Abstract
The HIV pandemic affects 36.7 million people worldwide, predominantly in resource-poor settings. Nucleic acid-based molecular detection of HIV plays a significant role in antiretroviral treatment monitoring for HIV patients, as well as diagnosis of HIV infection in infants. Currently available molecular diagnostic methods are complex, time-consuming and relatively expensive, thus limiting their use in resource-poor settings. Recent advances in microfluidics technology have made possible low-cost integrated miniaturized devices for molecular detection and quantification of HIV at the point of care. We review recent technical advances in molecular testing of HIV using microfluidic technology, with a focus on assays based on isothermal nucleic acid amplification. Microfluidic components for sample preparation, isothermal amplification and result detection are discussed and compared. We also discuss the challenges and future directions for developing an integrated “sample-to-result” microfluidic platform for HIV molecular detection.
Graphical Abstract
We review recent technical advances in molecular testing of HIV using microfluidic technology, including sample preparation, isothermal amplification and detection.

1. Introduction
According to the Joint United Nations Programme on HIV/AIDS (UNAIDS) [1], there are currently 36.7 million people living with HIV globally, about 70 percent of whom are in sub-Saharan Africa. Only 46% of infected adults have access to antiretroviral treatment. HIV has become the leading cause of human death from a single infectious agent, emphasizing the importance of developing convenient tests for HIV infection. Although lateral flow (LF) immunoassays for anti-HIV antibodies [2], such as OraQuick™ Advance HIV 1/2, enable rapid detection of established HIV infection, they cannot quantify HIV virus (i.e., copies of viral RNA/mL of plasma). These assays are not sufficiently sensitive to detect the disease during seroconversion. Unfortunately, when HIV-infected individuals are most contagious, they lack sufficient levels of anti-HIV antibodies for detection.
Nucleic acid-based molecular detection (i.e., HIV viral load testing) plays a critical role in monitoring antiretroviral treatment, in diagnosing HIV infection in newborns under 18 months of age, and in early detection of newly-infected individuals during the period before anti-HIV antibodies develop [3–6]. The World Health Organization (WHO) strongly recommends routine monitoring of antiretroviral therapy (ART) effectiveness using routine viral load testing at 6 months and every 12 months, to monitor treatment adherence and minimize failure [7,8]. Although viral load monitoring is the gold-standard method in resource-rich settings, its availability in resource-limited settings is hindered due to the lack of laboratory infrastructure, complex specimen collection, lack of infrastructure for preservation and transport, and high cost [9,10]. Lack of simple low-cost molecular diagnostic tools has contributed to high morbidity and mortality among HIV patients globally, especially in resource-poor settings [11]. Moreover, simpler alternatives to nucleic acid based viral load testing, such as CD4 cell counting, have limitations as surrogates for direct viral load testing [12]. CD4 cell count changes are neither sufficiently sensitive nor specific for effective patient monitoring [13]. Thus, there is an urgent need for point of care (POC) molecular diagnostic assays that allow rapid and low cost real time viral load measurements that do not rely on centralized laboratories, highly trained personnel, and refrigeration.
WHO has outlined a framework of the ASSURED (Affordable, Sensitive, Specific, User-friendly, Rapid and robust, Equipment-free and Deliverable) criteria for evaluating HIV POC devices for resource-limited settings (Table 1) [14]. WHO has surveyed partially automated portable detection platforms for measuring HIV such as GeneXpert® HIV-1 Viral Load Test (Cepheid), Alere™ q system (Alere™), cobas® Liat™ System (Roche), and EOSCAPE-HIV™ HIV Rapid RNA Assay system (Wave 80 Biosciences) [15]. Most of these systems merely replace conventional real-time PCR machines with portable thermal cyclers, and include little or no sample processing capabilities. These instruments can be better described as semi-automated benchtop instruments rather than POC diagnostic systems. Most of the above-listed diagnostic devices rely on relatively expensive processors (often in the $10,000s per unit) and a reagent cold chain. These instruments process one or a few samples at a time, which is incompatible with needs in many developing countries, where a medical team visiting a village would ideally process dozens of samples within a few hours. The systems mentioned above are most suitable for district hospitals and diagnostic centers, but not point of care settings such as home care and rural clinics with little infrastructure. According to WHO [16], the expected annual demand for HIV testing by 2018 is 15–30 million tests for viral load, 0.9–3.0 million for early infant diagnosis, and 160 million for rapid tests generally.
