Abstract
Background and Objectives:
The most common serotype of enterohaemorrhagic Esherichia coli group or Shiga-toxin-producing E. coli is O157:H7. Domestic and wild ruminants are regarded as the main natural reservoirs. O157:H7 serotype is the major cause of gastrointestinal infections in developed countries. In this study was conducted to survey on the toxigenic E. coli O157: H7 strains in milk of industrial dairy farms.
Materials and Methods:
A total number of 150 milk samples were collected from dairy industry in Khuzestan, over a period of 6 months and were evaluated by cultivation in selective media (CT-SMAC) and multiplex PCR.
Results:
Two isolates were identified as E. coli using biochemical tests, none of them were toxigenic E. coli O157:H7 as determined by multiplex PCR. Using direct PCR on milk samples, 45 samples contained at least one gene of the studied genes in this investigation (rfb, flic, stx1, stx2). With direct PCR, 2 milk samples were positive for toxigenic O157:H7.
Conclusion:
E. coli O157:H7 is present in this region and so the necessity for strict compliance of health standards is recommended. This is the first study on O157: H7 E. coli milk contamination in Khuzestan province. Based on these results, direct PCR is more accurate than indirect PCR.
Keywords: Milk, Escherichia coli O157:H7, PCR
INTRODUCTION
Serotype of enterohaemorrhagic E. coli is one of the most harmful food borne pathogenic bacteria and is responsible for many cases of infection and deaths worldwide (1). Infection with serotype is usually self-limiting, but the bacterium can cause hemorrhagic colitis (HC), hemolytic uremic syndrome (HUS) and thrombotic thrombocytopenic purpura in children and the immunocompromised patients, which are life-threatening complications (2).
Shiga toxin 1 and 2, which respectively encoded by the stx1 and stx2 genes (two phage-encoded cytotoxins), the intimin protein (encoded by the chromosomal gene eae), and enterohaemolysin (encoded by the ehxA gene), are the most commonly assayed virulence factors of Shiga-toxin-producing E. coli (3). The bacterium has a locus of enterocyte effacement (LEE), a pathogenicity island (4), encodes a type III secretion system which includes a translocated intimin receptor (Tir) (5) three enterohaemorrhagic E. coli secreted proteins A, B and D (EspA, EspB, and EspD) which are important in signal transduction of mammalian host cells as well as attaching and effacing lesion formation (6). Cattle are the major reservoir of this bacterium for human, although E. coli O157:H7 has also been isolated from sheep, goat, horses, dogs, deer, birds and flies (7). Consumption of unpasteurized contaminated milk and under-cooked contaminated meat with fecal material are the main transmission routes in human. Different methods such as culture in special media, serological and molecular assays have been used for detection of this serotype in food, environmental and clinical samples. Sorbitol-MacConkey agar (SMAC) supplemented with cefixime and potassium tellurite, is one of the most sensitive media and improves selection of E. coli O157:H7 from other serotypes of E. coli and non sorbitol fermenter bacteria such as Morganella, Aeromonas, Providencia, and Plesiomonas (8, 9).
Since the conventional methods take time, many studies have been designed based on molecular techniques such as PCR (10). Primers in many PCR assays are used for investigation of specific genes, but combining these primers in a single reaction would be a fast and reliable method for detection of the O157:H7 E. coli. The presence of these organisms in milk has been studied in different countries (11) but in Iran most studies have been done on the meat and another dairy products in shopping center or bulk milk of farms.
This study was designed to investigate the presence of E. coli O157:H7 in raw cattle milk in Khuzestan province, south west of Iran, using both bacterial culture method and PCR.
MATERIALS AND METHODS
Sample collection.
In total, 150 fresh milk samples were obtained from six dairy farms in different parts of Khuzestan Province (Ahvaz, Izeh, Baghmalek, Behbahan, Dezful and Shadegan) from March to September, 2013. The clinically healthy Holstein cows with normal physical characteristics of milk were selected for this study. The California Mastitis Test (CMT) was used to detect mastitic milks. Samples were in normal distribution and 10% of dairy farms in each sample were assayed. All samples were collected in sterile 50 ml containers, in total volume of 30–40 ml, aseptically, and were transported to the laboratory at 4°C within a < 6 h after sampling. For the isolation of E. coli O157:H7 from milk, the samples were centrifuged and the sediments were plated on selective media (referred to as the “milk pellet enrichment-direct plating method”, or MPEDP method).
