ABSTRACT
The putative contribution of natural killer (NK) cells to immunosurveillance in non-small cell lung cancer (NSCLC) has been an ongoing conundrum. Here, we used a readily standardizable quantitative real time polymerase chain reaction (qRT-PCR) to measure the expression of NK cell receptors in total peripheral blood mononuclear cells (PBMC) from healthy volunteers (HV), patients with gastrointestinal stromal tumors (GIST), neuroblastoma (NB), melanoma or NSCLC. We quantified NCR1 (which codes for NKp46) and NCR3 (which codes for NKp30), as well as that of three NCR3 splice variants (which give rise to immunostimulatory NKp30A and NKp30B, as well as to immunosuppressive NKp30C). NSCLC patients expressed lower levels of NCR1 than did HV. Remarkably, NCR3 was lower in NSCLC patients than in HV as well as in all other malignancies. Moreover, a discrete proportion of NSCLC patients exhibited a particular low ratio between NKp30B and NKp30C (ΔBC). In the overall cohort, low expression of NCR3 correlated with poor overall and progression-free survival (PFS). When patients were stratified according to the level of PD-L1 expression by NSCLC cells, within the PD-L1high category (>5% positive tumors), the sole parameter that affected prognosis was the expression of NCR1. However, in patients bearing tumors with negative PD-L1 expression on tumor or tumor-infiltrating stromal cells, the ΔBClow patients exhibited a dismal prognosis. Altogether, these results strongly suggest that NK cells mediate immunosurveillance against NSCLC and that measuring NK cell receptor expression by blood cells can yield useful biomarkers for patient stratification.
KEYWORDS: Biomarker, NKp30, NKp46, NK cells, NSCLC, PD-L1
Introduction
Although unsuccessful attempts have been made to treat NSCLC with autologous NK cells,1,2 the role of NK cells in natural immunosurveillance against NSCLC has been debated.
NK cells that infiltrate established NSCLC lesions are functionally deficient,3,4 and this notion of a functional NK cell defect has been extended to patients with advanced NSCLC.5 Intratumoral NK cells express low amounts of the NK receptors NKp30, NKp46, DNAM-1, CD16 and ILT2, while the expression of CD69 and NKp44 activation markers is increased.6 Moreover, intratumoral NK cells exhibit profound defects in their ability to activate degranulation and to produce IFNγ.7 The frequency of tumor-infiltrating NK cells with an immature CD11b− CD27− phenotype correlated with the tumor stage and tumor size in a mouse preclinical model.8 Thus, it appears that NK cells fail to exert their immunosurveillance function in NSCLC, perhaps as a result of intratumoral immunosuppression or exhaustion. No clear correlation has been established between the intratumoral expression level of the NK receptor NKp30 and that of its ligand B7-H6 and clinical pathological features or survival of NSCLC patients yet.9 This contrasts with the fact that T lymphocytes have clearly been shown to mediate anticancer effects in NSCLC, meaning that the presence of cytotoxic T lymphocytes within the tumor bed does have positive prognostic feature.10-12 Accordingly, checkpoint blockade by antibodies neutralizing PD-1 or PD-L1 has positive (and likely T-cell-mediated) effects on a fraction of NSCLC patients.13,14
In contrast to NSCLC, other human malignancies appear to be under strong NK-mediated immunosurveillance. This applies for example to breast,15,16 colorectal,17 renal18 and ovarian carcinoma,19 GIST,20,21 melanoma22-24 and chronic myeloid leukemia.25 Recently, we developed a relatively simple test that can be performed on PBMC to measure the absolute or relative abundance of the mRNAs coding for major NK cell receptors (such as NCR1, which codes for NKp46, and NCR3 which codes for NKp30), as well as the three splice variants of NKp30 that differ in their C-terminus (and hence in their signaling capabilities), NKp30A, NKp30B and NKp30C. Whereas the NKp30A and NKp30B isoforms stimulate cytotoxicity and T helper 1 cytokine secretion, respectively, the NKp30C isoform preferentially promotes the release of the immunosuppressive factor, interleukin-10.26 The relative expression of NKp30A, NKp30B and NKp30C is a stable trait that is not influenced by the location of NK cells (with no difference between intratumoral and peripheral expression patterns), nor by the progression/cure of cancer,26 infectious27 or autoimmune disease.28 Both genetic variants (such as single nucleotide polymorphisms in the NCR3 gene) and epigenetic factors have been invoked to explain the stable proportion of NKp30A, NKp30B and NKp30C in distinct individuals.26 Irrespective of the precise causes of the NKp30 isoform expression pattern, several studies established that a high abundance of the NKp30C or a low abundance of NKp30B isoforms (which results in a reduction in the ratio between NKp30B and NKp30C, ΔBC) has a negative impact on the prognosis of patients with GIST26 or NB,29 respectively.
Driven by these considerations, we decided to investigate the expression of NK cell receptors and their isoforms in the blood from NSCLC patients. As reported below, we found that the absolute and relative expression of NK cell receptors does have important prognostic features in two cohorts of advanced NSCLC patients.
Materials and methods
Patients and specimens
TIME study
We first considered the control arm of the TIME study (sponsored by Transgene, France) aimed at combining cis-platinum-based chemotherapy with a recombinant (MUC-1-IL-2) poxvirus30,31 and next, validated the results using the TG4010 arm of this protocol. Patient characteristics are depicted in Table S1. Patients were eligible for study inclusion in several European and American countries if they had an histologically confirmed, stage IV according to UICC, non-small-cell lung cancer without known activating EGFR mutation but MUC-1 expression on at least 50% tumor cells. Patients had to be treatment-naive, in good general status (PS 0 or 1 according to ECOG), with adequate hematological and biochemical parameters. The study was conducted under the oversight of an independent data monitoring committee, in accordance with the principles of the declaration of Helsinki and the Good Clinical Practice guidelines of the International Conference on Harmonization. The study was approved in each country by the appropriate regulatory and ethical committees. Patients provided a written informed consent before entering the screening process. The study was designed by the sponsor (Transgene). Monitoring, management and analysis of the data were performed by service providers under the supervision of the sponsor. Heparinized blood was drawn from patients at the time of clinical consultation, prior to chemotherapy. Clinical responses were assessed by computed tomography (CT) scan and the responses were classified according to RECIST.32 Other malignancies and controls HV were used as controls (EFS, Créteil and Besançon, France). Melanoma patients, GIST patients and NB-bearing children were already described.20,23,29,33
Peripheral blood mononuclear cells (PBMC)
PBMC were obtained by Ficoll-Hypaque density gradient (PAA Laboratories), washed twice in Dulbecco's phosphate-buffered saline (PBS, GIBCO Invitrogen), re-suspended in RLT buffer containing β−2 mercaptoethanol (Qiagen) to preserve the RNA quality, and stored at −80°C.
qRT-PCR
Total cellular RNA was isolated from PBMC with the RNeasy kit (Qiagen) following the manufacturer's recommendations. First strand cDNA was synthesized from 1 µg of total RNA using SuperScript™ III Reverse Transcriptase (Invitrogen) and random primers (Promega) according to the manufacturer's recommendations.
