiology. 1990;1:239–246. doi: 10.1097/00001648-199005000-00010. [DOI] [PubMed] [Google Scholar]
- 11.Greenland S. Basic methods for sensitivity analysis of biases. Int J Epidemiol. 1996;25:1107–1116. [PubMed] [Google Scholar]
- 12.Robins J, Rotnitzky A, Scharfstein D. Sensitivity analysis for selection bias and unmeasured confounding in missing data and causal inference models. In: Halloran E, Berry D, editors. Statistical Models in Epidemiiology, the Environment, and Clinical Trials. New York: Springer; 1999. [Google Scholar]
- 13.Phillips C, LaPole L. Quantifying errors without random sampling. BMC Med Res Methodol. 2003;3:9. doi: 10.1186/1471-2288-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenland S. Multiple-bias modelling for analysis of observational data. J R Stat Soc Ser A. 2005;168:267–306. [Google Scholar]
- 15.Greenland S. Bayesian perspectives for epidemiologic research: III. Bias analysis via missing-data methods. Int J Epidemiol. 2009;38:1662–1673. doi: 10.1093/ije/dyp278. [DOI] [PubMed] [Google Scholar]
- 16.Lash T, Schmidt M, Jenson A, et al. Methods to apply probabilistic bias analysis to summary estimates of association. Pharmacoepidemiol Drug Saf. 19:638–644. doi: 10.1002/pds.1938. [DOI] [PubMed] [Google Scholar]
- 17.Schneeweiss S, Avorn J. A review of uses of health care utilization databases for epidemiologic research on therapeutics. J Clin Epidemiol. 2005;58:323–37. doi: 10.1016/j.jclinepi.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Lash T, Fink A. Semi-automated sensitivity analysis to assess systematic errors in observational data. Epidemiology. 2003;14:451–458. doi: 10.1097/01.EDE.0000071419.41011.cf. [DOI] [PubMed] [Google Scholar]
- 19.Lash T, Abrams B, Bodnar L. Comparison of bias analysis strategies applied to a large data set. Epidemiology. 2014;25:576–582. doi: 10.1097/EDE.0000000000000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenland S. The impact of prior distributions for uncontrolled confounding and response bias: a case study of the relation of wire codes and magnetic fields to childhood leukemia. J Am Stat Assoc. 2003;98:47–54. [Google Scholar]
- 21.Greenland S. Sensitivity analysis, Monte Carlo risk analysis, and Bayesian uncertainty assessment. Risk Anal. 2001;21:579–583. doi: 10.1111/0272-4332.214136. [DOI] [PubMed] [Google Scholar]
- 22.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern. 2009;151:264–269. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 23.Schneeweiss S. Sensitivity analysis and external adjustment for unmeasured confounders in epidemiologic database studies of therapeutics. Pharmacoepidemiol Drug Saf. 2006;15:291–303. doi: 10.1002/pds.1200. [DOI] [PubMed] [Google Scholar]
- 24.Schlesselman J. Assessing effects of confounding variables. Am J Epidemiol. 1978;108:3–8. [PubMed] [Google Scholar]
- 25.Miettinen OS. Components of the crude risk ratio. Am J Epidemiol. 1972;96:168–72. doi: 10.1093/oxfordjournals.aje.a121443. [DOI] [PubMed] [Google Scholar]
- 26.Greenland S. Quantitative methods in the review of epidemiologic literature. Epidemiol Rev. 1987;9:1–30. doi: 10.1093/oxfordjournals.epirev.a036298. [DOI] [PubMed] [Google Scholar]
- 27.Corrao G, Nicotra F, Parodi A, et al. External adjustment for unmeasured confounders improved drug-outcome association estimates based on health care utilization data. J Clin Epidemiol. 2012;65:1190–1199. doi: 10.1016/j.jclinepi.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Jurek AM, Maldonado G, Greenland S. Adjusting for outcome misclassification: the importance of accounting for case-control sampling and other forms of outcome-related selection. Annals of Epidemiology. 2013;23:129–135. doi: 10.1016/j.annepidem.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 29.Corrao G, Nicotra F, Parodi A, et al. Cardiovascular protection by initial and subsequent combination of antihypertensive drugs in daily life practice. Hypertension. 2011;58:566–572. doi: 10.1161/HYPERTENSIONAHA.111.177592. [DOI] [PubMed] [Google Scholar]
- 30.Albert J, Pan I, Shih Y, et al. Effectiveness of radiation for prevention of mastectomy in older breast cancer patients treated with conservative surgery. Cancer. 2012;118:4642–4651. doi: 10.1002/cncr.27457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bannister-Tyrrell M, Ford JB, Morris JM, et al. Epidural analgesia in labour and risk of caesarean delivery. Paediatr Perinat Epidemiol. 2014;28:400–411. doi: 10.1111/ppe.12139. [DOI] [PubMed] [Google Scholar]
