Abstract
Across western North America, Mimulus guttatus exists as many local populations adapted to site-specific environmental challenges. Gene flow between locally adapted populations will affect genetic diversity both within demes and across the larger meta-population. Here, we analyze 34 whole genome sequences from the intensively studied Iron Mountain population (IM) in conjunction with sequences from 22 Mimulus individuals sampled from across western North America. Three striking features of these data address hypotheses about migration and selection in a locally adapted population. First, we find very high levels of intra-population polymorphism (synonymous π = 0.033). Variation outside of genes is likely even higher but difficult to estimate because excessive divergence reduces the efficiency of read mapping. Second, IM exhibits a significantly positive genome-wide average for Tajima’s D. This indicates allele frequencies are typically more intermediate than expected from neutrality, opposite the pattern observed in many other species. Third, IM exhibits a distinctive haplotype structure with a genome-wide excess of positive associations between rarer alleles at linked loci. This suggests an important effect of gene flow from other Mimulus populations, although a residual effect of population founding might also contribute. The combination of multiple analyses, including a novel tree-based analytic method, illustrate how the balance of local selection, limited dispersal, and meta-population dynamics manifests across the genome. The overall genomic pattern of sequence diversity suggests successful gene flow of divergent immigrant genotypes into IM. However, many loci show patterns indicative of local adaptation, particularly at SNPs associated with chromosomal inversions.
Keywords: population genomics, evolution, migration, local selection, Mimulus, inversions
INTRODUCTION
A fundamental question in evolutionary biology is how genetic variation is maintained in the face of selection. Here we consider this question as it relates to a species with many locally adapted, but subdivided demes (Slatkin 1987; Wade 2016; Wright 1932). Considering a species as a meta-population, as opposed to a homogenous unit, is necessary when local populations are genetically distinct. Local differentiation is more the rule than the exception in plant species. Govindaraju (1988) summarized studies of allozyme variation within and among populations of 102 plant species: On average, 26% of allelic variation (measured as FST or Gst; (Nei 1973)) is distributed among populations and this percentage is highly variable among species (see also (Hamrick & Godt 1996; Loveless & Hamrick 1984)). High mean FST has been corroborated by more recent surveys (Duminil et al. 2009; Nybom 2004) expanded to include DNA-based markers and many additional plant species. The nature and extent of differentiation has critical implications not only for the process of evolution, but how we study that process. In molecular population genetics for example, evolutionary inferences are usually based on summary statistics calculated from samples of gene sequences (Charlesworth et al. 1997; Tajima 1989; Watterson 1975). In a meta-population, the interpretation of these statistics depends entirely on the scale of sampling. Sampling of individuals at both local and geographical scale is essential to address questions about balance of evolutionary forces in a meta-population (Lack et al. 2015; Roesti et al. 2015; Ross-Ibarra et al. 2008).
In this paper, we combine whole genome sequencing with a multi-level sampling approach to investigate the maintenance of variation in the wildflower Mimulus guttatus. We examine a single focal population in conjunction with genomic data from individuals across the entire species complex. We use these data in a series of analyses to test basic predictions of the evolutionary theory of migration-selection balance (Charlesworth et al. 1997; Wright 1931; Yeaman & Whitlock 2011). We first confirm that previous marker based and single gene studies of M. guttatus, which indicated both high differentiation among populations and high intra-population (local) variation, are fully supported by genome-wide data. Second, we test the prediction that genomic regions subject to local selection will be less permeable to incoming haplotypes. Loci under local selection should have lower intra-population nucleotide diversity (π), but also higher FST and elevated absolute divergence between populations (Dxy), if gene flow occurs in other regions of the genome (Beaumont & Nichols 1996; Charlesworth et al. 1997; Cruickshank & Hahn 2014; Lewontin & Krakauer 1973). Third, we test the prediction that loci under local selection should exhibit distinct patterns of linkage disequilibria from the rest of the genome (Charlesworth 2006; Jacobs et al. 2016; Storz & Kelly 2008; Strobeck 1983). Finally, theory predicts that population genetic signatures of selection should be most pronounced when recombination is reduced (Begun & Aquadro 1992; Charlesworth et al. 1993; Kaplan et al. 1989). We test this prediction by comparing patterns of polymorphism, inter-population divergence, and linkage disequilibrium between genomic regions that harbor recombination-suppressing chromosomal inversion with the remainder of the genome.
An interesting and underappreciated prediction of migration-selection balance concerns the genome-wide pattern of linkage disequilibrium. Local selection and limited dispersal will allow populations to become differentiated in allele frequencies. As a consequence, when successful gene flow does (occasionally) occur, it can introduce divergent haplotypes into a population. Alleles that are rare in the focal population (or previously absent) are introduced in combinations. In this way, migration can generate positive associations between (locally) rare alleles at linked Single Nucleotide Polymorphisms (SNPs). To evaluate this prediction, we calculate “polarized” linkage disequilibrium (D) measuring the association of the minor alleles (the less common base at each contrasted SNP pair). Positive D indicates that minor alleles are positively associated (Langley & Crow 1974). To measure this signal, we compare the observed distribution of polarized D estimates between nearby SNPs with the predicted distribution under mutation-recombination-drift balance.
Our focal population for these studies, Iron Mountain (IM), has been the subject of intense evolutionary and ecological research for the past 30 years (Fishman & Kelly 2015; Flagel et al. 2014; Scoville et al. 2011; Willis 1993, 1996). In IM, plant lifespan is strictly limited by water availability. During the short window between the spring snow melt and summer drought (routinely 6–10 weeks), seedlings must grow, flower, mate, and set seed. These abiotic pressures impose strong selection (Mojica & Kelly 2010; Willis 1996) and the population exhibits adaption to local conditions (Hall & Willis 2006). Despite this, IM retains high internal variability in both molecular and quantitative genetic traits (Kelly & Willis 1998; Kelly & Arathi 2003). Chromosomal inversions segregate both within IM (Lee et al. 2016; Scoville et al. 2009) and also between IM and other populations (Holeski et al. 2014; Lowry & Willis 2010; Twyford & Friedman 2015).
IM is annual population within the M. guttatus species complex, an enormous collection of localized populations. Some of these populations are recognized as distinct taxa (e.g. M. nasutus) (Fenster & Ritland 1994; Fishman et al. 2002) or ecotypes (e.g. annual and perennial ecotypes of M. guttatus) (Lowry & Willis 2010). The M. guttatus complex occurs across western North America and has adapted to a wide range of habitats including serpentine barrens, heavy-metal-rich mine tailings, huge elevation ranges, and oceanic salt spray (Hall & Willis 2006; Kelly 2003; Lowry et al. 2009; Mojica et al. 2012). Populations within the complex are often inter-fertile to varying degrees and gene flow occurs between populations, including those named as distinct species (Brandvain et al. 2014). However, potentially strong selection against immigrant genotypes allows substantial genetic differentiation (Friedman et al. 2015; Hall et al. 2010; Hall & Willis 2006; Kooyers et al. 2015). The level of genetic differentiation increases with distance among populations in M. guttatus. Lowry and Willis (2010) estimated FST = 0.48 across a set of 30 populations spanning a latitudinal range from 35–45 degrees. Similarly, (Twyford & Friedman 2015) obtained an FST of 0.46 for populations sampled across a large extent of M. guttatus’ native range (latitudinal range: 31.2 – 53.8). FST is slightly lower (0.43) in a more geographically limited sampling of M. guttatus populations across Oregon, including IM (V. Koelling and J. K. Kelly, unpublished results from whole-genome sequencing study). If IM is compared to populations at much smaller distances (3 and 6 km, respectively), FST declines to 0.07 and 0.13, respectively (Monnahan et al. 2015). The geographic genetic data indicate limits on gene flow. Local adaptation should elevate differentiation relative to the level predicted by migration/drift balance.
In this study, we analyze genome sequences from 34 IM individuals in conjunction with 22 individuals from other populations across the species complex. We use a combination of techniques in two stages of analysis to test the migration-selection balance predictions outlined above. First, we characterize patterns of polymorphism and linkage disequilibria within IM. These analyses yield several striking results including a remarkably high level of synonymous site diversity, a tilt of the site frequency spectrum towards intermediate allele frequencies, and a distinctive haplotype structure in which minor alleles are positively associated at linked sites. In the second phase of the analysis, we compare IM sequences to individuals from allopatric populations within the species complex; estimating divergence relative to polymorphism. By reconstructing distance-based trees for thousands of intervals across the entire genome, we evaluate patterns of relatedness and the possibility of gene flow into IM. The varying structure of polymorphism within IM relative to divergence from other M. guttatus populations indicates an overall pattern of successful gene flow of divergent immigrant genotypes into IM. However, many loci show patterns indicative of local adaptation, particularly at SNPs associated with chromosomal inversions.
