ABSTRACT
The phosphatase Wip1 attenuates the DNA damage response (DDR) by removing phosphorylation marks from a number of DDR proteins (p53, MDM2, Chk1/2, p38). Wip1 also dephosphorylates and inactivates RelA. Notably, LZAP, a putative tumor suppressor, has been linked to dephosphorylation of several of these substrates, including RelA, p38, Chk1, and Chk2. LZAP has no known catalytic activity or functional motifs, suggesting that it exerts its effects through interaction with other proteins. Here we show that LZAP binds Wip1 and stimulates its phosphatase activity. LZAP had been previously shown to bind many Wip1 substrates (RelA, p38, Chk1/2), and our results show that LZAP also binds the previously identified Wip1 substrate, MDM2. This work identifies 2 novel Wip1 substrates, ERK1 and HuR, and demonstrates that HuR is a binding partner of LZAP. Pleasingly, LZAP potentiated Wip1 catalytic activity toward each substrate tested, regardless of whether full-length substrates or phosphopeptides were utilized. Since this effect was observed on ERK1, which does not bind LZAP, as well as for each of 7 peptides tested, we hypothesize that LZAP binding to the substrate is not required for this effect and that LZAP directly binds Wip1 to augment its phosphatase activity.
KEYWORDS: CDK5RAP3, C53, LZAP, phosphatase regulator, PPM1D, PP25; Wip1
Introduction
The wild-type p53-induced phosphatase 1 (Wip1), also known as protein phosphatase Mg2+/Mn2+-dependent 1D (PPM1D), or protein phosphatase 2Cδ (PP2Cδ), is an oncogenic nuclear serine/threonine phosphatase of the PP2C family. Wip1 was originally identified in a screen for p53-upregulated genes following DNA damage.1 As a negative regulator of the DNA damage response (DDR) pathway, Wip1 removes activating phosphorylation marks (mostly within pTXY and p(S/T)Q motifs) from a number of proteins active in the DDR.,2,3 The motifs recognized by Wip1 are identical to those recognized by the critical DDR kinases ATM, ATR, Chk1, and Chk2. The first described Wip1 substrate was pThr180 of MAPK p38α (hereafter p38), which is rapidly phosphorylated in response to cellular stress, including DNA damage.4 In addition to p38, several other proteins that are dephosphorylated by Wip1 have been described, including p53 (Ser15),5 checkpoint kinase 1 (Chk1) (Ser345),5 Chk2 (Thr68),6 ATM (Ser1981),7 γH2AX (Ser139),89 and Bax (Thr172, 174, 186),10 as well as 2 major negative regulators of p53, MDM2 (Ser345)11 and MDMX (Ser403).12 Hence, Wip1 is ideally positioned and plays a critical role for restoring the baseline state after DNA damage.
Given that p53 is dephosphorylated and inhibited by Wip1, the role of Wip1 as an oncogene was experimentally confirmed when Bulavin et al. showed cooperation between Wip1 and other oncogenic drivers to transform primary fibroblasts.13 Wip1 was further validated as an oncogene using mouse models that demonstrated a reduction in tumor occurrence and size following protein depletion or chemical inhibition of Wip1.13-16 In human tumors, amplification of PPM1D (17q22-q23) and subsequent increases in Wip1 protein levels were found in human breast cancer cell lines and primary tumors at an incidence rate of approximately 15%.13,17,18 Importantly, PPM1D amplification has since been identified in carcinomas of the ovary,19 pancreas,20 and stomach,21 as well as in medulloblastomas22 and neuroblastomas.23 More recently, Wip1 protein-truncating mutations have been identified within exon 6 of PPM1D in a small number (< 1%) of patients with breast,24 ovarian,24 lung,25 and colorectal carcinomas,26 as well as brainstem gliomas.27 Interestingly, these mutations confer a gain-of-function phenotype by encoding a hyperstable, truncated Wip1 protein that retains all phosphatase activity, phenotypically similar to amplification of PPM1D. Importantly, regardless of the mechanism, elevated levels of Wip1 predispose patients to cancer and correlate with tumor progression, invasion, metastasis, and poor patient prognosis.18-24
In contrast to accepted oncogenic activities of Wip1, it has also been found to have potential tumor suppressor activity through dephosphorylation of RelA (Ser536), a modification required for full transactivation of the NF-κB (p50/RelA) heterodimer. Wip1 activity toward RelA has been linked to negative regulation of canonical NF-κB signaling and dampening of the inflammatory response.28,29 Given this seemingly counterintuitive finding, the role of Wip1 in tumorigenesis may be complex, contributing both oncogenic and tumor suppressor functions depending on cellular context. Like other PPM phosphatases, Wip1 is active as a monomer, suggesting that in addition to regulation of Wip1 protein expression, other levels of regulation likely exist, potentially some mediated by binding partners of Wip1.30
LZAP (LXXLL/leucine zipper-containing ARF-binding protein), also known as CDK5RAP3, C53, IC53, and HSF-27, was initially identified as a binding partner of the Cdk5 activator p35.31 Our laboratory furthered insight into the activity of LZAP by showing that it binds alternative reading frame (ARF, p14ARF in human and p19ARF in mice) to activate p53, arrest cellular proliferation, and inhibit clonogenic growth.32 Evidence of a tumor suppressor role for LZAP was bolstered when LZAP protein levels were found to be markedly decreased in head and neck squamous cell carcinoma (HNSCC), where its loss inversely correlates with expression of NF-κB target genes.33 Further, LZAP inhibits cellular transformation, as well as growth and vascularization of xenograft tumors, through its ability to bind and inhibit RelA.33 A binding partner of LZAP, Ufl1, stabilizes LZAP protein levels, and its loss results in LZAP downregulation and enhanced NF-κB signaling and cell invasion in osteosarcoma and hepatocellular carcinoma cells.34,35 Moreover, in cancer cells treated with DNA damaging agents, LZAP binds and inhibits checkpoint kinases 1 and 2 (Chk1 and Chk2), compromising the G2/M checkpoint and resulting in subsequent apoptosis.36,37 Collectively, these data are consistent with a role for LZAP in tumor suppression.
Although activation of Chk1/2 serves as a barrier to cell death induced by radiotherapy and chemotherapeutic agents, basal checkpoint kinase activity is an essential component of the tumor surveillance network of healthy cells. Hence, in unstimulated cells, LZAP may compromise the ability of cells to arrest in response to DNA damage by promoting premature dephosphorylation of Chk1 and Chk2, contributing to genomic instability and subsequent tumorigenesis. We recently reported that LZAP inhibits p38 phosphorylation and activity by facilitating p38 association with Wip1.38 Considering that LZAP antagonizes p38 activity and that the role of p38 in cancers can vary from growth inhibitory to growth promoting, LZAP activities in tumors may depend on cellular context.
Although the evolutionally conserved LZAP plays critical roles in both development and cancer, it has no known functional motifs, catalytic activity, or described homologs suggesting that LZAP may exert its activity through interaction with other protein(s).33,39 Data from our laboratory and others link LZAP to a decrease in phosphorylation of some, but not all, of its binding partners, including RelA, p38, and Chk1/2.33,36-38 It is striking that each of these proteins is a target of the Wip1 phosphatase, suggesting that LZAP activity may be tightly linked to Wip1. Given this observation, as well as our finding that Wip1 is, at least partially, required for the LZAP-mediated reduction in phosphorylation and subsequent inactivation of p38, we hypothesized that LZAP may be a regulator of Wip1 activity.38 Here we report that LZAP directly binds Wip1 and enhances dephosphorylation of a number of its substrates.
Results
LZAP interacts with Wip1
We previously demonstrated that LZAP binds p38 MAPK and attenuates its phosphorylation and subsequent kinase activation. Interestingly, LZAP does not alter phosphorylation of upstream p38-activating kinases, instead mediating its inhibitory effect toward p38 through the major p38 phosphatase, Wip1.38 LZAP also decreases phosphorylation of RelA at Ser536,33 a mark targeted for dephosphorylation by Wip1.28 Additionally, LZAP dampens DNA damage-induced activation of Chk1 and Chk2 by reducing levels of phosphorylated Ser345 and Thr68, respectively, both of which are Wip1 targets.36,37 These findings led us to hypothesize that LZAP may directly bind Wip1 to regulate its catalytic activity.