Upon arrival at the laboratory, the sampling containers (containing 25 ml of milk) were centrifuged (Eppendorf, Germany) at 3000 rpm for 20 min at 4°C. The supernatant was discarded and the pellets were dissolved in1 ml of milk. One milliliter was added in 14 ml of Tryptone Soya Broth containing 20 mg/l novobiocin (mTSB-n) in 50 ml tubes. After 18h incubated at 37°C, the enriched cultures were centrifuged same before and loopful of pellet cells were used to streak on supplemented sorbitol MacConkey agar supplemented with cefixime (2.5 mg/l) and tellurite (0.05 mg/l) (CT-SMAC, Merck, Germany) and incubated for 24h at 37 °C. Three to five colonies that had the characteristic of E. coli morphology (clear and colorless) were chosen from each plates and cultured on blood Agar (BA). At the first step, every selected suspicious colony were streaked onto plates containing eosin methylene blue agar (EMB,Merck, Germany) and incubated at 37°C. After 24 h, biochemical tests including conventional indole reduction, methyl red, voges proskauer, citrate utilization and lysine decarboxylase tests was done for isolates with typical E. coli metallic sheen on EMB. Certain E. coli isolates were stored at −70°C in TSB with glycerol (20%).
Multiplex-PCR assay.
Every isolates of E. coli were screened by PCR for the presence of shiga-like toxins (stx1 and stx2), O157:H7 serotype (O157 and H7 genes). Standard strain of E. coli O157:H7 (ATCC 43894) and sterile distilled water were used as a positive and negative control, respectively.
After prepared of suspension of every E. coli colony in sterile TE (Tris-EDTA) buffer with 2% 2-mercaptoethanol, by heating the bacterial suspension for 10 min in boiling water temperature the bacteria was lysed. The lysate was spun at 13000 rpm for 3 min to pellet the cellular debris.
Supernatant was stored at −20°C as template for amplification by m-PCR. The primer sequences used were: flic H7 and rfb O157 which encoded the flagellar and somatic antigens respectively (12). Certain isolates as E. coli O157:H7, were examined by second m-PCR assay and using stx1 and stx2 genes specific primers (13) (Table 1). The amplification conditions and reagents for the m-PCR assays were those described by Berenjchi et al. (2010) with total volume of 25 μl in each m-PCR reaction and amplification mixture consisting of 12.5 μl 2X mastermix (Sinagen, Iran), 1μl of each primer (0.5 μM), and 5 μl of template. The thermocycler (Eppendorf, Germany) PCR program was started with initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 60 sec, annealing at 52°C for 30 sec and elongation at 72°C for 60 sec, and final extention at 72°C for 10 min (14). The PCR products were electrophoresed in TAE (Tris- Acetic acid- EDTA) buffer containing 1% agarose, visualized by safe- staining (Sinagen, Iran), illuminated by UV-transilluminator apparatus. As a DNA marker, 100 bp DNA ladder was used.
Table 1.
Primers sequence and expected sizes of studied genes
| Target gene | Primer sequence | Size (bp) |
|---|---|---|
| rfb | F: 5′- CGG ACA TCC ATG TGA TAT GG-3′ R: 5′- TTG CCT ATG TAC AGC TAA TCC -3′ |
259 |
| flic | F: 5′- GCG CTG TCG AGT TCT ATC GAG-3′ R: 5′- CAA CGG TGA CTT TAT CGC CAT TCC-3′ |
625 |
| stx1 | F: 5′- ACA CTG GAT GAT CTC AGT GG-3′ R: 5′- CTG AAT CCC CCT CCA TTA TG-3′ |
614 |
| stx2 | F: 5′- CCA TGA CAA CGG ACA GCA GTT-3′ R: 5′- CCT GTC AAC TGA GCA CTT TG-3′ |
779 |
Direct multiplex-PCR.
Direct PCR was done on enriched milk samples in N-TSB. For this purpose 1ml enriched milk sample in N-TSB from the first stage was defrosted and centrifuged with 13000 rpm for 3 min. Sediment was resolved in 1ml TE buffer and diluted 1:20 in TE buffer again. Bacterial suspension was used in DNA extraction like the previous protocol and was stored at −20°C. The second stage of PCR was done similar to first stage.