For qRT-PCR, 5 μL of first-strand cDNA was mixed with 12.5 μL of 2X TaqMan Gene Expression Master Mix (Applied Biosystems). For NKp30 isoforms A, B and C, and NKp46: 0.75 μL primers (10 μM) and probe (5 μM); and for the β−2-microglobulin housekeeping gene (β2M): 0.5 μL of primers (10 μM) and probe (5 μM), were used in a final volume of 25 μL. A StepOnePlus System (Applied Biosystems) was used for temperature cycling and real time fluorescence measurement. The PCR conditions were as follows: initial incubation at 50°C for 2 min, denaturation at 95°C for 10 min, followed by 45 cycles at 95°C for 15 sec, and 60°C (for NKp30a, b and c, and β2m transcripts) for 1 min. PCR primers and TaqMan probes are listed in Table S2. The relative expression of the gene of interest was calculated with the 2-ΔCt method34 normalized to the expression level of β2M, with the following formula: 2-ΔCt = 2(Ct (gene of interest) − Ct (housekeeping gene)) × 106. The level of expression of the distinct NKp30 isoforms compared with each other (delta, Δ) was determined using the following formula ΔNKp30x NKp30y = CtNKp30y − CtNKp30x, with Ct = Cycle threshold. The median of each NKp30 isoform pair delta (Δ) was used for the survival curve analysis.
PD-L1 staining in immunohistochemistry
Immunohistochemistry was performed on 4-µm sections from formalin-fixed paraffin-embedded tumor tissue, using a Ventana Discovery Ultra platform (Ventana, Tucson, AZ). Briefly, epitope retrieval was performed in CC1 buffer (Tris-based buffer, pH 8.0) during 90 min at 95°C. Anti-PD-L1 primary antibody clone E1L3N (Cell Signaling Technology, Danvers, MA) was incubated at 1 µg/mL during 1 h at room temperature. An anti-rabbit HQ amplification kit was used for detection with DAB as a chromogen. PD-L1 expression on tumor cells was scored semi-quantitatively based on the percentage of cells harboring a membranous staining of moderate or strong intensity. Membranous staining on alveolar macrophages was used as an internal positive control.
Statistical analyses
Kruskal–Wallis test with Dunn's multiple comparison test was used for comparison of the different groups, as indicated in the figure legends, using Prism 5 software (GraphPad San Diego, CA, USA). Correlations were analyzed by Spearman correlation test. Survival curves were plotted according to the Kaplan–Meier method and compared by log-rank test. In addition, Hazard Ratios (HRs) and corresponding 95% Confidence Interval (95% CI) were estimated using a Cox proportional hazards model. These analyses were performed in patients treated with placebo in TIME study and patients from experimental arm were used to validate the results with one-sided log-rank test. These analyses were performed using SAS 9.4 Software (SAS Institute, Cary, NC) and p value <0.05 was considered statistically significant. As these analyses were considered as exploratory, no multiplicity correction was applied.
Results and discussion
Reduced expression of NCR1/NKp46 and NCR3/NKp30 in PBMC from NSCLC patients
qRT-PCR was used to detect the relative abundance (compared to that of B2M which codes for β2-microglobulin) of the expression of NCR1 (which codes for NKp46) and NCR3 (which codes for NKp30) in total PBMC from HV, patients with GIST, NB, melanoma or NSCLC. The expression of NCR1 and NCR3 by PBMC from NSCLC patients was lower than that found in control cells from HV (Fig. 1A, B). Moreover, the abundance of the NCR3 mRNA was significantly lower in NSCLC than in any other of the malignant pathologies studied here (Fig. 1B). Of note, there was no obvious correlation between the total frequency of circulating NK cells and the expression of NCR1 and NCR3 (Fig. 1C, D), suggesting that the reduced abundance of mRNA species coding for NK cell receptors cannot be simply attributed to a relative NK lymphopenia.
Figure 1.
Relative expression of NCR1 and NCR3 in cancer patients (A–B): The relative expression of NCR1 (A) and NCR3 (B) normalized with respect to β2-microglobulin was determined by qRT-PCR from PBMC of healthy volunteers (HV) as well as from patients with gastrointestinal stromal tumors (GIST), neuroblastoma (NB), melanoma or non-small cell lung cancer (NSCLC). Each dot represents one patient. p values were calculated by means of the one-way ANOVA: Kruskal–Wallis test with Dunn's multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.001. (C–D): NCR1 (C) and NCR3 (D) relative expression in relation to the percentage of NK cells within PBMC samples from melanoma patients (n = 28). p values were calculated with the Spearman correlation test.
Altered NKp30 isoform expression in PBMC from NSCLC patients
Intrigued by the low overall abundance of NCR3 transcripts, we determined the relative expression of each of the NCR3-encoded NKp30 isoforms (NKp30A, B, C), which arise from alternative splicing. Again, the expression of all three mRNA species coding for the three distinct NKp30 isoforms was less abundant in PBMC from NSCLC patients than in control PBMC from HV and actually tended to be significantly lower than in patients with GIST, NB or melanoma (Fig. 2A, B and C). Moreover, there was a specific NSCLC-associated shift in the relative proportion of NKp30 isoforms (Fig. 2D). PBMC from NSCLC patients exhibited significantly elevated ΔAB (Fig. 3A) and ΔAC values (Fig. 3B) but reduced ΔBC values (Fig. 3C) as compared to HV or patients with GIST, NB or melanoma. However, these Δ values (ΔAB, ΔAC and ΔBC) did not correlate with the overall expression of NCR1 or NCR3 (which do correlate among each other, Fig. 3D).
Figure 2.
Absolute NKp30 isoforms expression in distinct cancer patients (A–C). The absolute expression of NKp30A (A), NKp30B (B) and NKp30C (C) normalized to β2m was assessed by qRT-PCR from healthy volunteers (HV, N = 106), metastatic GIST (N = 126), NB (N = 199), melanoma (N = 151) and NSCLC (N = 76) cancer patients. One-way ANOVA: Kruskall–Wallis test with Dunn's multiple comparison test. (D). Correlation matrix between the relative expression of NKp30 A, B and C isoforms, NCR3 and NCR1 gene in NSCLC cancer patients using the Spearman correlation test. *p < 0.05, **p < 0.01, ***p < 0.001.