MATERIALS AND METHODS
Focal population and plant samples
All sequences in this study (IM and allopatric) are based on samples from natural populations. IM is located in the cascade mountains of central Oregon (44.402217 N, –122.153317W) at an elevation of approximately 1400 meters. The population is predominantly outcrossing (Willis 1993), but plants are self-compatible. IM exhibits minimal internal spatial structure: The genetic relatedness of neighboring plants, typically separated by less than 1 cm, is essentially zero at micro-satellite loci (Sweigart et al. 1999). We propagated approximately 1200 independent lines of M. guttatus by single-seed descent (self-fertilization) for 5–13 generations. Each line was founded from the seed set of a separate field-collected plant sampled from the IM (Willis 1999a). As expected, the inbred lines are almost completely homozygous at microsatellite loci with different lines fixed for different alleles (Kelly 2003). Some novel mutations may have been introduced over the course of line formation, but the number of such mutations should be miniscule relative to standing variation (see RESULTS). It is likely that recessive alleles causing lethality or sterility (under greenhouse conditions) were lost during line formation (Willis 1999b). DNA from 39 of these lines was newly extracted and sequenced.
After eliminating some lines as redundant (see below), we combined these data with 9 previously sequenced IM lines (Flagel et al. 2014) and 21 allopatric samples (individuals from other populations or species in the complex (Supplemental Table 1); reads downloaded from the JGI Short Read Archive). For all samples (both IM and allopatric individuals), average read depth after filtering (as determined from VCF file using vcftools --depth) ranged from 2.6–24.4 (mean = 6.7; calculated for genotyped bases on chromosome 1, no indels included, Supplemental Table 1). We also newly sequenced an additional allopatric individual, hereafter called Iron Mtn. Perennial (IMP). This perennial population occurs in close proximity to the annual IM population. IMP might be taxonomically classified as Mimulus decorus on the basis of its distinctive morphology, including numerous long and thin underground stems (stolons), although the results of this study suggest substantial genetic similarity to IM. Like the IM samples, the sequenced allopatric individuals are inbred lines; descendants of wild plants propagated through multiple generations of self-fertilization in the greenhouse.
DNA extraction, library preparation, and sequencing
We collected and froze leaf tissue and extracted DNA using the Epicentre Leaf MasterPure kit (Epicentre, USA). Libraries for Illumina sequencing were made using the Illumina Nextera DNA kit (Illumina, USA). Individual barcodes were added during library preparation to facilitate multiplexing. Libraries were pooled in equal molar amounts based on concentrations measured using the Qubit high-sensitivity DNA assay and insert size distributions obtained from a Agilent bioanalyzer (HS-DNA chip, Agilent Technologies, USA). Up to 24 libraries were pooled in a single Illumina HiSeq 2500 Rapid-Run sequencing run generating 150-bp paired-end reads.
Alignment, Genotype Calling, and Residual Heterozygosity
After sequencing, we demultiplexed reads into individual samples and mapped them independently. Reads were aligned to the unmasked Mimulus guttatus v2.0 reference genome (http://www.phytozome.net/) (assembled length of 321Mb) using bowtie2 (Langmead & Salzberg 2012). Next, we converted SAM alignment files to binary format using samtools (Li et al. 2009) and then processed alignments with Picardtools (http://broadinstitute.github.io/picard; Commands: FixMates, MarkDuplicates, and AddReadGroups). The Picard processing validated read pairing, removed duplicate reads, and added read groups for analysis in the Genome Analysis Toolkit (GATK) (DePristo et al. 2011). We called genotypes using GATK UnifiedGenotyper (details in supplement). Genotype VCF files were converted to tabbed format using vcftools (vcf-to-tab) (Danecek et al. 2011).
After the initial genotyping, we masked putative SNPs that were excessively heterozygous (in more than 25% of lines) as these are likely due to mis-mapped reads. Mis-mapping is likely in regions of the reference genome where paralogs are incorrectly collapsed into a single gene. We further suppressed entire genomic intervals where, across lines, the mean heterozygosity divided by the average expected heterozygosity (given by the Hardy-Weinberg proportions) exceeds 0.5. Finally, for each individual, we calculated the ratio of observed to expected heterozygosity within 500 SNP windows across the genome. Within each line, we called a region heterozygous if the average was elevated across 10 successive windows. We identified a total of 429 residually heterozygous regions across all lines/chromosomes. This corresponds to 1.29% of the sequence in total. The Mendelian prediction for residual heterozygosity with single seed descent is 1.56% after 6 generations and 0.78% after 7 generations. The size distribution of putative residual heterozygous regions is consistent with the number of generations of selfing, the size of the genome 450–500 mB, and map length of about 125 cM per chromosome (Supplemental Figure 1).
Identification of related lines
After the genotype filtering described above, we constructed a similarity matrix for all IM lines using the Emboss fdnadist program with the Jukes-Cantor substitution matrix. A total of 4.1 million SNPs were called in 43 or more IM lines. Based on this approach, we identified lines that were excessively similar (Supplemental Figure 2). A distribution of pairwise similarity between IM lines shows clear outliers (Supplemental Figure 2a). For instance, IM777 is 0.997 similar to IM323. We thus determined these lines to be relatives and eliminated the IM323 from subsequent analyses. For each pair of IM lines with greater than 0.98 similarity, one line was removed from analysis. After filtering relatives, the following 34 lines were included for all subsequent tests: 62, 106, 109, 115, 116, 138, 170, 179, 238, 239, 266, 275, 359, 412, 479, 502, 549, 624, 657, 667, 693, 709, 742, 767, 777, 785, 835, 886, 909, 922, 1054, 1145, 1152, 1192 (Supplemental Figure 3).
Relationship of missing data and divergence
We delineated windows containing 500 SNPs, and within each window of each line, calculated (1) the number of called and uncalled sites and (2) the number of SNPs called for the reference allele as opposed to the alternative allele. The fraction missing data was calculated from (1), and the window divergence (fraction of calls to alternate) from (2). We performed a logistic regression in R with fraction missing as the response and divergence as the predictor: glm(formula = logit1$frac.missing ~ logit1$divergence, family = binomial). This revealed a strong relationship between missing data and divergence (fraction of called SNPs that differ from the reference genome; Supplemental Figure 4A). Given this relationship, we opted to focus our analyses where data was most complete using two complementary approaches. First, based on the fact that the fraction of missing data is much lower in coding regions (Supplemental Figure 4B) we conducted a series of gene-based analyses (e.g. synonymous versus non-synonymous diversity). The mean fraction of called bases in coding regions, calculated for each line, ranged from 0.63 to 0.86 (Supplemental Table 2). Second, we identified genomic windows (genic and inter-genic DNA) each consisting of 10,000 genotyped bases (monomorphic and polymorphic sites both count as genotyped bases). To qualify as a genotyped base, a site had to be scored in 30 of the 34 unrelated IM lines. The resulting windows ranged from 10,000–3,427,432 bases (of the reference genome) with a mean and median of 39,044 and 18,478 bases, respectively. Allowing 1,000 genotyped base overlapping steps between windows, a total of 74,445 windows span the 14 chromosomes. For these windows, we calculated population genetic and tree-based statistics.
Nucleotide diversity within genes (synonymous and non-synonymous π)
We converted filtered genotype files to fasta format for the entire genome for each separate line. When recreating line specific fasta files, missing data was not imputed, and indels and heterozygous sites were suppressed. We extracted coding sequences using Gffread (Langmead & Salzberg 2012). Each gene was individually extracted from the line specific coding sequences libraries and combined into a single fasta file containing 34 individual coding sequences for each gene. We calculated synonymous and non-synonymous diversity through pairwise comparisons of all lines using the KaKs_Calculator (Nei and Gojobori model) (Zhang et al. 2006). Only diversity measurements derived from genes with alignment lengths greater than 1000 bases were included (N=29,421). A πnon-syn and πsyn value was computed for each gene and was used to calculate genome-wide mean Ka and Ks.
Window analyses
We calculated statistics of polymorphism, divergence, and genealogy within windows of 10,000 genotyped bases. We then created a phylogenetic tree for every window using EMBOSS fdnadist (Rice et al. 2000) to calculate a nucleotide distance matrix (Jukes-Cantor substitution model). Trees were inferred from the distance matrix using EMBOSS fneighbor (Rice et al. 2000) and rooted using Mimulus dentilobius. Of the 74,445 windows, fneighbor failed to parse the distance matrix for only 28 windows, which we excluded from subsequent analysis. Next, for each tree we determined whether IM formed a monophyletic clade or was polyphyletic using a custom perl script (Vos 2015) dependent on the Bio::Phylo toolkit (Talevich et al. 2012). In cases that IM was polyphyletic, we determined how many allopatric samples had to be removed to restore IM monophyly using the perl monophyletic output (Vos 2015) and custom perl scripts.
