Abstract
Transient, multi-protein complexes are important facilitators of cellular functions. This includes the chaperome, an abundant protein family comprising chaperones, co-chaperones, adaptors, and folding enzymes—dynamic complexes of which regulate cellular homeostasis together with the protein degradation machinery1–6. Numerous studies have addressed the role of chaperome members in isolation, yet little is known about their relationships regarding how they interact and function together in malignancy7–17. As function is probably highly dependent on endogenous conditions found in native tumours, chaperomes have resisted investigation, mainly due to the limitations of methods needed to disrupt or engineer the cellular environment to facilitate analysis. Such limitations have led to a bottleneck in our understanding of chaperome-related disease biology and in the development of chaperome-targeted cancer treatment. Here we examined the chaperome complexes in a large set of tumour specimens. The methods used maintained the endogenous native state of tumours and we exploited this to investigate the molecular characteristics and composition of the chaperome in cancer, the molecular factors that drive chaperome networks to crosstalk in tumours, the distinguishing factors of the chaperome in tumours sensitive to pharmacologic inhibition, and the characteristics of tumours that may benefit from chaperome therapy. We find that under conditions of stress, such as malignant transformation fuelled by MYC, the chaperome becomes biochemically ‘rewired’ to form a network of stable, survival-facilitating, high-molecular-weight complexes. The chaperones heat shock protein 90 (HSP90) and heat shock cognate protein 70 (HSC70) are nucleating sites for these physically and functionally integrated complexes. The results indicate that these tightly integrated chaperome units, here termed the epichaperome, can function as a network to enhance cellular survival, irrespective of tissue of origin or genetic background. The epichaperome, present in over half of all cancers tested, has implications for diagnostics and also provides potential vulnerability as a target for drug intervention.
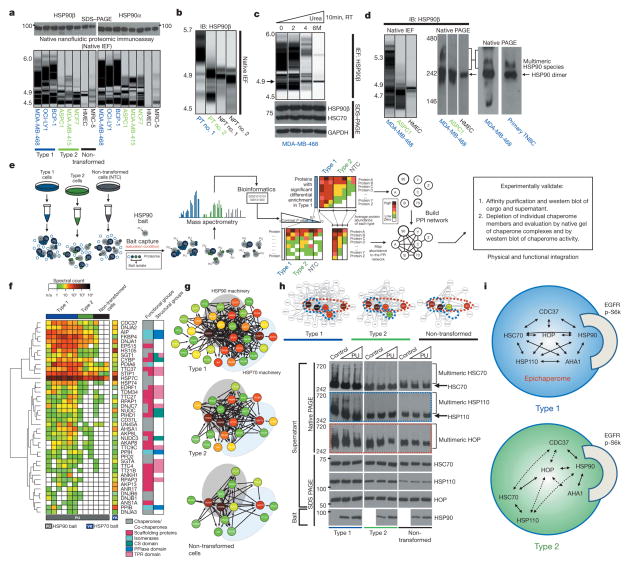
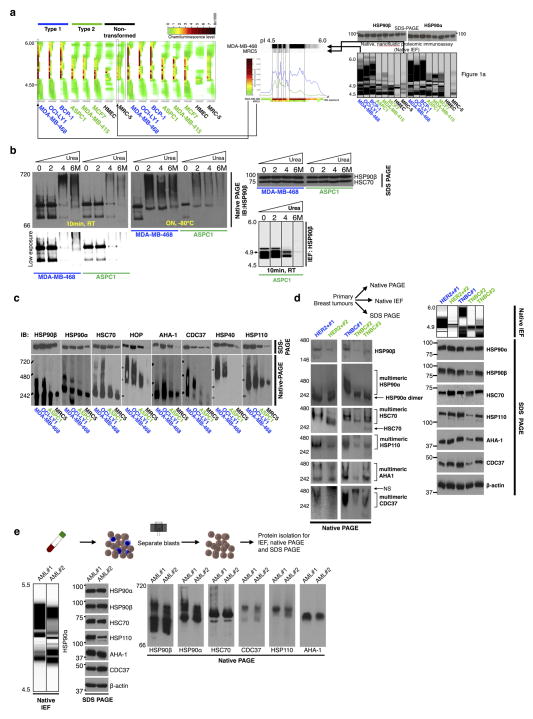
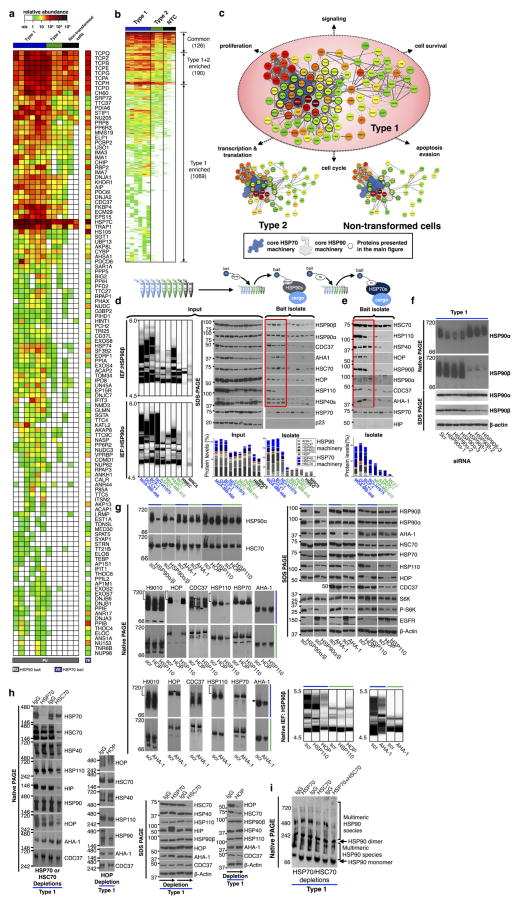
To investigate the chaperome in tumours we first analysed HSP90, the most abundant chaperome member in human cells1,2. In cultured non-transformed cells and in normal primary breast tissue (NPT, the normal tissue surrounding or adjacent to the corresponding primary tumour) (Fig. 1a, b), HSP90 focused primarily as a single species at the predicted isoelectric point (pI) of 4.9. However, cancer cell lines analysed by this method contained a complex mixture of HSP90 species spanning a pI range of 4.5 to 6; HSP90α and HSP90β isoforms were part of these complexes. Furthermore, although all cancer cell lines contained a number of HSP90 complexes with pI < 4.9, a subset was enriched in HSP90 complexes with the unusual pI of ≥5, herein referred to as ‘type 1’ cells. We refer to cancer cell lines that contained mainly complexes with pI < 4.9 as ‘type 2’ cells. This distinction in HSP90 complexes was also evident in primary tumours (Fig. 1b). The total levels of HSP90 were essentially identical among all analysed samples, irrespective of whether they were type 1 or type 2 (Fig. 1a; see further analyses).
Figure 1. A subset of cancer cells are enriched in stable multimeric chaperome complexes.
a–d, The biochemical profile of indicated chaperome members in cell lines and primary specimens. IB, immunoblotting; TNBC, triple-negative breast cancer; NPT, the normal tissue surrounding or adjacent to the corresponding primary tumour; PT, primary tumour; RT, room temperature. The gel representation of the chromatogram is shown for IEF. See also Extended Data Fig. 2a, b. e, Workflow used to identify the chaperome components and establish their interconnectivity in cells. f, Heat map illustrating core HSP90 chaperome members enriched (P < 0.1) in type 1 tumours. Last lane, HSP70-interacting chaperome. g, Networks showing interactions between chaperome proteins. See also Extended Data Fig. 5. h, Changes in multimeric chaperome complexes and total chaperome levels in cell homogenates challenged with control or increasing concentrations of the HSP90-directed bait. All data were repeated independently twice with representative images shown. For uncropped gel data, see Supplementary Fig. 1. i, In both type 1 and 2 tumours, the HSP90 machinery is functional and regulates its onco-client proteins such as EGFR and p-S6K, but only type 1 but not type 2 tumours are characterized by stable, multimeric chaperome complexes that physically and functionally integrate the HSP90 and HSP70 machinery components.
Under denaturing conditions, HSP90 in type 1 tumours focused mainly at the pI of ~4.9 (Fig. 1c). We therefore directed our attention on proteins interacting with HSP90 as the main instrument for pI change in type 1 tumours. HSP90 is known to interact with several co-chaperones including activator of HSP90 ATPase homologue 1 (AHA1, also known as AHSA1), cell division cycle 37 (CDC37), and HSP70–HSP90 organizing protein (HOP, also known as stress-inducible phosphoprotein 1 (STIP1)) which links HSP90 to the HSP70 machinery. Each of these co-chaperones has a distinct role. CDC37 facilitates activation of kinases, AHA1 augments HSP90 ATPase activity, and HSP70 and HOP participate in the chaperoning of proteins2–5,13. We observed that cultured cells and primary tumours enriched in the high pI HSP90 species were also enriched in high-molecular-weight, multimeric forms of HSP90 and of other essential chaperome members (Fig. 1d and Extended Data Fig. 2c–e).
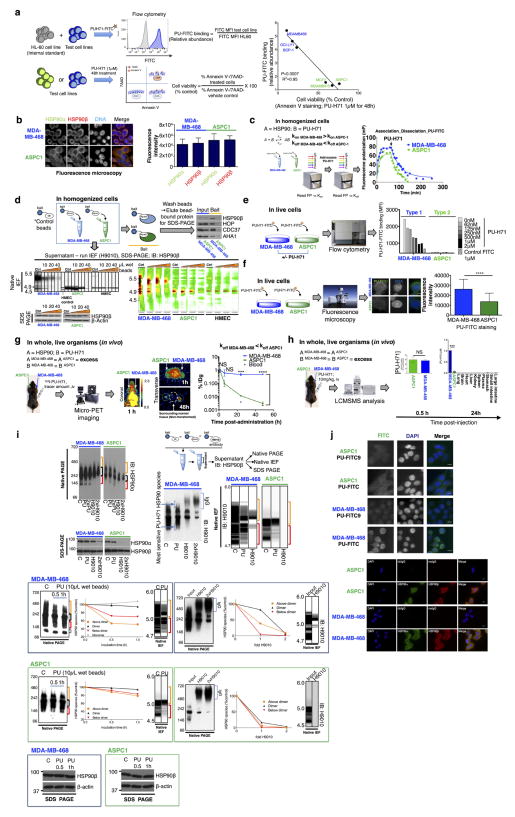
We found that PU-H71, an HSP90 inhibitor that binds to HSP90 more strongly when HSP90 is complexed with co-chaperones and onco-client proteins7,18,19, also bound HSP90 more tightly in type 1 than in type 2 cells (Extended Data Fig. 3a–j). This was independent of chaperome expression or intracellular ATP levels (as PU-H71 is an ATP competitor) (Extended Data Fig. 4). At the molecular level, and unlike the anti-HSP90 antibody H9010, the small fraction of cellular HSP90 that was part of the high-molecular-weight species enriched in type 1 tumours was most sensitive to PU-H71 (Extended Data Fig. 3c, h).
Our data suggested that a biochemically altered chaperome exists in type 1 tumours, so we investigated its composition (Fig. 1e–h). HSP90 protein isolates in type 1 tumours contained a significant enrichment of a number of chaperome proteins known to function as chaperone, co-chaperone, scaffolding, adaptor, interface mediators, foldase and isomerase proteins. Surprisingly, they also incorporated a large number of HSP70 chaperone regulators, in addition to the expected and known HSP90 regulators (Supplementary Discussion). Similarly, an HSP70- directed bait isolated numerous HSP90 regulators in type 1 tumours (Fig. 1f and Extended Data Fig. 5a–e). Multiple connectivity networks that integrate the HSP90 and HSP70 machineries and expand their functional reach through participating scaffolding proteins were characteristic of type 1 but not of type 2 tumours or non-transformed cells (Fig. 1g). High-molecular-weight complexes that incorporate HSP90, HOP, HSC70 (heat shock cognate 70 kDa protein, the constitutively expressed HSP70 paralogue also known as HSPA8)2 and its co-chaperone HSP110 (heat shock 105 kDa/110 kDa protein 1)2, were present in type 1 but not in type 2 or non-transformed cells. The HSP90 bait readily depleted these multimeric species but left the non-bound HSC70 and HSP110 species unaltered (Fig. 1h). Similarly, dual knock-down of HSP90α and HSP90β or AHA1 modulated the high molecular HSC70 complexes only in type 1 tumours; knockdown of HSP110 modulated the multimeric HSP90 complexes only in type 1 tumours. Only the HOP knockdown modulated HSP90 and HSP70 complexes in both tumour types (Extended Data Fig. 5f, g). HSP90 was functional in both tumour types (chaperoned the kinases EGFR and p-S6K) and knockdown of HSP90 and AHA1 inhibited this activity in both tumour types (Extended Data Fig. 5g). Both the HSP90α and HSP90β paralogues, but mainly HSC70 and not HSP70 (the inducible HSP70 paralogue also known as HSP72 or HSP70-1)2, participated in the reconfiguration of the chaperome in type 1 tumours (Extended Data Fig. 5h, i). Substantial reconfiguration of the chaperome organization only modestly affected the total chaperome levels (Extended Data Fig. 5g, h).