Figure 3.

Relative NKp30 isoform Δ ratios in cancer patients (A–C). The NKp30 ΔAB (A), ΔAC (B) and ΔBC ratios (C) were calculated for healthy volunteers (HV, n = 106), metastatic GIST patients (n = 126), NB (N = 199), melanoma (N = 151) and NSCLC (N = 76) cancer patients are shown and analyzed by one-way ANOVA: Kruskall–Wallis test with Dunn's multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.001. (D). Correlation matrix between the relative expression of NKp30 ΔAB, ΔAC, ΔBC, NCR3 and NCR1 gene was assessed in NSCLC patients using the Spearman correlation test.
Prognostic impact of NCR1/NKp46, NCR3/NKp30 and NKp30 isoform expression in NSCLC
First, we analyzed the prognostic value of NCR1/NKp46 expression levels by stratifying the NSCLC patients' cohort according to the median of transcripts considered as a cut-off value. No significant difference was found between the two subgroups of expression of NCR1 transcripts with PFS or OS, the hazard ratio (HR=0.72 and 0.67, and p = 0.17 and 0.13, respectively) (Table 1).
Table 1.
Impact of NK receptor expression on NSCLC patient survival.
| <Cut-off | >Cut-off | |||||
|---|---|---|---|---|---|---|
| Nb pts | Median (95% CI) | Nb pts | Median (95% CI) | HR (95% CI) | p value | |
| PFS – NKp30a | 41/39 | 4.3 [2.7–5.6] mos | 35/31 | 6.9 [4.1–8.3] mos | 0.56 [95% CI: 0.34–0.91] | 0.0157 |
| OS – NKp30a | 41/30 | 10.3 [8.2–14.5] mos | 35/20 | 17.3 [10.0-ne] mos | 0.63 [95% CI: 0.36–1.11] | 0.1073 |
| PFS – NKp30b | 41/39 | 5.1 [2.8–5.9] mos | 35/31 | 4.9 [4.0–6.9] mos | 0.96 [95% CI: 0.60–1.55] | 0.8649 |
| OS – NKp30b | 41/28 | 10.0 [8.3–15.2] mos | 35/22 | 14.5 [10.3–18.9] mos | 0.74 [95% CI: 0.42–1.30] | 0.2902 |
| PFS – NKp30c | 41/39 | 4.3 [2.8–5.5] mos | 35/31 | 6.6 [4.1–8.1] mos | 0.68 [95% CI: 0.42–1.11] | 0.1186 |
| OS – NKp30c | 41/28 | 10.3 [8.4–18.9] mos | 35/22 | 14.1 [9.5–20.8] mos | 0.76 [95% CI: 0.43–1.33] | 0.3282 |
| PFS – dAB | 40/36 | 4.9 [4.2–6.8] mos | 36/33 | 4.3 [2.7–6.9] mos | 0.99 [95% CI: 0.61–1.59] | 0.9680 |
| OS – dAB | 40/24 | 13.7 [9.8–20.8] mos | 36/26 | 10.4 [8.1–18.7] mos | 1.47 [95% CI: 0.84–2.56] | 0.1727 |
| PFS – dBC | 38/35 | 5.5 [3.9–6.9] mos | 38/35 | 4.6 [3.9–6.0] mos | 1.31 [95% CI: 0.81–2.13] | 0.2667 |
| OS – dBC | 38/26 | 10.4 [8.1–20.1] mos | 38/24 | 13.7 [10.0–18.9] mos | 0.84 [95% CI: 0.48–1.47] | 0.5509 |
| PFS – dAC | 36/33 | 4.7 [4.1–6.0] mos | 40/36 | 5.5 [3.0–7.0] mos | 0.91 [95% CI: 0.57–1.47] | 0.7037 |
| OS – dAC | 36/24 | 9.8 [8.1–17.4] mos | 40/26 | 13.5 [10.0–20.1] mos | 0.87 [95% CI: 0.50–1.51] | 0.6131 |
| PFS – NCR3 | 40/39 | 4.2 [2.7–5.5] mos | 36/30 | 6.9 [4.2–8.3] mos | 0.53 [95% CI: 0.33–0.87] | 0.0101 |
| OS – NCR3 | 40/30 | 9.8 [6.4–13.5] mos | 36/20 | 18.7 [12.1-ne] mos | 0.53 [95% CI: 0.30–0.93] | 0.0250 |
| PFS – NCR1 | 40/38 | 4.4 [3.0–5.6] mos | 36/31 | 6.0 [4.0–7.0] mos | 0.72 [95% CI: 0.44–1.16] | 0.1737 |
| OS – NCR1 | 40/28 | 10.0 [8.1–13.7] mos | 36/22 | 17.4 [10.3–20.1] mos | 0.65 [95% CI: 0.37–1.14] | 0.1287 |
| PFS – dAB <−0.2 vs. >−0.2 | 38/34 | 4.6 [4.1–6.0] mos | 38/35 | 5.1 [3.9–6.9] mos | 0.94 [95% CI: 0.58–1.50] | 0.7798 |
| OS – dAB <−0.2 vs. >−0.2 | 38/24 | 13.5 [9.5–18.9] mos | 38/26 | 10.8 [8.4–20.1] mos | 1.17 [95% CI: 0.67–2.04] | 0.5780 |
| PFS – dBC <−1.3 vs. >−1.3 | 13/13 | 4.3 [1.6–6.6] mos | 63/56 | 4.9 [4.1–6.8] mos | 0.65 [95% CI: 0.35–1.21] | 0.1731 |
| OS – dBC <−1.3 vs >−1.3 | 13/11 | 9.4 [2.7–10.8] mos | 63/39 | 14.1 [10.0–18.9] mos | 0.50 [95% CI: 0.25–0.98] | 0.0400 |
| PFS – dBC 1.5 | 26/23 | 4.6 [3.0–6.0] mos | 37/33 | 5.8 [4.0–7.0] mos | 0.82 [95% CI: 0.48–1.40] | 0.4547 |
| OS – dBC 1.5 | 26/16 | 13.3 [6.4–18.9] mos | 37/23 | 17.3 [9.8–20.1] mos | 0.85 [95% CI: 0.45–1.62] | 0.6209 |
| PFS – dBC <−1.3 vs. >1.5 | 13/13 | 4.3 [1.6–6.6] mos | 26/23 | 4.6 [3.0–6.0] mos | 0.76 [95% CI: 0.38–1.52] | 0.4241 |
| OS – dBC <−1.3 vs. >1.5 | 13/11 | 9.4 [2.7−10.8] mos | 26/16 | 13.3 [6.4–18.9] mos | 0.51 [95% CI: 0.23–1.11] | 0.0820 |
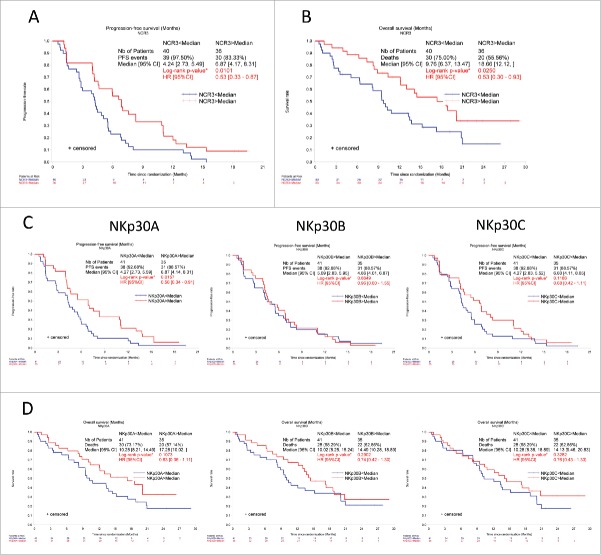
Next, we stratified NSCLC lung patients according to the expression level of NCR3 and each NKp30 isoform and observed that low expression of all transcripts encoded by NCR3, as well as low expression of NKp30A (but not that of NKp30B or NKp30C), had a negative prognostic impact on PFS and overall survival (OS) of NSCLC patients (Fig. 4).