We calculated S (the number of polymorphisms), π (nucleotide diversity), Tajima’s D (Tajima 1989), and LD statistics in each window using custom python scripts. For the linkage disequilibrium (D), we estimated the association of the minor alleles (less common base) at each contrasted SNP pair. Positive D indicates that minor alleles are positively associated (Langley & Crow 1974). We also standardize D as the correlation coefficient, r = D/ Sqrt[p(1−p)q(1−q)], and from that, calculate r2 (the ZnS test for selection is r2 conditioned on S (Kelly 1997)). We calculated r and r2 for SNP pairs across each chromosome to estimate the long-range pattern of LD. For comparison to observed LD, we performed neutral simulations using calibrated, empirical estimates for 4Nu (from nucleotide diversity) and 4Nr (from LDhelmet as described below) by updating the programs used in Storz et al (Storz et al. 2012). Absolute nucleotide divergence, Dxy, between IM annuals and all allopatric individuals was calculated using a perl script (LaMariposa) dependent on BioPerl::PopGen modules (Dxy is equivalent to πXY (Nei & Li 1979)). Dxy was calculated on a single base increment for all sites that had at least one IM and one allopatric individual genotyped. Next, using these values, an average Dxy value was calculated for the same 10,000 genotyped base windows used for other population genetic statistics.
Recombination rates within IM
We used LDhelmet (Chan et al. 2012) to estimate fine-scale recombination rates with recalled genomes in fasta format as inputs. First, using the “find_confs” command, 50 SNP windows were used to scan the genome and create a haplotype configuration file. Next, a likelihood lookup table and Pade coefficients were generated using a population scaled mutation rate of 0.015 (this was based on a preliminary estimate for genome-wide π within IM). In the final step, the “rjmcmc” command was run using the previously generated haplotype configuration, likelihood table, and Pade coefficients and a Jukes-Cantor mutation matrix to estimate recombination rates. Exon specific recombination rates were calculated. Only pairs of SNPs contained within exons were used. The genome-wide mean estimate for 4Nr was used to calibrate the neutral simulator described above.
Species-wide statistics
As a contrast to the population genetic estimates within IM, we constructed a sample with a single IM line (IM767) and 21 of the allopatric individuals (excluding M. dentilobius as it is outside of the M. guttatus complex). For this species-wide sample, we calculated nucleotide diversity within genes and other statistics (S, Tajima’s D, LD) genome-wide, requiring that SNP be called in at least 15 of 21 lines for inclusion.
RESULTS
Nucleotide diversity within IM and divergence from allopatric populations
Within genes, mean synonymous nucleotide diversity (πsyn = 0.033) is five-fold greater than mean non-synonymous diversity (πnon-syn = 0.006) within IM (Supplemental Figure 5). Nucleotide diversity (genic and non-genic) varied across windows with a mean value of πGenome = 0.014. The variance in pairwise π among the 561 contrasts between 34 IM lines within each genomic window also exhibits many localized peaks across the genome (Supplemental Figure 10). The mean of Var[π] is 0.0000831, and this statistic is positively correlated with nucleotide diversity in the window and with LD measured as r or r2 (Supplemental Figures 8F and 10, 11).
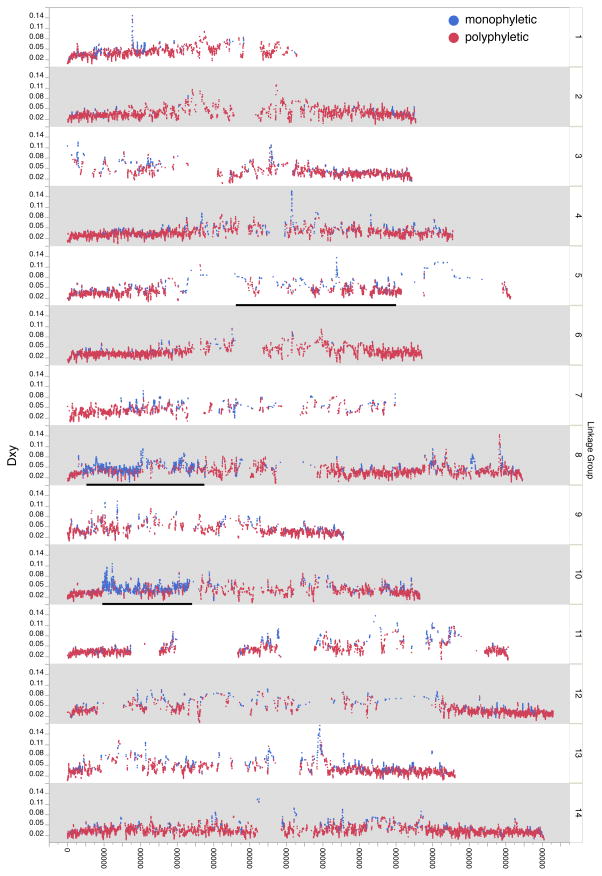
Diversity within IM was lower than divergence of IM sequences from allopatric individuals (Dxy): Mean Dxy = 0.038, range 0.002–0.166 (Supplemental Figure 8E) with several clear peaks of high Dxy (Figure 2). Dxy is the IM versus allopatric sequences analog of πGenome (IM versus IM) and thus the estimated ratio of divergence to polymorphism is about 2.7.
Figure 2.
Genomic distribution of absolute divergence (Dxy). Blue dots = monophyletic windows; red dots = polyphyletic windows. Bars on chromosomes5, 8, and 10 denote approximate locations of chromosomal inversions.
We delineated each of the three inversions mapped in the IMxPR RIL population (Holeski et al. 2014) by locating genetic markers used in the cross to locations in the v2 genome build (bars in Figure 2). This is a cross between annual (IM) and perennial (PR) genotypes. We did not analyze variation within the chromosome 6 inversion that segregates within IM because it has been largely purged from the sequenced lines (Lee et al. 2016). Sequence divergence (Dxy) is significantly elevated within the inversion regions on chromosomes 5, 8, and 10 relative to genome-wide averages: DxyGenome = 0.037, Dxyinversion(8) = 0.044 (p<0.0001), Dxyinversion(5) = 0.068 (p<0.0001), and Dxyinversion(10) = 0.042 (p<0.0001). The chromosome 8 and 10 inversions shows significantly lower overall nucleotide diversity within IM while the chromosome 5 is not statistically different from genome-wide levels: πGenome=0.014, πinversion(10) = 0.010 (p<0.0001), πinversion(8) = 0.012 (p<0.0001), and πinversion(5) = 0.014 (p=0.66). Interestingly, Var[π] is statistically elevated in the chromosome 5 inversion but statistically lower in the regions on chromosomes 8 and 10: Var[π]Genome= 0.000085, Var[π]inversion(5)=0.000109 (p=0.0002), Var[π]inversion(10) =0.000060 (p<0.0001), and Var[π]inversion(8) = 0.000053 (p<0.0001).
Linkage disequilibrium and recombination rate
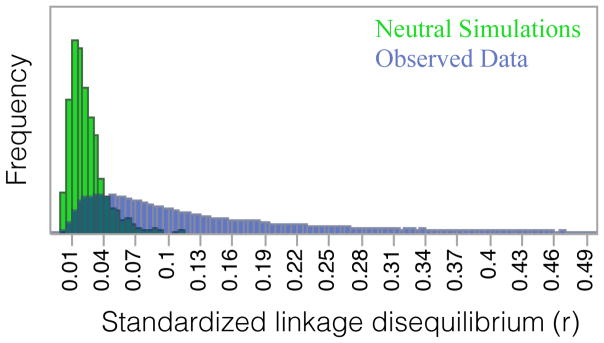
Chromosomal segments that have recently entered the IM population by migration from other differentiated demes are likely to consist of SNP alleles that at are rare within IM. Such migration should generate patterns of positive association between rare alleles at linked SNPs. To investigate this possibility, we estimated LD using a consistent coding for minor alleles at each SNP. This “polarized” measure of LD indicates a striking excess of positive associations (Figure 1) relative to the level predicted with neutrality. If an IM line harbors the less frequent base at a SNP, it is much more likely to have the less frequent base at neighboring SNPs. The neutral distribution of Figure 1 was obtained using the average, genome-wide ρ (4Nr) of 0.0042 obtained from LDhelmet (Chan et al. 2012). When measured as r2, linkage disequilibrium is high at short distances (~100bp) and shows a rapid decay with sequence distance (Supplemental Figure 12). It should be noted that the pattern of long-range LD differs among chromosomes (Supplemental Figure 12). The average genic ρ is 0.0052 and recombination hotspots are clearly evident (Supplemental Figure 13).
Figure 1.
Genome-wide distribution of r in IM annuals shows an excess of positive associations between rare alleles relative to neutral simulations.