Table 1.
ASSURED Characteristics by WHO for HIV POC Tests [14]
| Characteristic | Target Specification |
|---|---|
| Affordable | Less than $500 per instrument, less than $10 per test |
| Sensitivity, Specificity | Lower limit of detection: 500 HIV RNA copies per ml of blood |
| User-friendly | 1–2 days training, easy to use |
| Rapid and robust | < 30 minutes for diagnosis; Shelf-life > 1 year at room temperature. |
| Equipment-free | Compact, battery-powered, on-site data analysis, easy disposal, easy sample handling, no cold chain |
| Deliverable | Portable, hand-held |
Over the past decade, microfluidic technologies--“lab on chip”--have emerged as an attractive approach for nucleic acid-based testing at the point of care. Lab on a chip strategies offer lower cost, shorter test time, smaller sample and reagent volumes, reduced contamination, and better containment of infectious materials. These strategies also allow potential automation from sample preparation to detection in inexpensive portable devices [17–21]. In recent years, progress has been made on microfluidic devices for sample preparation, nucleic acid amplification and product detection.
Recently, efforts by us and others have been directed towards developing an integrated “sample-to-result” microfluidic platform for HIV molecular detection [22,23]. Here, we review advances on microfluidic devices for inexpensive POC molecular diagnostics of HIV proviral DNA and viral RNA. We introduce various microfluidic components and discuss isothermal amplification assays useful for molecular diagnostic of HIV. We also discuss current challenges and future prospects for developing integrated “sample-to-result” microfluidic devices that can be used by minimally-trained personnel in resource-limited settings.
2. Sample Preparation
The purity, integrity and concentration of target pathogen nucleic acids strongly influences downstream amplification and detection. Sample preparation represents a significant challenge for POC molecular detection since it involves manipulations such as plasma separation and nucleic acid extraction, by either manual operations or separate dedicated instruments (i.e., centrifuges) [24]. Blood is the most widely used sample type to monitor disease progression and the response to antiretroviral therapy in HIV infection. However, blood is a complex biological analyte comprised of white blood cells, red blood cells, proteins, platelets, and plasma. There are numerous technical challenges that need to be overcome when preparing high quality nucleic acid sample from blood with chip-compatible formats. In recent years, a variety of microfluidic devices have been developed to perform rapid sample preparation from blood for medical diagnostics [25].
2.1 Plasma Separation
Although plasma separation is not required for molecular diagnostics of HIV, plasma separation is essential for HIV viral load testing used in monitoring antiretroviral therapy, since the inclusion of lymphocytes in the blood sample would overestimate viral load due to admixture of proviral DNA [26,27]. In clinical laboratories, serum and plasma are typically separated from whole blood by centrifugation. However, centrifugation is not optimal for POC diagnostic assays, and FDA guidelines for simple ‘out-of-laboratory’ tests suggest avoiding the use of centrifugation [28]. Over the past decade, various plasma separation microdevices have been designed and developed based on different separation mechanisms, such as capillary imbibition [29,30], blood cell sedimentation [31,32], cross-flow filtration [33,34], and on-chip centrifugation [35]. However, many of these methods are not suitable for HIV viral load testing because they either produce too small plasma volume (i.e., ≤1 μL) or require high blood dilution.