Figure 4.
High NCR3 and NKp30A absolute expression levels are positive prognostic markers in NSCLC patients: first cohort. Progression-free survival (A, C) and overall survival (B, D) of NSCLC patients according to the median relative expression of NCR3 (A, B), and NKp30 isoforms A, B and C (C, D) was assessed at diagnosis, prior to therapy by univariate analysis using the Kaplan–Meier (Mantel–cox) test. *p < 0.05, **p < 0.01, ***p < 0.001.
Indeed, the hazard ratio (HR, with a 95% CI) was 0.53 (0.33–0.87, p = 0.0101) and 0.53 (0.30–0.93, p = 0.0250) for high vs. low NCR3 expression for PFS and OS, respectively. These results were confirmed in the validation set with HR at 0.66 (p = 0.036) and 0.63 (p = 0.040) (Fig. 5). These values were comparable for high vs. low NKp30A expression, with a HR = 0.56 (0.34–0.91, p = 0.016) for PFS, although NKp30A expression did not significantly affect OS with a HR = 0.63 (0.36–1.11, p = 0.11). Similar results were observed in the validation cohort with HR = 0.70 (p = 0.067) and 0.69 (p = 0.078). In contrast to the expression of NCR3 and individual NKp30 isoforms, which distributed in a roughly Gaussian mode, the ΔAB and ΔAC values distributed in a bimodal and trimodal fashion, respectively (Fig. S1). However, none of the Δ values (ΔAB, ΔAC and ΔBC) did affect PFS or OS of NSCLC patients in a significant fashion when the patients were stratified according to the median values or when cut-offs were calculated to fit multimodal distributions (Table 1). Hence, it appears that both the overall expression of NCR3 and the specific level of NKp30A (but not that of the other NKp30 isoforms nor the Δ values) have a prognostic impact in NSCLC patients.
Figure 5.
High NCR3 and NKp30A absolute expression levels are positive prognostic markers in NSCLC patients: validation cohort. Progression-free survival (A, C) and overall survival (B, D) of NSCLC patients according to the median relative expression of NCR3 (A, B), and NKp30 isoforms A, B and C (C, D) was assessed at diagnosis, prior to therapy by univariate analysis using the one-side Log-rank test. *p < 0.05, **p < 0.01, ***p < 0.001.
Interaction between PD-L1 expression by tumor cells and NK receptors
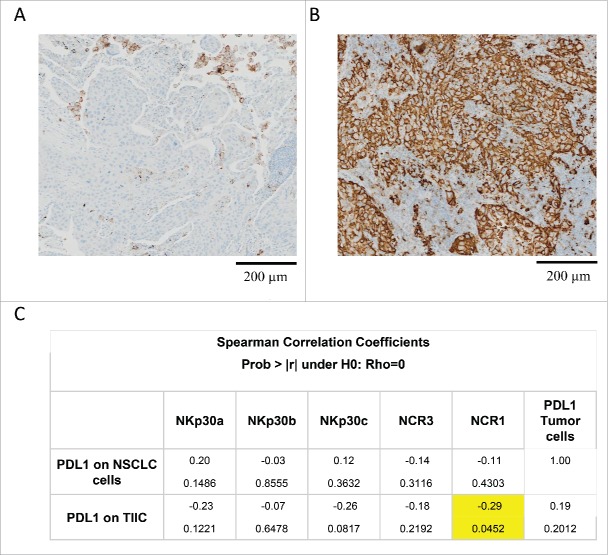
A validated immunohistochemical method was used to classify NSCLC specimens into those with low PD-L1 expression (“PD-L1-negative”) or those expressing PD-L1 on >5% of neoplastic cells (“PD-L1-positive”) (Fig. 6A, B), as well as into cancers in which stromal cells including tumor-infiltrating immune cells (TIIC) are PD-L1-negative (TIIC−) or PD-L1-positive (TIIC+). Of note, PD-L1 expression by NSCLC cells or TIIC was not correlated to the expression of NCR3 or any of the NKp30 isoforms. However, PD-L1 expression by TIIC was negatively correlated to that of NCR1 (Fig. 6C). Although PD-L1 expression on tumor cells had no significant impact on patient survival on its own (not shown), we investigated possible interactions between tumor cell PD-L1 expression and NK receptor expression with respect to patient prognosis (Fig. 7). When NSCLC fell into the PD-L1-positive category, the sole parameter that correlated with OS and PFS was the expression of NCR1 (but not NCR3) (Figs. 7, 8A–B vs. C–D), meaning that high NCR1 expression protected the host when tumors expressed PD-L1. However, none of the absolute or relative values of NKp30 isoforms affected prognosis (Fig. 7). When NSCLC fell into the PD-L1-negative category, the sole parameter that correlated with OS (but not PFS) was the ΔBC value (Fig. 9A, B). Indeed, patients with a low ΔBC value (<1.3) had a dismal prognosis as compared to the rest of patients (with a ΔBC value >1 .3) or the subgroup of patients with a high ΔBC value (>1 .5). Confirming these data, similar conclusions were drawn when examining the prognosis value of ΔBC in cancers with negative/low PD-L1 expression on stromal tumor-infiltrating cells (Fig. 9C, D). Hence, a relative increase of the inhibitory NKp30C isoform apparently has negative consequences for patient survival, in particular if the tumor is PD-L1-negative. However, in the second cohort receiving the TG4010 vaccine, neither NCR1 nor ΔBC value had predictive value for PFS nor OS in the PD-L1 positive or PD-L1 negative subset (not shown).