Together, these results lend support for the existence in type 1 tumours of HSP90- and HSP70-centric complexes that incorporate the co-chaperones of both machineries and integrate the chaperome into a large functional and physical network (Fig. 1h). Through scaffolding, adaptor, and interface modulator proteins, they bridge the chaperome to numerous cellular processes vital for tumour cell function. We refer to this highly integrated chaperome network of type 1 tumours as the epichaperome. Only a fraction of the entire chaperome pool participates in the chaperome rewiring of type 1 tumours. In contrast to the integrated epichaperome found in type 1 cells, no such integration is found in normal cells and type 2 tumours. In those cases, the HSP90 machinery only loosely interacts with the HSP70 machinery, mainly through the ubiquitous HSP90–HOP–HSP70 connection. In type 2 tumours, the two major chaperome machineries co-exist as insular chaperome communities that are only partially connected to each other.
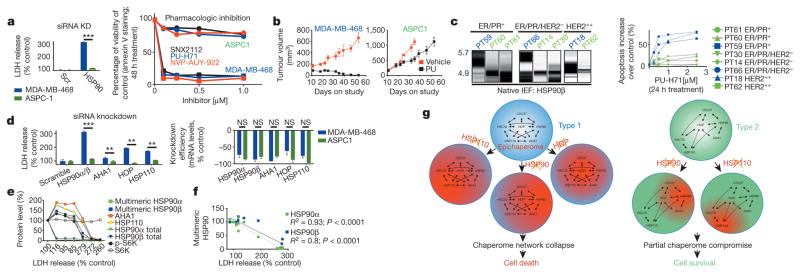
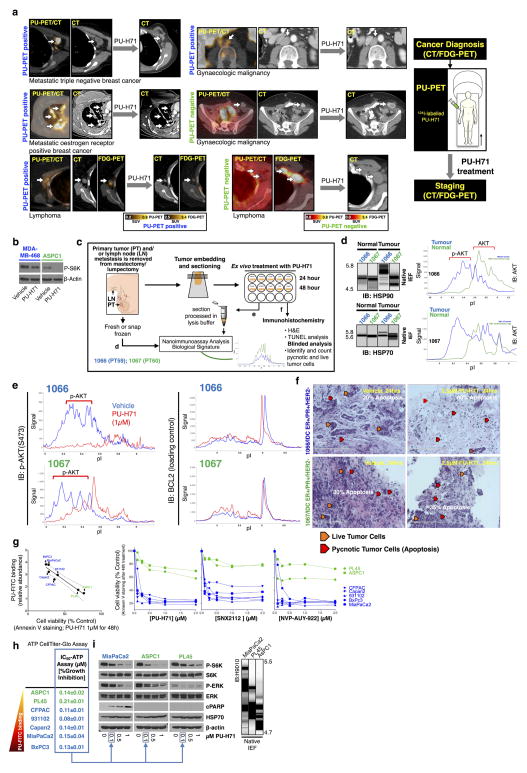
Next, we investigated the functional relevance of the integrated epichaperome as compared to that of individual chaperome members or individual chaperome machineries. First, we investigated the reliance of type 1 and type 2 tumours on individual chaperome members (Fig. 2). In cells with essentially identical HSP90 levels, targeting of both HSP90α and HSP90β was toxic to type 1 but not type 2 tumours (Fig. 2a). We confirmed this in mice bearing xenograft tumours (Fig. 2b), in primary specimens ex vivo (Fig. 2c), and in human patients (Extended Data Fig. 6a). By contrast, while being toxic only to type 1 tumours, targeting of HSP90 inactivated its chaperone activity in both tumour types (see inhibition and/or degradation of HSP90 regulated proteins and pathways, such as EGFR and PI3K/AKT, and cell growth inhibition with similar half-maximum inhibitory concentration (IC50) potencies; Extended Data Fig. 6b–i). As observed for HSP90, downregulation of other individual chaperome members led to cell death in type 1 but not in type 2 cells (Fig. 2d). When we interfered with epichaperome formation by reducing the levels of one of its components, AHA1, cells became less amenable to killing by PU-H71 (Extended Data Fig. 7a). From these observations, we propose that the epichaperome has a role as the survival facilitator of type 1 tumours. In type 1 tumours, we observed that a striking decline (>95% at protein level, Fig. 2e and Extended Data Fig. 7b–f) in overall total HSP90 levels was initially paralleled by an increase in the epichaperome. This occurred by an increased production of chaperome members that presumably sequestered the remaining HSP90 into the high-molecular-weight complexes. Under these conditions of low total HSP90 but high epichaperome levels, no cell death was observed. HSP90 function was also not impaired (see EGFR and p-S6K). As HSP90 levels continued to drop, however, an inflection was observed. At this point, the epichaperome levels dropped, chaperome members were depleted, a sudden drop in HSP90 function occurred, and cell death ensued. Epichaperome expression significantly correlated with cell viability (Fig. 2f). In contrast, in type 2 cells, a similar drastic reduction of HSP90 levels halted its activity, but failed to re-wire the chaperome into the epichaperome and did not result in cell death (Extended Data Fig. 7g). Together, these findings are consistent with the formation of the epichaperome as a survival mechanism for type 1 tumours. When the epichaperome is dismantled by ablation of a chaperome component, the network collapses and leads to cell death. In type 2 tumours in which the integration of the chaperome is only partial and most chaperome members function as insular communities, depletion of a chaperome member only ‘locally’ compromises the chaperome, while overall cellular survival is maintained (Fig. 2g).
Figure 2. The epichaperome facilitates cancer cell survival.
a, Changes in cell viability upon HSP90α and HSP90β knockdown (mean ± s.d., unpaired t-test, n = 6) or pharmacologic inhibition (mean, n = 3), as indicated. ***P < 0.001. b, Tumour volume in mice (n = 5) treated for the indicated time with PU-H71 or vehicle. Error bars show mean ± s.d. c, Multimeric HSP90 complexes in primary breast cancer specimens (n = 8), clustered by biologic subtype, and their ex vivo sensitivity to PU-H71. d, Cytotoxicity upon siRNA knockdown of key chaperome members. For knockdown efficiency see messenger RNA (right) and protein levels (see Extended Data Fig. 5g). Error bars show mean ± s.d., unpaired t-test, n = 6. **P < 0.01; ***P < 0.001. LDH, lactate dehydrogenase. e, Epichaperome, total chaperome levels, chaperome activity and cell viability of type 1 cells in which several concentrations of siRNAs (n = 7) against HSP90α and HSP90β were titrated in. f, Correlative analysis between epichaperome levels and cell viability for data in e (Pearson’s r, two-tailed, n = 14). See also Extended Data Fig. 7. For uncropped gel data, see Supplementary Fig. 1. g, Summary schematic.
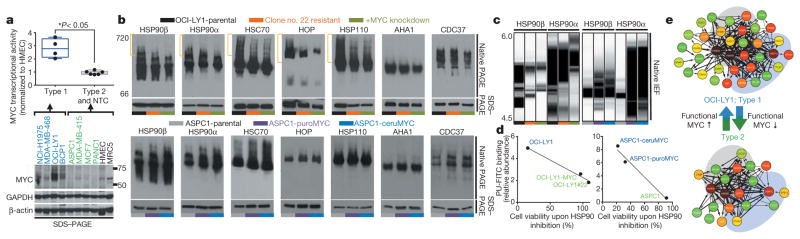
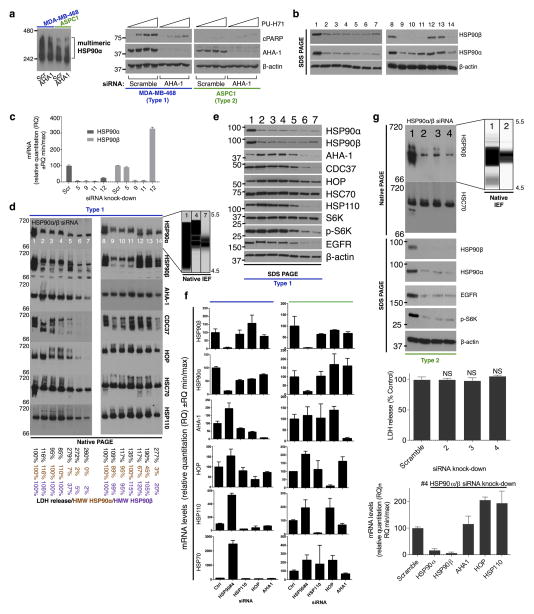
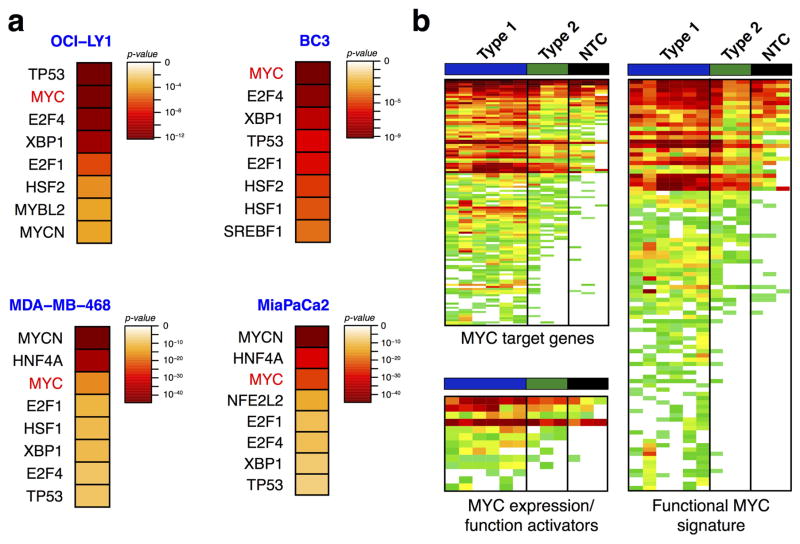
To understand the molecular mechanisms leading to epichaperome formation, we investigated the HSP90 interactome in type 1 and type 2 cells (Extended Data Fig. 5b). We identified MYC as a transcription factor that could most probably explain the protein signature observed in type 1 tumours. MYC target genes were significantly enriched in type 1 tumours, as was a MYC transcriptional signature and positive regulators of MYC expression/function (Extended Data Fig. 8). We experimentally confirmed a significantly higher MYC transcriptional activity in type 1 versus type 2 cancer cells (Fig. 3a). Knockdown of MYC re-wired type 1 cells into type 2; this was reflected in the composition of the chaperome complexes, the binding to PU-H71 and reduced sensitivity to HSP90 inhibition (Fig. 3b–d). We also observed a decrease in MYC mRNA and protein levels in type 1 cells that became resistant to HSP90 inhibitors after long-term treatment with suboptimal HSP90 inhibitor concentrations, and demonstrated their rewiring into type 2 cells (Fig. 3b–d and Extended Data Fig. 9a–e). However, the introduction of a functional MYC gene into a type 2 cancer cell rewired it to become type 1 (Fig. 3b–d and Extended Data Fig. 9f–l). Oncogenes that require single chaperome machinery activity for sufficient transformative power (for example, vSRC and mutated MET kinase require HSP90 (ref. 3)) were unable to induce the formation of the epichaperome, nor was epichaperome formation necessary to buffer their oncogenic function (Extended Data Fig. 9m–p). Together these findings suggest that the transcription factor, MYC, at least in part, causes the molecular rewiring of the chaperome into the epichaperome as observed in type 1 tumours (Fig. 3e).
Figure 3. MYC is a driver of chaperome rewiring into the epichaperome.
a, MYC transcriptional activity (top) and protein levels (bottom) in the indicated cells. β-actin and GAPDH, loading controls (unpaired t-test, each data point is the mean of two technical replicates and represents a cell line). HMEC, human mammary epithelial cells. b–d, Changes in multimeric chaperome complexes (top) and total protein (bottom) in the indicated homogenates of cells where MYC levels were modulated as indicated. Data were repeated independently twice with representative data shown. In d each data point is the mean from two independent experiments and represents a cell line. For uncropped gel data, see Supplementary Fig. 1. e, Summary schematic showing MYC as the cellular switch for epichaperome assembly and disassembly.