Distribution of monophyletic IM clusters across the genome
In order to compare the genomic patterns of diversity in IM with divergence among populations, we constructed a phylogenetic tree for every window of 10,000 genotyped bases in the genome including both IM and allopatric samples. The monophyly of IM samples was evaluated individually for each tree. A minority (10,504) phylogenetic trees were monophyletic for IM while the majority (64,913) showed IM as polyphyletic; in those genomic regions some IM lines were more similar to allopatric sequences than to other IM lines (Supplemental Table 4). Genome-wide, monphyletic windows were found both in gene dense and gene sparse regions. For each polyphyletic IM window, we calculated the number of allopatric samples that would have to be removed from the tree for IM to be monphyletic. For 6,958 of the polyphyletic trees (11%), only one allopatric sequence would have to be removed to restore IM monophyly (Supplemental Table 5). For 48% of these 6,958 trees, the geographically proximate Iron Mtn. Perennial (IMP) was responsible for IM polyphyly (Supplemental Table 5). This pattern is evident across all linkage groups but the incidence is highest for chromosome 8 where 65% of polyphyletic trees where IM monophyly is broken by a single individual are due to IMP (Supplemental Table 5). Table 1 summarizes relationship between population genetic statistics, IM mono/poly-phyly, and the location of structural polymorphisms. Nucleotide diversity (π), Var[π], polarized LD (r), the number of segregating sites, and Tajima’s D were all significantly elevated in polyphyletic windows when compared to monophyletic windows (Table 1). Dxy was significantly lower in polyphyletic windows relative to monophyletic windows (Table 1).
Table 1.
Contrast of population genetic statistics between monophyletic and polyphyletic windows. With these groups we classified windows as within one of the mapped inversions or elsewhere in genome. N=number of windows. The difference between monophyletic and polyphyletic windows is highly significant for all statistics.
| Tree Status | Genomic Location | N | π | Tajima’s D | Var[π] | r | Dxy |
|---|---|---|---|---|---|---|---|
| IM monophyletic | Inversion-5 | 35 | 0.0139 | −0.3277 | 3.59E-05 | 0.0782 | 0.0907 |
| Inversion-8 | 1455 | 0.0117 | 0.1091 | 3.62E-05 | 0.0553 | 0.0465 | |
| Inversion-10 | 700 | 0.0091 | 0.2108 | 3.33E-05 | 0.1082 | 0.0458 | |
| Rest of genome | 8314 | 0.0119 | 0.0793 | 4.81E-05 | 0.0871 | 0.0499 | |
| IM polyphyletic | Inversion-5 | 42 | 0.0136 | −0.2747 | 17.06E-05 | 0.2769 | 0.0482 |
| Inversion-8 | 925 | 0.0135 | 0.2818 | 7.99E-05 | 0.1213 | 0.0399 | |
| Inversion-10 | 630 | 0.0110 | 0.2820 | 8.93E-05 | 0.2474 | 0.0370 | |
| Rest of genome | 62316 | 0.0143 | 0.1498 | 8.94E-05 | 0.1339 | 0.0356 |
Relationship of Var[π] and tree topology
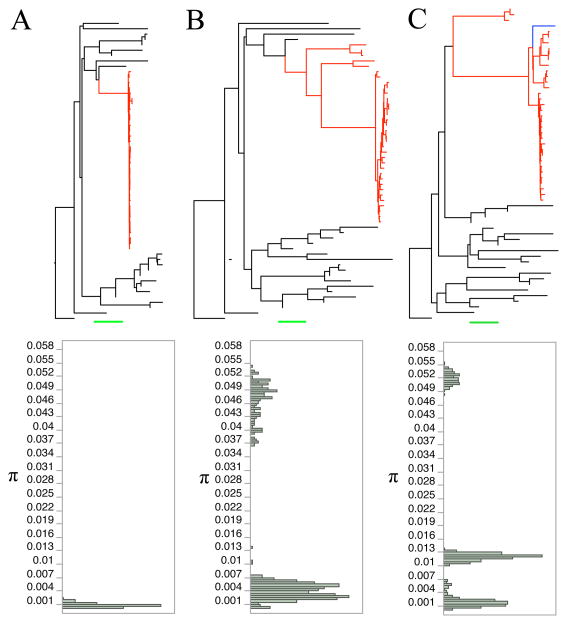
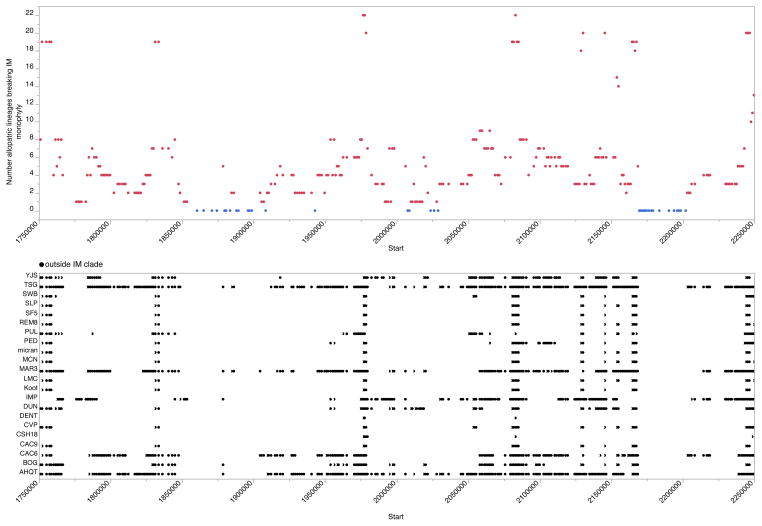
Combining Var[π] and tree topology allows for identification of genomic regions that have experienced a selective sweep or introgression event (Figure 3 and Supplemental Figure 15). On average, we expect monophyletic regions of the genome to have lower Var[π], a trend we observe (Table 1). First, we selected the lowest Var[π] window and the topology suggests a selective sweep – short branches within a monophyletic IM clade (IM individuals within this block possess nearly the exact same haplotype; Figure 3A). Next, we picked the highest Var[π] region for all monophyletic windows (Figure 3B). Here, the IM population contains several highly diverged sequences. Introgression may well have been the original source of the divergent lineages in Figure 3B, although locally they may be maintained by selective processes within IM. It is also possible to identify the genomic effects of introgression from individual allopatric sequences. To illustrate this, we extracted all regions of the genome where IMP is solely responsible for breaking IM monophyly and extracted high Var[π] regions. Tree and π distributions from these windows show evidence of introgression and segregation of very divergent haplotypes (long branches within IM and multimodal π distribution; Figure 3C, Supplemental Figure 15). Two key features of the tree-inference analysis are indicated by a specific (but typical) interval on chromosome 5 (Figure 4). Most windows are paraphyletic, but the number and identity allopatric individuals that break monophyly change along a chromosome. Second, genomic windows where IM sequences are monophyletic are distinctly clustered, such as around the 1.875mb and 2.175mb locations on chromosome 5 (Figure 4A).
Figure 3.
Relationship of tree topology and distribution of pairwise π values within IM annuals. Highlighted red branches are IM annuals. Black bars are allopatric individuals. Green scale bar equals 0.01 for panels A, B and C. Pairwise π values between all 34 IM annuals were calculated (total 561 comparisons). Var[π] was calculated using these values. (A) Lowest Var[π] region for all monophyletic windows shows evidence of selective sweep. Top: tree shows that all IM annuals are very similar. Bottom: Distribution of raw π values for within IM comparisons shows that all IM annuals possess almost the exact same haplotype. (B) Highest Var[π] region for all monophyletic windows shows evidence of multiple distinct haplotypes within IM. (C) Second highest Var[π] region of all trees where IMP alone ruins IM monophyly shows evidence of introgression event including IMP and multiple distinct segregating haplotypes. Blue branch=IMP.
Figure 4.
(A) The number of allopatric individuals that break IM monophyly is depicted as a function of window location along chromosome 5. Monophyletic windows are blue dots. (B) The identity of allopatric individuals breaking IM monophyly is indicated for the same genomic region.
Statistics in the species-wide sample
Across the species complex, nucleotide diversity within genes is very high: πsyn= 0.063 (SE= 0.0001), πnon-syn= 0.0099 (SE= 0.00005). Across genotyped bases (genic and non-genic), there are approximately twice as many SNPs in the species-wide sample than in the IM sample (S = 11,715,727 vs. 5,676,399). However, differences in the missing data pattern between these samples impedes comparisons based on the windows used for IM only. Thus, we calculated Tajima’s D and LD for each in consecutive non-overlapping 50 SNP windows. For the 113,519 windows of IM sample, the mean for Tajima’s D was 0.159 (SE=0.003; SD = 1.082) and the mean for r was 0.245 (SE=0.001, SD=0.194). For the 234,308 windows of the species-wide sample, the mean Tajima’s D was −0.906 (SE=0.001, SD = 0.472) and the mean for r was 0.036 (SE=0.0001, SD=0.043).