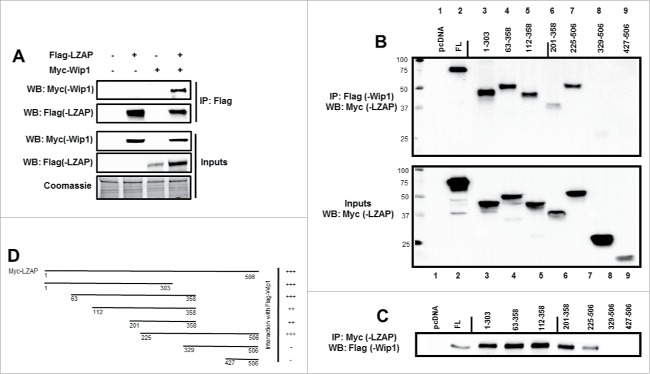
To begin exploring interaction between LZAP and Wip1, U2OS cells were transiently transfected with constructs encoding Flag-LZAP and Myc-Wip1, alone or in concert, prior to immunoprecipitation with Flag antibody-conjugated agarose. Myc-Wip1 was detected in Flag immunoprecipitates only in the presence of Flag-LZAP (Fig. 1A, lane 4 vs. lane 3). Empty vector was used as a negative control (lanes 1–3), and inputs confirm expression of both tagged constructs (bottom panels).
Figure 1.
LZAP binds to Wip1. (A) Ectopically expressed LZAP and Wip1 co-immunoprecipitate. U2OS cells were transfected with indicated plasmids encoding tagged Flag-LZAP or Myc-Wip1. Immunoprecipitates were prepared using Flag affinity matrix to pull down LZAP, resolved on SDS-PAGE, and immunoblotted by antibodies recognizing Myc(-Wip1) or Flag(-LZAP). Expression of LZAP and Wip1 was confirmed by immunoblotting with Flag or Myc antibodies, respectively. (B and C) A central region of LZAP likely mediates its interaction with Wip1. U2OS cells were transfected with indicated plasmids encoding Flag-Wip1 and Myc-tagged truncation mutants of LZAP. Expression of Myc-LZAP truncation mutants was confirmed by immunoblotting with an antibody recognizing Myc (B, lower panel). Immunoprecipitates were prepared using either Flag (B, upper panel) or Myc (C) affinity matrix to pull down Wip1 or LZAP, respectively, resolved on SDS-PAGE, and immunoblotted by antibodies recognizing Myc(-LZAP) (B) or Flag(-Wip1) (C). In each panel, all lanes were run on the same gel; solid line indicates where images were cropped. Binding of LZAP truncations to full-length Wip1 is schematically summarized (D).
To further confirm this interaction, as well as define regions of LZAP that mediate its interaction with Wip1, the converse co-immunoprecipitation was performed in U2OS cells using full-length Flag-Wip1 and Myc-LZAP, as well as various Myc-LZAP truncations. Expression of Myc-LZAP truncations was confirmed by immunoblot of whole cell lysate (Fig. 1B, bottom panel). Empty vector was used as a negative control (Fig. 1B and 1C, lane 1), and the interaction between full-length overexpressed full-length LZAP and Wip1 presented in Fig. 1A was confirmed (Fig. 1B, top panel, lane 2, and Fig. 1C, lane 2).
Results generated by immunoprecipitation of either Wip1 (Fig. 1B) or LZAP (Fig. 1C) indicate that an extended region of the N-terminus of LZAP, αα (amino acids) 1–303, is sufficient for interation with Wip1 (lanes 3+4). LZAP fragments including αα 112–358, 201–358, and 225–506 also bound Wip1 in these assays (lanes 5, 6, and 7, respectively). Interestingly, further truncation to generate constructs αα 329–506 and 427–506 abrogated Wip1 interaction (lanes 8 and 9). Together, these data show that the carboxy-terminus of LZAP is dispensable for Wip1 binding, and LZAP mediates its interaction with Wip1 through its N-terminus. Binding activity of LZAP truncations is schematically summarized in Fig. 1D.
LZAP promotes Wip1-catalyzed dephosphorylation of its full-length substrates
Our laboratory previously showed that LZAP binds p38, and that increasing amounts of LZAP potentiate dephosphorylation of p38. Conversely, knockdown of LZAP increases p38 phosphorylation and its kinase activation. LZAP did not alter activation of the primary kinases upstream of p38, MAP kinase kinases 3 and 6 (MKK3 and MKK6), but instead promoted association of p38 with its primary phosphatase, Wip1. Moreover, knockdown of Wip1 attenuated the ability of LZAP to promote dephosphorylation of p38.38 These results, combined with binding between LZAP and Wip1, suggest that LZAP may directly or indirectly stimulate Wip1-catalyzed dephosphorylation.
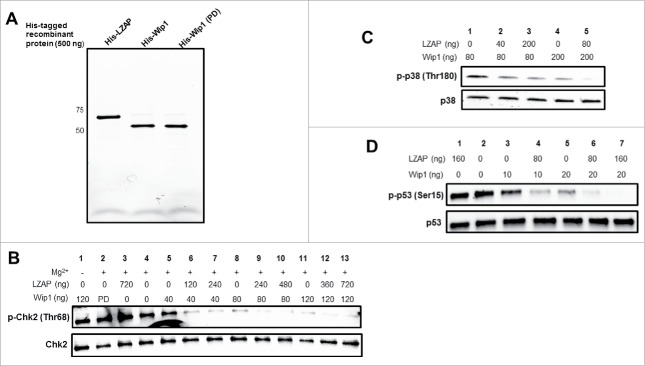
To begin exploring these possibilities, we performed in vitro phosphatase assays using full-length phosphorylated substrates of Wip1. These substrates were generated by transfection of 293T cells with Flag-tagged constructs, treatment with mitomycin C to enrich for phosphorylated species, immunoprecipitation with Flag antibody-conjugated agarose, and elution with 3x Flag peptide. These phospho-proteins were then incubated with bacterially expressed His-Wip1 (wild-type and phosphatase-dead) in the presence or absence of His-LZAP (see Fig. 2A for image of recombinant proteins).
Figure 2.
LZAP potentiates Wip1-catalyzed dephosphorylation of full-length phosphoproteins. (A) 500 ng of purified His-LZAP (lane 1), His-Wip1 (wild-type, lane 2), and His-Wip1 (phosphatase dead, lane 3) were resolved via SDS-PAGE and imaged. (B-D) Flag-tagged, full-length, phosphorylated Chk2 (B), p38 (C), and p53 (D) were used as substrates for in vitro Wip1 phosphatase assays. Substrates were incubated with the indicated amounts of bacterially-produced His-Wip1 and His-LZAP in phosphatase buffer with or without magnesium for 30 minutes at 30 degrees. Reactions were resolved by SDS-PAGE and immunoblotted using antibodies recognizing phosphorylated substrates (top panels) or total substrates (bottom panels).
Wip1 has been shown to dephosphorylate Chk2 at Thr68, resulting in its inactivation.6 Therefore, we tested whether LZAP could potentiate Wip1 phosphatase activity toward this mark. As expected, addition of Wip1 decreased Chk2 phosphorylation at Thr68 in a dose-dependent manner (Fig. 2B, lane 4 vs. lanes 5, 8, and 11). Wip1 is a magnesium-dependent phosphatase, and exclusion of required magnesium (lane 1) or use of a point mutant Wip1 lacking phosphatase activity (lane 2) did not result in Chk2 dephosphorylation, suggesting the Wip1 phosphatase activity was responsible for observed decreased Chk2 phosphorylation.
Of interest, the addition of LZAP to the reaction strongly increased Wip1 dephosphorylation of full-length Chk2 at Thr68, for each of 3 different doses of Wip1 tested (lanes 6 and 7 vs. lane 5; lanes 9 and 10 vs. lane 8; and lanes 12 and 13 vs. lane 11). Consistent with the absence of functional motifs in LZAP, addition of LZAP without Wip1 did not alter Chk2 phosphorylation (lane 3).
To determine if the effect of LZAP to enhance Wip1 dephosphorylation of substrates was restricted to Chk2, 2 additional substrates, p38 and p53, were tested using the same in vitro Wip1 phosphatase assay. P38 (Thr180) was the first described Wip1 substrate,4 and Wip1 activity toward p53 (Ser15) was discovered shortly thereafter.5 Consistent with results obtained using p-Chk2 (Thr68), LZAP potentiated Wip1 activity toward p38 (Thr180) (Fig. 2C, lanes 2 and 3 vs. lane 1, and lane 5 vs. lane 4).