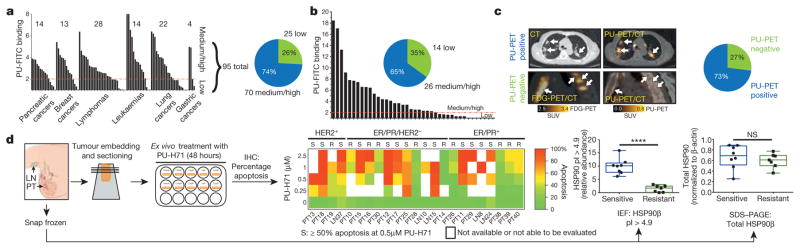
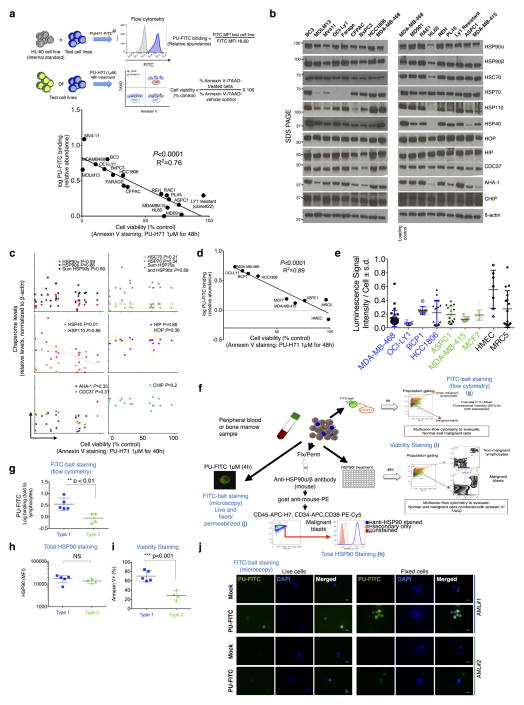
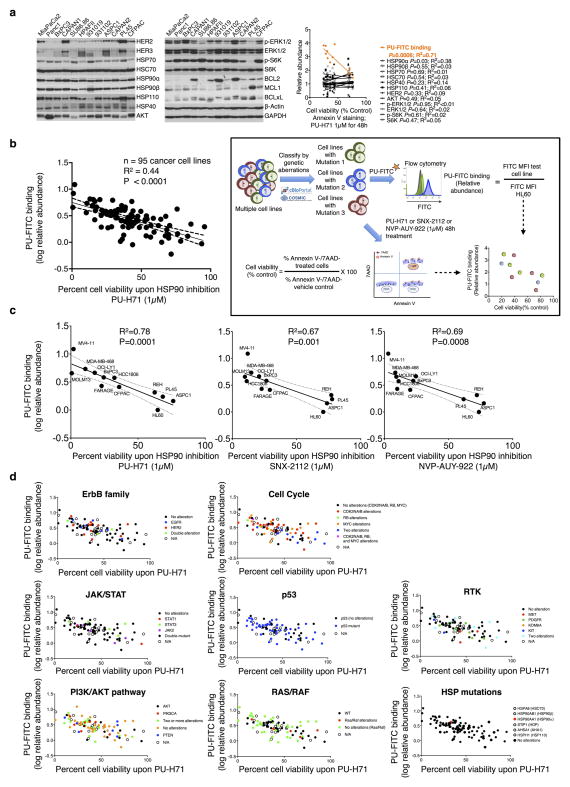
The data point to the epichaperome and not the individual chaperome members as potential targets of chaperome-directed intervention in cancer. We thus assessed the prevalence of tumours expressing the epichaperome complexes. In probing cancer cell lines representing pancreatic, gastric, lung, and breast cancers, as well as lymphomas and leukaemias, we found that approximately 60–70% presented medium to high levels of epichaperome complexes (Fig. 4a). Similar results were obtained with primary liquid tumours (Fig. 4b) and solid tumours including lymphomas (Fig. 4c). This establishes that over half of tumours tested use the epichaperome irrespective of their subtype, provenance or genetic background. Toxicity to HSP90 inhibition correlated with the presence of the epichaperome (P = 0.0006; R2 = 0.71) but was independent of the levels of chaperome members, HSP90 client proteins, anti-apoptotic proteins and genetic alterations. This correlation held in over 90 cell lines encompassing breast cancer, lung cancer, pancreatic and gastric cancers, leukaemia and lymphomas (P < 0.0001; R2 = 0.44) and was true for several HSP90 inhibitors19,20 (Extended Data Fig. 10). The data thus indicate that it is the abundance of the epichaperome, and not merely its existence, that is indicative of the reliance of tumours on the epichaperome. If patients were to be selected for epichaperome therapy, not only the existence of this species but also its abundance should be measured. To further confirm these findings, we collected primary breast cancer specimens (n = 40) obtained from surgery. Of these, 23 specimens were suitable samples to be evaluated for PU-H71 sensitivity and/or epichaperome abundance by isoelectric focusing (IEF) (that is, HSP90 of a pI > 4.9) and total HSP90 by SDS–PAGE (Fig. 4d). In these specimens, we found a spectrum of sensitivities, ranging from 0% to 100% for apoptotic response, with 56% undergoing at least 50% apoptosis when challenged with 0.5 μM PU-H71. Abundance of the epichaperome but not of total HSP90 significantly correlated with sensitivity (Fig. 4d, P < 0.0001).
Figure 4. More than half of all tumours tested express the epichaperome.
a–c, Epichaperome measurement (abundance measured by PU-FITC, see Methods) in a panel of 95 cancer cell lines (a), 40 primary acute myeloid leukaemias (AMLs) (b), and epichaperome detection (by PU-PET, see Methods) in 51 solid tumours and lymphomas, in patients (c). Each bar represents a cell line; data are the mean from two independent experiments. For PU-PET, cross-sectional CT and PU-PET images of representative tumours are shown each at the same transaxial plane. Location of the tumours is indicated by arrows. Scale bars, PET window display intensity scales, with upper and lower standardized uptake value (SUV) thresholds. d, Ex vivo apoptotic sensitivity of primary breast tumours to PU-H71 (n = 23) was compared to epichaperome abundance and total HSP90 levels. PT, primary tumour; LN, lymph node (error bars represent mean ± s.d., unpaired t-test, n = 15). ****P < 0.0001.
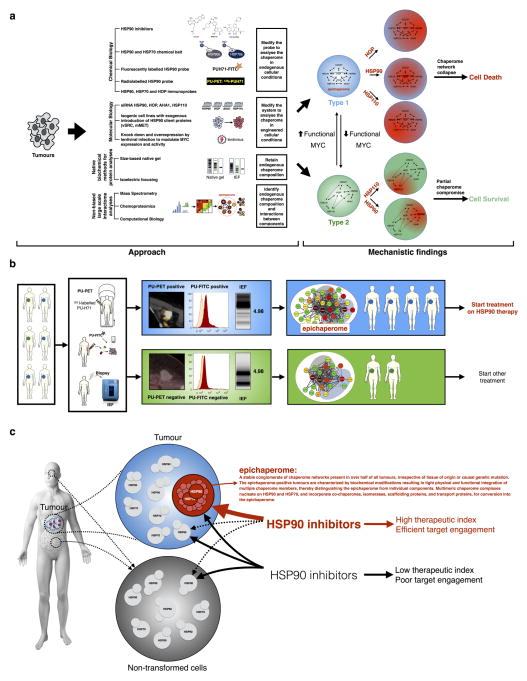
Here we report the discovery of a new mechanism of tumour regulation. Our study unveils a novel usage of the chaperome in epichaperome networks for cancer cell survival. The epichaperome results from changes in the chaperome that are driven by a change in the cellular milieu, that is, activation of MYC, rather than defects in the chaperome members, composition, number or structure. It manifests as an enhanced physical integration of the HSP90 and HSP70 machineries, resulting in the utilization of their capacities in the tumour cell environment, and thereby also presenting a vulnerability that might possibly be exploited therapeutically with pharmacological modulators. Our results offer a blueprint for the future development of therapeutic inhibitors specific for multimeric chaperome complexes, and might encourage further drug developments and advances in innovative companion diagnostics (Extended Data Fig. 1).
Methods
Reagents
HSP90 inhibitors used in this study including PU-H71, PU-DZ13, NVP-AUY922, and SNX-2112 were synthesized as previously reported7,19. 17-DMAG was purchased from Sigma. HSP90 bait (PU-H71 beads)21, HSP70 bait (YK beads)22, biotinylated YK (YK-biotin)22, fluorescently labelled PU-H71 (PU-FITC)23, the control derivatives PU-TEG and PU-FITC9 (ref. 24), and the radiolabelled PU-H71-derivative 124I-PU-H71 (ref. 25) were generated as previously described. The specificity of PU-H71 for HSP90 and over other proteins was extensively analysed7. Thus binding of PU-H71 in cell homogenates, live cells and organisms denotes binding to HSP90 species characteristic of each analysed tumour or tissue. Combined with the findings that PU-H71 binds more tightly to HSP90 in type 1 than in type 2 cells, an observation true for cell homogenates, live cells, and in vivo, at the organismal level, we propose that labelled versions of PU-H71 are reliable tools to perturb, identify and measure the expression of the high-molecular-weight, multimeric HSP90 complexes in tumours. The specificity of YK probes for HSP70 was previously reported22,26–28.
Cell lines
Cell lines were obtained from laboratories at WCMC or MSKCC, or were purchased from the American Type Culture Collection (ATCC) or Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ). Cells were cultured as per the providers’ recommended culture conditions. Cells were authenticated using short tandem repeat profiling and tested for mycoplasma. The pancreatic cancer cell lines include: ASPC-1 (CRL-1682), PL45 (CRL-2558), MiaPaCa2 (CRL-1420), SU.86.86 (CRL-1837), CFPAC (CRL-1918), Capan-2 (HTB-80), BxPc-3 (CRL-1687), HPAFII (CRL-1997), Capan-1 (HTB-79), Panc-1 (CRL-1469), Panc05.04 (CRL-2557) and Hs766t (HTB-134) (purchased from the ATCC); 931102 and 931019 are patient derived cell lines provided by Y. Janjigian, MSKCC. Breast cancer cell lines were obtained from ATCC and include MDA-MB-468 (HTB-132), HCC1806 (CRL-2335), MDA-MB-231 (CRM-HTB- 26), MDA-MB-415 (HTB-128), MCF-7 (HTB-22), BT-474 (HTB-20), BT-20 (HTB-19), MDA-MB-361 (HTB-27), SK-Br-3 (HTB-30), MDA-MB-453 (HTB- 131), T-47D (HTB-133), AU565 (CRL-2351), ZR-75-30 (CRL-1504), ZR-75-1 (CRL-1500). Lymphoma cell lines include: Akata1, Mutu-1 and Rae-1 (provided by W. Tam, WCMC); BCP-1 (CRL-2294), Daudi (CCL-213), EB1 (HTB-60), NAMALWA (CRL-1432), P3HR-1 (HTB-62), SU-DHL-6 (CRL-2959), Farage (CRL-2630), Toledo (CRL-2631) and Pfeiffer (CRL-2632) (obtained from ATCC); HBL-1, MD901 and U2932 (kindly provided by J. Angel Martinez-Climent, Centre for Applied Medical Research, Pamplona, Spain); Karpas422 (ACC-32), RCK8 (ACC-561) and SU-DHL-4 (ACC-495) (obtained from the DSMZ); OCI-LY1, OCI-LY3, OCI-LY4, OCI-LY7 and OCI-LY10 (obtained from the Ontario Cancer Institute); TMD8 (kindly provided by L. M. Staudt, NIH); BC-1 (derived from an AIDS-related primary effusion lymphoma); IBL-1 and IBL-4 (derived from an AIDS-related immunoblastic lymphoma) and BC3 (derived from a non-HIV primary effusion lymphoma). Leukaemia cell lines include: REH (CRL-8286), HL-60 (CCL-240), KASUMI-1 (CRL-2724), KASUMI-4 (CRL-2726), TF-1 (CRL-2003), KG-1 (CCL-246), K562 (CCL-243), TUR (CRL-2367), THP-1 (TIB-202), U937 (CRL-1593.2), MV4-11 (CRL-9591) (obtained from ATCC); KCL-22 (ACC-519), OCI-AML3 (ACC-582) and MOLM-13 (ACC-554) (obtained from DSMZ). The lung cancer cell lines include: NCI-H3122, NCI-H299 (provided by M. Moore, MSKCC); EBC1 (provided by Dr Mellinghoff, MSKCC); PC9 (kindly provided by D. Scheinberg, MSKCC), HCC15 (ACC-496) (DSMZ), HCC827 (CRL-2868), NCI-H2228 (CRL-5935), NCI-H1395 (CRL-5868), NCI-H1975 (CRL-5908), NCI-H1437 (CRL-5872), NCI-H1838 (CRL-5899), NCI-H1373 (CRL-5866), NCI-H526 (CRL-5811), SK-MES-1 (HTB-58), A549 (CCL-185), NCI-H647 (CRL-5834), Calu-6 (HTB-56), NCI-H522 (CRL-5810), NCI-H1299 (CRL-5803), NCI-H1666 (CRL-5885) and NCI-H1703 (CRL-5889) (obtained from ATCC). The gastric cancer cell lines include: MKN74 (obtained from G. Schwarz, Columbia University), SNU-1 (CRL-5971) and NCI-N87 (CRL-5822) (obtained from ATCC), OE19 (ACC-700) (DSMZ). The non-transformed cell lines MRC-5 (CCL-171), human lung fibroblast and HMEC (PCS-600-010), human mammary epithelial cells were obtained from ATCC. NIH-3T3, and NIH-3T3 cell lines stably expressing either mutant MET (Y1248H) or vSRC, were provided by L. Neckers, National Cancer Institute (NCI), USA, and were previously reported29,30.
Primary breast cancer specimens
Patient tissue was obtained with informed consent and authorized through institutional review board (IRB)-approved bio-specimen protocol number 09-121 at Memorial Sloan Kettering Cancer Centre (New York, New York). Specimens were treated for 24 h or 48 h with the indicated concentrations of PU-H71 as previously described31. Following treatment, slices were fixed in 4% formalin solution for 1 h, then stored in 70% ethanol. For tissue analysis, slices were embedded in paraffin, sectioned, slide-mounted, and stained with haematoxylin and eosin (H&E). Apoptosis and necrosis of the tumour cells (as percentage) was assessed by reviewing all the H&E slides of the case (controls and treated ones) in toto, blindly, allowing for better estimation of the overall treatment effect to the tumour. In addition, any effects to precursor lesions (if present) and any off-target effects to benign surrounding tissue, were analysed. Tissue slides were assessed blindly by a breast cancer pathologist who determined the apoptotic events in the tumour, as well as any effect on adjacent normal tissue31.
Primary acute myeloid leukaemia (AML)
Cryopreserved primary AML samples were obtained with informed consent and Weill Cornell Medical College IRB approval (IRB number 0910010677 and IRB number 0909010629). Samples were thawed and cultured for in vitro treatment as described previously32.
Clinical trials
The microdose 124I-PU-H71 PET-CT (Dunphy, M. PET imaging of cancer patients using 124I-PUH71: a pilot study available from: http://clinicaltrials.gov; NCT01269593) and phase I PU-H71 therapeutic (Gerecitano, J. The first-in-human phase I trial of PU-H71 in patients with advanced malignancies available from: http://clinicaltrials.gov; NCT01393509) studies were approved by the institutional review board (protocols 10-139 and 11-041, respectively), and conducted under an exploratory investigational new drug (IND) application approved by the US Food and Drug Administration. Patients provided signed informed consent before participation. 124I-PU-H71 tracer was synthesized in-house by the institutional cyclotron core facility at high specific activity.