DISCUSSION
Traditionally, sequencing efforts in evolutionary genomics have focused on sampling a single individual from each of multiple populations distributed across the full range of a species. Only recently have evolutionary biologists begun generating whole-genome sequence datasets specific to demes, populations of individuals connected by mating in recent time, e.g. (Burri et al. 2015; Kubota et al. 2015; Mackay et al. 2012). This study uses a combination of intensive within and across population whole-genome sequencing to test hypotheses about the balance between evolutionary forces acting in a meta-population. Within the Iron Mountain (IM) population, we observe very high sequence diversity and atypically intermediate allele frequencies. Our data support the prediction that genomic regions subject to local selection are less permeable to introgressing haplotypes introduced by immigrant pollen or seed. Estimates for polarized linkage disequilibria (LD) provide evidence that migration does introduce alleles into IM, at least within genomic regions where gene flow is not impeded by local selection. Finally, the data provide further support to the growing body of examples that chromosomal inversions are an important component of local adaptation (Balanyà et al. 2003; Cheng et al. 2012; Coluzzi et al. 2002; Fang et al. 2012; Feder et al. 2003; Gilburn & Day 1999; Hoffmann & Rieseberg 2008; Jones et al. 2012; Krimbas & Powell 1992; Lowry & Willis 2010). Owing to recombination suppression, these inversions can have pronounced effects on gene sequence evolution (Figure 2).
Diversity within IM
Variation is extremely high for a single population (πsyn = 0.033, πnon-syn = 0.006, πGenome = 0.014), consistent with a previous study of several autosomal genes within M. guttatus (πsyn= 0.061 in (Puzey & Vallejo-Marín 2014)). The estimate for non-synonymous diversity is much lower, reiterating the usual pattern of purifying selection on amino acid changes that is observed in most species. Our genomic estimate (πGenome = 0.014), which includes both genic and non-coding sequence, is almost certainly an underestimate. There is a strong tendency for missing data to increases with divergence from the reference genome (Supplemental Figure 4). The consequence is a downward bias in π due to ascertainment; we are less likely to map (and thus analyze) sequences that are most divergent. This is not surprising, but to our knowledge, has not been demonstrated previously. We expect this to be a general phenomenon extending across most studies of this kind. High levels of insertion/deletion variation (Flagel et al. 2014) is likely contributing to incomplete read mapping and subsequent underestimation of nucleotide diversity.
The high diversity within IM is notable for a single local population. Leffler (Leffler et al. 2012) recently summarized nucleotide diversity across a wide-range of species, classifying estimates by sampling strategy (one population or multiple populations), site type (synonymous, non-synonymous, etc.), and chromosome type (autosome or sex). Considering single populations (N=9, species from five phyla: Arthropoda, Chlorophyta, Chordata, Mollusca, and Pinophyta), the mean and median of autosomal πsyn were 0.014 and 0.011, respectively (range 0.001–0.033). Across multiple populations (N=50) the mean and median were 0.010 and 0.006, respectively. Mimulus guttatus thus represents one of the most variable species yet described, excepting the hyper-diverse nematode recently reported by Dey et al. (2013).
The species-wide FST of ~0.5 for M. guttatus (see Introduction) implies that only half of allelic variation resides within demes, on average. This high FST implies that migration, if successful, should introduce novel haplotypes into IM. This may help to explain the pattern of Linkage Disequilibrium (LD) observed in IM, where rarer alleles at proximate SNP pairs are positively associated across the genome (Figure 1). Most sequencing studies do not specify associations between SNPs in terms of features of alternative alleles, and as a consequence, the direction of LD estimates is meaningless. Indeed, direction is lost when calculating r2, which is commonly used to measure the strength of association between SNPs (e.g. Supplemental Figure 12). Here, we polarize bases (minor vs major) based on allele frequency so that the absolute value of LD can inform questions about evolutionary process. Langley and Crow (Langley & Crow 1974) developed an epistatic selection model to explain the negative LD observed in allozyme data. In IM, LD is highly variable in both direction and magnitude. However, we suggest that the positive mean for LD is due, at least in part, to migration. Immigrants from divergent populations tend to generate positive LD by introducing novel alleles in combinations.
The third striking feature of intra-IM variation, the positive average value for Tajima’s D (Table 1), also requires a careful consideration of both genomic and spatial scale. At the scale of individual loci, this statistic is routinely used as a test for selection. Negative Tajima’s D (extreme allele frequencies) can result from strong purifying selection or a recent selective sweep, while positive values (intermediate allele frequencies) suggest balancing selection (Tajima 1989). Different genomic windows of our survey illustrate each of these outcomes. The window on chromosome 8 at position 8.25 Mb exhibits the topology predicted by a recent selective sweep within IM (Supplemental Figure 16) and Tajima’s D = −2.31 across 191 SNPs. In contrast, the meiotic drive locus on chromosome 11 exhibits positive Tajima’s D over a large genomic interval: Mean value = 0.46 across 561 genomic windows from position 9.5 Mb to 11.7 Mb. Previous study of this region indicates a balanced polymorphism owing to meiotic drive. The Drive allele maintained at a population frequency of approximately 35% owing to balancing benefits and costs (Fishman & Kelly 2015; Fishman & Saunders 2008).
Demographic arguments are typically invoked if the genomic average of Tajima’s D is significantly non-zero; population expansion to explain negative values, population contraction for positive values (Tajima 1989). However, these interpretations depend on the spatial scale of sampling. A species that is expanding through many local populations, each founded from a limited propagule, can exhibit positive Tajima’s D within demes (Ross-Ibarra et al. 2008), even if the statistic is negative in a species wide sample. To illustrate, consider a population founded by a single seed that rapidly expands to large population size. At the beginning, all SNPs in this population will be at loci that were heterozygous in the founder. Such SNPs will have initial population frequencies of 0.5 and thus produce the highest possible values for Tajima’s D. The statistic will be reduced by subsequent drift, pushing allele frequencies away from 0.5, and novel mutations that necessarily start at low frequency (1/2N). However, simulations indicate a substantial time persistence of this founding effect (Ross-Ibarra et al. 2008) and local demes of many species are likely to be quite young on the “coalescent” time scale.
The comparison of genetic diversity statistics between our IM sample and our species-wide sample further indicate the need to interpret population genetic statistics in a meta-population context. Consistent with the high species-wide FST, the amount variation in our complex-wide sample is about double that within IM in terms of the number of polymorphic sites. However, the nature variation with respect to allele frequencies is quite different. While the mean Tajima’s D is positive within IM, it is very significantly negative (mean = −0.90) in the complex-wide sample. This is similar to what has been observed in several Drosophila species (Fabian et al. 2012; Mackay et al. 2012; Nolte et al. 2013), as well as in Arabidopsis (Schmid et al. 2005). It is not surprising that lineages within the species complex have acquired lineage-specific mutations (adaptive, neutral, or deleterious), and by definition, such mutation will be rare in the complex-wide sample. In the next section, we describe results from our second phase of analysis where we compare IM sequences to 22 genomes from “allopatric individuals” sampled from across the M. guttatus species complex.
Genome-wide effects of migration and localized signatures of selection
Most genomic windows exhibit polyphyly of the IM samples, i.e. some IM lines are more similar to other populations than to other IM lines. The high frequency of polyphyly is not caused by a few divergent IM lines repeatedly breaking monophyly. Instead, most IM lines exhibit similarity to allopatric sequences within portions of their genomes; the identity of lines that break monophyly changing across the genome. This is expected given previous evidence that IM is an internally well-mixed, outbred population (Sweigart et al. 1999). IM polyphyly could be due to either ancestral polymorphism that is continuing to segregate or introgression from neighboring populations. These are difficult to distinguish and both are likely important. However, several observations suggest low, but non-trivial, immigration of genotypes into IM. First, the previously noted positive LD pattern (Figure 1) is significantly elevated in polyphyletic windows (Table 1). Second, the geographically proximate IMP is the most frequent cause of IM polyphyly (when a single allopatric is the cause; Supplemental Table 5) consistent with migration from this source. Third, the number and identity of allopatric sequences breaking IM monophyly turn over rapidly as one moves along a chromosome (Figure 4) suggesting a diverse set of contributors to IM.
If there is gene flow into IM, we expect its molecular signal to vary across the genome. In particular, introgression should be reduced at loci subject to local adaptation (Beaumont & Nichols 1996; Lewontin & Krakauer 1973). Despite the general tendency towards polyphyly, many genomic windows exhibit a distinct pattern suggesting local adaptation. The clearest signature of selection is a genomically localized reduction in nucleotide diversity coupled with increased divergence of IM from other populations of M. guttatus (Figure 2) (Nosil et al. 2009). One of the most striking observations from Table 1 is the elevated Dxy, depressed π and Var[π] in monophyletic windows. Reduced π is a one signal of directional selection while elevated Dxy is indicative of locally beneficial variants.