Similar results were obtained using p-p53 (Ser15) as a substrate. As expected, Wip1 potentiated dephosphorylation of Ser15 in a dose-dependent manner (Fig. 2D, lanes 3 and 5 vs. lane 2). Addition of LZAP enhanced Wip1-mediated dephosphorylation for 2 different doses of Wip1 tested (lane 4 vs. lane 3 and lanes 6 and 7 vs. lane 5). Moreover, incubation with LZAP alone had no effect on p-Ser15 levels (lane 1 vs. 2). Together, these data suggest that LZAP enhances Wip1 dephosphorylation of several of its substrates in the absence of other factors or proteins.
LZAP enhances the intrinsic catalytic activity of Wip1
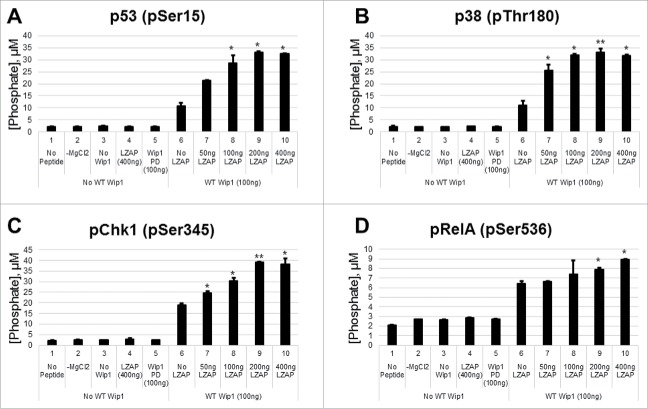
Our data and that of others reveal that LZAP binds and alters phosphorylation of a wide spectrum of Wip1 full-length substrates, including Chk2 (Fig. 2B),37 p38 (Fig. 2C),38 p53 (Fig. 2D), and RelA.33 Because each of these Wip1 substrates has increased dephosphorylation with expression of LZAP, 2 non-exclusive mechanisms of LZAP activity are possible. LZAP may potentiate Wip1-catalyzed dephosphorylation through binding both substrate and phosphatase to facilitate their association, or alternatively, LZAP association with Wip1 could enhance its intrinsic catalytic activity. The phosphatase assays shown in Fig. 2 using full length proteins as substrate do not distinguish these possibilities. Hence, we performed phosphatase assays using short phospho-peptides that are unlikely to bind to LZAP. In addition, we tested phospho-peptides containing consensus Wip1 dephosphorylation sites derived from proteins whose association with Wip1 and LZAP were unknown.
As expected and previously described, Wip1 dephosphorylated peptides corresponding to p53 (pSer15),5 p38 (pThr180),4 and Chk1 (pSer345)5 (Figs. 3A-3C, lanes 6 vs. lanes 1–5). Importantly, pre-incubation of His-Wip1 with His-LZAP increased Wip1 phosphatase activity toward each of these peptides in a dose-dependent manner (Figs. 3A-3C, lanes 7–9 vs. lane 6). As expected, no phosphatase activity was detected in the absence of peptide (lanes 1), Mg2+ (lanes 2), or Wip1 (lanes 3), or in the presence of LZAP alone (lanes 4). Phosphatase-dead His-Wip1 (lanes 5) also lacked detectable phosphatase activity.
Figure 3.
LZAP enhances the intrinsic catalytic activity of Wip1 toward phosphopeptides of LZAP target proteins. Phosphorylated peptides derived from p53 (A), p38 (B), Chk1 (C), and RelA (D) were used as substrates for in vitro Wip1 phosphatase assays. Substrates were incubated with the indicated amounts of bacterially-produced His-Wip1 and His-LZAP in phosphatase buffer with or without magnesium for 30 minutes at 30 degrees. Phosphatase reactions were mixed with the malachite green substrate, incubated for 30 minutes at room temperature, and OD620 was measured. Absorbance values were converted to levels of released phosphate using a standard curve. Data presented are the mean of at least 2 independent experiments, and error bars represent standard deviation. * and ** indicate p < 0.05 and p < 0.01, respectively, as calculated by 2-tailed student t-test.
Phosphorylation of RelA at serine 536 is required for full transcriptional activity of the p50/RelA NF-κB heterodimer. Since improper regulation of NF-κB activity is linked to many human malignancies, addition and removal of this modification is tightly regulated by a number of proteins. In accordance with previous reports identifying Wip1 as a phosphatase for RelA (pSer536),28 Wip1 dephosphorylated a short phospho-peptide corresponding to this mark (Fig. 3D, lane 6 vs. lanes 1–5). Importantly, LZAP potentiated Wip1 activity toward RelA (lanes 7–10 vs. lane 4), providing a potential mechanism for the previously reported association between LZAP and RelA dephosphorylation.33
Although many kinases have been identified that can conjugate this moiety (casein kinase II, NF-κB-inducing kinase (NIK), TRAF family member-associated NF-κB activator (TANK)-binding kinase 1 (TBK1), and Rho-associated protein kinase 1 (ROCK1)),40 the canonical NF-κB signaling pathway is activated by IκB kinase complex (IKK).41 Positive regulators of IKK activity toward Ser536 include upstream kinases Akt and ERK1/242 and the adaptor protein Rap1.43 In contrast, LZAP antagonizes phosphorylation of pSer536.33 Therefore, we found it plausible that LZAP and Rap1 interact and/or compete to govern the phosphorylation status of Ser536.
To examine a potential interaction between LZAP and Rap1, in vitro studies were performed using recombinant GST, GST-Rap1, and His-LZAP (Supplementary Fig. 1A). First, recombinant His-LZAP was immunoprecipitated using LZAP antiserum in the presence of GST or GST-Rap1, and complexes were resolved via SDS-PAGE (Supplementary Fig. 1B, left). LZAP was efficiently immunoprecipitated (middle panel), and neither GST (bottom panel) nor GST-Rap1 (top panel) was detected. Next, GST or GST-Rap1 was incubated with a Rap1 antibody in the presence of His-LZAP. GST-Rap1, but not GST or His-LZAP, was detected in Rap1 immunoprecipitates. 10% inputs demonstrate specificity of antibodies used (right panels). Taken together, we conclude that LZAP and Rap1 do not interact in vitro.
Additionally, we tested whether Rap1 can modulate RelA dephosphorylation by Wip1 via competition with LZAP. We found that in the presence of LZAP and Wip1, addition of 2 different doses of Rap1 to the phosphatase assay had no effect on substrate dephosphorylation (Supplementary Fig. 1C, lanes 5 and 6 vs. lane 4). Therefore, it is unlikely that Rap1 modulates RelA phosphorylation through competition with LZAP, in addition to its widely accepted mechanism of increasing RelA phosphorylation through its interaction with the IKK complex.43
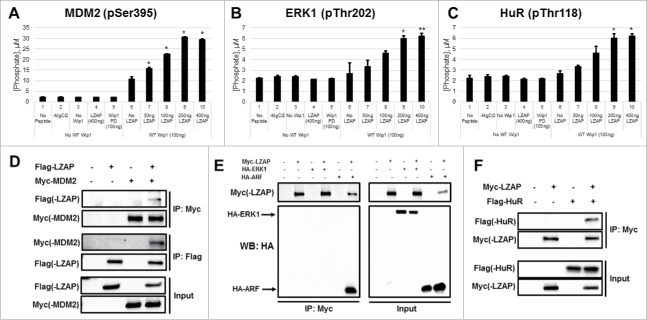
To further examine the breadth of this LZAP effect on Wip1 phosphatase activity, we tested Wip1 substrates that have not been previously associated with LZAP activity. Wip1 removes an inhibitory phosphorylation mark from MDM2 at Ser395.11 Although Wip1 activity to ERK1 and the RNA-binding protein HuR has not been previously described, both contain consensus Wip1 sites. HuR is phosphorylated at residues Ser88 (Chk2), Ser100 (Chk2), and Thr118 (Chk2 and p38),44 resulting in increased HuR stability and enhanced binding to some of its target mRNA sequences. MEK phosphorylates ERK1 at Thr202 as a prerequisite to its activation.45 Peptides were synthesized that corresponded to each of these 3 phosphorylation sites within HuR, as well as the single sites within ERK1 and MDM2. Importantly, His-Wip1 showed activity toward MDM2 (Ser395) and ERK1 (Thr202) (Figs. 4A and 4B, lane 6). Of the 3 HuR peptides tested, Wip1 only displayed robust activity toward Thr118 (Fig. 4C, lane 6, and data not shown). Pleasingly, a similar pattern of enhanced phosphatase activity in the presence of LZAP was observed when compare with Wip1 alone (compare lanes 7–10 to lane 6).