Figure 6.
Correlation between PD-L1 expression and NK cell receptors. (A–B). Example of PD-L1 staining by immunohistochemistry of a negative (A) and a positive (B) case of NSCLC sample. Membranous staining of alveolar macrophages is observed (positive control). Scale bar: 200 µm. (C) Correlation matrix between the relative expression of NKp30 isoforms A, B, C, NCR3, NCR1 and PD-L1 expression on tumor cells and tumor-infiltrating immune cells (TIIR).
Figure 7.
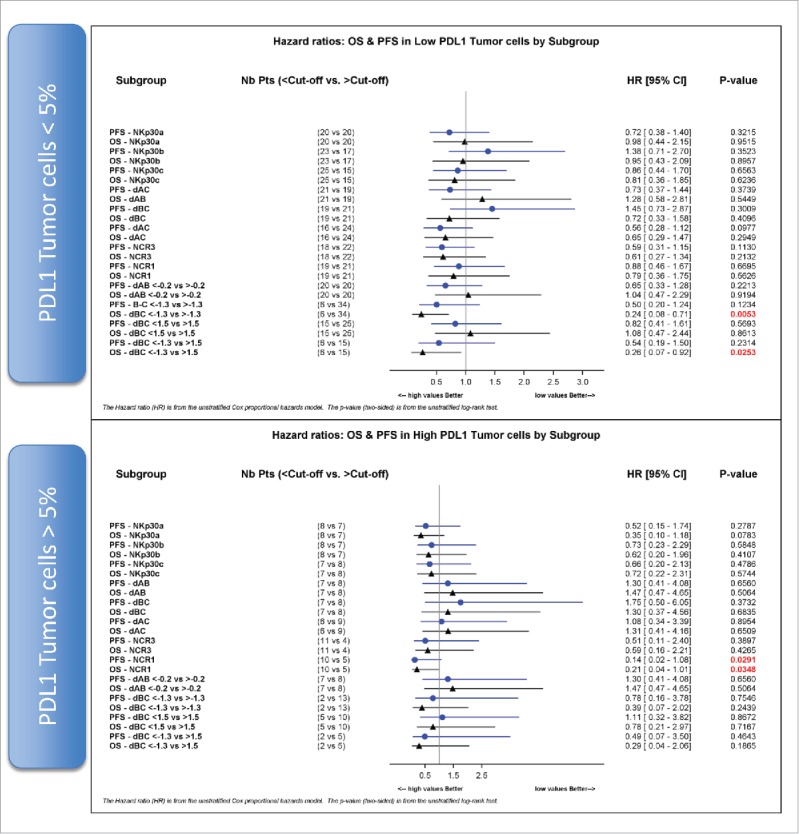
NCR1 is a prognostic factor in PD-L1 positive NSCLC. Hazard ratios of progression-free survival (PFS) and overall survival (OS) according to the median values, unless otherwise specified, of absolute expression of NKp30 isoforms A, B, C and relative expression of NKp30 Δ ratios, NCR3 and NCR1 are shown for PD-L1-negative (upper panel) and PD-L1 positive (lower panel) tumor cells (cut-off value of 5%).
Figure 8.

NCR1 absolute expression predicts PFS and OS of PD-L1 positive NSCLC cancer patients. Progression-free survival (A, C) and overall survival (B, D) of NSCLC cancer patients according to the median relative expression of NCR1 was assessed in PD-L1 negative (A, B) and PD-L1 positive (C, D) tumors by univariate analysis using the Kaplan–Meier method *p < 0.05 by Log-rank test. HR and corresponding 95% CI were estimated using a Cox regression model.
Figure 9.
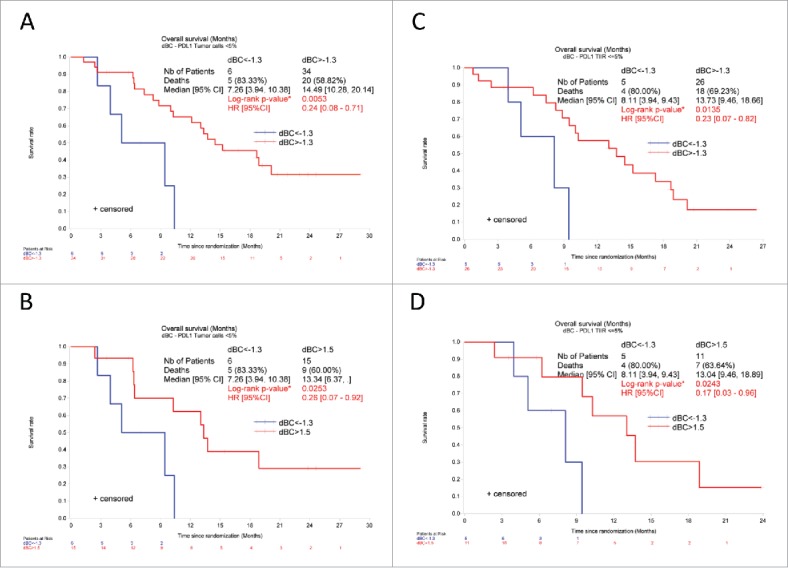
Low NKp30 ΔBC ratio affects the overall survival of NSCLC patients with PD-L1 negative tumors. Kaplan–Meier curve of overall survival of NSCLC patients according to the median (A, C, cut-off 1.3) or to the adjusted value (B, D, <1.3 vs. > 1.5) of NKp30 ΔBC ratio in patients with PD-L1 low (<5%) tumor cells (A, B) or TIIC (C, D). *p < 0 .05 by Log-rank test. HR and corresponding 95% CI were estimated using a Cox regression model.