RESULTS
Although 14 colonies of non-sorbitol fermenting (NSF) were isolated from 150 milk samples, after enrichment and selective plating only two isolates were identification as E. coli by biochemical tests. In m-PCR assay, using specific primers for rfb and flic genes, the isolate confirmed serotypes other than E. coli O157:H7 and the second m-PCR assay, using specific primers for stx1 and stx2 genes, showed that the isolates were harboring none of the toxin genes (stx1, stx2). In the second stage of research by direct PCR on 150 enriched milk samples, different patterns of studied genes were observed in 45 samples. Column chart of primary and secondary PCR results is shown below. Twenty (33.3%) of 150 samples were positive for E. coli O157 (carrier for rfb gene), eighteen isolates of which (90%) were non carrier for H7 gene (O157) and only two (10%) were motile (O157:H7). E. coli O157 was found with the highest level in milk samples of Behbahan and Eizeh counties. In this research the number of stx2 producing bacteria was more than stx1 producing bacteria (Fig. 1).
Fig. 1.
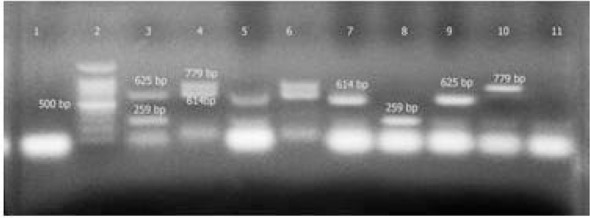
Results of direct mPCR; line1: negative control, line2: marker 100 bp, line3: positive control for rfb (259bp) and flic (625bp), line4: positive control for stx1 (614bp) and stx2 (779bp), lines 5 and 6:positive samples for all genes studied, lines 7, 8, 9, 10: four positive samples for every one of the genes studied, line 11: negative sample.
DISCUSSION
Due to the consumption of unpasteurized and traditional dairy product in Khuzestan province, we decided to survey on the presence of E. coli O157: H7 in the collected bovine milk from dairy farms. In this study, 2 isolates of E. coli O157:H7 (1.3%) were detected from 150 milk samples by direct PCR assay, but by conventional culture method and multiplex-PCR none of the samples were positive from this serotype (O157 and H7) and the virulence genes studied (stx1, stx2). Our findings do not differ greatly from those reported abroad from raw cow’s milk. Studies in Egypt and Austria, were reported that 6% and 3% of raw cow’s milk samples, were contaminated with E. coli O157:H7 respectively (15), but generally the prevalence of this serotype is various from 1 to 13% in European countries (16). Similarities and differences in the various regions may be due to similarities and differences in climate and hygiene of the area. Although according to our finding the incidence of this serotype of E. coli in milk is low, considering the low infective dose, the presence of this pathogen in raw cow’s milk is important in this area of Iran. The same result was observed in the study conducted by Rahimi et al. in 3 provinces (Isfahan, Charmahal va Bakhtiari and Khuzestan) of Iran. From 201 traditional dairy products samples, non-O157 E. coli in 14 (7%), O157:NM E. coli in 3 (1.5%) and O157:H7
E. coli in 1(0.49%), were recognized. All the O157:H7/NM E. coli isolates were positive for eaeA and stx1 and/or stx2, and one isolate was positive for EhlyA. In stx positive isolates, 1 and 2 isolates had stx1 and stx2, respectively (17). As well as in studies carried out on ground beef hamburger and donor kabab samples in Khuzestan and parts of Iran, presence of E. coli O157:H7 was determined based on culture and m-PCR (18). Several studies have been conducted around the world to determine the presence of this serotype of E. coli in various foods such as meat (19). Comparison of various studies results is difficult because of difference in methodologies, such as isolation procedures, the type of improved enrichment, differences in sample size, and the type of sample and how and when it was collected. Culture in selective media and PCR used in this study is a conventional and available method for isolation of O157:H7 and a sensitive method for detection of virulence genes, respectively. Although using rainbow agar (a new chromogenic medium for the detection of O157: H7 E. coli) has been found to be more sensitive than CTSMAC, but the difference is not significant (20). In multiplex PCR, the use combination primers for the detection of several genes eliminates the possibility of false positives, which may occur if non-O157:H7 strains were to acquire an O157:H7 specific gene. Although in some strains despite the encoding H7 flagella antigen gene within the genome, immunore-active H7 flagella antigen is not expressed, and may lead to false negative results (21). Direct m-PCR assay was evaluated as a very suitable method for detection of toxigenic O157:H7 Taking this approach not only decreases medium used, but also increased precision in milk samples (20) because of decrease in false negative results due to viable but non-culturable bacteria (VBNC) which can be present in foods under unsuitable conditions. Also, we used primers specific for flagellar and somatic antigens genes (confirmed by serotyping), and then for virulence factors such as shiga-like toxin 1(SLT1) and shiga-like toxin 2 (SLT2). The ability to detect rough isolates or the masked O antigen isolates is main advantage of the employed m-PCR method (22). By direct PCR in this research, stx1 and stx2 were detected in two infected samples by E. coli O157:H7, but in another (43 samples) frequency of stx2 (5.3%) was more than stx1 (4%). In previous studies in USA, European countries and Japan have been reported that the stx2 gene was more common than the stx1 (23).