Figure 4.
LZAP enhances the intrinsic catalytic activity of Wip1 toward phosphopeptides and binds MDM2 and HuR. (A-C) Phosphorylated peptides derived from MDM2 (A), ERK1 (B), and HuR (C) were used as substrates for in vitro Wip1 phosphatase assays. Substrates were incubated with the indicated amounts of bacterially-produced His-Wip1 and His-LZAP in phosphatase buffer with or without magnesium for 30 minutes at 30 degrees. Phosphatase reactions were mixed with the malachite green substrate, incubated for 30 minutes at room temperature, and OD620 was measured. Absorbance values were converted to levels of released phosphate using a standard curve. Data presented are the mean of at least 2 independent experiments, and error bars represent standard deviation. * and ** indicate p<0.05 and p<0.01, respectively, as calculated by 2-tailed Student t-test. (D) LZAP and MDM2 interact in mammalian cells. U2OS cells were transfected with indicated plasmids encoding tagged Flag-LZAP or Myc-MDM2, and ectopic expression of these proteins was confirmed by immunoblot of whole cell lysate (bottom panels). Immunoprecipitates were prepared using Myc affinity matrix (top panels) to pull down MDM2 or Flag affinity matrix to pull down LZAP (middle panels), resolved on SDS-PAGE, and immunoblotted by antibodies recognizing Myc(-MDM2) or Flag(-LZAP). (E) LZAP and ERK1 do not interact in mammalian cells. U2OS cells were transfected with indicated plasmids encoding tagged Myc-LZAP, HA-ERK1, or HA-ARF, and ectopic expression of these proteins was confirmed (right panels). Immunoprecipitates were prepared using Myc affinity matrix to pull down LZAP (left panels), resolved on SDS-PAGE, and immunoblotted by antibodies recognizing HA(-ERK1 and ARF) and Myc(-LZAP). (F) LZAP and HuR interact in mammalian cells. U2OS cells were transfected with indicated plasmids encoding tagged Myc-LZAP or Flag-HuR, and ectopic expression of these proteins was confirmed (bottom panels). Immunoprecipitates were prepared using Myc affinity matrix to pull down LZAP (top panels), resolved on SDS-PAGE, and immunoblotted by antibodies recognizing Flag(-HuR) or Myc(-LZAP).
As LZAP binds to many Wip1 substrates, we explored possible LZAP interactions with MDM2, HuR, and ERK1. Unexpectedly, a novel, specific interaction between LZAP and MDM2 was identified by co-immunoprecipitation of expressed Myc-MDM2 and Flag-LZAP. Myc-MDM2 was detected in Flag-LZAP immunoprecipitates and vice versa (Fig. 4D, top and middle panels). Protein expression was confirmed by immunoblot of whole cell lysate (Fig. 4D, bottom panels). A similar approach demonstrated specific binding between Myc-LZAP and Flag-HuR (Fig. 4F). Interestingly, however, no association was found between LZAP and ERK1 (Fig. 4E). An HA-tagged construct of ARF, a known LZAP binding protein,32 was included as a positive control. Given that ERK1 is not a binding partner of LZAP, yet LZAP still augments Wip1 phosphatase activity toward its phospho-peptide, this suggests that LZAP binding to the target is not required for this effect. Positive results in each of 7 short peptides tested also support this hypothesis, as it is highly unlikely that LZAP specifically interacts with each of these. Although we cannot rule out contributing effects of LZAP's association with the substrates toward potentiating their dephosphorylation, these data suggest that these interactions are not required, and that LZAP binds Wip1 to enhance its intrinsic catalytic activity.
Discussion
Here, we show that LZAP bound directly to Wip1 and activated Wip1 phosphatase activity to all targets tested. Although many cellular targets for Wip1 have been identified, an abundance of proteins dephosphorylated by Wip1 have activity in the DNA damage response (e.g. Chk1, Chk2, ATM, p53, H2AX), and it has been proposed that the major cellular role of Wip1 is to restore homeostasis after DNA damage is repaired.30 ERK1 and HuR, 2 novel Wip1 targets identified in this study (Fig. 4), add another level of complexity into cellular DNA damage response network. The biological outcome of Wip1-dependent de-phosphorylation of these proteins deserves further investigation.
Premature termination of the DDR or ablation of the cell's tumor surveillance networks due to spurious activation of Wip1 can have negative consequences for the cell and organism, requiring tight regulation of Wip1 activity. Previous research has described regulation of Wip1 activity almost exclusively through modulation of Wip1 expression or degradation. Transcription of PPM1D is tightly controlled by a number of transcription factors, including p53, and PPM1D mRNA stability is governed by both microRNAs and RNA binding proteins.46 In unstressed cells, Wip1 protein levels are constitutively maintained at a low level by HIPK2-mediated phosphorylation and subsequent proteasomal degradation.47 However, mechanisms governing the catalytic activity of Wip1 independently of alterations in its protein levels remained unknown until recently.
Phosphorylation can mark proteins for ubiquitination and proteasomal destruction, as described above for Wip1, but also can enhance or inhibit protein activities. During mitosis, Wip1 activity is downregulated not only by its proteasomal degradation, but also by phosphorylation of 7 sites within its phosphatase domain. Although kinase(s) responsible for all phosphorylation events are not currently known, phosphomimetic mutations of 7 phospho-acceptor sites within Wip1 completely abolished its phosphatase activity.48 This work increased understanding of Wip1 regulation and provided insight that regulation of Wip1 phosphatase activity utilizes mechanisms independent of changes in its protein levels.
A recent study identified Tax, the transforming protein encoded by human T cell leukemia virus type 1 (HTLV-1), as a novel regulator of Wip1 phosphatase activity. The oncogenic Tax protein is critical for transformation and has many activities including activation of NF-κB with resultant cytokine production and signaling. Tax expression also results in mitogenic stress at least partially through activation of G1 cyclin-dependent kinases that ultimately leads to DNA damage.49 In response to DNA lesions, cells expressing Tax initiate DDR; however, the DNA damage-induced G1/S cell cycle arrest is compromised allowing cells containing unrepaired DNA lesions to enter S phase.50 Cell cycle progression is enabled by muted recognition of DNA lesions as marked by decreased γ-H2AX foci in Tax expressing cells. Mechanistically, Tax decreases γ-H2AX foci through binding and stimulating activity of the γ-H2AX phosphatase, Wip1. Stimulation of Wip1 phosphatase activity by Tax is not limited to γ-H2AX but applies to all Wip1 targets tested including p38 and UNG. The Tax viral protein is the first identified protein that binds and enhances Wip1 activity. In the context of HTLV-1 infection, activation of Wip1 by Tax may contribute to incomplete DNA damage repair and cell cycle progression with unrepaired DNA therefore contributing to carcinogenesis.
P53 is the most commonly altered tumor suppressor gene in human cancer, and NF-κB is an oncogene that is activated in many tumor types.51 Wip1 dephosphorylates and inhibits both p53 and RelA to dampen the DNA damage response as well as canonical NF-κB signaling, suggesting that Wip1 has both oncogenic and tumor suppressor-like activities. The balance between NF-κB and p53 inhibition by the phosphatase activity of Wip1 could tip cellular outcomes either promoting or protecting from cancer.
However, extensive cross-talk between p53 and NF-κB, both antagonistic and cooperative, has been well-described and adds another potential level of complexity to the role of Wip1 in tumorigenesis.52,53 For example, in response to moderate levels of DNA damage, p53 promotes ribosomal S6 kinase 1 (RSK1) activity toward RelA (Ser536), activating pro-survival pathways that promote re-entry into the cell cycle following DNA repair.54,55 In this context, RelA inactivation by Wip1 may be required for restoring homeostasis post-DDR. Moreover, in cooperation with wild-type p53, NF-κB has been reported to induce expression of the TP53 gene in colon cancer cells.56 In contrast to these examples of positive cross-talk, NF-κB has been shown to decrease p53 levels in response to chemotherapy via upregulation of MDM2, providing a potential mechanism for drug resistance.57 Moreover, mutual antagonization between p53 and NF-κB has been reported through sequestration of transcriptional coactivators CBP and p300.53 In lung cancer cells, Nutlin, an MDM2 antagonist that blocks interaction between p53 and MDM2, inhibits NF-κB signaling in a p53-dependent manner.58 Given the complexity of p53 and NF-κB interactions, Wip1 activity toward RelA may indirectly affect levels and activity of p53, and vice versa, potentially further promoting or inhibiting tumor initiation and progression.