Wang et al. [36] developed a microfluidic-based plasma separation device with a 2-μm pore size membrane filter, which separated plasma and HIV virus from pre-diluted whole blood with virus recovery efficiencies ranging from 73.1% to 82.5%. However, the pre-dilution of blood reduces the ultimate limit of detection, which is critical in HIV viral load quantification. To meet stringent limit-of-detection specifications, Liu et al. [37] reported a membrane-based, sedimentation-assisted plasma separator (Fig. 1A) that was able to separate a 275 μL of plasma from 1.8 mL of whole blood. The separation process utilizes both size exclusion-based filtration and gravitational sedimentation of cells. The HIV-laden plasma recovered from the device was successfully subjected to isothermal amplification in a custom-made molecular diagnostic chip, demonstrating that the plasma was compatible with downstream nucleic acid amplification. However, to obtain more than a milliliter of whole blood typically requires venipuncture sampling, which is not ideal for POC diagnostic applications. Finger or heel-prick sampling is relatively simple, minimally invasive and more convenient [38,39], and has been validated against standard phlebotomy in HIV testing [40]. To this end, Liu et al. [41] developed a superhydrophobic clamshell plasma separator (Fig. 1B) incorporating superhydrophobic coatings to reduce non-specific absorption and achieve high efficiency plasma separation. The device is capable of extracting 65 μL of plasma from 200 μL of undiluted finger-prick blood (Maximum volume of finger-prick blood is 250–500 μL [42]). The plasma separated by the superhydrophobic separator was used for the detection of cell-free Schistosoma mansoni DNA [41] and HIV virus.
Fig. 1.
(A) A photograph of a membrane-based, sedimentation-assisted, plasma separator that is able to separate a relatively large volume of plasma from few milliliters of venipuncture blood. Reproduced with permission from ref. 37. Copyright 2013 American Chemical Society. (B) A photograph of a clamshell, superhydrophobic plasma separator for high-efficiency plasma separation from several hundred microliters of whole blood (finger-prick blood volume). Reproduced with permission from ref. 41. Copyright 2016 Royal Society of Chemistry. (C) Two photographs of the integrated sample preservation device that is capable of separating plasma and preserving sample. Reproduced with permission from ref. 48. Copyright 2013 Royal Society of Chemistry.
2.2 Dry Sample Preservation
Rather than carry out the test and deliver an immediate result, an alternative for resource-limited settings is to collect, preserve and ship clinical samples to a diagnostic laboratory. For example, blood samples can be preserved by blotting onto cellulose-based filter papers (i.e., dried blood spots (DBS)) for subsequent viral load testing [43], early infant diagnosis [44] and drug resistance genotyping [45]. Pathogens in dried blood spots are deactivated due to the inclusion of appropriate chemicals in the paper [46]. Such dried samples can be shipped through regular mail safely, reducing the burden and cost of sample transport to centralized laboratories. However, the use of DBS or whole blood generally has limitations, providing poor specificity at low viral loads (i.e., <5,000 copies/ml) and overestimating HIV viral load due to contamination with proviral DNA and intracellular RNA [47]. To this end, Begolo et al.[48] described a SlipChip-based microfluidic device (Fig. 1C) for on-chip plasma separation and dry plasma preservation using chemical stabilization matrices. This device is portable, compact, and self-contained and can be operated by minimally trained personnel in resource-limited settings. The on-chip dry plasma samples can be rehydrated and recovered for analysis in a clinical laboratory. The authors demonstrated that the dry preserved HIV-1 RNA in their device was stable at 50 °C for over a 5-week period. Nevertheless, the use of such sample preparation and transport approaches forgoes an important advantage of POC tests: namely, test results available within the time frame of a patient visit.
2.3 Nucleic Acid Extraction
Extraction, purification and concentration of nucleic acids from clinical samples (i.e., plasma, saliva, blood) for enzymatic amplification are critical because inhibitors in the samples can affect polymerase reaction efficiency and the accuracy of detection results. Although nucleic acid extraction-free methods have been reported for HIV detection in whole blood and plasma [49], they are limited to the very small sample volume (a small fraction of the reaction mix) and suffer from low sensitivity and poor reproducibility. Currently, on-chip nucleic acid extraction is the main bottleneck in developing an integrated microfluidic-based, molecular diagnostic platform. In general, conventional nucleic acids are extracted from complex samples, using centrifugation, ion exchange, liquid-liquid extraction (i.e., phenol chloroform), ethanol precipitation, and silica solid-phase extraction (i.e., spin-column). In recent years, several nucleic acid extraction microfluidic devices based on different mechanisms have been developed for HIV detection at the point of care.