Epichaperome detection by PU-PET (124I-PU-H71 positron emission tomography)
For PU-PET, research PET-CT was performed using an integrated PET-CT scanner (Discovery DSTE, General Electric). CT scans for attenuation correction and anatomic coregistration were performed before tracer injection. Patients received 185 megabecquerel (MBq) of 124I-PU-H71 by peripheral vein over two minutes. PET data were reconstructed using a standard ordered subset expected maximization iterative algorithm. Emission data were corrected for scatter, attenuation, and decay. 124I-PU-H71 scans (PU-PET) were performed at 24 h after tracer administration. Each picture shown in Fig. 4c and Extended Fig. 6a is a scan taken of an individual patient. PET window display intensity scales for FDG and PU-PET fusion PET-CT images are given for both PU-PET and FDG-PET. Numbers in the scale bar indicate upper and lower SUV thresholds that define pixel intensity on PET images. The phase I trial included patients with solid tumours and lymphomas who had undergone prior treatment and currently had no curative treatment options. Patient cohorts were treated with PU-H71 at escalating dose levels determined by a modified continuous reassessment model. Each patient was treated with his or her assigned dose of PU-H71 on day 1, 4, 8, and 11 of each 21-day cycle.
Neurons
Human embryonic stem cells (hESCs) were differentiated with a modified dual-SMAD inhibition protocol towards floor plate-based midbrain dopaminergic (mDA) neurons as described previously33. hESCs were maintained on mouse embryonic fibroblasts and passaged with Dispase (STEMCELL Technologies). For each differentiation, hESCs were harvested with Accutase (Innovative Cell Technology). At day 30 of differentiation, hESC-derived mDA neurons were replated and maintained on dishes precoated with polyornithine (PO; 15 μg ml−1), laminin (1 μg ml−1), and fibronectin (2 μg ml−1) in Neurobasal/ B27/l-glutamine-containing medium (NB/B27; Life Technologies) supplemented with 10 μM Y-27632 (until day 32) and with BDNF (brain-derived neurotrophic factor, 20 ng ml−1; R&D), ascorbic acid (AA; 0.2 mM, Sigma), GDNF (glial cell line-derived neurotrophic factor, 20 ng ml−1; R&D), TGFβ 3 (transforming growth factor type β 3, 1 ng ml−1; R&D), dibutyryl cAMP (0.5 mM; Sigma), and DAPT (10 nM; Tocris). Two days after replating, mDA neurons were treated with 1 μg ml−1 mitomycin C (Tocris) for 1 h to kill any remaining non-post mitotic contaminants. Assays were performed at day 65 of neuron differentiation.
Epichaperome abundance measurement using the PU-FITC flow cytometry assay
The PU-FITC assay was performed as previously described7,23. Briefly, cells were incubated with 1 μM PU-FITC at 37 °C for 4 h. Then cells were washed twice with FACS buffer (PBS/0.5% FBS), and resuspended in FACS buffer containing 1 μg ml−1 DAPI. HL-60 cells were used as internal control to calculate fold binding for all cell lines tested. The mean fluorescence intensity (MFI) of PU-FITC in treated viable cells (DAPI negative) was evaluated by flow cytometry. For primary AML specimens, cells were also stained with anti-CD45-APC-H7, to identify blasts and lymphocyte populations (BD biosciences). Blasts and lymphocyte populations were gated based on SSC versus CD45. The fold PU-FITC binding of leukaemic blasts (CD45dim) was calculated relative to lymphocytes (CD45hiSSClow). The FITC derivative FITC9 was used as a negative control.
PU-FITC microscopy
Cells were seeded on coverslips in 6-well plate and cultured overnight. Cells were treated with 1 μM PU-FITC or negative control (PU-FITC9, an HSP90 inert PU-H71 derivative labelled with FITC). At 4 h post-treatment, cells were fixed with 4% formaldehyde at room temperature for 30 min, and the coverslips were mounted on slides with DAPI-Fluoromount-G Mounting Media (Southern Biotech). The images were captured using EVOS FL Auto imaging system (ThermoFisher Scientific) or a confocal microscope (Zeiss LSM5).
HSP90 immunofluorescence staining
Cells were seeded on coverslips and cultured overnight. Cells were fixed with 4% formaldehyde at room temperature for 30 min, washed three times with PBS, and permeabilized with 0.2% Triton X-100 in blocking buffer (PBS/5% BSA) for 10 min. Cells were incubated in blocking buffer for 30 min, and then incubated with rabbit anti-human HSP90α antibody (1:500, Abcam 2928) and mouse anti-human HSP90β (1:500, Stressmarq H9010), or rabbit and mouse normal IgG, in blocking buffer for 1 h. Cells were washed three times with PBS, and incubated with goat anti-mouse Alexa Fluor 568 and goat anti-rabbit Alexa Fluor 488 (1:1,000, ThermoFisher Scientific) in blocking buffer in the dark for 1 h. Cells were then washed three times with PBS, and the coverslips were removed from the plate, and mounted on slides with DAPI-Fluoromount-G Mounting Media (Southern Biotech). The images were captured using EVOS FL Auto imaging system (ThermoFisher Scientific) or a confocal microscope (Zeiss LSM5). Fluorescence intensity was quantified by the integrated density algorithm as implemented in ImageJ.
PU-FITC or GM-cy3B binding to HSP90 in cell homogenates
Assays were carried out in black 96-well microplates (Greiner Microlon Fluotrac 200). A stock of 10 μM PU-FITC (or GM-cy3B34) was prepared in DMSO and diluted with Felts buffer (20 mM Hepes (K), pH 7.3, 50 mM KCl, 2 mM DTT, 5 mM MgCl2, 20 mM Na2MoO4, and 0.01% NP40 with 0.1 mg ml−1 BGG). To each well was added the fluorescent dye-labelled HSP90 ligand (3 nM PU-FITC or 6 nM GM-cy3B), and cell lysates (7.5 μg) in a final volume of 100 μl Felts buffer. For each assay, background wells (buffer only), and tracer controls (PU-FITC only) were included on assay plate. To determine the equilibrium binding of GM-cy3b, increasing amounts of lysate (up to 20 μg of total protein) were incubated with tracer. The assay plate was placed on a shaker at room temperature for 60 min and the FP values in mP were measured every 5 min. At time t = 60 min, dissociation of fluorescent ligand was initiated by adding 1 μM PU-H71 in Felts buffer to each well and then placing the assay plate on a shaker at room temperature and measuring the FP values in mP every 5 min. The assay window was calculated as the difference between the FP value recorded for the bound fluorescent tracer and the FP value recorded for the free fluorescent tracer (defined as mP – mPf). Measurements were performed on a Molecular Devices SpectraMax Paradigm instrument (Molecular Devices, Sunnyvale, CA), and data were imported into SoftMaxPro6 and analysed in GraphPad Prism 5.
Protein analysis by the NanoPro capillary-based immunoassay platform
To identify and separate chaperome complexes in tumours, and to overcome the limitations of classical protein chromatography methods for resolving complexes of similar composition and size, we took advantage of a capillary-based platform that combines isoelectric focusing (IEF) with immunoblotting capabilities35. This methodology uses an immobilized pH gradient to separate native multimeric protein complexes based on their isoelectric point (pI), and allows for subsequent probing of immobilized complexes with specific antibodies. The method uses only minute amounts of sample, thus enabling the interrogation of primary specimens. Cultured cells were lysed in 20 mM HEPES pH 7.5, 50 mM KCl, 5 mM MgCl2, 0.01% NP40, 20 mM Na2MoO4 buffer, containing protease and phosphatase inhibitors. Primary specimens were lysed in either Bicine-Chaps or RIPA buffers (ProteinSimple). Total protein assay was performed on an automated system, NanoPro 1000 Simple Western (ProteinSimple), for charge-based separation. Briefly, total cell lysates were diluted to a final protein concentration of 250 ng μl−1 using a master mix containing 1× Premix G2 pH 3-10 separation gradient (Protein simple) and 1×isoelectric point standard ladders (ProteinSimple). Samples diluted in this manner maintained their native charge state, and were loaded into capillaries (ProteinSimple) and separated based on their isoelectric points at a constant power of 21,000 μ Watts for 40 min. Immobilization was performed by UV-light embedded in the Simple Western system, followed by incubations with anti- HSP90β (SMC-107A, StressMarq Biosciences), anti-HSP90α (ab2928, Abcam), anti-HSP70 (SPA-810, Enzo), AKT (4691), P-AKT (9271) or BCL2 (2872) from Cell Signaling Technology and subsequently with HRP-conjugated anti-Mouse IgG (1030-05, SouthernBiotech) or with HRP-conjugated anti-Rabbit IgG (4010-05, SouthernBiotech). Protein signals were quantitated by chemiluminescence using SuperSignal West Dura Extended Duration Substrate (Thermo Scientific), and digital imaging and associated software (Compass) in the Simple Western system, resulting in a gel-like representation of the chromatogram. This representation is shown for each figure.
Western blotting
Protein was extracted from cultured cells in 20 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40 buffer with protease and phosphatase inhibitors added (Complete tablets and PhosSTOP EASYpack, Roche). Ten to fifty μg of total protein was subjected to SDS–PAGE, transferred onto nitrocellulose membrane, and incubated with indicated antibodies. HSP90β (SMC-107) and HSP110 (SPC-195) antibodies were purchased from Stressmarq; HER2 (28-0004) from Zymed; HSP70 (SPA-810), HSC70 (SPA-815), HIP (SPA-766), HOP (SRA-1500), and HSP40 (SPA- 400) from Enzo; HSP90β (ab2927), HSP90α (ab2928), p23 (ab2814), GAPDH (ab8245) and AHA1 (ab56721) from Abcam; cleaved PARP (G734A) from Promega; CDC37 (4793), CHIP (2080), EGFR (4267), S6K (2217), phospho-S6K (S235/236) (4858), P-AKT (S473) (9271), AKT (4691), P-ERK (T202/Y204) (4377), ERK (4695), MCL1 (5453), Bcl-XL (2764), BCL2 (2872), c-MYC (5605) and HER3 (4754) from Cell Signaling Technology; and β-actin (A1978) from Sigma-Aldrich. The blots were washed with TBS/0.1% Tween 20 and incubated with appropriate HRP-conjugated secondary antibodies. Chemiluminescent signal was detected with Enhanced Chemiluminescence Detection System (GE Healthcare) following the manufacturer’s instructions.
Native-cognate antibodies
We screened a panel of anti-chaperome antibodies for those that interacted with the target protein in its native form. We reasoned that these antibodies were more likely to capture stable multimeric forms of the chaperome members. These native-cognate antibodies were used in native-PAGE and IEF analyses of chaperome complexes. HSP90β (SMC-107) and HSP110 (SPC-195) antibodies were purchased from Stressmarq; HSP70 (SPA-810), HSC70 (SPA-815), HOP (SRA-1500), and HSP40 (SPA-400) from Enzo; HSP90β (ab2927), HSP90α (ab2928), and AHA1 (ab56721) from Abcam; CDC37 (4793) from Cell Signaling Technology.
Native gel electrophoresis
Cells were lysed in 20 mM Tris pH 7.4, 20 mM KCl, 5 mM MgCl2, 0.01% NP40, and 10% glycerol buffer by a freeze-thaw procedure. Primary samples were lysed in either Bicine-Chaps or RIPA buffers (ProteinSimple). Twenty-five to one hundred μg of protein was loaded onto 4–10% native gradient gel and resolved at 4 °C. The gels were immunoblotted as described above following either incubation in Tris-Glycine-SDS running buffer for 15 min before transfer in regular transfer buffer for 1 h, or directly transferred in 0.1% SDS-containing transfer buffer for 1 h.
siRNA knockdown
Cells were plated at 1 × 106 per 6 well-plate and transfected with an siRNA against human AHA1 (AHSA1; 5′-TTCAAATTGGTCCACGGATAA-3′), HSP90α (HSP90AA1; no. 1 5′-ATGGCATGACAACTACTTTAA-3′; no. 2 5′-AACC CTGACCATTCCATTATT-3′; no.3 5′-TGCACTGTAAGACGTATGTAA-3′), HSP90β (HSP90AB1; no., 5′-CAAGAATGATAAGGCAGTTAA-3′; no. 5′-TACGTT GCTCACTATTACGTA-3′; no.3 5′-CAGAAGACAAGGAGAATTACA-3′) HSP90α /β (no.1 5′-CAGAATGAAGGAGAACCAGAA-3′, no.2 5′-CACAACGA TGATGAACAGTAT-3′), HSP110 (HSPH1; 5′-AGGCCGCTTTGTAGTTC AGAA-3′) from Qiagen or HOP (STIP1) (Dharmacon; M-019802-01), or a negative control (scramble; 5′-CAGGGTATCGACGATTACAAA-3′) with Lipofectamine RNAiMAX reagent (Invitrogen), incubated for 72 h and subjected to further analysis.
qRT–PCR
Total mRNA was isolated using TRIzol Reagent (Invitrogen) following the manufacturer’s recommended protocol. Reverse transcription of mRNA into cDNA was performed using QuantiTect Reverse Transcription Kit (Qiagen). qRT–PCR was performed using PerfeCTa SYBR (Quanta Bioscience), 10 nM AHSA1 (forward: 5′-GCGGCCGCTTCTAGTAGTTT-3′ and reverse: 5′-CATCTCTCTCCGTCCAGTGC-3′) and GAPDH (forward: 5′-CAAAGGCACAGTCAAGGCTGA-3′ and reverse: 5′-TGGTGAAGACG CCAGTAGATT-3′) primers, or 1× QuantiTect Primers for HSP110 (HSPH1), HSP90α (HSP90AA1), HSP90β (HSP90AB1), HSP70 (HSPA1A), HOP (STIP1) (Qiagen) following recommended PCR cycling conditions. Melting curve analysis was performed to ensure product uniformity.