It is interesting that both Tax and LZAP are associated with increased Wip1 activity, yet LZAP and Tax have opposite effects on NF-κB and p53. Whereas Tax expressing cells display decreased p53 activity and strong activation of NF-κB,49 the opposite is true for LZAP, where LZAP expression is associated with activation of p53 and inhibition of NF-κB. By attenuating NF-κB activation, LZAP hinders in vitro invasion and transformation, as well as in vivo xenograft growth and tumor vascularity.33 Moreover, we have shown that LZAP has both ARF-dependent and ARF-independent activities that contribute to p53-dependent cell cycle arrest and inhibition of clonogenic growth.32 The fact that LZAP helps to remove inhibitory phosphorylation at Ser395 on the p53 destructor MDM2 (Fig. 4A) and stimulates Wip1 phosphatase activity toward p53 Ser15 (Figs. 2D and 3A), a phospho-site that is involved in p53 stabilization and activation, suggest that LZAP also affects p53 through Wip1-independent pathway(s). Differential effects of LZAP and Tax on the critical tumor suppressor, p53, and the oncogene, NF-κB, may at least partially explain different roles of Tax and LZAP in tumorigenesis.
Although we and others have ascribed many tumor suppressor activities to LZAP, its contributions toward tumorigenesis may be complicated and depend on cellular context. Its role in hepatocellular carcinoma remains controversial, as one study demonstrated that LZAP overexpression promotes hepatocellular carcinoma metastasis,59 whereas others have demonstrated a tumor suppressor role in these cancers.34,60 Additionally, LZAP has been shown to promote colorectal xenograft growth and has been proposed as a marker for colon cancer progression.61 LZAP is also overexpressed in a portion of lung adenocarcinoma, where its levels correlate with poor prognosis.62 The role of LZAP in tumorigenesis or maintenance likely depends on cellular context.
Mass spectrometry performed in 293T cells following overexpression of Flag-LZAP identified 2 Wip1 family members, PPM1A and PPM1B, as LZAP-interacting proteins (data not shown). Previously published results from our laboratory identified PPM1A as a novel RelA phosphatase with tumor-suppressor activity.63 Importantly, we showed that LZAP cooperates with PPM1A to dephosphorylate RelA at serine 536 (data not shown), suggesting that the ability of LZAP to modulate phosphatase activity may extend to other members of the PP2C family of phosphatases. Notably, while Wip1/PPM1D is a widely accepted oncogene, PPM1A acts as an important tumor suppressor. Therefore, differential activation of these 2 phosphatases by LZAP may also contribute to its seemingly conflicting roles in cancer initiation and progression.
LZAP has no known catalytic activity, suggesting that its activity likely depends on interaction with other proteins. Here we show that LZAP directly interacts with Wip1 and enhances its catalytic activity toward a number of its substrates. We speculate that LZAP acts similarly to Tax, by binding Wip1 and inducing a conformational change to bolster its phosphatase activity. Data presented here highlight a novel cellular mechanism for moderating Wip1 activity through identification of LZAP as a positive regulator of Wip1 phosphatase activity.
Materials and methods
Cell lines and transfection
Human cell lines U2OS (osteosarcoma) and HEK 293T (transformed embryonic kidney epithelia) were cultured in complete growth media recommended by the American Type Culture Collection (ATCC) at 37°C in 5% CO2. Transfections of plasmid DNA were performed using Lipofectamine 2000 (Thermo Fisher, https://www.thermofisher.com/us/en/home/brands/product-brand/lipofectamine/lipofectamine-2000.html) (293T) or FuGENE 6 (Promega, https://www.promega.com/products/reporter-assays-and-transfection/transfection-reagents/fugene-6-low-toxicity-simple-transfection/fugene-6-transfection-reagent/) (U2OS) as per manufacturer's protocol. The total amount of DNA transfected was kept equal by adding appropriate amounts of empty vector (pcDNA3.1).
Expression constructs
pCMV-Neo-Bam-Flag-Wip1 and pET-23a-His-Wip1-ΔExon6, both wild-type (WT) and phosphatase-dead (PD, D314A), were a gift of Dr. Lawrence Donehower (Baylor University). The sequence encoding full-length Wip1 (1–605) was amplified from pCMV-Neo-Bam-Flag-Wip1 by PCR and sub-cloned into pcDNA3-Myc3. pcDNA3-Flag-LZAP,32 pET-3E-His-LZAP,33 pcDNA3-Myc3-LZAP, and all Myc-LZAP truncations38 were cloned as previously described. pcDNA3-Flag-p38α was provided by Dr. Jiahuai Han.64 All constructs were verified by DNA sequencing.
Antibodies and reagents
Primary antibodies for immunoblotting include Flag (Sigma, http://www.sigmaaldrich.com/catalog/product/sigma/f3165?lang = enandregion = US); phospho-p38 (Thr180/Tyr182) (Cell Signaling, https://www.cellsignal.com/products/primary-antibodies/phospho-p38-mapk-thr180-tyr182–28b10-mouse-mab/9216), phospho-p53 (Ser15) (https://www.cellsignal.com/products/primary-antibodies/phospho-p53-ser15-antibody/9284?N = 4294956287&Ntt = p53+ser+15andfromPage = plp), phospho-Chk2 (Thr68) (https://www.cellsignal.com/products/primary-antibodies/phospho- chk2-thr68-antibody/2661), p38 (https://www.cellsignal.com/products/primary-antibodies/p38a-mapk-antibody/9218), and Chk2 (https://www.cellsignal.com/products/primary-antibodies/chk2–1c12-mouse-mab/3440); and HA (Santa Cruz, https://www3.scbt.com/scbt/ product/ha-probe-antibody-f-7), Myc (https://www3.scbt.com/scbt/product/c-myc-antibody-9e10), p53 (https://www3.scbt.com/scbt/product/p53-antibody-do-1), and Rap1 (. Other reagents include HRP-conjugated secondary antibodies (Promega, https://www.promega.com/products/imaging-and-immunological-detection/elisas-and-antibodies/horseradish-peroxidase_conjugated-antibodies/), goat anti-mouse IgG (H+L) secondary antibody (Dylight 550 conjugate) (Thermo Fisher, https://www.thermofisher.com/antibody/product/Goat-anti-Mouse-IgG-H-L-Secondary-Antibody-Polyclonal/84540), goat anti-rabbit IgG (H+L) secondary antibody (Dylight 650 conjugate) (https://www.thermofisher.com/antibody/product/Goat-anti-Rabbit-IgG-H-L-Secondary-Antibody-Polyclonal/84546), mitomycin C (Sigma, http://www.sigmaaldrich.com/catalog/product/sigma/m4287?lang = enandregion = US), 3x Flag peptide (http://www.sigmaaldrich.com/catalog/product/sigma/f4799?lang = enandregion = US), and recombinant GST (http://www.sigmaaldrich.com/catalog/product/sigma/srp5348?lang = enandregion = US) and GST-Rap1 (https://www.novusbio.com/products/terf2ip-recombinant-protein_h00054386-p01).