2.3.1 Filter Membrane-based Extraction
Spin-column methods with chaotropic agents and silica-based nucleic acid binding phases have been widely used on microfluidic chips. These have used multiple nucleic acid binding matricies, such as porous isolation membranes [50,51], silica-coated microchannels [52], monolithic sol-gels [53], and chitosan-coated silica beads [54]. Liu et al. [55] reported a multifunctional amplification reactor chip (Fig. 2A) incorporating a flow-through, Flinders Technology Associates (Whatman FTA®) membrane (inset of Fig. 2A) for the isolation, purification, concentration and detection of HIV RNA in saliva. The HIV RNA captured by the isolation membrane could be used directly as template for isothermal amplification without further elution, thus simplifying flow control. In addition to nucleic acid purification, the flow-through isolation membrane served to concentrate the nucleic acids. The amount of sample lysate loaded on the membrane can be more than 100 times larger than the volume of the reaction chamber, decoupling sample volume from amplification volume. This amplification reactor chip allowed detection of less than 10 HIV particles per sample. In another approach, these authors [37] also immobilized a Qiagen® Silica membrane in a microfluidic device and achieved a sensitivity of ~300 copies/mL HIV RNA in plasma.
Fig. 2.
Microfluidic-based nucleic acid extraction of HIV. (A) A multifunctional amplification reactor chip integrated with a nucleic acid isolation membrane for HIV virus detection in saliva. Top inset is a side view of the multifunctional amplification reactor with a flow-through isolation membrane. Reproduced with permission from ref. 55. Copyright 2011 Royal Society of Chemistry. (B) Top: schematic of the immiscible phase filter for wash-free nucleic acid extraction. Bottom: Photograph of immiscible phase filter-based cartridge containing lysis/binding buffer, elution buffer, and a red colored liquid wax. Reproduced with permission from ref. 56. Copyright 2010 Elsevier. (C) Photograph of wax-based IFAST device for nucleic acid extraction. Reproduced with permission from ref. 57. Copyright 2014 Elsevier. (D) A schematic of AirJump operation for high throughput nucleic acid extraction: (1) an elution plate is placed above a sample plate loaded with PMPs. (2) Upon application of a magnet, PMPs-bound nucleic acids “jump” across the air gap and are deposited in the elution plate. Reproduced with permission from ref. 58. Copyright 2016 American Chemical Society.
2.3.2 Magnetic Beads based Extraction
Sur et al. [56] developed an immiscible phase strategy for wash-free nucleic acid extraction of HIV in a microfluidic device. The paramagnetic particles (PMPs) bind the target nucleic acids and are magnetically transported by an externally-applied magnetic field through a channel filled with hydrophobic liquid wax that connects the lysis chamber to the elution chamber (Fig. 2B). The hydrophobic liquid wax acts as an immiscible phase filter and barrier, which prevents migration of lysate from the lysis chamber to the elution chamber and blocks potential inhibitors (i.e., ethanol, chaotropic salts). Transporting PMPs through the immiscible phase yielded pure nucleic acids that were compatible with downstream nucleic acid amplification. A sensitivity of 60 copies per plasma sample was demonstrated with the extracted HIV RNA. Based on the idea of immiscible phase filter-based extraction, Berry et al. [57] reported a wax-based microfluidic device for nucleic acid extraction known as immiscible filtration-assisted by surface tension (IFAST) (Fig. 2C). They used viral-like particles (VLPs) to simulate HIV virus, and spiked VLPs into fetal bovine serum to simulate whole blood samples infected with HIV. The IFAST device was capable of detecting VLPs in mock samples with a sensitivity of 50 copies/mL of sample. The authors also validated that RNA extracted by the IFAST device could be stored at 37°C for 1 week without significant loss. In another example, they recently reported an “AirJump” device (Fig. 2D) for high-throughput nucleic acid extraction by relying on an air gap, instead of immiscible phase filters [58]. In the “AirJump” device, the PMPs-bound nucleic acids are drawn directly out of a 96-well plate and through a gap of air via a magnetic. The “AirJump” device allowed quantification of HIV virus down to a sensitivity of 10 copies per sample. The simple, rapid and high-through nucleic acid extraction ability of the “AirJump” device is potentially useful in microfluidic-based nucleic acid extraction by miniaturizing the plate.