Selection effects should be most pronounced when recombination is suppressed. Chromosomal inversions suppress recombination within heterozygotes over broad genomic scales and a number of inversions have been identified in M. guttatus (Holeski et al. 2014; Lowry & Willis 2010; Twyford & Friedman 2015). Three of these, on chromosomes 5, 8, and 10 respectively, are located approximately to the genome sequence in Figure 2. Consistent with a migration-selection balance model for these loci (Guerrero et al. 2012), we find that sequence divergence (Dxy) is significantly elevated within all three inversion regions relative to genome-wide observations (Figure 2). The inversion regions on chromosomes 8 and 10 show significantly lower overall nucleotide diversity within IM, while the chromosome 5 inversion is on not statistically different from genome-wide levels.
Direct evidence for local selection on the chromosome 8 inversion come from experiments mapping QTL for important for life history traits and flowering time to this locus (Hall et al. 2006; Hall et al. 2010). Reciprocal transplant experiments have directly shown local selection on inversion-8 (Lowry & Willis 2010). In fact, these previous experiments predict the current findings of increased IM monophyly (61% of genomic windows are monophyletic in the inversion-8 region relative to 14% for the genome as a whole 14%), and increased Dxy within this region. This region also shows slightly lower nucleotide diversity (0.012) than the genome average (0.14), consistent with increased monophyly (Table 1). Also, while the general pattern appears to be resistance to introgression, there is evidence for gene exchange via recombination or gene conversion between IM and IMP within inversion-8. IMP is the sole cause of IM polyphyly far more frequently within inversion-8 than genome-wide. IMP is responsible for 409 of 925 (44%) polyphyletic windows, while outside the inversion, IMP is solely responsible for only 2923 of 62,988 (4.6%) polyphyletic windows. This collection of estimates is interesting given that inversion-8 is the genomic region most closely association life-history variation the species range (Lowry & Willis 2010).
The phenotypic and fitness effects of the inversions on chromosomes 5 and 10 remain to be investigated, although the recent genome scan by (Twyford & Friedman 2015) suggests that inversion-5 may be associated with life-history differences among populations. Like inversion-8, inversion-10 exhibits reduced intra-IM variation but increased divergence. In contrast, intra-IM π within inversion-5 is comparable to the genome-wide average. The very elevated Dxy, high Var[π], but moderate π within the inversion-5 could be explained by reduced gene flow with allopatric populations and balancing local selection. The proportion of monophyletic windows within the inversion was substantially elevated: 35 of 77 (45%) of windows are monophyletic. The proportion of monophyletic windows within inversion-10 is even higher (700 of 1330, 52%). Interestingly, IMP is not solely responsible for breaking IM monophyly in any of the 42 polyphyletic chromosome 5 windows while IMP is the solely responsible for breaking IM monophyly in 241 of 630 chromosome 10 windows. The variable patterns of ancestry across inverted regions is perhaps not too surprising. Many different population/species of the M. guttatus complex are potential contributors to IM, and they may differ in the whether they have the IM orientation for a particular inversion.
Candidate genes for local adaptation
Haasland Payseur (2016) recently reviewed the literature on genome scans for natural selection and concluded that “the ability to detect individual instances of selection can decrease as the fraction of genome affected by linked selection grows.” This can be true for a number of reasons, but our study certainly one illustrates one them. Perhaps our most compelling evidence of selection is the pattern of sequence data in regions with suppressed recombination owing to inversions. This suppression produces the broad signal that we detect (many sites affected) but it also hinders discrimination of the specific genetic changes that are affecting fitness. However, we do see more localized patterns of polymorphism and divergence that are potential signatures of selection. We report these as tentative candidates, worthy of further study.
As a first step to identify specific genes potentially involved in local adaptation, we calculated outlier residuals from a Dxy vs. π contrast from all monophyletic windows. Supplemental File 1 reports the 2.5% most negative windows for IM π (controlling for Dxy). A total of 882 genes were located in these outlier windows, and they have significantly lower πsyn (πsyn outlier=0.014, πsyn=0.035, p<0.0001; for genes with alignment length >= 200 and p-value <=0.05, see methods) and lower non-synonymous diversity (πnon-syn outlier=0.002, πnon-syn=0.006, p<0.0001). Genes involved in the flowering pathway, germination timing, stress responses, and trichome development are present in this outlier class. Several very intriguing candidates are immediately worth follow-up functional work. DELAY OF GERMINATION1 (DOG1), a gene involved in timing of germination and flowering in Arabidopsis thaliana (Bentsink et al. 2006; Chiang et al. 2013), as well as several genes involved in flowering time in A. thaliana, including Short Vegetative Phase (SVP) (Lee et al. 2013) and ATMBD9 (Peng et al. 2006b), are contained within the outlier windows. To complement the Dxy vs. π residual analysis, we selected monophyletic windows whose Var[π] and π values are below the 10% minimum for all monophyletic windows (Var[π] <=0.00001 and π<=0.00591). For the low Var[π] and π windows, a total of 690 windows were identified (Supplemental File 2; interval list of outlier windows). Genes within this outlier group include a possible ortholog of A. thaliana gene AtMBD9 (Arabidopsis METHYL-CpG BINDING DOMAIN 9), which is an important regulator of flowering time through interactions with FLC (Peng et al. 2006a), as well as a possible ortholog of Incurvata2 (ICU2) whose Arabidopsis mutant exhibits early flowering (Barrero et al. 2007). The fact that genes in both outlier groups regulate phenological transitions in A. thaliana and that phenology is critical for IM fitness suggests that research following up on these candidate loci may move us a step closer to a gene-level understanding of local adaptation.
Gene genealogies in a meta-population
A component of our analysis is the construction of distance-based trees relating IM and allopatric sequences for thousands of intervals across the entire genome. These trees estimate the genealogy of sequences subject to the important caveat that historical recombination within a locus will make our estimate a compromise among multiple true genealogies. Recognizing this, we do not use the trees for formal hypothesis testing, e.g. to provide a p-value on the null hypothesis that a genomic region is selectively neutral. Instead, the trees provide a compelling visual illustration of the relationship between molecular summary statistics, such as Tajima’s D or r2, and hypothesized evolutionary events, such as selective sweeps or introgression (Figure 3, Supplemental Figures 15–16). Second, the trees can provide classifiers, e.g. Is IM monophyletic or polyphyletic? This classification is useful for the analysis and interpretation of population genetic statistics that more formally characterize intra-population polymorphism, inter-population divergence, and linkage disequilibria (Table 1). The relative occurrence of these classes (monophyletic versus polyphyletic) is clearly affected by major evolutionary events such as chromosomal inversion (Figure 2).
The trees also provide an avenue for thinking about the genealogical process in a structured population with recombination. The observed phylogenetic relationship between sequences changes as one moves along a chromosome of M. guttatus (Figure 4), which is to be expected in an outbred species with recombination. Beyond that, several features of the data suggest that unlike selection, lineage coalescence is perhaps rarely a local phenomenon. First, the number of reproductive adults in IM (far exceeding 100,000 in most years) is at least an order of magnitude greater than the number of generations since the population was founded (likely less than 10,000 and perhaps much less). Second, the high frequency of polyphyly is predicted if lineages are coalescing outside of IM within the larger meta-population. Third, even genomic windows where IM sequences are monophyletic do not imply a Most Recent Common Ancestor (MRCA) within IM. Sequences that coalesce within IM should differ only at sites that experienced mutation within the clade. We would not expect the same nucleotide positions to be polymorphic in neighboring M. guttatus populations (except due to occasional independent, parallel mutation). In fact, pooled population sequencing data indicates that IM polymorphisms are usually shared between populations with the same alternative bases segregating (Monnahan et al. 2015). Perhaps most important is the simple fact that any two IM sequences are likely to exhibit a large number of nucleotide differences (high π) in a given genomic window. This suggests a substantial time since their MRCA. If the neutral mutation rate per base pair is 10−8 or 10−9, the high π observed even in monophyletic windows (Table 1) implies numbers of generations to the MRCA that are far greater than the age of IM (Hudson 1990; Watterson 1975). In aggregate, these observations suggest that the genealogy of sequences within IM is determined more by population founding, migration, and natural selection (hitch-hiking effects associated with linkage), than by the standard coalescent process of genetic drift within IM.
An important open question is the extent to which population-level processes, such as selection generated by local environmental conditions, influence the amount and distribution of genetic variation species-wide. Local adaptation not only influences the specific loci that are targets of selection, but also the “effective migration rate” via its effect on the relative fitness of immigrant individuals (or gametes). As described in the INTRODUCTION, there is a long history of spatial population genetics in plant biology. Species vary enormously in the proportion of genetic variation that exists within relative to among demes. Genome sequencing can now provide a much more thorough characterization, quantifying absolute levels of variation within and among populations, and providing novel information from haplotype structure (LD). This study of M. guttatus illustrates that high differentiation among populations does not imply low intra-population variation. Hierarchical studies of other species, e.g. (Long et al. 2013), are required to determine the generality of this pattern.