Protein depletion
To investigate which of the two HSP70 paralogues is involved in epichaperome formation we performed immunodepletions with HSP70 and HSC70 antibodies. Protein lysates were immunoprecipitated consecutively three times with either an HSP70 (Enzo, SPA-810), HSC70 (Enzo, SPA-815) or HOP (kindly provided by M. B. Cox, University of Texas at El Paso), or with the same species normal antibody as a negative control (Santa Cruz). The resulting supernatant was collected and run on a native or a denaturing gel.
Native gel electrophoresis and isoelectric focusing (IEF) under denaturing conditions
Tumour lysates were mixed with 10 M urea (dissolved in Felts buffer) to reach the indicated final concentrations of 2 M, 4 M and 6 M. After incubation for 10 min at room temperature or frozen overnight at −80 °C, the lysates were loaded onto 4–10% native gradient gel and resolved at 4 °C or applied to the IEF capillary. The HSP90β bands were detected by using antibody purchased from Stressmarq (SMC-107).
MYC knockdown by lentiviral-delivered shRNA
A lentiviral vector expressing the MYC shRNA, as previously described36, was requested from Addgene (Plasmid 29435, c-MYC shRNA sequence: GACGAGAACAGTTGAAACA). Viruses were prepared by co-transfecting the shRNA vector, the packaging plasmid psPAX2 and the envelop plasmid pMD2.G into HEK293 cells. OCI-LY1 cells were then infected with lentiviral supernatants in the presence of 4 μg ml−1 polybrene for 24 h. Following flow cytometry selection for positive cells, cells were expanded for further experiments. The MYC protein level was confirmed at 10 days post-infection by western blot using the anti-MYC antibody (Cell Signaling Technology, 5605).
Exogenous MYC expression
Viruses were prepared by co-transfection of the lentiviral vector expressing the MYC shRNA with pLM-mCerulean-2A-cMyc (Addgene, 23244) or pCDH-puro-cMYC (Addgene, 46970), the packaging plasmid psPAX2, and the envelope plasmid pMD2.G into HEK293 cells. ASPC1 cells were then infected with lentiviral supernatants in the presence of 4 μg ml−1 polybrene for 24 h and sorted for mCerulean positive cells or selected with puromycin treatment. Changes in cell size after infection were monitored by analysing the forward scatter (FSC) of intact cells via flow cytometry. MYC protein levels were analysed at 4 days post-infection by western blot.
MYC transcription factor binding activity assay
Whole cell extracts were prepared by homogenizing cells in RIPA buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 1% NP40, 0.25% sodium deoxycholate, 10% glycerol, protease inhibitors). MYC activity was determined using the TransAM c-Myc Kit (Active Motif, 43396), following the manufacturer’s instructions.
Cell viability assessment ATP assay
Cell viability was assessed using CellTiter-Glo luminescent Cell Viability Assay (Promega) after a 72 h PU-H71 treatment. The method determines the number of viable cells in culture based on quantification of the ATP present, which signals the presence of metabolically active cells, and was performed as previously reported37. For the annexin V staining, cells were labelled with Annexin V-PE and 7AAD after PU-H71 treatment for 48 h, as previously reported38. The necrotic cells were defined as annexin V+/7AAD+, and the early apoptotic cells were defined as annexin V+/7AAD−. For the LDH assay the release of lactate dehydrogenase (LDH) into the culture medium only occurs upon cell death. Following indicated treatment, the culture medium was collected and centrifuged to remove living cells and cell debris. The collected medium was incubated at room temperature for 30 min with the Cytotox-96 Non-radioactive Assay kit (Promega) LDH substrate.
In vivo studies
All animal studies were conducted in compliance with MSKCC’s Institutional Animal Care and Use Committee (IACUC) guidelines. Female athymic nu/nu mice (NCRNU-M, 20–25 g, 6 weeks old) were obtained from Harlan Laboratories and were allowed to acclimatize at the MSKCC vivarium for 1 week before implanting tumours. Mice were provided with food and water ad libitum. Tumour xenografts were established on the forelimbs for PET imaging and on the flank for efficacy studies. Tumours were initiated by sub-cutaneous injection of 1 × 107 cells for MDA-MB-468 and 5 × 106 for ASPC1 in a 200 μl cell suspension of a 1:1 v/v mixture of PBS with reconstituted basement membrane (BD Matrigel, Collaborative Biomedical Products). Before administration, a solution of PU-H71 was formulated in citrate buffer. Sample size was chosen empirically based on published data39. No statistical methods were used to predetermine sample size. Animals were randomly assigned to groups. Studies were not conducted blinded.
Small-animal PET imaging
Imaging was performed with a dedicated small-animal PET scanner (Focus 120 microPET; Concorde Microsystems, Knoxville, TN). Mice were maintained under 2% isoflurane (Baxter Healthcare, Deerfield, IL) anaesthesia in oxygen at 2 litres per min during the entire scanning period. To reduce the thyroid uptake of free iodide arising from metabolism of tracer, mice received 0.01% potassium iodide solution in their drinking water starting 48 h before tracer administration. For PET imaging, each mouse was administered 9.25 MBq (250 μCi) of 124I-PU-H71 via the tail vein. List-mode data (10 to 30 min acquisitions) were obtained for each animal at various time points post-tracer administration. An energy window of 420–580 keV and a coincidence timing window of 6 ns were used. The resulting list-mode data were sorted into 2-dimensional histograms by Fourier rebinning; transverse images were reconstructed by filtered back projection (FBP). The image data were corrected for non-uniformity of scanner response, dead-time count losses, and physical decay to the time of injection. There was no correction applied for attenuation, scatter, or partial-volume averaging. The measured reconstructed spatial resolution of the Focus 120 is 1.6-mm FWHM at the centre of the field of view. Region of interest (ROI) analysis of the reconstructed images was performed using ASIPro software (Concorde Microsystems, Knoxville, TN), and the maximum pixel value was recorded for each tissue/organ ROI. A system calibration factor (that is, μCi per ml per cps per voxel) that was derived from reconstructed images of a mouse-size water-filled cylinder containing 18F was used to convert the 124I voxel count rates to activity concentrations (after adjustment for the 124I positron branching ratio). The resulting image data were then normalized to the administered activity to parameterize the micro- PET images in terms of per cent injected dose per gram (%ID per g) ( corrected for decay of 124I to the time of injection). Post-reconstruction smoothing was applied only for visual representation of images in the figures. Upon euthanasia, radio-activity (124I) was measured in a gamma-counter (Perkin Elmer 1480 Wizard 3 Auto Gamma counter) using a 400–600 keV energy window. Count data were background- and decay-corrected to the time of injection, and the percent injected dose per gram (%ID per g) for each tumour sample was calculated using a calibration curve to convert counts to radioactivity, followed by normalization to the total activity injected.
Efficacy studies
Mice (n = 5) bearing MDA-MB-468 or ASPC1 tumours reaching a volume of 100–150 mm3 were treated i.p. using PU-H71 (75mg per kg) or vehicle, on a 3 times per week schedule, as indicated. Tumour volume (in mm3) was determined by measurement with Vernier calipers, and was calculated as the product of its length × width2 × 0.5. Tumour volume was expressed on indicated days as the median tumour volume ± s.d. indicated for groups of mice. Mice were euthanized after similar PU-H71 treatment periods, and at a time before tumours reached a size that resulted in discomfort or difficulty in physiological functions of mice in the individual treatment group, in accordance with our IUCAC protocol.
LC-MS/MS analyses
Frozen tissue was dried and weighed before homogenization in acetonitrile/H2O (3:7). PU-H71 was extracted in methylene chloride, and the organic layer was separated and dried under vacuum. Samples were reconstituted in mobile phase. The concentrations of PU-H71 in tissue or plasma were determined by high-performance LC-MS/MS. PU-H71-d6 was added as the internal standard40. Compound analysis was performed on the 6410 LC-MS/MS system (Agilent Technologies) in multiple reaction monitoring mode using positive-ion electrospray ionization. For tissue samples, a Zorbax Eclipse XDB-C18 column (2.1 × 50 mm, 3.5 μm) was used for the LC separation, and the analyte was eluted under an isocratic condition (80% H2O + 0.1% HCOOH: 20% CH3CN) for 3 min at a flow rate of 0.4 ml min−1. For plasma samples, a Zorbax Eclipse XDB-C18 column (4.6 × 50 mm, 5 μm) was used for the LC separation, and the analyte was eluted under a gradient condition (H2O + 0.1% HCOOH:CH3CN, 95:5 to 70:30) at a flow rate of 0.35 ml min−1.
Chemical bait precipitation and proteomics
Protein extracts were prepared either in 20 mM HEPES pH 7.5, 50 mM KCl, 5 mM MgCl2, 1% NP40, and 20 mM Na2MoO4 for PU-H71 beads pull-down, or in 20 mM Tris pH 7.4, 150 mM NaCl, and 1% NP40 for YK beads pull-down. Samples were incubated with the PU-H71 beads (HSP90 bait) for 3–4 h or with the YK beads (HSP70 bait, for chemical precipitation) overnight, at 4 °C, then washed and subjected to SDS–PAGE with subsequent immunoblotting and western blot analysis. For HSP70 proteomic analyses, cells were incubated with a biotinylated YK-derivative, YK-biotin. Briefly, MDA-MB-468 cells were treated for 4 h with 100 μM biotin-YK5 or d-biotin as a negative control. Cells were collected and lysed in 20 mM Tris pH 7.4, 150 mM NaCl, and 1% NP40 buffer. Protein extracts were incubated with streptavidin agarose beads (Thermo Scientific) for 1 h at 4 °C, washed with 20 mM Tris pH 7.4, 150 mM NaCl, and 0.1% NP40 buffer and applied onto SDS– PAGE. The gels were stained with SimplyBlue Coomassie stain (Invitrogen Life Science Technologies). Proteomic analyses were performed using the published protocol7,18,22. Control beads contained an inert molecule as previously described7,18,22.
Protein identification by nano-liquid chromatography coupled to tandem mass spectrometry (LC–MS/MS) analysis
Affinity-purified protein complexes from type 1 tumours (n = 6; NCI-H1975, MDA-MB-468, OCI-LY1, Daudi, IBL1, BC3), type 2 tumours (n = 3; ASPC1, OCI-LY4, Ramos) and from non-transformed cells (n = 3; MRC5, HMEC and neurons) were resolved using SDS-polyacrylamide gel electrophoresis, followed by staining with colloidal, SimplyBlue Coomassie stain (Invitrogen Life Science Technologies) and excision of the separated protein bands. Control beads that contained an inert molecule were subjected to the same steps as PU-H71 and YK beads and served as a control experiment. To ensure that we captured a majority of the HSP90 complexes in each cell type, we performed these studies under conditions of HSP90-bait saturation. The number of gel sections per lane averaged to be 14. In situ trypsin digestion of gel bound proteins, purification of the generated peptides and LC–MS/MS analysis were performed using our published protocols7,18,22. After the acquisition of raw files, Proteowizard (version 3.0.3650)41 was used to create a Mascot Generic Format (mgf) file containing accurate mass for each peak and its corresponding ms2 ions. Each mgf was then subjected to search a human segment of Uniprot protein database (20,273 sequences, European Bioinformatics Institute, Swiss Institute of Bioinformatics and Protein Information Resource) using Mascot (Matrix Science; version 2.5.0; http://www.matrixscience.com). Decoy proteins were added to the search to allow for the calculation of false discovery rates (FDR). The search parameters were as follows: (i) two missed cleavage tryptic sites were allowed; (ii) precursor ion mass tolerance = 10 p.p.m.; (iii) fragment ion mass tolerance = 0.8 Da; and (iv) variable protein modifications were allowed for methionine oxidation, deamidation of asparagine and glutamines, cysteine acrylamide derivatization and protein N-terminal acetylation. MudPit scoring was typically applied using significance threshold score P < 0.01. Decoy database search was always activated and, in general, for merged LS–MS/MS analysis of a gel lane with P < 0.01, false discovery rate averaged around 1%. The Mascot search result was finally imported into Scaffold (Proteome Software, Inc.; version 4_4_1) to further analyse tandem mass spectrometry (MS/MS) based protein and peptide identifications. X! Tandem (The GPM, http://thegpm.org; version CYCLONE (2010.12.01.1) was then performed and its results are merged with those from Mascot. The two search engine results were combined and displayed at 1% FDR. Protein and peptide probability was set at 95% with a minimum peptide requirement of 1. Protein identifications were expressed as Exclusive Spectrum Counts that identified each protein listed. Primary data, such as raw mass spectrometry files, Mascot generic format files and proteomics data files created by Scaffold have been deposited onto the Massive site (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp; MassIVE Accession ID: MSV000079877). In each of the Scaffold files that validate and import Mascot searched files, peptide matches, scoring information (Mascot, as well as X! Tandem search scores) for peptide and protein identifications, MS/MS spectra, protein views with sequence coverage and more, can be easily accessed. To read the Scaffold files, free viewer software can be found at (http://www.proteome-software.com/products/free-viewer/). Peptide matches and scoring information that demonstrate the data processing are available in Supplementary Table 1f–q.