Immunoprecipitation and immunoblotting
For immunoprecipitation of ectopically expressed proteins, cells were lysed in RIPA buffer (Sigma, http://www.sigmaaldrich.com/catalog/product/sigma/r0278?lang = enandregion = US) supplemented with Complete Mini EDTA-free Protease Inhibitor cocktail (Roche, http://www.sigmaaldrich.com/catalog/product/roche/11836170001?lang = enandregion = US) and PhosStop (Roche, https://www.lifescience.roche.com/webapp/wcs/stores/servlet/ProductDisplay?catalogId = 10001andpartNumber = 3.2.7.8.1.1). 200 µg of lysates were pre-cleared for 30 minutes using 8 µg normal rabbit IgG (Santa Cruz, https://www3.scbt.com/scbt/product/normal-rabbit-igg) or normal mouse IgG (https://www3.scbt.com/scbt/product/normal-mouse-igg) and 20µL protein A/G beads (https://www3.scbt.com/scbt/product/protein-a-g-plus-agarose) prior to incubation with 15 µL agarose beads conjugated to antibodies recognizing Flag (Sigma, http://www.sigmaaldrich.com/catalog/product/sigma/a2220?lang = enandregion = US) or Myc (http://www.sigmaaldrich.com/catalog/product/sigma/a7470?lang = enandregion = US). Immunoprecipitation of 100 ng His-LZAP, GST-Rap1, or GST was performed in the presence of 2% BSA in TBST, using LZAP antiserum or an antibody recognizing Rap1 (Santa Cruz, https://www.scbt.com/scbt/product/rap1-antibody-4c8–1) and protein A/G beads. Immunoblotting was performed as described previously.33
Expression and purification of recombinant proteins
E. coli BL21(DE3) cells (Promega, https://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/bl21_de3_plyss-competent-cells/) were transformed with pET-3E-His-LZAP or pET-23-His-Wip1-ΔExon6 (wild-type or D314A), grown to an OD600 of 0.6, induced with 1mM IPTG for 4 hours, and pelleted. Pellets were lysed in lysis buffer (50 mM Tris, pH 8.0, 300 mM NaCl, 10 mM imidazole, 1% NP-40, 300mM NaCl, and 1 mg/mL lysozyme). Lysates were sonicated 10 times with 10 second pulses, clarified by centrifugation, and incubated with Ni2+-NTA agarose (Qiagen, https://www.qiagen.com/us/shop/sample-technologies/protein/ni-nta-agarose/#orderinginformation) for one hour. Unbound lysate was allowed to flow through, and beads were washed 3 times (50 mM Tris, pH 8.0, 300 mM NaCl, 20 mM imidazole, 1% NP-40, and 300 mM NaCl). Proteins were eluted into 50 mM Tris, pH 8.0, 300 mM NaCl, 250 mM imidazole, 1% NP-40, and 300 mM NaCl. Eluates were concentrated and desalted using Amicon Ultra-0.5 Centrifugal Filter Devices (Millipore, http://www.emdmillipore.com/US/en/product/Amicon-Ultra-0.5%C2%A0mL-Centrifugal-Filters-for-DNA-and-Protein-Purification-and-Concentration,MM_NF-C82301) prior to snap-freeze in single-use aliquots. All buffers were supplemented with EDTA-Free Protease Inhibitors (Roche).
In vitro malachite green phosphatase assay
Prior to addition of substrate, His-Wip1 (WT or PD) was incubated with varying amounts of His-LZAP, or Rap1, in phosphatase assay buffer (125 mM Tris, pH 7.5, 0.25 mM EDTA, 0.05% BME, and 62.5 mM MgCl2) on ice for 20 minutes. These complexes were then incubated with 750µg of each phospho-peptide (LifeTein, http://www.lifetein.com/peptide_synthesis_services.html) for 30 minutes at 30°C in a total volume of 50 µL. This reaction was then incubated with the Malachite Green substrate (BioAssay Systems, https://www.bioassaysys.com/Phosphate-Assay-Kit-(POMG-25H).html) for 30 minutes at room temperature prior to measuring OD620. A standard curve was prepared using standards provided with the kit, and absorbance values were converted to liberated phosphate (µg). See Table 1 for peptide sequences used.
Table 1.
Phosphorylated peptide sequences.
| Protein | Residue | Sequence |
|---|---|---|
| p53 | Ser15 | VEPPLpSQETFS |
| Chk1 | Ser345 | QGISFpSQPTCPD |
| p38 | Thr180 | TDDEMpTGYVAT |
| MDM2 | Ser395 | ESEDYpSQPSTS |
| RelA | Ser536 | GDEDFSpSIADMD |
| ERK1 | Thr202 | DHTGFLpTEYVATR |
| HuR | Thr118 | GLPRTMpTQKDVED |
Amino acid sequences of phosphorylated peptides utilized in the malachite green phosphatase assays outlined in Figs. 3 and 4.
In vitro full-length phosphatase assays
293T cells were transfected with constructs encoding Flag-tagged p53, p38, and Chk2. Twenty hours prior to harvesting, cells were stimulated with mitomycin C (2.5 µg/mL) to enrich for the phosphorylated species of these proteins. Flag-tagged phosphorylated proteins were immunoprecipitated as described above and eluted using 3x Flag peptide (100 µg/mL). Phosphatase assays were performed exactly as described above, using Flag-phospho-p53, p38, and Chk2 as substrates. Reactions were resolved using SDS-PAGE, transferred, and probed using phospho-specific or pan antibodies.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Institute of Dental and Craniofacial Research under R01 DE013173.
Author contributions
Conceived the experiments: JJW NI HA XL LD WGY. Provided reagents: LD. Conducted the experiments: JJW NI. Analyzed the data: JJW NI WGY. Prepared the figures: JJW. Wrote the manuscript: JJW NI WGY. All authors reviewed the manuscript.
References
- [1].Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Vande Woude GF, O'Connor PM, Appella E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci U S A 1997; 94:6048-53; PMID:9177166; http://dx.doi.org/ 10.1073/pnas.94.12.6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yamaguchi H, Durell SR, Chatterjee DK, Anderson CW, Appella E. The Wip1 phosphatase PPM1D dephosphorylates SQ/TQ motifs in checkpoint substrates phosphorylated by PI3K-like kinases. Biochemistry 2007; 46:12594-603; PMID:17939684; http://dx.doi.org/ 10.1021/bi701096s [DOI] [PubMed] [Google Scholar]
- [3].Yamaguchi H, Minopoli G, Demidov ON, Chatterjee DK, Anderson CW, Durell SR, Appella E. Substrate specificity of the human protein phosphatase 2Cdelta, Wip1. Biochemistry 2005; 44:5285-94; PMID:15807522; http://dx.doi.org/ 10.1021/bi0476634 [DOI] [PubMed] [Google Scholar]
- [4].Takekawa M, Adachi M, Nakahata A, Nakayama I, Itoh F, Tsukuda H, Taya Y, Imai K. p53-inducible Wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. The EMBO journal 2000; 19:6517-26; PMID:11101524; http://dx.doi.org/ 10.1093/emboj/19.23.6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev 2005; 19:1162-74; PMID:15870257; http://dx.doi.org/ 10.1101/gad.1291305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Oliva-Trastoy M, Berthonaud V, Chevalier A, Ducrot C, Marsolier-Kergoat MC, Mann C, Leteurtre F. The Wip1 phosphatase (PPM1D) antagonizes activation of the Chk2 tumour suppressor kinase. Oncogene 2007; 26:1449-58; PMID:16936775; http://dx.doi.org/ 10.1038/sj.onc.1209927 [DOI] [PubMed] [Google Scholar]
- [7].Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, et al.. Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol Cell 2006; 23:757-64; PMID:16949371; http://dx.doi.org/ 10.1016/j.molcel.2006.07.010 [DOI] [PubMed] [Google Scholar]
- [8].Cha H, Lowe JM, Li H, Lee J-S, Belova GI, Bulavin DV, Fornace AJ. Wip1 directly dephosphorylates gamma-H2AX and attenuates the DNA damage response. Cancer Res 2010; 70:4112-22; PMID:20460517; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Moon S-H, Lin L, Zhang X, Nguyen T-A, Darlington Y, Waldman AS, Lu X, Donehower LA. Wild-type p53-induced phosphatase 1 dephosphorylates histone variant gamma-H2AX and suppresses DNA double strand break repair. J Biol Chem 2010; 285:12935-47; PMID:20118229; http://dx.doi.org/ 10.1074/jbc.M109.071696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Song JY, Ryu SH, Cho YM, Kim YS, Lee BM, Lee SW, Choi J. Wip1 suppresses apoptotic cell death through direct dephosphorylation of BAX in response to γ-radiation. Cell Death Dis 2013; 4:e744-e; PMID:23907458; http://dx.doi.org/ 10.1038/cddis.2013.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lu X, Ma O, Nguyen T-A, Jones SN, Oren M, Donehower LA. The Wip1 Phosphatase Acts as a Gatekeeper in the p53-Mdm2 Autoregulatory Loop. Cancer Cell 2007; 12:342-54; PMID:17936559; http://dx.doi.org/ 10.1016/j.ccr.2007.08.033 [DOI] [PubMed] [Google Scholar]