Supplementary Material
Acknowledgments
We would like to thank Stephen Wright, Patrick Monnahan and Jenn Coughlan for advice on the content and/or comments on this manuscript. This work was supported by grants from the National Institutes of Health to J.K. and J.W. (R01 GM073990) and the National Science Foundation to J.R.P. (NPGI-IOS-1202778).
Footnotes
Data Availability: All sequence data generated here is available on the Short Read Archive. SRA numbers: SAMN05852485-SAMN05852522.
Author Contributions: JP, JW, and JK designed this experiment. JP made the libraries and directed sequencing. JP and JK performed all genomic analyses and wrote the paper.
Cited
- Balanyà J, Serra L, Gilchrist GW, Huey RB. Evolutionary pace of chromosomal polymorphism in colonizing populations of Drosophila subobscura: an evolutionary time series. Evolution. 2003;57:1837–1845. doi: 10.1111/j.0014-3820.2003.tb00591.x. [DOI] [PubMed] [Google Scholar]
- Barrero JM, González-Bayón R, del Pozo JC, Ponce MR, Micol JL. INCURVATA2 Encodes the Catalytic Subunit of DNA Polymerase α and Interacts with Genes Involved in Chromatin-Mediated Cellular Memory in Arabidopsis thaliana. The Plant Cell. 2007;19:2822–2838. doi: 10.1105/tpc.107.054130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society of London Series B-Biological Sciences. 1996;263:1619–1626. [Google Scholar]
- Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proceedings of the National Academy of Sciences. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y, Kenney AM, Flagel L, Coop G, Sweigart AL. Speciation and Introgression between Mimulus nasutus and Mimulus guttatus. PLoS Genet. 2014;10:e1004410. doi: 10.1371/journal.pgen.1004410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri R, Nater A, Kawakami T, et al. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Research. 2015;25:1656–1665. doi: 10.1101/gr.196485.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AH, Jenkins PA, Song YS. Genome-wide fine-scale recombination rate variation in Drosophila melanogaster. PLoS genetics. 2012;8:e1003090. doi: 10.1371/journal.pgen.1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. The Effect Of Deleterious Mutations On Neutral Molecular Variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Nordborg M, Charlesworth D. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genetical Research. 1997;70:155–174. doi: 10.1017/s0016672397002954. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. Balancing Selection and Its Effects on Sequences in Nearby Genome Regions. PLoS Genet. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, White BJ, Kamdem C, et al. Ecological Genomics of Anopheles gambiae Along a Latitudinal Cline: A Population-Resequencing Approach. Genetics. 2012;190:1417–1432. doi: 10.1534/genetics.111.137794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang GC, Barua D, Dittmar E, et al. Pleiotropy in the wild: the dormancy gene DOG1 exerts cascading control on life cycles. Evolution. 2013;67:883–893. doi: 10.1111/j.1558-5646.2012.01828.x. [DOI] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. A Polytene Chromosome Analysis of the Anopheles gambiae Species Complex. Science. 2002;298:1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Molecular Ecology. 2014;23:3133–3157. doi: 10.1111/mec.12796. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Chan CKW, Thomas CG, Cutter AD. Molecular hyperdiversity defines populations of the nematode Caenorhabditis brenneri. PNAS. 2013;110:11056–11060. doi: 10.1073/pnas.1303057110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duminil J, Hardy OJ, Petit RJ. Plant traits correlated with generation time directly affect inbreeding depression and mating system and indirectly genetic structure. BMC Evolutionary Biology. 2009;9:1–14. doi: 10.1186/1471-2148-9-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian DK, Kapun M, Nolte V, et al. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Molecular ecology. 2012;21:4748–4769. doi: 10.1111/j.1365-294X.2012.05731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Pyhäjärvi T, Weber AL, et al. Megabase-Scale Inversion Polymorphism in the Wild Ancestor of Maize. Genetics. 2012;191:883–894. doi: 10.1534/genetics.112.138578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Roethele JB, Filchak K, Niedbalski J, Romero-Severson J. Evidence for Inversion Polymorphism Related to Sympatric Host Race Formation in the Apple Maggot Fly, Rhagoletis pomonella. Genetics. 2003;163:939–953. doi: 10.1093/genetics/163.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster CB, Ritland K. Quantitative Genetics Of Mating System Divergence In the Yellow Monkeyflower Species Complex. Heredity. 1994;73:422–435. [Google Scholar]
- Fishman L, Kelly AJ, Willis JH. Minor quantitiative trait loci underlie floral traits associated with mating system divergence in Mimulus. Evolution. 2002;56:2138–2155. doi: 10.1111/j.0014-3820.2002.tb00139.x. [DOI] [PubMed] [Google Scholar]
- Fishman L, Kelly JK. Centromere-associated meiotic drive and female fitness variation in Mimulus. Evolution. 2015;69:1208–1218. doi: 10.1111/evo.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Saunders A. Centromere-Associated Female Meiotic Drive Entails Male Fitness Costs in Monkeyflowers. Science. 2008;322:1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- Flagel LE, Willis JH, Vision TJ. The standing pool of genomic structural variation in a natural population of Mimulus guttatus. Genome biology and evolution. 2014;6:53–64. doi: 10.1093/gbe/evt199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Twyford AD, Willis JH, Blackman BK. The extent and genetic basis of phenotypic divergence in life history traits in Mimulus guttatus. Molecular Ecology. 2015;24:111–122. doi: 10.1111/mec.13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilburn AS, Day TH. Female mating behaviour, sexual selection and chromosome I inversion karyotype in the seaweed fly, Coelopa frigida. Heredity. 1999;82:276–281. doi: 10.1038/sj.hdy.6884830. [DOI] [PubMed] [Google Scholar]
- Govindaraju DR. Mating Systems and the Oportunity for Group Selection in Plants. Evolutionary Trends in Plants. 1988;2:99–106. [Google Scholar]
- Guerrero RF, Rousset F, Kirkpatrick M. Coalescent patterns for chromosomal inversions in divergent populations. Philosophical Transactions of the Royal Society B: Biological Sciences. 2012;367:430–438. doi: 10.1098/rstb.2011.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasl RJ, Payseur BA. Fifteen years of genomewide scans for selection: trends, lessons and unaddressed genetic sources of complication. Molecular Ecology. 2016;25:5–23. doi: 10.1111/mec.13339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Basten CJ, Willis JH. Pleiotropic Quantitative Trait Loci Contribute to Population Divergence in Traits Associated With Life-History Variation in Mimulus guttatus. Genetics. 2006;172:1829–1844. doi: 10.1534/genetics.105.051227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Lowry DB, Willis JH. Is local adaptation in Mimulus guttatus caused by trade-offs at individual loci? Molecular Ecology. 2010;19:2739–2753. doi: 10.1111/j.1365-294X.2010.04680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Willis JH. Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution. 2006;60:2466–2477. [PubMed] [Google Scholar]
- Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. Phil Trans Roy Soc London Biol Sci. 1996;351:1291–1298. [Google Scholar]
- Hoffmann AA, Rieseberg LH. Revisiting the Impact of Inversions in Evolution: From Population Genetic Markers to Drivers of Adaptive Shifts and Speciation? 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holeski L, Monnahan P, Koseva B, et al. A High-Resolution Genetic Map of Yellow Monkeyflower Identifies Chemical Defense QTLs and Recombination Rate Variation. G3: Genes/Genomes/Genetics. 2014;4:813–821. doi: 10.1534/g3.113.010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Gene genealogies and the coalescent process. Oxford surveys in Evolutionary biology. 1990;7:1–44. [Google Scholar]
- Jacobs GS, Sluckin TJ, Kivisild T. Refining the Use of Linkage Disequilibrium as a Robust Signature of Selective Sweeps. Genetics. 2016;203:1807–1825. doi: 10.1534/genetics.115.185900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan NL, Hudson RR, Langley CH. The “hitchhiking effect” revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AJ, Willis JH. Polymorphic microsatellite loci in Mimulus guttatus and related species. Mol Ecol. 1998;7:769–774. [Google Scholar]
- Kelly JK. A test of neutrality based on interlocus associations. Genetics. 1997;146:1197–1206. doi: 10.1093/genetics/146.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JK. Deleterious mutations and the genetic variance of male fitness components in Mimulus guttatus. Genetics. 2003;164:1071–1085. doi: 10.1093/genetics/164.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JK, Arathi HS. Inbreeding and the genetic variance of floral traits in Mimulus guttatus. Heredity. 2003;90:77–83. doi: 10.1038/sj.hdy.6800181. [DOI] [PubMed] [Google Scholar]
- Kooyers NJ, Greenlee AB, Colicchio JM, Oh M, Blackman BK. Replicate altitudinal clines reveal that evolutionary flexibility underlies adaptation to drought stress in annual Mimulus guttatus. New Phytologist. 2015;206:152–165. doi: 10.1111/nph.13153. [DOI] [PubMed] [Google Scholar]
- Krimbas CB, Powell JR. Drosophila Inversion Polymorphism. CRC Press; Boca Raton, FL: 1992. [Google Scholar]
- Kubota S, Iwasaki T, Hanada K, et al. A Genome Scan for Genes Underlying Microgeographic-Scale Local Adaptation in a Wild Arabidopsis Species. PLoS Genet. 2015;11:e1005361. doi: 10.1371/journal.pgen.1005361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack JB, Cardeno CM, Crepeau MW, et al. The Drosophila Genome Nexus: A Population Genomic Resource of 623 Drosophila melanogaster Genomes, Including 197 from a Single Ancestral Range Population. Genetics. 2015;199:1229–1241. doi: 10.1534/genetics.115.174664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMariposa GitHub: popgen_scripts. https://github.com/LaMariposa/popgen_scripts.