Bioinformatics analyses
The exclusive spectrum count values, an alternative for quantitative proteomic measurements42, were used for protein analyses. CHIP and PP5 were examined and used as internal quality controls among the samples. Statistics were performed using R (version 3.1.3) limma package43,44. For entries with zero spectral counts, and to enable further analyses, we assigned an arbitrary small number of 0.1. The data were then transformed into logarithmic base 10 for analysis. Linear models were fit to the transformed data and moderated standard errors were calculated using empirical Bayesian methods. For Fig. 1f and Extended Data Fig. 5a, a moderated t-statistic was used to compare protein enrichment between type 1 cells and combined type 2 and non-transformed cells45. For Extended Data Fig. 5b, the t-statistic was performed to compare protein enrichment among type 1 cells, type 2 cells and non-transformed cells (see Supplementary Table 1). Heat maps were created to display the selected proteins using the package “gplots” and “lattice”46,47. See Supplementary Table 1 in which the table tab ‘a’ corresponds to Fig. 1f and contains core chaperome networks in type 1, type 2 and non-transformed cells; the table tab ‘b’ corresponds to Extended Data Fig. 5a and contains comprehensive chaperome networks in type 1, type 2 and non-transformed cells; the table tab ‘c’ corresponds to Extended Data Fig. 5b and Extended Data Fig. 8b and contains the HSP90 interactome as isolated by the HSP90 bait in type 1, type 2 and non-transformed cells; the table tab ‘d’ corresponds to Extended Data Fig. 8a and contains upstream transcriptional regulators that explain the protein signature of type1 tumours and the table tab ‘e’ contains metastasis-related proteins characteristic of type 1 tumours.
The protein–protein interaction (PPI) network and upstream transcriptional regulators
To understand the physical and functional protein-interaction properties of the HSP90-interacting chaperome proteins enriched in type 1 tumours, we used the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database48. Proteins displayed in the heat map were uploaded in STRING database to generate the PPI networks. STRING builds functional protein-association networks based on compiled available experimental evidence. The thickness of the edges represents the confidence score of a functional association. The score was calculated based on four criteria: co-expression, experimental and biochemical validation, association in curated databases, and co-mentioning in PubMed abstracts48. Proteins with no adjacent interactions were not shown. The colour scale in nodes indicates the average enrichment of the protein (measured as exclusive spectral counts) in type 1, type 2, and non-transformed cells, respectively. The network layout for type 1 tumours was generated using edge-weighted spring-electric layout in Cytoscape with slight adjustments of marginal nodes for better visualization49. The layout for type 2 and non-transformed cells retains that of type 1 for better comparison. Proteins with average relative abundance values less than 1 were deleted from analyses. The biological processes in which they participate and the functionality of proteins enriched in type 1 tumours were assigned based on gene ontology terms and based on their designated interactome from UniProtKB, STRING, and/or I2D databases48,50–53. The Upstream Regulator analytic, as implemented in Ingenuity Pathways Analysis (IPA, QIAGEN Redwood City, http://www.qiagen.com/ingenuity), was used to identify the cascade of upstream transcriptional regulators that can explain the observed protein expression changes in type 1 tumours. The analysis is based on prior knowledge of expected effects between transcriptional regulators and their target genes stored in the Ingenuity Knowledge Base. The analysis examines how many known targets of each transcription regulator are present in the data set, and calculates an overlap P value for upstream regulators based on significant overlap between dataset genes and known targets regulated by a transcription regulator. For Extended Data Fig. 8b, proteins were selected based on 3 pre-curated lists (MYC target genes based on the analysis report from INGENUITY, MYC signature genes based on the reported list provided in ref. 54 and MYC expression/ function activators were manually curated from UniProt and GeneCards databases).
Sequencing data
Cell lines with information available in the cBioPortal for cancer genomics (http://www.cbioportal.org) were evaluated for mutations in pathways implicated in cancer: P53, RAS, RAF, PTEN, PIK3CA, AKT, EGFR, HER2, CDK2NA/B, RB, MYC, STAT1, STAT3, JAK2, MET, PDGFR, KDM6A, KIT. Mutations in major chaperome members (HSP90AA1, HSP90AB1, HSPH1, HSPA8, STIP1, AHSA1) were also evaluated.
Statistical analysis
Data were visualized and statistical analyses performed using GraphPad Prism (version 6; GraphPad Software) or R statistical package. In each group of data, estimate variation was taken into account and is indicated in each figure as s.d. or s.e.m. If a single panel is presented, data are representative of 2 or 3 biological or technical replicates, as indicated. P values for unpaired comparisons between two groups with comparable variance were calculated by two-tailed Student’s t-test. Pearson’s tests were used to identify correlations among variables. Significance for all statistical tests was shown in figures for not significant (NS), * P < 0.05, * * P < 0.01, * * * P < 0.001 and * * * * P < 0.0001. No samples or animals were excluded from analysis, and sample size estimates were not used. Animals were randomly assigned to groups. Studies were not conducted blinded, with the exception of all patient specimen histological analyses.
Extended Data
Extended Data Figure 1. Summary of the experimental design and findings.
a, Schematic of the experimental approach to address four key questions concerning the chaperome in cancer: (1) what are the molecular characteristics and composition of the chaperome in cancer; (2) what molecular factors drive chaperome networks to crosstalk in tumours; (3) what distinguishes the chaperomes of tumours that are sensitive to pharmacologic inhibition from those that are not; and (4) what are the characteristics of tumours that may benefit from chaperome therapy? To retain the endogenous proteome/chaperome make-up and function, we applied a variety of chemical biology tools and biochemical methods that retain native protein conformations and complexes. This approach minimally interferes with the system it interrogates, thus providing answers closer to the reality of disease. When applicable, data were validated by alternative and complementary methods, as indicated. This approach led to the discovery of a novel mechanism of tumour regulation. Specifically, we have identified and characterized the epichaperome, a modified chaperome network. Our data demonstrate that heterogeneous and stable, multimeric chaperome complexes nucleating on HSP90 and HSP70, and incorporating co-chaperones, isomerases, scaffolding proteins, and transport proteins, bring about the effective physical and functional integration of the chaperome machinery into the epichaperome. Chaperome rewiring into the epichaperome is fuelled by powerful transcription activators such as MYC. Only under conditions in which the chaperome becomes tightly integrated both functionally and physically to form the epichaperome are tumours addicted to individual chaperome members. The epichaperome is the survival mechanism for type 1 tumours; when the epichaperome is dismantled by ablation of a component chaperome, the chaperome network collapses leading to cell death. In contrast, in type 2 tumours in which the integration of the chaperome is only partial and most chaperome members function as insular communities, depletion of chaperome members only ‘locally’ compromises the chaperome, maintaining overall cellular survival. b, c, Therapeutic and diagnostic implications of the findings. We propose the epichaperome as a biomarker to stratify patients for chaperome therapy, such as HSP90 inhibitors. This work also provides several ways to measure the epichaperome in clinic, that is, a non-invasive imaging assay (PU-PET) for solid tumours, a flow cytometry assay based on PU-FITC for liquid tumours and a native protein separation and analysis for minute biopsy specimens (isoelectric focusing; NanoPro technique) (b). We also propose that HSP90 is a cancer target when integrated into the epichaperome. Thus, HSP90 inhibitors that are specific for HSP90 when part of the epichaperome would be preferred for clinical use. Non-discriminate pharmacological agents that target chaperome members regardless of whether they are in the epichaperome or are part of dynamic complexes, such as in normal cells, could lead to toxicities and a low therapeutic index. For example, GI and ocular toxicities have been associated with some HSP90 inhibitors and not others due to chronic HSP90 inhibition in these normal tissues.
Extended Data Figure 2. Biochemical profile of chaperome members in cancer cell lines and primary specimens.
a, The biochemical profile of HSP90 in cell lines was analysed by native capillary isoelectric focusing. The ‘heat map’ representation shows snapshots of HSP90 complexes as detected under different antibody blotting exposure times. See also Fig. 1a. b, The biochemical profile of HSP90 in samples denatured with urea. RT, room temperature; ON, overnight. Data were repeated independently twice with representative data shown. c–e, The biochemical profile of indicated chaperome members in cell lines (c, n = 4) and primary specimens (breast cancer, n = 5 (d), and acute myeloid leukaemia (AML), n = 2 (e)) was analysed by native capillary isoelectric focusing (IEF), native-PAGE and SDS–PAGE. The schematic for the isolation and separation of AML blasts for biochemical analyses is shown in e, top. TNBC, triple negative breast cancer; HER2+, HER2-overexpressing breast cancer. For uncropped gel data, see Supplementary Fig. 1.
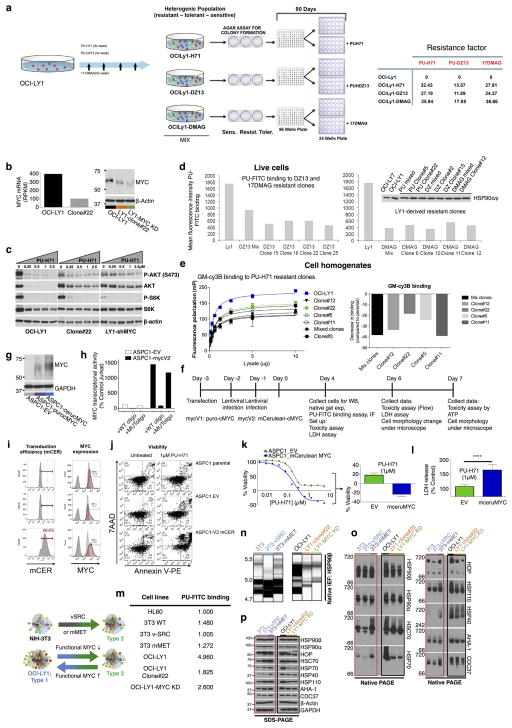
Extended Data Figure 3. PU-H71 and its labelled versions are reliable tools to perturb, identify and measure the expression of the high-molecular-weight, multimeric HSP90 complexes in tumours.
a, Correlative analysis between binding of a fluorescently (FITC) labelled PU-H71 (PU-FITC) to the panel of cancer cells shown in Fig. 1a (n = 6) and their apoptotic sensitivity to HSP90 inhibition (Pearson’s r, two-tailed). Each data point represents a cell line. b, MDA-MB-468 (type 1) and ASPC1 (type 2) contain similar levels of HSP90 but only MDA-MB-468 expresses the high-molecular-weight chaperome species (see also Fig. 1a). HSP90α and HSP90β levels were quantified by fluorescence microscopy (n = 50; mean ± s.d.; unpaired t-test, NS). Scale bar, 10 μm. c, Association and dissociation of PU-FITC (a FITC labelled PU-H71) from HSP90 was probed in cell homogenates by fluorescence polarization. Average from technical duplicates is shown on the graph. The experiment was carried out twice with similar results. d, Association and dissociation of PU-H71-bait (a solid-support immobilised PU-H71) from HSP90 was probed in cell homogenates by chemical precipitation followed by analyses of HSP90 in the supernatant and of HSP90 isolated on the solid support. A solid-support containing immobilized PU-H71 and an HSP90-inert molecule (control bait) were incubated with cell homogenates for 2 h. The bait-captured cargo was isolated and analysed by western blot (bait). The HSP90 species in the supernatant were separated and analysed as indicated. For isoelectric focusing, both the gel (for experimental duplicates) and the heat map representations of different exposure times are shown for each experimental condition. HMEC cells are shown for reference. Data were repeated independently twice with representative data shown. e, f, Association and dissociation of PU-H71 from type 1 and 2 tumours, measured in cells. Binding of PU-FITC to live cells was analysed by flow cytometry and fluorescence microscopy, as indicated. PU-FITC (1 μM for flow and 5 μM for microscopy) was added to cells and incubated for 4 h before fluorescence signal detection. To show specificity of binding, the signal was competed off in a dose-dependent manner with unlabelled PU-H71. Control FITC, a triethylene glycol labelled FITC. e, Mean from two technical replicates; f, mean ± s.d., n = 50 individual cells, unpaired t-test, * * * * P < 0.0001. The fluorescence intensity of PU-FITC staining was quantified by ImageJ. Scale bar, 10 μm. g, Association and dissociation of PU-H71 from type 1 and 2 tumours, measured in vivo. Biodistribution of 124I-PU-H71 (a 124I radiolabelled version of PU-H71) was monitored live in tumour-bearing mice. Each mouse bears one xenografted MDA-MB-468 and one ASPC1 tumour, of similar volume, as indicated. Following intraveneous (iv) injection of a tracer amount of the 124I-PU-H71 agent, mice were monitored by micro-positron emission tomography (PET) imaging. Representative images taken at the indicated times post-injection are shown. Note that immediately after injection (1 h timepoint image), the agent is widely and uniformly distributed throughout the body and in each tumour. The off rate from type 1 tumours is slower compared to type 2 and non-transformed tissues (that is, distinct koff from type 1 tumours versus type 2 tumours). The image is representative of five individual mice. In an independent experiment, radioactivity was measured in a gamma-counter upon mouse euthanasia and data were graphed to monitor the time-dependent distribution of PU-H71. Graph; radioactivity, measured as %IDg; injected dose per gram, versus time upon euthanasia (mean ± s.d., n = 8, ASPC1; n = 34, MDA-MB-468, pooled experiments of mice bearing individual tumours). Means were compared by unpaired t-tests between MDA-MB-468 and ASPC1 at each time point (NS, not significant; * * * P < 0.001; * * * * P < 0.0001). h, Same as in g for a therapeutic dose of injected PU-H71, as indicated. Levels of intact PU-H71 in the indicated tumours, tissues and plasma were determined by liquid chromatography tandem mass spectrometry (LC-MS/MS) in mice (n = 5) euthanized at the indicated times post-PU-H71 injection. Graph; mean ± s.d., unpaired t-tests between MDA-MB-468 and ASPC1 (NS, not significant; * * * P < 0.001). i, Dose- and time-dependent binding of PU-H71 and H9010 (an anti-HSP90 antibody) to HSP90 species expressed in type 1 and type 2 tumour cells. C, control beads containing an HSP90-inert molecule. PU, 10 μl PU-H71 wet beads; 2× PU, 20 μl PU-H71 wet beads; H9010; 2 μl antibody immobilized on agarose beads. Because the IgG interferes with the HSP90 signal (see the high molecular smear in the native gels), native lysates were used for a control (input). Graph shows quantification of time-dependent changes in HSP90 species. j, Representative fluorescence microscopy images of live cells stained with PU-FITC (top) as compared to antibodies specific for HSP90 (bottom). rbtIgG, rabbit IgG control, msIgG, mouse IgG control. Scale bar, 10 μm. Micrograph is representative of four captured images. For uncropped gel data, see Supplementary Fig. 1.