Concluding remarks
The present study reveals a probable aberration of NK cell function in NSCLC. This aberration consists in a constitutively low expression at the mRNA level of the two major NK cell receptors, NCR1/NKp46 and NCR3/NKp30, as well as a reduced expression of all major NKp30 isoforms when compared to patients with other malignancies or healthy controls. The reasons for this low expression of NK cell receptors are elusive. There is no general numeric NK cell defect in NSCLC patients,6-8 hence ruling out the (trivial) explanation that a reduction in the amount of circulating NK cells relative to other leukocyte subset would account for reduced NCR1/NKp46 and NCR3/NKp30 expression. Previous longitudinal studies have shown that NKp30 isoforms are stably expressed over time, before and after curative treatment of GIST or NB.26,29 Such longitudinal information is missing in this study, in which NK cell receptor expression was only determined before the initiation of NSCLC treatment, meaning that it cannot be determined whether tumor burden affects the levels of NCR1/NKp46 and NCR3/NKp30 in circulating blood cells or whether this parameter is constitutively perturbed, irrespective to the absence or presence of the cancer.
Importantly, NSCLC patients exhibited significant changes in the ratio of activating NKp30 isoforms (NKp30A, NKp30B) and the inhibitory isoform (NKp30C). We found among NSCLC patients (but not among HV nor among patients with other malignancies such as GIST, NB or melanoma) a discrete group of samples with rather low ΔBC values (<1.3). However, these ΔBC values did not correlate with the overall expression of NCR1 or NCR3, suggesting that the mechanisms that lead to the downregulation of overall NK cell receptor expression and those involved in the regulation of NKp30 isoform expression are not the same. It is tempting to hypothesize, yet remains to be evaluated, that NK cells from patients with low (<1.3) ΔBC values are defective in their immunosurveillance function, hence shifting from an anticancer effector function to an immunosuppressive one.
In a highly speculative scenario, NSCLC might be under dual immunosurveillance by T lymphocytes (which mostly detect tumor-specific antigenic peptides presented by MHC class I molecules) and NK cells (which mostly detect the absence of MHC class I expression).35-38 The expression of PD-L1 allowed us to categorize patients into two groups with rather low (<5%) PD-L1 expression by tumor cells or a high (>5%) expression level. PD-L1 expression is thought to constitute an active mechanism of immunosubversion, allowing the malignant cells to paralyze or “exhaust” T lymphocytes.39 Interestingly, it is only among the patients bearing PD-L1-negative (<5%) NSCLC, either at the level of tumor cells or at the stromal tumor-infiltrating lymphocytes that low (<1 .3) ΔBC values had a negative impact on survival. However, in cases of high PD-L1 tumor expression, the expression of NCR1 appears crucial to dictate PFS and OS. Although very few data point to a regulatory role of the PD-L1/PD-1 axis on the NK cell arm of anticancer immunity,40,41 it is tempting to speculate that the ménage à trois) between DC, NK and T cells, which is supposedly regulated by NCR3 and PD1,42 is coordinated to orchestrate the priming arm of tumor immunosurveillance while other interactions (tumor cells, NK and T cells) depending mostly on NCR1 may mostly affect the effector arm of immunity. Thus, the combined failure of T and NK cell-mediated immunosurveillance may be particularly negative in its consequences.
These observations are based on a relatively small number of patients. However, when examining the second arm of the Phase 2b Transgene clinical trial “Time”, the arm receiving the poxvirus recombinant for MUC-1 and IL-2 (TG4010) vaccine, the prognostic value of NCR3 remained significant. However, the prognostic value of NCR1 or the ΔBC ratio in cancers with positive or low PD-L1 expression, respectively were not verified. Our NK cell parameters may reflect the NK cell status of these patients, known to influence the outcome of the TG4010 vaccine. When TG4010 was co-administered with chemotherapy in non-operable NSCLC, the response rates, the PFS and OS were all significantly improved.31 The authors confirmed the negative predictive value of an NK parameter, i.e a high-baseline percentage of circulating activated NK cells (CD16+CD56+CD69+) (TrPAL) (observed in 25% cases) for all these clinical metrics relevant for the bioactivity of the product. However, the NCR1, NCR3 and NCR3 isoforms were outperformed by the TrPAL parameter to predict the long term benefit of the TG4010 vaccine.
Irrespective of these limitations, it appears highly plausible that NK cell function impacts on NSCLC progression and that a comparatively simple (qRT-PCR-based) blood test allowing to measure the expression of NK cell receptor isoforms can yield clinically useful information. Future will tell whether such a diagnostic procedure will improve the clinical management of NSCLC with immunotherapies.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
LZ is supported by Transgene, as well as ARC, LIGUE, INCA, ISREC and Swiss Bridge Foundation. GK is supported by the Ligue contre le Cancer (équipe labelisée), Agence National de la Recherche (ANR) – Projets blancs, ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases, Association pour la recherche sur le cancer (ARC), Cancéropôle Ile-de-France, Institut National du Cancer (INCa), Institut Universitaire de France, Fondation pour la Recherche Médicale (FRM), the European Commission (ArtForce), the European Research Council (ERC), the LabEx Immuno-Oncology, the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE), the SIRIC Cancer Research and Personalized Medicine (CARPEM) and the Paris Alliance of Cancer Research Institutes (PACRI).