In this study all milk samples were collected in hot seasons (spring to fall), which is consistent with the findings from previous studies that showed, summer and early fall are peak prevalence of infection (24). Different distribution of E. coli O157:H7 as the season changes has been reported previously (25), with the highest prevalence in summer and the lowest in winter, so it is possible that the contamination rate is variable.
CONCLUSION
This is the first study on contamination of raw milk with O157:H7 E. coli in Khuzestan province of Iran. Comparison of the two methods results showed that direct PCR for pre-enrichment milk samples is more accurate and because of culture process deletion, is more economical and faster. The percentage of positive raw milk samples of the O157:H7 E. coli isolates reported herein are the same as what has been reported previously from different dairy products of several provinces in Iran. Although this results, showed that a low percentage of raw milk in Khuzestan is contaminated with this serotype of E. coli which is potentially pathogenic for humans, but because of high consumption of raw dairy products in the province, further research is essential in this area for detection of other serotypes of O157:H7. Although finding effective pre-harvest control measures is not easy, but it is necessary to apply some approaches for preventing and reducing the incidence of STEC in animal reservoirs in primary production, finally reducing the pathogens’ in foods and water, and on consumer education.
ACKNOWLEDGEMENT
Authors would like to thank the research council of Shahid Chamran University of Ahvaz for their financial support and Dr Firuzi in Shiraz University and Dr. Nofouzi in Tabriz University for prepared of control positive strain of E. coli.
REFERENCES
- 1. Gilbert C, Winters D, O’Leary A, Slavik M. Development of a triplex PCR assay for the specific detection of Campylobacter jejuni, Salmonella spp. and Escherichia coli O157:H7. Mol Cell Probes 2003;17: 135– 138 . [DOI] [PubMed] [Google Scholar]
- 2. Gryko R, Sobieszczanska BM, Stopa PJ, Bartoszcze MA. Comparison of multiplex PCR, and immunochromatographic method sensitivity for the detection of Escherichia coli O157:H7 in minced beef. Acta Microbiol Pol 2002; 51; 121–129. [PubMed] [Google Scholar]
- 3. Law D. Virulence factors of Escherichia coli O157 and other Shiga toxin producing E. coli. J Appl Microbiol 2000; 88, 729–745. [DOI] [PubMed] [Google Scholar]
- 4. Amani J, Mousavi SL, Rafati S, Salmanian AH. Immunogenicity of a plant-derived edible chimeric EspA, Intimin and Tir of Escherichia coli O157:H7 in mice. Plant Science 2011; 180: 620– 627 . [DOI] [PubMed] [Google Scholar]
- 5. Clark SC, Haigh RD, Freestone PPE, Williams PH. Virulence of enteropathogenic Escherichia coli, a global pathogen. Clin Microbiol Rev 2003; 16 (3): 365– 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Elliott SJ, Wainwright LA, McDaniel TK, Jarvis KG, Deng YK, Lai LC, et al. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol 1998; 28: 1– 4 . [DOI] [PubMed] [Google Scholar]
- 7. Hancock DD, Besser TE, Rice DH, Ebel ED, Herriott DE, Carpenter LV. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the Northwestern USA. Prev Vet Med 1998; 35: 11– 19 . [DOI] [PubMed] [Google Scholar]
- 8. March SB, Ratnam S. Sorbitol- MacConkey medium for detection of Escherichia coli O157:H7 associated with hemorrhagic colitis. J Clin Microbiol 1986; 23: 869– 872 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heuvelink AE, Zwartkruis-Nahuis JTM, De Boer E. Evaluation of media and test kits for the detection and isolation of Escherichia coli O157 from minced beef. J Food Protect 1997; 60: 817– 824 . [DOI] [PubMed] [Google Scholar]
- 10. Bai J, Shi X, Nagaraja TG. A multiplex PCR procedure for the detection of six major virulence genes in Escherichia coli O157:H7. J Microbiol Methods 2010; 82: 85– 89 . [DOI] [PubMed] [Google Scholar]