- 32.Corrao G, Parodi A, Nicotra F, et al. Better compliance to antihypertensive medications reduces cardiovascular risk. J Hypertens. 2011;29:610–618. doi: 10.1097/HJH.0b013e328342ca97. [DOI] [PubMed] [Google Scholar]
- 33.Corrao G, Ibrahim B, Nicotra F, et al. Statins and the risk of diabetes: evidence from a large population-based cohort study. Diabetes Care. 2014;37:2225–2232. doi: 10.2337/dc13-2215. [DOI] [PubMed] [Google Scholar]
- 34.Corrao G, Soranna D, Arfè A, et al. Are generic and brand-name statins clinically equivalent? Evidence from a real data-base. Eur J Intern Med. 2014;25:745–750. doi: 10.1016/j.ejim.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Corrao G, Ibrahim B, Nicotra F, et al. Long-term use of statins reduces the risk of hospitalization for dementia. Atherosclerosis. 2013:171–176. doi: 10.1016/j.atherosclerosis.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Ghirardi A, Bari Di M, Zambon A, et al. Effectiveness of oral bisphosphonates for primary prevention of osteoporotic fractures: evidence from the AIFA-BEST observational study. Eur J Clin Pharmacol. 2014;70:1129–1137. doi: 10.1007/s00228-014-1708-8. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt M, Johansen MB, Lash TL, et al. Antiplatelet drugs and risk of subarachnoid hemorrhage: a population-based case-control study. J Thromb Haemost. 2010;8:1468–1474. doi: 10.1111/j.1538-7836.2010.03856.x. [DOI] [PubMed] [Google Scholar]
- 38.Ahrens K, Lash TL, Louik C, et al. Correcting for exposure misclassification using survival analysis with a time-varying exposure. Ann Epidemiol. 2012;22:799–806. doi: 10.1016/j.annepidem.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huybrechts K, Palmsten K, Avorn J, et al. Antidepressant use in pregnancy and the risk of cardiac defects. N Engl J Med. 2014;370:2397–2407. doi: 10.1056/NEJMoa1312828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmsten K, Huybrechts KF, Micheis KB, et al. Antidepressant use and risk for preeclampsia. Epidemiology. 2013;24:682–691. doi: 10.1097/EDE.0b013e31829e0aaa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barron TI, Cahir C, Sharp L, et al. A nested case-control study of adjuvant hormonal therapy persistence and compliance, and early breast cancer recurrence in women with stage I-III breast cancer. Br J Cancer. 2013;109:1513–1521. doi: 10.1038/bjc.2013.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahmud S, Franco E, Turner D, et al. Use of non-steroidal anti-inflammatory drugs and prostate cancer risk: a population-based nested case-control study. PLoS One. 2011;28:e16412. doi: 10.1371/journal.pone.0016412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brunet L, Moodie E, Cox J, et al. Opioid use and risk of liver fibrosis in HIV/hepatitis C virus-coinfected patients in Canada. HIV Med. 2015 doi: 10.1111/hiv.12279. epub 1–10. [DOI] [PubMed] [Google Scholar]
- 44.MacLehose RF, Gustafson P. Is probabilistic bias analysis approximately Bayesian? Epidemiology. 2012;23:151–8. doi: 10.1097/EDE.0b013e31823b539c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steenland K, Greenland S. Monte Carlo sensitivity analysis and Bayesian analysis of smoking as an unmeasured confounder in a study of silica and lung cancer. Am J Epidemiol. 2004;160:384–92. doi: 10.1093/aje/kwh211. [DOI] [PubMed] [Google Scholar]
- 46.Gustafson P, Greenland S. The performance of random coefficient regression in accounting for residual confounding. Biometrics. 2006;62:760–8. doi: 10.1111/j.1541-0420.2005.00510.x. [DOI] [PubMed] [Google Scholar]
- 47.Morrissey M, Spiegelman D. Matrix methods for estimating odds ratios with misclassified exposure data: extensions and comparisons. Biometrics. 1999;55:338–344. doi: 10.1111/j.0006-341x.1999.00338.x. [DOI] [PubMed] [Google Scholar]
- 48.Spiegelman D, Rosner B, Logan R. Estimation and inference for logistic regression with covariate misclassification and measurement error in main study/validation study designs. J Am Stat Assoc. 2000;95:51–61. [Google Scholar]
- 49.Zucker D, Spiegelman D. Corrected score estimation in the proportional hazards model with misclassified discrete covariates. Stat Med. 2008;27:1911–1933. doi: 10.1002/sim.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gustafson P. Measurement error and misclassification in statistics and epidemiology: impacts and Bayesian adjustments. CRC Press; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.