Extended Data Figure 4. Binding affinity of PU-H71 for cellular HSP90 is independent of the expression of HSP90 and other chaperome members, and is not affected by intracellular ATP concentration variations.
a, Correlative analysis for PU-H71-sensitive HSP90 species abundance, as measured by PU-FITC capture, and cell viability upon a 48 h treatment with PU-H71 (1 μM), as measured by annexin V staining (Pearson’s r, two-tailed). Each data point represents a cell line (n = 17); data points are the mean from two biological replicates ran in duplicate or triplicate. To account for intercellular background signal variability, HL60 cells were spiked in and used as internal control for each cell line; thus binding is presented as a ratio of the signal obtained in the analysed cell over that in HL60 cells. y axis, log values of the binding ratios. b, Cell lines analysed in a were lysed and total levels of the indicated chaperome members were determined by western blot. β-actin; protein loading control. c, A correlative analysis was performed between total chaperome levels, as obtained in b, and cell viability values, as determined in a; no significant and/or robust relationship was observed (Pearson’s r, two-tailed). d, e, In a panel of type 1, type 2 and non-transformed cells (n = 9), binding to PU-H71 and cell killing by PU-H71 (d) was compared to intracellular ATP levels (e). d, Correlation, Pearson’s r, two-tailed; e, mean ± s.d., each symbol represents an experimental replicate (MDA-MB-468, n = 23; OCI-LY1, n = 15; BCP-1, n = 8; HCC-1806, n = 16; ASPC1, n = 15; MDA-MB-415, n = 8; MCF7, n = 8; HMEC, n = 7; MRC5, n = 16). f, Schematic showing the experimental design for the isolation and analysis of primary AML samples. g, AML samples were stained with PU-FITC, and blasts (malignant) and lymphocytes (normal) were separated and analysed by flow cytometry. The signal in blasts over lymphocytes (used as internal standard) was graphed to classify clones as type 1 (> 2.1 PU-FITC binding ratio of signal in blast versus lymphocytes) and type 2 (< 2.1 PU-FITC binding ratio) (mean ± s.d., n = 9, unpaired t-test, * * P < 0.01). h, Total HSP90 levels were measured by staining with an HSP90 antibody after cell fixation and permeabilization (mean ± s.d., n = 9, unpaired t-test, NS, not significant). i, Viability of blasts following exposure to PU-H71 (1 μM) for 48 h was measured by annexin V/7AAD staining (mean ± s.d., n = 9, unpaired t-test, * * * P < 0.001). j, PU-FITC staining of live and fixed/permeabilized unfractionated AMLs was visualized by fluorescence microscopy. Scale bar, 100 μm. Micrograph is representative of two captured images. The biochemical profile of AML no. 1 and AML no. 2 is presented in Extended Data Fig. 2e. For uncropped gel data, see Supplementary Fig. 1.
Extended Data Figure 5. Chaperome networks in type 1, type 2 and non-transformed cells.
a, b, Heat maps illustrating all chaperome members (a) and the interactome of HSP90 (b) isolated by the HSP90 bait and identified upon mass spectrometry and bioinformatics analyses enriched (P < 0.1) in type 1 tumours over type 2 tumours and non-transformed cells. Protein sorting was based on hierarchical clustering. Last lane of the heat map in a shows the enrichment of these proteins on the HSP70 bait. c, Network illustrating the connectivity of proteins isolated by the HSP90 bait and identified upon mass spectrometry and bioinformatics analyses; chaperome members and proteins with scaffolding, adaptor, protein interface modulator roles and significantly enriched (P < 0.1) in type 1 tumours over type 2 tumours and non-transformed cells are shown. The thickness of the edges (connection lines) represents the robustness of the functional interaction. The colour of nodes represents protein abundance. For comparison, type 2 and non-transformed cells are also show. Core interactions are shown in Fig. 1f. d, e, The cargo or interacting proteins of HSP90 (d) and HSP70 (e) isolated by the PU-H71 and YK-chemical baits from the indicated cell homogenates. Protein levels in individual cell homogenates (input) were analysed by IEF and SDS–PAGE, as indicated. Proteins precipitated on the chemical bait were analysed by SDS–PAGE. Protein levels from each experimental condition were quantified and graphed (bottom). Data were repeated independently twice with representative data shown. f–i, Changes in multimeric chaperone complexes in cells challenged with multiple siRNAs against HSP90α or HSP90β (f), HSP90α /β , AHA1, HOP or HSP110 (g) and in cell homogenates challenged with antibodies specific for the indicated HSP70 paralogues and for HOP (h, i), as indicated. Levels of proteins in the homogenate were probed by SDS–PAGE or native- PAGE, as indicated. All data were repeated independently twice with representative data shown. For uncropped gels, see Supplementary Fig. 1.
Extended Data Figure 6. HSP90 is functional and susceptible to exogenous inhibitors in type 2 as well as in type 1 cells, but only inhibition of HSP90 in type 1 cells is toxic to the cell.
a, The response of type 1 and 2 tumours classified by PU-PET avidity, to PU-H71 treatment, is shown. Patients were treated as part of the NCT01393509 clinical study. Each picture is a scan of data taken of an individual patient. PU-PET images were taken at 24 h after 124I-PU-H71 tracer administration. Scale bars (bottom of panel); PET window display intensity scales for FDG and PU-PET fusion PET-CT images. Numbers in the scale bars indicate upper and lower SUV thresholds that define pixel intensity on PET images. b, Changes in HSP90 machinery function upon pharmacologic inhibition (PU-H71, 1 μM for 24 h). Inhibition of PI3K/AKT activity was monitored; see p-S6K surrogate for AKT activity in cell lines and p-AKT in primary specimens (below). Data in cell lines were repeated independently twice with representative data shown. For HSP90α /β knockdown data, see Extended Data Fig. 5g. c–f, Treatment schematic and representative examples of primary breast cancer specimens (n = 2) treated ex vivo with PU-H71. c, Workflow for the analysis of the primary specimens. d, Molecular signature of tumour and adjacent normal tissue of the surgical specimen as analysed by native, nanofluidic proteomic assay (NanoPro; native IEF), for HSP90 and HSP70 (gel representation), and AKT (chromatogram representation). e, Molecular response of tumour sections treated for 24 h ex vivo with PU-H71 (1 μM). AKT (an HSP90 client) activity was probed with the indicated antibody. BCL2 was chosen as a loading standard; this protein is insensitive to HSP90 inhibition in the analysed primary breast specimens (native IEF, chromatogram representation). f, Apoptotic response of the indicated tumour specimens to ex vivo treatment with PU-H71 or vehicle. Apoptosis and necrosis of the tumour cells (as percentage) is assessed by reviewing all the haematoxylin and eosin (H&E) slides of the case (controls and treated ones) in toto, blindly, allowing for better estimation of the overall treatment effect to the tumour. Image representative of the entire specimen section. g–j, Response profile of a panel of pancreatic cancer cells to HSP90 inhibition. g, Changes in cell viability following HSP90 pharmacologic inhibition by three chemically distinct agents, as indicated. Mean from two to three technical replicates is shown. Subclassification of the analysed cell lines by PU-FITC binding is shown on the left. h, The effect of PU-H71 on cell growth was measured with an assay that analyses intracellular ATP levels. Cells were treated for 72 h with PU-H71 and the half maximal inhibitory growth concentration (IC50) was determined. Mean ± s.d.; n = 6. i, Representative examples of type 1 and type 2 cells treated for 24 h with the indicated concentrations of PU-H71. Inhibition of HSP90 is demonstrated by a decrease in HSP90 client function (p-S6K and p-ERK) and by HSP70 induction, and evidenced in both type 1 and 2 tumour cells. Induction of apoptosis, as demonstrated by the appearance of cleaved PARP (cPARP), is however, specific to type 1 tumour cells. β-actin, protein loading control. The HSP90 biochemical signature of the select cells is shown on the right. The blue arrows indicate the close relationship between the growth inhibitory IC50 values and HSP90 function inhibition, suggesting that HSP90 inactivation is sufficient to inhibit growth (that is, have a static effect) in both type 1 and 2 tumours. In contrast, substantial induction of apoptosis is specific to type 1 tumours. Thus, HSP90 is functional in type 2 and is engaged by the HSP90 inhibitors—the resistance phenotype of type 2 tumours cannot be explained by an inability of the HSP90 inhibitor to engage HSP90. Data are representative of two independent experiments. For uncropped gel data, see Supplementary Fig. 1.
Extended Data Figure 7. The epichaperome facilitates cancer cell survival.
The expression of the epichaperome was altered by epichaperome components knockdown (a) or by titrating into cells siRNAs that targeted both the HSP90α and the HSP90β paralogues (b–g) to test whether the epichaperome facilitates survival in type 1 tumours. a, Epichaperome levels were altered by AHA1 siRNA knockdown or a control (scramble, Scr) siRNA (left panel) and cell viability, as measured by PARP cleavage, was determined in cells treated for 24 h with increasing concentrations of PU-H71 (0, 0.5, 1 and 2 μM) (right panel). See also Extended Data Fig. 5g for biochemical signature of cells after AHA1 knockdown. Data are representative of two independent experiments. b–e, Total protein (b, e), mRNA (c) and multimeric species of indicated chaperome members (d) levels were monitored in MDA-MB-468 type 1 cells in which several concentrations of siRNAs against HSP90α /β (n = 14) were titrated in. 1 and 8 are control scramble; 2, 3, 4, 9, 10, 11 are 0.915, 1.83, 3.66, 0.366, 1.83 and 14.64 nM of siRNA no. 1, respectively; and 5, 6, 7, 12, 13, 14 are 0.366, 0.915, 3.66, 0.0229, 0.0915 and 0.366nM of siRNA no. 2, respectively. Cell viability in each condition was monitored by LDH release. Values for each experimental condition (as percentage control scramble) were quantified and are noted under the native gels in a. HMW, high molecular weight. For gels, experiments were repeated independently twice with representative data shown. For graphs, mean ± s.d., n = 6. f, Changes in chaperome members (mRNA) were monitored following siRNA knockdown of the indicated individual chaperome members in MDA-MB-468 and ASPC1 cells. Error bars show mean ± s.d., n = 6. g, Same as for b–d in ASPC1, type 2 cells (1 through 4 siRNA concentrations, as in d. Error bars represent mean ± s.d., n = 6, unpaired t-test, NS, not significant). For uncropped gel data, see Supplementary Fig. 1.
Extended Data Figure 8. MYC is a driver of chaperome rewiring into the epichaperome.
a, The top eight transcriptional regulators ranked by significant overlap between dataset genes and known targets regulated by the indicated transcription regulators in type 1 tumours. b, Heat map illustrating the enrichment in type 1 tumours of MYC target genes, MYC transcriptional signature and of proteins that regulate MYC function and/ or expression.
Extended Data Figure 9. Chaperome rewiring into the epichaperome is fuelled by powerful transcription activators such as MYC.
a–e, Establishment and characterization of HSP90-inhibitor-resistant cells. a, Schematic detailing the establishment and separation of clones cross-resistant to PU-H71, PU-DZ13 and 17-DMAG. These compounds are chemically distinct HSP90 inhibitors. b, RNA-seq and western blot analyses of clone no. 22 indicate that cellular resistance to HSP90 inhibitors is associated with MYC downregulation. RPKM, reads per kilobase of exon per million mapped reads. Western blot data are representative of two independent experiments. c, HSP90 remains functional in the HSP90-inhibitor resistant clones, as well as in cancer cells in which MYC expression is reduced by shRNA knockdown. Cells were treated for 24 h with the indicated concentrations of PU-H71, and HSP90 client protein function (p-AKT and p-S6K levels) was analysed by western blot. For gels, experiments were repeated independently twice with a representative gel shown. d, PU-FITC binding to the indicated resistant clones (n = 10) presented as relative, mean fluorescence intensity values, was measured by flow cytometry. The parental OCI-LY1 (type 1 tumour cell) is shown for comparison. Inset shows total HSP90 levels measured by western blot in the indicated clones (n = 8). IB; anti-HSP90 (F8) sc-13119. e, Binding of a fluorescently labelled geldanamycin derivative (GM-cy3B) to the indicated cell homogenates was measured by fluorescence polarization. Graph shows mean from three technical replicates. f, Experimental design for the creation and characterization of MYC-expressing ASPC1 cells. g, Levels of MYC and HSP90 were analysed by western blot in the indicated infection conditions (day 4 post-lentiviral infection). Data were repeated independently twice with a representative blot shown. h, Transcriptional activity of MYC in infected cells was measured using the TransAM c-Myc Transcription Factor ELISA. Mean from three technical replicates is shown. i, Flow cytometry confirmed the expression of MYC in infected ASPC1 cells. mCerulean and MYC were co-expressed with a 2A peptide linker, which was self-cleaved after protein translation. Data were repeated independently twice with representative data shown. j–l, Viability of ASPC1 cells infected with either empty vector or MYC was assessed using an assay that quantifies annexin V/7AAD-stained cells (j), ATP levels (k), LDH release (l) following treatment with PU-H71, as indicated. k, Mean of four technical replicates. l, Error bars show mean ± s.d., n = 6, unpaired t-test, * * * * P < 0.0001. m–p, HSP90 oncogenic kinase clients do not require the epichaperome for cell transforming activity. m, PU-FITC binding to the indicated live cells presented as a ratio (fluorescent signal of measured cells over signal in HL60 cells; HL60, internal standard). n–p, Changes in multimeric chaperome complexes (n, o) and total protein (p) in the indicated conditions. All data were repeated independently twice with representative data shown. OCI-LY1 (type 1 cells) and OCI-LY1 rewired to type 2 following MYC loss are presented for direct comparison purposes. For uncropped gels, see Supplementary Fig. 1.