3. Isothermal Nucleic Acids Amplification Methods
Although several amplification-free, nucleic acid-based testing methods (i.e., DNA biosensor) [59,60] have been reported for HIV detection, nucleic acid amplification testing is still the most widely used technology for HIV molecular diagnostics and viral load testing. PCR/RT-PCR is the most commonly used method. Most of commercially available portable molecular diagnostic systems, such as Cepheid GeneXpert, IQuum LIAT analyzer, utilize real-time RT-PCR assay to quantify HIV virus due to the high sensitivity, specificity and reproducibility. Early microfluidic-based molecular diagnostic devices used PCR technology to detect HIV [56,61]. However, PCR technology requires precise temperature control and rapid thermal cycling between 55 and 95°C, which is cumbersome to implement in resource-limited settings.
Nucleic acid-based molecular diagnostics have been simplified with the advent of isothermal amplification methods such as loop-mediated isothermal amplification (LAMP), reverse transcription loop-mediated isothermal amplification (RT-LAMP) [62,63], nucleic acid sequence based amplification (NASBA) [64,65], recombinase polymerase amplification (RPA) [66,67], and helicase dependent amplification (HDA) [68]. Isothermal amplification assays are very suitable for chip-based POC diagnostic applications due to their simple operation, high amplification efficiency, and easy detection. Moreover, isothermal amplification techniques are capable of combining reverse transcription, thus allowing detection of RNA targets such as HIV genomes. Additionally, the amplification ‘cocktails’ for isothermal amplification reactions can be lyophilized or preserved in dry form, eliminating the need for a cold-chain [69]. Isothermal amplification-based microfluidic technologies have recently been reviewed [70]. Here, we focus on several isothermal nucleic acid amplification assays (Table 1) that have recently been used for molecular detection of HIV, including assays of HIV proviral DNA and viral RNA.
3.1 Loop-Mediated Isothermal Amplification (LAMP)
LAMP is a rapid and sensitive sequence-specific isothermal nucleic acid amplification technique with a reaction temperature of 60–65 °C. It employs a strand-displacing Bst DNA polymerase and 4 or 6 primers. Among the isothermal amplification methods currently available, it appears that LAMP is the most widely used for nucleic acid-based detection of pathogens. Multiple LAMP primer sets have been developed to target conserved HIV-1 gene regions.
Curtis et al. [71] first reported a simple LAMP/ RT-LAMP assay for the detection of HIV-1 DNA and RNA using LAMP primers targeting conserved sequences located within the protease (PR) and capsid (p24) coding regions. Sensitivity of 10 and 100 copies per reaction was achieved for detection of HIV-1 DNA and RNA, respectively. The authors also demonstrated the feasibility of direct detection of HIV-1 nucleic acid in plasma and whole blood samples without need of prior nucleic acid extraction. Further, they designed various HIV-1 RT-LAMP primer sets targeting other conserved regions, such as the Reverse transcriptase (RT) [49] and Integrase (IN) coding regions [72]. However, due to the variation of HIV-1 genomic sequences [73,74], many of these LAMP primers showed high sensitivity and specificity only for HIV-1 subtype B, the most common HIV-1 subtype in the developed world [71,72,75], but often performed poorly with other subtypes (there are at least 9 genetically distinct subtypes recognized so far among the HIV-1 strains responsible for the AIDS epidemic, identified A–K). HIV-1 subtype C has infected the greatest number of people worldwide, particularly in Sub-Saharan Africa [76,77].