References
- 1.Krause SW, Gastpar R, Andreesen R, Gross C, Ullrich H, Thonigs G, Pfister K, Multhoff G. Treatment of colon and lung cancer patients with ex vivo heat shock protein 70-peptide-activated, autologous natural killer cells: a clinical phase i trial. Clin Cancer Res 2004; 10:3699-707; PMID:15173076; http://dx.doi.org/ 10.1158/1078-0432.CCR-03-0683 [DOI] [PubMed] [Google Scholar]
- 2.Yang YJ, Park JC, Kim HK, Kang JH, Park SY. A trial of autologous ex vivo-expanded NK cell-enriched lymphocytes with docetaxel in patients with advanced non-small cell lung cancer as second- or third-line treatment: phase IIa study. Anti Cancer Res 2013; 33:2115-22; PMID:2364576316237071 [PubMed] [Google Scholar]
- 3.Le Maux Chansac B, Moretta A, Vergnon I, Opolon P, Lecluse Y, Grunenwald D, Kubin M, Soria JC, Chouaib S, Mami-Chouaib F. NK cells infiltrating a MHC class I-deficient lung adenocarcinoma display impaired cytotoxic activity toward autologous tumor cells associated with altered NK cell-triggering receptors. J Immunol 2005; 175:5790-8; PMID:16237071; http://dx.doi.org/ 10.4049/jimmunol.175.9.5790 [DOI] [PubMed] [Google Scholar]
- 4.Carrega P, Morandi B, Costa R, Frumento G, Forte G, Altavilla G, Ratto GB, Mingari MC, Moretta L, Ferlazzo G. Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56 bright CD16(-) cells and display an impaired capability to kill tumor cells. Cancer 2008; 112:863-75; PMID:18203207; http://dx.doi.org/ 10.1002/cncr.23239 [DOI] [PubMed] [Google Scholar]
- 5.Ciszak L, Kosmaczewska A, Werynska B, Szteblich A, Jankowska R, Frydecka I. Impaired zeta chain expression and IFN-gamma production in peripheral blood T and NK cells of patients with advanced lung cancer. Oncology Reports 2009; 21:173-84; PMID:19082459; http://dx.doi.org/ 10.3892/or_00000205 [DOI] [PubMed] [Google Scholar]
- 6.Cremer I, Fridman WH, Sautes-Fridman C. Tumor microenvironment in NSCLC suppresses NK cells function. Onco Immunol 2012; 1:244-6; PMID:22720258; http://dx.doi.org/21708957 10.4161/onci.1.2.18309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Platonova S, Cherfils-Vicini J, Damotte D, Crozet L, Vieillard V, Validire P, Andre P, Dieu-Nosjean MC, Alifano M, Regnard JF, Fridman WH et al.. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res 2011; 71:5412-22; PMID:21708957; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-4179 [DOI] [PubMed] [Google Scholar]
- 8.Jin J, Fu B, Mei X, Yue T, Sun R, Tian Z, Wei H. CD11b(-)CD27(-) NK cells are associated with the progression of lung carcinoma. PloS One 2013; 8:e61024; PMID:23565296; http://dx.doi.org/ 10.1371/journal.pone.0061024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Zhang G, Qin Y, Bai R, Huang J. B7-H6 expression in non-small cell lung cancers. Int J Clin Exp Pathol 2014; 7:6936-42; PMID:25400778 [PMC free article] [PubMed] [Google Scholar]
- 10.de Chaisemartin L, Goc J, Damotte D, Validire P, Magdeleinat P, Alifano M, Cremer I, Fridman WH, Sautes-Fridman C, Dieu-Nosjean MC. Characterization of chemokines and adhesion molecules associated with T cell presence in tertiary lymphoid structures in human lung cancer. Cancer Res 2011; 71:6391-9; PMID:21900403; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-0952 [DOI] [PubMed] [Google Scholar]
- 11.Remark R, Becker C, Gomez JE, Damotte D, Dieu-Nosjean MC, Sautes-Fridman C, Fridman WH, Powell CA, Altorki NK, Merad M et al.. The non-small cell lung cancer immune contexture. A major determinant of tumor characteristics and patient outcome. Am J Respiratory Critical Care Med 2015; 191:377-90; http://dx.doi.org/ 10.1164/rccm.201409-1671PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sariban E, Mitchell T, Kufe D. Expression of the c-raf protooncogene in human hematopoietic cells and cell lines. Blood 1987; 69:1437-40; PMID:2436688 [PubMed] [Google Scholar]
- 13.Taube JM. Unleashing the immune system: PD-1 and PD-Ls in the pre-treatment tumor microenvironment and correlation with response to PD-1/PD-L1 blockade. Oncoimmunol 2014; 3:e963413; PMID:25914862; http://dx.doi.org/23592754 10.4161/21624011.2014.963413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buque A, Bloy N, Aranda F, Castoldi F, Eggermont A, Cremer I, Fridman WH, Fucikova J, Galon J, Marabelle A et al.. Trial Watch: Immunomodulatory monoclonal antibodies for oncological indications. Oncoimmunol 2015; 4:e1008814; PMID:26137403; http://dx.doi.org/23592754 10.1080/2162402X.2015.1008814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mamessier E, Bourgin C, Olive D. When breast cancer cells start to fend the educational process of NK cells off. Oncoimmunol 2013; 2:e26688; PMID:24498553; http://dx.doi.org/23592754 10.4161/onci.26688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu D, Geschwind JF, Karthikeyan S, Miller E, Kunjithapatham R, Wang Z, Ganapathy-Kanniappan S. Metabolic perturbation sensitizes human breast cancer to NK cell-mediated cytotoxicity by increasing the expression of MHC class I chain-related A/B. Oncoimmunol 2015; 4:e991228; PMID:25949910; http://dx.doi.org/23592754 10.4161/2162402X.2014.991228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sconocchia G, Eppenberger S, Spagnoli GC, Tornillo L, Droeser R, Caratelli S, Ferrelli F, Coppola A, Arriga R, Lauro D et al.. NK cells and T cells cooperate during the clinical course of colorectal cancer. Oncoimmunol 2014; 3:e952197; PMID:25610741; http://dx.doi.org/23592754 10.4161/21624011.2014.952197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geissler K, Fornara P, Lautenschlager C, Holzhausen HJ, Seliger B, Riemann D. Immune signature of tumor infiltrating immune cells in renal cancer. Oncoimmunol 2015; 4:e985082; PMID:25949868; http://dx.doi.org/23592754 10.4161/2162402X.2014.985082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pesce S, Tabellini G, Cantoni C, Patrizi O, Coltrini D, Rampinelli F, Matta J, Vivier E, Moretta A, Parolini S et al.. B7-H6-mediated downregulation of NKp30 in NK cells contributes to ovarian carcinoma immune escape. Oncoimmunol 2015; 4:e1001224; PMID:25610741; http://dx.doi.org/23592754 10.1080/2162402X.2014.1001224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rusakiewicz S, Semeraro M, Sarabi M, Desbois M, Locher C, Mendez R, Vimond N, Concha A, Garrido F, Isambert N et al.. Immune infiltrates are prognostic factors in localized gastrointestinal stromal tumors. Cancer Res 2013; 73:3499-510; PMID:23592754; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-0371 [DOI] [PubMed] [Google Scholar]