Extended Data Figure 10. The apoptotic response profile of a panel of cancer cells following HSP90 inhibition is independent of levels of chaperome members, HSP90 client proteins and anti-apoptotic molecules, tissue of origin or causal genetic mutations.
a, Total levels of the indicated chaperome members, HSP90 client proteins and anti-apoptotic molecules were analysed by western blot in a panel of pancreatic cancer cells (n = 12). GAPDH and β-actin, protein loading controls. Protein levels were quantified and graphed against the viability of these cells upon HSP90 inhibition. A correlative analysis was performed (Pearson’s r, two-tailed). Each data point represents a cell line. PU-FITC binding is shown for comparison. b, Correlative analysis of epichaperome abundance, as measured by PU-FITC staining, and cell viability upon a 48 h treatment with PU-H71 (1 μM), as measured by annexin V staining (Pearson’s r, two-tailed). Each data point represents a cell line (n = 95); data are the mean from two orthree biological replicates. Cells representing pancreatic, gastric, lung, and breast cancers, along with lymphomas and leukaemias were chosen for analysis. c, Same as above for the treatment of cancer cells (n = 12) with three chemically distinct HSP90 inhibitors. d, Same as b, but for each cell line, known genetic lesions were added. No specific genetic alteration could be found distinguishing the two tumour types; whereas p53-, Ras-, Myc-, HER-, PI3K/AKT-, and JAK- cell cycle-related defects were found in tumours that were sensitive to PU-H71, they were also evident in PU-H71 resistant cells. We found genetic defects in major chaperome members to be rare, with BCP-1 cells only carrying an HSP90AA1 missense mutation (P596S). No mutations in HSP90AB1, HSPH1, HSPA8, STIP1 and AHSA1 were reported in this large panel of cell lines. These were obtained from the cBioPortal for Cancer Genomics website (http://www.cbioportal.org/).
Acknowledgments
We thank D. Gewirth, A. Younes, J. Young and E. Devaney for reading the manuscript. This work was supported by the NIH (R01 CA172546, R01 CA155226, P01 CA186866, P30 CA08748, P50 CA86438, DP2 OD007399), the Jane H. Gordon Breast Cancer Research Fund, the Breast Cancer Research Fund, Susan G. Komen for the Cure, W. H. Goodwin, A. Goodwin and the Commonwealth Foundation for Cancer Research, and the Experimental Therapeutics Center of MSKCC, the Hirshberg Foundation for Pancreatic Cancer, the Irma T. Hirschl Foundation and the Rubenstein Center for Pancreatic Research. A.R. was supported by the National Center for Advancing Translational Sciences of the NIH under award number UL1TR000457.
Footnotes
Author Information Primary data, such as raw mass spectrometry files, Mascot generic format files and proteomics data files created by Scaffold have been deposited in the MassIVE database (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp); MassIVE accession ID: MSV000079877
The authors declare competing financial interests: details are available in the online version of the paper. Readers are welcome to comment on the online version of the paper
Supplementary Information is available in the online version of the paper.
Author Contributions A.R., T.W., P.Y., E.D.G., M.P.S.D., M.R., J.F.G., M.A., N.P., A.C., H.Z., J.K. III, B.B., C.P., C.X., R.T., H.J.P., P.P., M.F.F., F.S., E.C.-L., A.B., S.K., C.Y., O.L. and H.E.-B. performed experiments. S.M., T.T., C.H., N.P., L.C., A.M., E.C., L.S., J.S.L. and G.J.R. provided reagents. N.P., A.R., E.D.G., T.W., P.Y., P.Z., B.B., R.L., A.M., S.M.L., J.S.L., M.P.S.D., R.I.P., F.C., J.F.G., H.E-B., G.C. and M.L.G. participated in the design and analysis of various experiments, S.P., R.I.P. and T.W. performed statistical analyses and G.C. and M.L.G. wrote the paper. All authors read the manuscript and/or discussed the experiments.
Reviewer Information Nature thanks J. W. Kelly and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
References
- 1.Brehme M, et al. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Reports. 2014;9:1135–1150. doi: 10.1016/j.celrep.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finka A, Goloubinoff P. Proteomic data from human cell cultures refine mechanisms of chaperone-mediated protein homeostasis. Cell Stress Chaperones. 2013;18:591–605. doi: 10.1007/s12192-013-0413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taipale M, et al. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014;158:434–448. doi: 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Echtenkamp FJ, Freeman BC. Expanding the cellular molecular chaperone network through the ubiquitous cochaperones. Biochim Biophys Acta. 2012;1823:668–673. doi: 10.1016/j.bbamcr.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Echeverría PC, Bernthaler A, Dupuis P, Mayer B, Picard D. An interaction network predicted from public data as a discovery tool: application to the Hsp90 molecular chaperone machine. PLoS One. 2011;6:e26044. doi: 10.1371/journal.pone.0026044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 7.Moulick K, et al. Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90. Nature Chem Biol. 2011;7:818–826. doi: 10.1038/nchembio.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodford MR, et al. Impact of posttranslational modifications on the anticancer activity of Hsp90 inhibitors. Adv Cancer Res. 2016;129:31–50. doi: 10.1016/bs.acr.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Kamal A, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 10.Alarcon SV, et al. Tumor-intrinsic and tumor-extrinsic factors impacting Hsp90-targeted therapy. Curr Mol Med. 2012;12:1125–1141. doi: 10.2174/156652412803306729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mollapour M, Neckers L. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim Biophys Acta. 2012;1823:648–655. doi: 10.1016/j.bbamcr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Assimon VA, Southworth DR, Gestwicki JE. Specific binding of tetratricopeptide repeat proteins to heat shock protein 70 (Hsp70) and heat shock protein 90 (Hsp90) is regulated by affinity and phosphorylation. Biochemistry. 2015;54:7120–7131. doi: 10.1021/acs.biochem.5b00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuehlke A, Johnson JL. Hsp90 and co-chaperones twist the functions of diverse client proteins. Biopolymers. 2010;93:211–217. doi: 10.1002/bip.21292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrestha L, Young JC. Function and chemotypes of human Hsp70 chaperones. Curr Top Med Chem. 2016;16:2812–2828. doi: 10.2174/1568026616666160413142028. [DOI] [PubMed] [Google Scholar]
- 15.Smith JR, et al. Restricting direct interaction of CDC37 with HSP90 does not compromise chaperoning of client proteins. Oncogene. 2015;34:15–26. doi: 10.1038/onc.2013.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radons J. The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperones. 2016;21:379–404. doi: 10.1007/s12192-016-0676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel PD, et al. Paralog-selective Hsp90 inhibitors define tumor-specific regulation of HER2. Nature Chem Biol. 2013;9:677–684. doi: 10.1038/nchembio.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nayar U, et al. Targeting the Hsp90-associated viral oncoproteome in gammaherpesvirus-associated malignancies. Blood. 2013;122:2837–2847. doi: 10.1182/blood-2013-01-479972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shrestha L, Patel HJ, Chiosis G. Chemical tools to investigate mechanisms associated with HSP90 and HSP70 in disease. Cell Chemical Biology. 2016;23:158–172. doi: 10.1016/j.chembiol.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taldone T, Ochiana SO, Patel PD, Chiosis G. Selective targeting of the stress chaperome as a therapeutic strategy. Trends Pharmacol Sci. 2014;35:592–603. doi: 10.1016/j.tips.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taldone T, et al. Design, synthesis, and evaluation of small molecule Hsp90 probes. Bioorg Med Chem. 2011;19:2603–2614. doi: 10.1016/j.bmc.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodina A, et al. Affinity purification probes of potential use to investigate the endogenous Hsp70 interactome in cancer. ACS Chem Biol. 2014;9:1698–1705. doi: 10.1021/cb500256u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taldone T, et al. Synthesis of purine-scaffold fluorescent probes for heat shock protein 90 with use in flow cytometry and fluorescence microscopy. Bioorg Med Chem Lett. 2011;21:5347–5352. doi: 10.1016/j.bmcl.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiosis G, et al. Uses of labeled hsp90 inhibitors. 20140242602 A1. US patent. 2014
- 25.Taldone T, et al. Radiosynthesis of the iodine-124 labeled Hsp90 inhibitor PU-H71. J Labelled Comp Radiopharm. 2016;59:129–132. doi: 10.1002/jlcr.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodina A, et al. Identification of an allosteric pocket on human Hsp70 reveals a mode of inhibition of this therapeutically important protein. Chem Biol. 2013;20:1469–1480. doi: 10.1016/j.chembiol.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taldone T, et al. Heat shock protein 70 inhibitors. 2. 2,5′-thiodipyrimidines, 5-(phenylthio)pyrimidines, 2-(pyridin-3-ylthio)pyrimidines, and 3-(phenylthio)pyridines as reversible binders to an allosteric site on heat shock protein 70. J Med Chem. 2014;57:1208–1224. doi: 10.1021/jm401552y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang Y, et al. Heat shock protein 70 inhibitors. 1. 2,5′-thiodipyrimidine and 5-(phenylthio)pyrimidine acrylamides as irreversible binders to an allosteric site on heat shock protein 70. J Med Chem. 2014;57:1188–1207. doi: 10.1021/jm401551n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyajima N, et al. The HSP90 inhibitor ganetespib synergizes with the MET kinase inhibitor crizotinib in both crizotinib-sensitive and -resistant MET-driven tumor models. Cancer Res. 2013;73:7022–7033. doi: 10.1158/0008-5472.CAN-13-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corben AD, et al. Ex vivo treatment response of primary tumors and/or associated metastases for preclinical and clinical development of therapeutics. J Vis Exp. 2014;92:e52157. doi: 10.3791/52157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hassane DC, et al. Chemical genomic screening reveals synergism between parthenolide and inhibitors of the PI-3 kinase and mTOR pathways. Blood. 2010;116:5983–5990. doi: 10.1182/blood-2010-04-278044. [DOI] [PubMed] [Google Scholar]
- 33.Miller JD, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013;13:691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moulick K, et al. Synthesis of a red-shifted fluorescence polarization probe for Hsp90. Bioorg Med Chem Lett. 2006;16:4515–4518. doi: 10.1016/j.bmcl.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 35.Fan AC, et al. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat Med. 2009;15:566–571. doi: 10.1038/nm.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28:3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodina A, et al. Selective compounds define Hsp90 as a major inhibitor of apoptosis in small-cell lung cancer. Nat Chem Biol. 2007;3:498–507. doi: 10.1038/nchembio.2007.10. [DOI] [PubMed] [Google Scholar]
- 38.Zong H, et al. A hyperactive signalosome in acute myeloid leukemia drives addiction to a tumor-specific Hsp90 species. Cell Reports. 2015;13:2159–2173. doi: 10.1016/j.celrep.2015.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caldas-Lopes E, et al. Hsp90 inhibitor PU-H71, a multimodal inhibitor of malignancy, induces complete responses in triple-negative breast cancer models. Proc Natl Acad Sci USA. 2009;106:8368–8373. doi: 10.1073/pnas.0903392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taldone T, Zatorska D, Kang Y, Chiosis G. A facile and efficient synthesis of d6-labeled PU-H71, a purine-scaffold Hsp90 inhibitor. J Labelled Comp Radiopharm. 2010;53:47–49. [Google Scholar]
- 41.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skarra DV, et al. Label-free quantitative proteomics and SAINT analysis enable interactome mapping for the human Ser/Thr protein phosphatase 5. Proteomics. 2011;11:1508–1516. doi: 10.1002/pmic.201000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- 44.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarkar D. Lattice: Multivariate Data Visualization with R. Springer; 2008. [Google Scholar]
- 47.Scales M, Jäger R, Migliorini G, Houlston RS, Henrion MY. visPIG—a web tool for producing multi-region, multi-track, multi-scale plots of genetic data. PLoS One. 2014;9:e107497. doi: 10.1371/journal.pone.0107497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Licata L, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown KR, Jurisica I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol. 2007;8:R95. doi: 10.1186/gb-2007-8-5-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21:2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- 54.Chandriani S, et al. A core MYC gene expression signature is prominent in basal-like breast cancer but only partially overlaps the core serum response. PLoS One. 2009;4:e6693. doi: 10.1371/journal.pone.0006693. [DOI] [PMC free article] [PubMed] [Google Scholar]