To this end, Ocwieja et al. [78] performed a bioinformatic analysis to identify regions highly conserved in all the HIV-1 subtypes that allowed development of RT-LAMP primers to detect multiple HIV-1 subtypes. The RT-LAMP primers have been demonstrated to detect HIV-1 subtypes A, B, C, D, and G with a sensitivity ranging from 500 to 5000 copies per reaction. Another study described RT-LAMP primer set optimized for the detection of HIV-1 variants (i.e., HIV-1 subtype B, C) circulating in China [79]. RT-LAMP assays have also been used for detection of the HIV-2 virus [80], which is much less prevalent and localized mainly to West Africa.
3.2 Recombinase Polymerase Amplification (RPA)
RPA is a novel isothermal nucleic acid amplification technique that uses a recombinase complex to insert primers into specific DNA sites and initiate the amplification reaction by using a strand displacing DNA polymerase. One of RPA’s major advantages is that it does not require precise temperature control and can amplify DNA to detectable levels in less than 30 min. This is advantageous for point of care diagnostic applications, where precise temperature control is not available. RPA allows for instrument-free amplification. In addition, RPA has a high tolerance to impure samples and can directly amplify DNA in clinical specimens. Unlike PCR and other isothermal amplification assays, the amplification mechanism of RPA assay is incompatible with current widely-used intercalating dyes and molecular beacons. To this end, TwistDX has developed TwistAmp® exo, real-time fluorescent detection kit that allows single tube fluorescent detection using TwistDx's proprietary fluorescent TwistAmp® exo Probe [82,83].
Boyle et al. [84] identified two sets of RPA primers that target two distinct regions of the HIV-1 genome, including the long terminal repeat (LTR) and pol gene, by scanning 63 HIV-1-specific primer and probe combinations. These pol and LTR primers have been demonstrated to amplify 98.6% and 93% of the diverse HIV-1 variants, respectively. They can detect 3 copies of proviral DNA with both fluorescence detection and lateral-flow assay. Rohrman et al. [85] used a set of RPA primers that target gag for DNA detection in a simple paper/plastic hybrid device that stores enzymes and facilitates mixing of reaction components. The device demonstrated the ability to detect 10 copies of HIV DNA in 15 min. In another effort, Crannell et al. [86] reported a real-time fluorescence quantitative RPA assay to quantify HIV-1 DNA. Lillis et al. [87] described a rapid and sensitive RT-RPA assay for the simultaneous detection of both HIV-1 RNA and DNA by adding reverse transcriptase as well. As of yet, RT-RPA has not been directly used to quantify viral RNA for HIV viral load testing.
3.3 Helicase dependent amplification (HDA)
HDA is an isothermal nucleic acid amplification method that utilizes a DNA helicase enzyme to separate double stranded DNA (dsDNA) into a single stranded DNA template. Tang et al. [88] reported an IsoAmp HIV-1 assay (BioHelix Corporation) for HIV-1 viral RNA detection using isothermal reverse-transcription helicase-dependent amplification (RT-HDA). In their assay, the samples were human plasma spiked with bacteriophage VLPs containing HIV RNA. RNA templates for RT-HDA were isolated by spin-columns on the bench. The HDA amplicons detection was carried out in a disposable cartridge (BioHelix Express Strip (BESt), BioHelix Corporation) with an embedded nucleic acid lateral flow strip. A sensitivity of ~50 copies was achieved for HIV RNA per assay. With a similar HDA protocol and the BioHelix cartridge, Jordan et al. [89] detected HIV-1 DNA that was manually extracted from DBS with a sensitivity of ~470 copies/ml of HIV-1 DNA.