- 21.Yoshimoto T, Mizoguchi I, Katagiri S, Tauchi T, Furusawa JI, Chiba Y, Mizuguchi J, Ohyashiki JH, Ohyashiki K. Immunosurveillance markers may predict patients who can discontinue imatinib therapy without relapse. Oncoimmunol 2014; 3:e28861; PMID:25057448; http://dx.doi.org/24225017 10.4161/onci.28861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Messaoudene M, Avril MF, Caignard A. When unity makes strength: Combinatorial NK cell-based immunotherapies against melanoma. Oncoimmunol 2014; 3:e28048; PMID:25340005; http://dx.doi.org/24225017 10.4161/onci.28048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messaoudene M, Fregni G, Fourmentraux-Neves E, Chanal J, Maubec E, Mazouz-Dorval S, Couturaud B, Girod A, Sastre-Garau X, Albert S et al.. Mature cytotoxic CD56(bright)/CD16(+) natural killer cells can infiltrate lymph nodes adjacent to metastatic melanoma. Cancer Res 2014; 74:81-92; PMID:24225017; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-1303 [DOI] [PubMed] [Google Scholar]
- 24.de Andrade LF, Ngiow SF, Martinet L, Smyth MJ. Natural Killer cell control of mutant melanoma during targeted therapy. Oncoimmunol 2015; 4:e998119; PMID:26137412; http://dx.doi.org/21552268 10.1080/2162402X.2014.998119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ilander M, Kreutzman A, Mustjoki S. IFNalpha induces prolonged remissions modeling curative immunologic responses in chronic myeloid leukemia. Oncoimmunol 2014; 3:e28781; PMID:25050224; http://dx.doi.org/21552268 10.4161/onci.28781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delahaye NF, Rusakiewicz S, Martins I, Menard C, Roux S, Lyonnet L, Paul P, Sarabi M, Chaput N, Semeraro M et al.. Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nat Med 2011; 17:700-7; PMID:21552268; http://dx.doi.org/ 10.1038/nm.2366 [DOI] [PubMed] [Google Scholar]
- 27.Prada N, Antoni G, Commo F, Rusakiewicz S, Semeraro M, Boufassa F, Lambotte O, Meyer L, Gougeon ML, Zitvogel L. Analysis of NKp30/NCR3 isoforms in untreated HIV-1-infected patients from the ANRS SEROCO cohort. Oncoimmunol 2013; 2:e23472; PMID:23802087; http://dx.doi.org/26207902 10.4161/onci.23472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rusakiewicz S, Nocturne G, Lazure T, Semeraro M, Flament C, Caillat-Zucman S, Sene D, Delahaye N, Vivier E, Chaba K et al.. NCR3/NKp30 contributes to pathogenesis in primary Sjogren's syndrome. Sci Translational Med 2013; 5:195ra96; PMID:23884468; http://dx.doi.org/26207902 10.1126/scitranslmed.3005727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semeraro M, Rusakiewicz S, Minard-Colin V, Delahaye NF, Enot D, Vely F, Marabelle A, Papoular B, Piperoglou C, Ponzoni M et al.. Clinical impact of the NKp30/B7-H6 axis in high-risk neuroblastoma patients. Sci Translational Med 2015; 7:283ra55; PMID:25877893; http://dx.doi.org/26207902 10.1126/scitranslmed.aaa2327 [DOI] [PubMed] [Google Scholar]
- 30.Rotonda C, Anota A, Mercier M, Bastien B, Lacoste G, Limacher JM, Quoix E, Bonnetain F. Impact of TG4010 vaccine on health-related quality of life in advanced non-small-cell lung cancer: results of a Phase IIB clinical trial. Plos One 2015; 10:e0132568; PMID:26207902; http://dx.doi.org/ 10.1371/journal.pone.0132568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quoix E, Lena H, Losonczy G, Forget F, Chouaid C, Papai Z, Gervais R, Ottensmeier C, Szczesna A, Kazarnowicz A et al.. TG4010 immunotherapy and first-line chemotherapy for advanced non-small-cell lung cancer (TIME): results from the phase 2b part of a randomised, double-blind, placebo-controlled, phase 2b/3 trial. Lancet Oncol 2016; 17:212-23; PMID:26727163; http://dx.doi.org/ 10.1016/S1470-2045(15)00483-0 [DOI] [PubMed] [Google Scholar]
- 32.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC et al.. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000; 92:205-16; PMID:10655437; http://dx.doi.org/ 10.1093/jnci/92.3.205 [DOI] [PubMed] [Google Scholar]
- 33.Romero AI, Chaput N, Poirier-Colame V, Rusakiewicz S, Jacquelot N, Chaba K, Mortier E, Jacques Y, Caillat-Zucman S, Flament C et al.. Regulation of CD4(+)NKG2D(+) Th1 cells in patients with metastatic melanoma treated with sorafenib: role of IL-15Ralpha and NKG2D triggering. Cancer Res 2014; 74:68-80; PMID:24197135; http://dx.doi.org/11846609 10.1158/0008-5472.CAN-13-1186 [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/ 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 35.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 2011; 331:1565-70; PMID:21436444; http://dx.doi.org/ 10.1126/science.1203486 [DOI] [PubMed] [Google Scholar]
- 36.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science 2011; 331:44-9; PMID:21212348; http://dx.doi.org/ 10.1126/science.1198687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zitvogel L, Galluzzi L, Smyth MJ, Kroemer G. Mechanism of action of conventional and targeted anticancer therapies: reinstating immunosurveillance. Immunity 2013; 39:74-88; PMID:23890065; http://dx.doi.org/ 10.1016/j.immuni.2013.06.014 [DOI] [PubMed] [Google Scholar]
- 38.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nature reviews. Immunol 2015; 15:405-14; PMID:26027717; http://dx.doi.org/21724589 10.1038/nri3845 [DOI] [PubMed] [Google Scholar]
- 39.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annual Rev Immunol 2008; 26:677-704; PMID:18173375; http://dx.doi.org/21724589 10.1146/annurev.immunol.26.021607.090331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terme M, Ullrich E, Aymeric L, Meinhardt K, Desbois M, Delahaye N, Viaud S, Ryffel B, Yagita H, Kaplanski G et al.. IL-18 induces PD-1-dependent immunosuppression in cancer. Cancer Res 2011; 71:5393-9; PMID:21724589; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-0993 [DOI] [PubMed] [Google Scholar]
- 41.Benson DM Jr, Bakan CE, Mishra A, Hofmeister CC, Efebera Y, Becknell B, Baiocchi RA, Zhang J, Yu J, Smith MK et al.. The PD-1/PD-L1 axis modulates the natural killer cell vs. multiple myeloma effect: a therapeutic target for CT-011, a novel monoclonal anti-PD-1 antibody. Blood 2010; 116:2286-94; PMID:20460501; http://dx.doi.org/ 10.1182/blood-2010-02-271874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferlazzo G, Tsang ML, Moretta L, Melioli G, Steinman RM, Munz C. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J Exp Med 2002; 195:343-51; PMID:11828009; http://dx.doi.org/ 10.1084/jem.20011149 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.