- [12].Zhang X, Lin L, Guo H, Yang J, Jones SN, Jochemsen A, Lu X. Phosphorylation and degradation of MdmX is inhibited by Wip1 phosphatase in the DNA damage response. Cancer Res 2009; 69:7960-8; PMID:19808970; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-0634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bulavin DV, Demidov ON, Si Saito, Kauraniemi P, Phillips C, Amundson SA, Ambrosino C, Sauter G, Nebreda AR, Anderson CW, et al.. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat Genet 2002; 31:210-5; PMID:12021785; http://dx.doi.org/ 10.1038/ng894 [DOI] [PubMed] [Google Scholar]
- [14].Belova GI, Demidov ON, Fornace AJ, Bulavin DV. Chemical inhibition of Wip1 phosphatase contributes to suppression of tumorigenesis. Cancer Biol Ther 2005; 4:1154-8; PMID:16258255; http://dx.doi.org/ 10.4161/cbt.4.10.2204 [DOI] [PubMed] [Google Scholar]
- [15].Bulavin DV, Phillips C, Nannenga B, Timofeev O, Donehower LA, Anderson CW, Appella E, Fornace AJ. Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16(Ink4a)-p19(Arf) pathway. Nat Genet 2004; 36:343-50; PMID:14991053; http://dx.doi.org/ 10.1038/ng1317 [DOI] [PubMed] [Google Scholar]
- [16].Nannenga B, Lu X, Dumble M, Van Maanen M, Nguyen T-A, Sutton R, Kumar TR, Donehower LA. Augmented cancer resistance and DNA damage response phenotypes in PPM1D null mice. Mol Carcinog 2006; 45:594-604; PMID:16652371; http://dx.doi.org/ 10.1002/mc.20195 [DOI] [PubMed] [Google Scholar]
- [17].Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven WG, Nguyen KCQ, Gabriele T, McCurrach ME, Marks JR, Hoey T, et al.. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet 2002; 31:133-4; PMID:12021784; http://dx.doi.org/ 10.1038/ng888 [DOI] [PubMed] [Google Scholar]
- [18].Rauta J, Alarmo E-L, Kauraniemi P, Karhu R, Kuukasjärvi T, Kallioniemi A. The serine-threonine protein phosphatase PPM1D is frequently activated through amplification in aggressive primary breast tumours. Breast Cancer Res Treat 2006; 95:257-63; PMID:16254685; http://dx.doi.org/ 10.1007/s10549-005-9017-7 [DOI] [PubMed] [Google Scholar]
- [19].Hirasawa A, Saito-Ohara F, Inoue J, Aoki D, Susumu N, Yokoyama T, Nozawa S, Inazawa J, Imoto I. Association of 17q21-q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin Cancer Res 2003; 9:1995-2004; PMID:12796361 [PubMed] [Google Scholar]
- [20].Loukopoulos P, Shibata T, Katoh H, Kokubu A, Sakamoto M, Yamazaki K, Kosuge T, Kanai Y, Hosoda F, Imoto I, et al.. Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: identification of genetic indicators that predict patient outcome. Cancer Sci 2007; 98:392-400; PMID:17233815; http://dx.doi.org/ 10.1111/j.1349-7006.2007.00395.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fuku T, Semba S, Yutori H, Yokozaki H. Increased wild-type p53-induced phosphatase 1 (Wip1 or PPM1D) expression correlated with downregulation of checkpoint kinase 2 in human gastric carcinoma. Pathol Int 2007; 57:566-71; PMID:17685927; http://dx.doi.org/ 10.1111/j.1440-1827.2007.02140.x [DOI] [PubMed] [Google Scholar]
- [22].Buss MC, Remke M, Lee J, Gandhi K, Schniederjan MJ, Kool M, Northcott PA, Pfister SM, Taylor MD, Castellino RC. The WIP1 oncogene promotes progression and invasion of aggressive medulloblastoma variants. Oncogene 2015; 34:1126-40; PMID:24632620; http://dx.doi.org/ 10.1038/onc.2014.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Saito-Ohara F, Imoto I, Inoue J, Hosoi H, Nakagawara A, Sugimoto T, Inazawa J. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res 2003; 63:1876-83; PMID:12702577 [PubMed] [Google Scholar]
- [24].Ruark E, Snape K, Humburg P, Loveday C, Bajrami I, Brough R, Rodrigues DN, Renwick A, Seal S, Ramsay E, et al.. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature 2013; 493:406-10; PMID:23242139; http://dx.doi.org/ 10.1038/nature11725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zajkowicz A, Butkiewicz D, Drosik A, Giglok M, Suwiński R, Rusin M. Truncating mutations of PPM1D are found in blood DNA samples of lung cancer patients. Br J Cancer 2015; 112:1114-20; PMID:25742468; http://dx.doi.org/ 10.1038/bjc.2015.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kleiblova P, Shaltiel IA, Benada J, Ševčík J, S Pecháčková, Pohlreich P, Voest EE, Dundr P, Bartek J, Kleibl Z, et al.. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. J Cell Biol 2013; 201:511-21; PMID:23649806; http://dx.doi.org/ 10.1083/jcb.201210031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang L, Chen LH, Wan H, Yang R, Wang Z, Feng J, Yang S, Jones S, Wang S, Zhou W, et al.. Exome sequencing identifies somatic gain-of-function PPM1D mutations in brainstem gliomas. Nat Genet 2014; 46:726-30; PMID:24880341; http://dx.doi.org/ 10.1038/ng.2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chew J, Biswas S, Shreeram S, Humaidi M, Wong ET, Dhillion MK, Teo H, Hazra A, Fang CC, López-Collazo E, et al.. WIP1 phosphatase is a negative regulator of NF-kappaB signalling. Nat Cell Biol 2009; 11:659-66; PMID:19377466; http://dx.doi.org/ 10.1038/ncb1873 [DOI] [PubMed] [Google Scholar]
- [29].Lu X, Yarbrough WG. Negative regulation of RelA phosphorylation: emerging players and their roles in cancer. Cytokine Growth Factor Rev 2015; 26:7-13; PMID:25438737; http://dx.doi.org/ 10.1016/j.cytogfr.2014.09.003 [DOI] [PubMed] [Google Scholar]
- [30].Lu X, Nguyen T-A, Moon S-H, Darlington Y, Sommer M, Donehower LA. The type 2C phosphatase Wip1: an oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer metastasis Rev 2008; 27:123-35; PMID:18265945; http://dx.doi.org/ 10.1007/s10555-008-9127-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ching YP, Qi Z, Wang JH. Cloning of three novel neuronal Cdk5 activator binding proteins. Gene 2000; 242:285-94; PMID:10721722; http://dx.doi.org/ 10.1016/S0378-1119(99)00499-0 [DOI] [PubMed] [Google Scholar]
- [32].Wang J, He X, Luo Y, Yarbrough WG. A novel ARF-binding protein (LZAP) alters ARF regulation of HDM2. Biochem J 2006; 393:489-501; PMID:16173922; http://dx.doi.org/ 10.1042/BJ20050960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang J, An H, Mayo MW, Baldwin AS, Yarbrough WG. LZAP, a putative tumor suppressor, selectively inhibits NF-kappaB. Cancer Cell 2007; 12:239-51; PMID:17785205; http://dx.doi.org/ 10.1016/j.ccr.2007.07.002 [DOI] [PubMed] [Google Scholar]
- [34].Kwon J, Cho HJ, Han SH, No JG, Kwon JY, Kim H. A novel LZAP-binding protein, NLBP, inhibits cell invasion. J Biol Chem 2010; 285:12232-40; PMID:20164180; http://dx.doi.org/ 10.1074/jbc.M109.065920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wu J, Lei G, Mei M, Tang Y, Li H. A novel C53/LZAP-interacting protein regulates stability of C53/LZAP and DDRGK domain-containing Protein 1 (DDRGK1) and modulates NF-kappaB signaling. J Biol Chem 2010; 285:15126-36; PMID:20228063; http://dx.doi.org/ 10.1074/jbc.M110.110619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jiang H, Luo S, Li H. Cdk5 activator-binding protein C53 regulates apoptosis induced by genotoxic stress via modulating the G2/M DNA damage checkpoint. J Biol Chem 2005; 280:20651-9; PMID:15790566; http://dx.doi.org/ 10.1074/jbc.M413431200 [DOI] [PubMed] [Google Scholar]