- Langley CH, Crow JF. The direction of linkage disequilibrium. Genetics. 1974;78:937–941. doi: 10.1093/genetics/78.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ryu H-S, Chung KS, et al. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science. 2013;342:628–632. doi: 10.1126/science.1241097. [DOI] [PubMed] [Google Scholar]
- Lee YW, Fishman L, Kelly JK, Willis JH. A Segregating Inversion Generates Fitness Variation in Yellow Monkeyflower (Mimulus guttatus) Genetics. 2016;202:1473–1484. doi: 10.1534/genetics.115.183566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler EM, Bullaughey K, Matute DR, et al. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS biology. 2012;10:e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC, Krakauer J. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Rabanal FA, Meng D, et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 2013;45:884–890. doi: 10.1038/ng.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveless MD, Hamrick JL. Ecological determinants of genetic structure in plant populations. Annual Review of Ecology and Systematics. 1984:15. [Google Scholar]
- Lowry DB, Hall MC, Salt DE, Willis JH. Genetic and physiological basis of adaptive salt tolerance divergence between coastal and inland Mimulus guttatus. New Phytologist. 2009;183:776–788. doi: 10.1111/j.1469-8137.2009.02901.x. [DOI] [PubMed] [Google Scholar]
- Lowry DB, Willis JH. A Widespread Chromosomal Inversion Polymorphism Contributes to a Major Life-History Transition, Local Adaptation, and Reproductive Isolation. PLoS Biol. 2010;8:e1000500. doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Richards S, Stone EA, et al. The Drosophila melanogaster genetic reference panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica JP, Kelly JK. Viability selection prior to trait expression is an essential component of natural selection. Proceedings of the Royal Society B-Biological Sciences. 2010;277:2945–2950. doi: 10.1098/rspb.2010.0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica JP, Lee YW, Willis JH, Kelly JK. Spatially and temporally varying selection on intrapopulation quantitative trait loci for a life history trade-off in Mimulus guttatus. Molecular ecology. 2012;21:3718–3728. doi: 10.1111/j.1365-294X.2012.05662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnahan PJ, Colicchio J, Kelly JK. A genomic selection component analysis characterizes migration-selection balance. Evolution. 2015;69:1713–1727. doi: 10.1111/evo.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Analysis of Gene Diversity in Subdivided Populations. Proceedings of the National Academy of Sciences. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte V, Pandey RV, Kofler R, Schlötterer C. Genome-wide patterns of natural variation reveal strong selective sweeps and ongoing genomic conflict in Drosophila mauritiana. Genome research. 2013;23:99–110. doi: 10.1101/gr.139873.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, ORTIZ-BARRIENTOS D. Divergent selection and heterogeneous genomic divergence. Molecular ecology. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Nybom H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology. 2004;13:1143–1155. doi: 10.1111/j.1365-294X.2004.02141.x. [DOI] [PubMed] [Google Scholar]
- Peng M, Cui Y, Bi Y-M, Rothstein SJ. AtMBD9: a protein with a methyl-CpG-binding domain regulates flowering time and shoot branching in Arabidopsis. The Plant Journal. 2006a;46:282–296. doi: 10.1111/j.1365-313X.2006.02691.x. [DOI] [PubMed] [Google Scholar]
- Peng M, Cui Y, Bi YM, Rothstein SJ. AtMBD9: a protein with a methyl-CpG-binding domain regulates flowering time and shoot branching in Arabidopsis. The Plant Journal. 2006b;46:282–296. doi: 10.1111/j.1365-313X.2006.02691.x. [DOI] [PubMed] [Google Scholar]
- Puzey J, Vallejo-Marín M. Genomics of invasion: diversity and selection in introduced populations of monkeyflowers (Mimulus guttatus) Molecular ecology. 2014;23:4472–4485. doi: 10.1111/mec.12875. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends in genetics. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Roesti M, Kueng B, Moser D, Berner D. The genomics of ecological vicariance in threespine stickleback fish. Nat Commun. 2015:6. doi: 10.1038/ncomms9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J, Wright SI, Foxe JP, et al. Patterns of Polymorphism and Demographic History in Natural Populations of Arabidopsis lyrata. PLoS ONE. 2008;3:e2411. doi: 10.1371/journal.pone.0002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid KJ, Ramos-Onsins S, Ringys-Beckstein H, Weisshaar B, Mitchell-Olds T. A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics. 2005;169:1601–1615. doi: 10.1534/genetics.104.033795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville A, Lee YW, Willis JH, Kelly JK. Contribution of chromosomal polymorphisms to the G-matrix of Mimulus guttatus. New Phytologist. 2009;183:803–815. doi: 10.1111/j.1469-8137.2009.02947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville AG, Lee YW, Willis JH, Kelly JK. Explaining the heritability of an ecologically significant trait in terms of individual QTLs. Biology letters. 2011;7:896–898. doi: 10.1098/rsbl.2011.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- Storz JF, Kelly JK. Effects of Spatially Varying Selection on Nucleotide Diversity and Linkage Disequilibrium: Insights From Deer Mouse Globin Genes. Genetics. 2008;180:367–379. doi: 10.1534/genetics.108.088732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Natarajan C, Cheviron ZA, Hoffmann FG, Kelly JK. Altitudinal variation at duplicated β-globin genes in deer mice: effects of selection, recombination, and gene conversion. Genetics. 2012;190:203–216. doi: 10.1534/genetics.111.134494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck C. Expected Linkage Disequilibrium For a Neutral Locus Linked to a Chromosomal Arrangement. Genetics. 1983;103:545–555. doi: 10.1093/genetics/103.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart A, Karoly K, Jones A, Willis JH. The distribution of individual inbreeding coefficients and pairwise relatedness in a population of Mimulus guttatus. Heredity. 1999;83:625–632. doi: 10.1038/sj.hdy.6886020. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talevich E, Invergo BM, Cock PJ, Chapman BA. Bio. Phylo: A unified toolkit for processing, analyzing and visualizing phylogenetic trees in Biopython. BMC bioinformatics. 2012;13:209. doi: 10.1186/1471-2105-13-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyford A, Friedman J. Adaptive divergence in the monkey flower Mimulus guttatus is maintained by a chromosomal inversion. Evolution. 2015;69:1476–1486. doi: 10.1111/evo.12663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos R. Monophylizer. 2015 https://github.com/naturalis/monophylizer/tree/v1.0.1.
- Wade MJ. Adaptation in Metapopulations: How Interaction Changes Evolution. University of Chicago Press; 2016. [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Pop Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Willis JH. Partial self fertilization and inbreeding depression in two populations of Mimulus guttatus. Heredity. 1993;71:145–154. [Google Scholar]
- Willis JH. Measures of phenotypic selection are biased by partial inbreeding. Evolution. 1996;50:1501–1511. doi: 10.1111/j.1558-5646.1996.tb03923.x. [DOI] [PubMed] [Google Scholar]
- Willis JH. Inbreeding load, average dominance, and the mutation rate for mildly deleterious alleles in Mimulus guttatus. Genetics. 1999a;153:1885–1898. doi: 10.1093/genetics/153.4.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JH. The role of genes of large effect on inbreeding depression in Mimulus guttatus. Evolution. 1999b;53:1678–1691. doi: 10.1111/j.1558-5646.1999.tb04553.x. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian population. Genetics. 1931:16. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. 1932:356–366. [Google Scholar]
- Yeaman S, Whitlock MC. THE GENETIC ARCHITECTURE OF ADAPTATION UNDER MIGRATION–SELECTION BALANCE. Evolution. 2011;65:1897–1911. doi: 10.1111/j.1558-5646.2011.01269.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao X-Q, et al. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics, proteomics & bioinformatics. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.