4. Portable Incubators
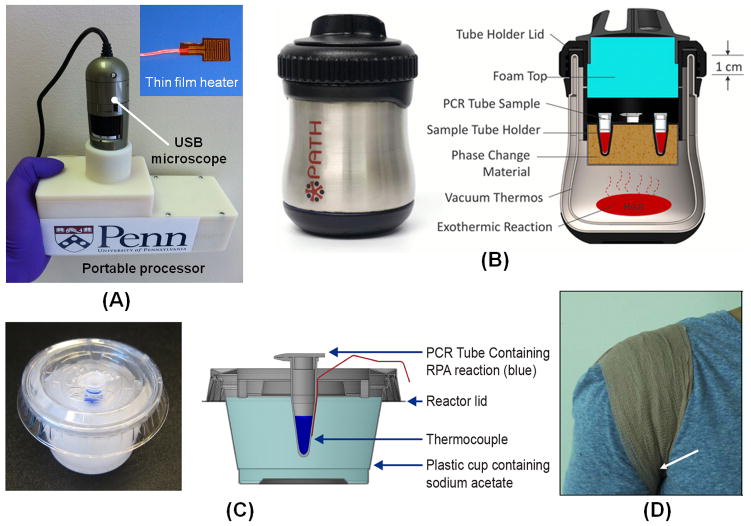
Enzymatic amplification, whether for PCR or isothermal amplification, typically requires an elevated temperature for optimal DNA synthesis. Several heating strategies have been developed for HIV detection at the point of care. Liu et al. [90] developed a portable electronic processor (Fig. 3A) for HIV viral load testing by using their nuclemeter chip platform. The processor consists of a flexible, polyimide-based, thin film heater, an electronic circuit board, and batteries. The processor is powered by four AAA batteries and is capable of maintaining the chip at a constant temperature (i.e. ~63ºC) for RT-LAMP amplification and can work continuously for more than 10 hours without replacing the batteries.
Fig. 3.
Inexpensive portable incubators for POC molecular detection of HIV. (A) A photograph of a handheld battery-powered processor for LAMP amplification and detection. Reproduced with permission from ref. 90. Copyright 2014 Nature. (B) Non-instrumented nucleic acid amplification (NINA) heater for LAMP amplification of HIV-1. Reproduced with permission from ref. 92. Copyright 2014 PloS One. (C) Sodium acetate trihydrate (SAT) heater used to incubate RPA reaction for the rapid detection of proviral HIV-1 DNA. Reproduced with permission from ref. 93. Copyright 2014 PloS One. (D) A photograph of RPA reaction setup for HIV-1 DNA detection in the axilla using a bandage. The arrow indicates the approximate position of the RPA tube. Reproduced with permission from ref. 86. Copyright 2014 PloS One.
In some developing countries, where HIV is widely prevalent, there is lack of reliable electrical power supply. A simple, electricity-free heater is thus important for low-cost nucleic acid amplification testing. Recently, various electricity-free heaters have been developed for isothermal amplification using exothermic chemical reactions, boiling water and body heat [86,91–93]. For example, Singleton et al. [92] developed a simple, affordable, and robust chemical heater for LAMP-based HIV detection by using Mg-Fe alloy as heating source. The temperature of the chemical heater was regulated by a phase change in the material. The heater can work at a wide range of ambient temperatures, from 16°C to 30°C, and the cost of reaction material is approximately US$0.06 per test. The authors, however, extracted the nucleic acids of HIV for isothermal amplification with benchtop spin-column. They combined the chemical heater with lateral flow-detection and demonstrated sensitive detection of HIV-1 within less than 80 minutes. However, the device works only at relatively temperatures (up to 30 °C) and may be a challenge to be used in Sub-Saharan Africa where ambient temperature is often higher.