- [37].Jiang H, Wu J, He C, Yang W, Li H. Tumor suppressor protein C53 antagonizes checkpoint kinases to promote cyclin-dependent kinase 1 activation. Cell Res 2009; 19:458-68; PMID:19223857; http://dx.doi.org/ 10.1038/cr.2009.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].An H, Lu X, Liu D, Yarbrough WG. LZAP inhibits p38 MAPK (p38) phosphorylation and activity by facilitating p38 association with the wild-type p53 induced phosphatase 1 (WIP1). PLoS One 2011; 6:e16427-e; PMID:21283629; http://dx.doi.org/ 10.1371/journal.pone.0016427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Liu D, Wang W-D, Melville DB, Cha YI, Yin Z, Issaeva N, Knapik EW, Yarbrough WG. Tumor suppressor Lzap regulates cell cycle progression, doming, and zebrafish epiboly. Developmental dynamics: an official publication of the American Association of Anatomists 2011; 240:1613-25; PMID:21523853; http://dx.doi.org/ 10.1002/dvdy.22644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tong L, Tergaonkar V. Rho protein GTPases and their interactions with NFkappaB: crossroads of inflammation and matrix biology. Biosci Rep 2014; 34; PMID:24877606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cildir G, Low KC, Tergaonkar V. Noncanonical NF-kappaB Signaling in Health and Disease. Trends Mol Med 2016; 22:414-29; PMID:27068135; http://dx.doi.org/ 10.1016/j.molmed.2016.03.002 [DOI] [PubMed] [Google Scholar]
- [42].Dey A, Wong E, Kua N, Teo HL, Tergaonkar V, Lane D. Hexamethylene bisacetamide (HMBA) simultaneously targets AKT and MAPK pathway and represses NF kappaB activity: implications for cancer therapy. Cell Cycle 2008; 7:3759-67; PMID:19029824; http://dx.doi.org/ 10.4161/cc.7.23.7213 [DOI] [PubMed] [Google Scholar]
- [43].Teo H, Ghosh S, Luesch H, Ghosh A, Wong ET, Malik N, Orth A, de Jesus P, Perry AS, Oliver JD, et al.. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat Cell Biol 2010; 12:758-67; PMID:20622870; http://dx.doi.org/ 10.1038/ncb2080 [DOI] [PubMed] [Google Scholar]
- [44].Kim HH, Abdelmohsen K, Gorospe M. Regulation of HuR by DNA Damage Response Kinases. J Nucleic Acids 2010; 2010; PMID:20798862; http://dx.doi.org/ 10.4061/2010/981487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Butch ER, Guan K-L. Characterization of ERK1 Activation Site Mutants and the Effect on Recognition by MEK1 and MEK2. J Biol Chem 1996; 271:4230-5; PMID:8626767; http://dx.doi.org/ 10.1074/jbc.271.8.4230 [DOI] [PubMed] [Google Scholar]
- [46].Lowe J, Cha H, Lee M-O, Mazur SJ, Appella E, Fornace AJ. Regulation of the Wip1 phosphatase and its effects on the stress response. Frontiers in bioscience (Landmark edition) 2012; 17:1480-98; PMID:22201816; http://dx.doi.org/ 10.2741/3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Choi DW, Na W, Kabir MH, Yi E, Kwon S, Yeom J, Ahn J-W, Choi H-H, Lee Y, Seo KW, et al.. WIP1, a homeostatic regulator of the DNA damage response, is targeted by HIPK2 for phosphorylation and degradation. Mol Cell 2013; 51:374-85; PMID:23871434; http://dx.doi.org/ 10.1016/j.molcel.2013.06.010 [DOI] [PubMed] [Google Scholar]
- [48].Macurek L, Benada J, Müllers E, Halim VA, Krejčíková K, Burdová K, Pecháčková S, Hodný Z, Lindqvist A, Medema RH, et al.. Downregulation of Wip1 phosphatase modulates the cellular threshold of DNA damage signaling in mitosis. Cell cycle (Georgetown, Tex) 2013; 12:251-62; PMID:23255129; http://dx.doi.org/ 10.4161/cc.23057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Grassmann R, Aboud M, Jeang K-T. Molecular mechanisms of cellular transformation by HTLV-1 Tax. Oncogene 2005; 24:5976-85; PMID:16155604; http://dx.doi.org/ 10.1038/sj.onc.1208978 [DOI] [PubMed] [Google Scholar]
- [50].Dayaram T, Lemoine FJ, Donehower LA, Marriott SJ. Activation of WIP1 phosphatase by HTLV-1 Tax mitigates the cellular response to DNA damage. PLoS One 2013; 8:e55989-e; PMID:23405243; http://dx.doi.org/ 10.1371/journal.pone.0055989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell 2008; 132:344-62; PMID:18267068; http://dx.doi.org/ 10.1016/j.cell.2008.01.020 [DOI] [PubMed] [Google Scholar]
- [52].Tergaonkar V. p53 and NFkappaB: fresh breath in the cross talk. Cell Res 2009; 19:1313-5; PMID:19956183; http://dx.doi.org/ 10.1038/cr.2009.132 [DOI] [PubMed] [Google Scholar]
- [53].Tergaonkar V, Perkins ND. p53 and NF-kappaB crosstalk: IKKalpha tips the balance. Mol Cell 2007; 26:158-9; PMID:17466617; http://dx.doi.org/ 10.1016/j.molcel.2007.04.006 [DOI] [PubMed] [Google Scholar]
- [54].Janssens S, Tschopp J. Signals from within: the DNA-damage-induced NF-kappaB response. Cell Death Differ 2006; 13:773-84; PMID:16410802; http://dx.doi.org/ 10.1038/sj.cdd.4401843 [DOI] [PubMed] [Google Scholar]
- [55].Bohuslav J, Chen LF, Kwon H, Mu Y, Greene WC. p53 induces NF-kappaB activation by an IkappaB kinase-independent mechanism involving phosphorylation of p65 by ribosomal S6 kinase 1. J Biol Chem 2004; 279:26115-25; PMID:15073170; http://dx.doi.org/ 10.1074/jbc.M313509200 [DOI] [PubMed] [Google Scholar]
- [56].Benoit V, Hellin AC, Huygen S, Gielen J, Bours V, Merville MP. Additive effect between NF-kappaB subunits and p53 protein for transcriptional activation of human p53 promoter. Oncogene 2000; 19:4787-94; PMID:11032029; http://dx.doi.org/ 10.1038/sj.onc.1203831 [DOI] [PubMed] [Google Scholar]
- [57].Tergaonkar V, Pando M, Vafa O, Wahl G, Verma I. p53 stabilization is decreased upon NFkappaB activation: a role for NFkappaB in acquisition of resistance to chemotherapy. Cancer Cell 2002; 1:493-503; PMID:12124178; http://dx.doi.org/ 10.1016/S1535-6108(02)00068-5 [DOI] [PubMed] [Google Scholar]
- [58].Dey A, Wong ET, Bist P, Tergaonkar V, Lane DP. Nutlin-3 inhibits the NFkappaB pathway in a p53-dependent manner: implications in lung cancer therapy. Cell Cycle 2007; 6:2178-85; PMID:17786042; http://dx.doi.org/ 10.4161/cc.6.17.4643 [DOI] [PubMed] [Google Scholar]
- [59].Mak GW-Y, Chan MM-L, Leong VY-L, Lee JM-F, Yau T-O, Ng IO-L, Ching Y-P. Overexpression of a novel activator of PAK4, the CDK5 kinase-associated protein CDK5RAP3, promotes hepatocellular carcinoma metastasis. Cancer Res 2011; 71:2949-58; PMID:21385901; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-4046 [DOI] [PubMed] [Google Scholar]
- [60].J-j Zhao, Pan K, J-j Li, Y-b Chen, J-g Chen, Lv L, Wang D-d, Pan Q-z, Chen M-s, Xia J-c. Identification of LZAP as a new candidate tumor suppressor in hepatocellular carcinoma. PLoS One 2011; 6:e26608-e; PMID:22028922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chen J, Shi Y, Li Z, Yu H, Han Y, Wang X, Sun K, Yang T, Lou K, Song Y, et al.. A functional variant of IC53 correlates with the late onset of colorectal cancer. Molecular medicine (Cambridge, Mass) 2011; 17:607-18; PMID:21394385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Stav D, Bar I, Sandbank J. Usefulness of CDK5RAP3, CCNB2, and RAGE genes for the diagnosis of lung adenocarcinoma. Int J Biol Markers 2007; 22:108-13; PMID:17549666 [DOI] [PubMed] [Google Scholar]
- [63].Lu X, An H, Jin R, Zou M, Guo Y, Su PF, Liu D, Shyr Y, Yarbrough WG. PPM1A is a RelA phosphatase with tumor suppressor-like activity. Oncogene 2014; 33:2918-27; PMID:23812431; http://dx.doi.org/ 10.1038/onc.2013.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Han J, Lee JD, Jiang Y, Li Z, Feng L, Ulevitch RJ. Characterization of the Structure and Function of a Novel MAP Kinase Kinase (MKK6). J Biol Chem 1996; 271:2886-91; PMID:8621675; http://dx.doi.org/ 10.1074/jbc.271.6.2886 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.