- 11. Solomakos N, Govaris A, Angelidis AS, Pournaras S, Burriel AR, Kritas SK, et al. Occurrence, virulence genes and antibiotic resistance of Escherichia coli O157 isolated from raw bovine, caprine and ovine milk in Greece. Food Microbiol 2009; 26):865–871. [DOI] [PubMed] [Google Scholar]
- 12. Philpott D, Ebel F. E. coli: shigatoxin methods and protocols. 1st EdnTotowa, New Jersey, Humana Press Inc; 2003; PP: 9–45. [Google Scholar]
- 13. Holland JL, Louie L, Simor AE, Louie M. PCR detection of Escherichia coli O157:H7 directly from stools: evaluation of commercial extraction methods for purifying fecal DNA. J Clin Microbiol 2000; 38: 4108– 4113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brenjchi M, Jamshidi A, Farzaneh N, Bassami MR. Identification of shiga toxin producing Escherichia coli O157:H7 in raw cow milk samples from dairy farms in Mashhad using multiplex PCR assay. Iran J Vet Res 2011; 12(2), Ser. No. 35 [Google Scholar]
- 15. Allerberger F, Dierich MP. (1996). Enterohemorrhagic Escherichia coli in Austria. VTEC’97, abstract V37/I p.4 3rd international symposium and workshop on shiga toxin (Verocytotoxin)-producing Escherichia coli infections. Baltimore, MD, USA: Lois Joy Galler Foundation for HUS, Melville, NY, USA. [Google Scholar]
- 16. Blanco J, Blanco M, Blanco JE, Mora A, Alonso MP, Gonzàles EA, et al. (2001). Epidemiology of verocytotoxigenic Escherichia coli (VTEC) in ruminants . In: Verocytotoxigenic Escherichia coli. Duffy G, Garvey P, McDowell DA. (eds). Food sciences & Nutrition Press, Trumbull, Connecticut: 113–148 [Google Scholar]
- 17. Rahimi R, Kazemeini HR, Salajegheh M. Escherichia coli O157:H7/NM prevalence in raw beef, camel, sheep, goat, and water buffalo meat in Fars and Khuzestan provinces, Iran. Vet Res Forum 2012; 3 : 15– 17 . [PMC free article] [PubMed] [Google Scholar]
- 18. Farajzadeh Sheikh A, Rostami S, Amin M, Abbaspour A, Goudarzi H, Hashemzadeh M. Isolation and identification of Escherichia coli O157:H7 from ground beef hamburgers in Khuzestan Province, Iran. Afr J Microbiol Res 2013; 7: 413– 417 . [Google Scholar]
- 19. Hussein HS. Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. J Anim Sci 2007; 85: E63– 72 . [DOI] [PubMed] [Google Scholar]
- 20. Cagney C, Crowley H, Duffy G, Sheridan JJ, O’Brien S, Carney E, et al. Prevalence and numbers of Escherichia coli O157:H7 in minced beef and beef burgers from butcher shops and supermarkets in the Republic of Ireland. Food Microbiol 2004; 21: 203– 212 . [Google Scholar]
- 21. Tutenel AV, Pierard D, Vandekerckhove D, Van Hoof J, De Zutter L. Sensitivity of methods for the isolation of Escherichia coli O157 from naturally infected bovine feces. Vet Microbiol 2003; l., 94:341–346. [DOI] [PubMed] [Google Scholar]
- 22. Al-Ajmi D, Padmanabha J, Denman SE, Gilbert RA, Al Jassim RAM, McSweeney CS. Evaluation of a PCR detection method for Escherichia coli O157: H7/h-bovine faecal samples. Lett Appl Microbiol 2006; 42: 386– 391 . [DOI] [PubMed] [Google Scholar]
- 23. Desmarchelier PM, Bilge SS, Fegan N, Mills L, Vary JC J, Tarr PI. A PCR specific for Escherichia coli O157:H7 based on rfb locus encoding O157 lipopolysaccharide. J Clin Microbiol 1998; 36: 1801– 1804 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keen EJ, Elder RO. Isolation of shiga-toxigenic Escherichia coli O157 from hide surface and the oral cavity of finished beef feedlot cattle. JAVMA 2002; 220: 756– 763 . [DOI] [PubMed] [Google Scholar]
- 25. Fedio WM, Jinneman KC, Yoshitomi KJ, Zapata R, Wendakoon CN, Browning P, et al. Detection of E. coli O157: H7 in raw ground beef by Pathatrix (TM) immunomagnetic-separation, real-time PCR and cultural methods. Int J Food Microbiol 2011; 148: 87– 92 . [DOI] [PubMed] [Google Scholar]