Lillis et al. [93] developed a simple sodium acetate trihydrate (SAT) heater (Fig. 3C) to incubate RPA assays for infant HIV diagnostics. These SAT heaters are rechargeable by heating at 80 °C for 20 minutes. The heat for resetting the SAT heaters can be provided by exothermic chemical reaction, such as water-activated flameless ration heater (Mg-Fe alloy/water) [92]. They have also demonstrated that RPA can work over a wide temperature range (25 °C to 43 °C), and does not require a precise incubation temperature control. In another effort, Crannell et al. [86] used human body heat (Fig. 3D) to incubate RPA reaction tubes for HIV-1 DNA detection. The axilla (armpit) was found to be the ideal site for RPA incubation.
5. Detection Technologies
5.1 Real time quantitative detection
Real-time fluorescence PCR is the most widely used method for quantifying nucleic acids in biological samples and has become the “gold standard” for HIV viral load testing. Signal detection in real-time fluorescence PCR or isothermal amplification is currently based on the fluorescence emitted by intercalating dyes or fluorescently-labeled probes. Detection typically relies on relatively expensive instrumentation. Simpler and cheaper instruments are more appropriate for real-time monitoring of amplification reactions at the point of care. The use of portable fluorescence detectors and smartphones offers a simple and inexpensive method of real-time amplification detection.
For example, Liu et al. [55] used a portable fluorescent reader to monitor the fluorescence emission of their isothermal amplification reactor for HIV viral load testing in real time. The reader consists of a light-emitting diode (LED) as the excitation light source and a photodiode for fluorescence detection. A portable USB microscope (Fig. 3A) [90] was used to monitor in real time the fluorescence emission of multiple isothermal amplification reactors. Damhorst et al. [94] used high-output blue LED to excite the fluorescent dye of their microfluidic chip and a smartphone camera to record fluorescence emission of the amplification reaction in real time. A smartphone flashlight could function as the excitation source to eliminate the need for a separate light source [95]. However, real-time fluorescence detection typically requires relatively expensive optical filters to select the appropriate excitation and emission light. Alternatively, BART (Bioluminescent Assay in Real-Time) uses a new reporter for isothermal amplification assays. The luciferase enzyme produces transient bioluminescence during DNA synthesis [96, 97]. BART-based detection eliminates the need for an excitation source and optical filters, and avoids potential interference due to autofluorescence of reagents and chip materials. Other methods for real-time quantitative nucleic acid detection include pH-sensing [98], turbidity-based detection [99], and surface plasmon resonance (SPR) biosensing [100].
5.2 Nucleic Acid Lateral Flow Strip Assay
Endpoint qualitative detection of amplicons produced during enzymatic amplification reaction is often performed in the laboratory with gel electrophoresis, which is not ideal for POC diagnostics. The lateral flow-based assay is a simple and rapid alternative technology. Recently nucleic acid lateral flow strips (Fig. 4A) were coupled with enzymatic amplification assays, including PCR, LAMP, HDA and RPA for rapid detection of amplicons. Since lateral flow strips detect amplicons, their inherent low sensitivity is of no concern. Rohrman et al [85] reported a simple and inexpensive paper-based device for RPA amplification of HIV DNA in combination with the lateral flow assay. RPA amplicons are labeled with 5’ FAM antigen and 3’ biotin during RPA amplification. The strips contain nanoparticles conjugated to anti-FAM antibodies and a detection line with anti-biotin antibodies. During the lateral flow process, the RPA amplicons bind to the nanoparticle conjugates and detection line for visual detection. The strip also contains a control line with anti-rabbit antibodies, which bind to free nanoparticles conjugates to indicate whether the strip is working properly. As an alternative to gold nanoparticles, upconverting phosphor (UCP) reporters have been used as label for nucleic acid lateral flow strip detection for PCR amplicons of HIV RNA [101]. The major advantages of the UCP reporters, compared to gold nanoparticle labels, include higher sensitivity, longer shelf life, and lower background autofluorescence.
Fig. 4.
Various detection methods for HIV molecular diagnostics. (A) Cartridge integrated with instrument-free lateral flow strip for detecting HIV amplicons. Reproduced with permission from ref. 92. Copyright 2014 PloS One. (B) Photograph of a digital PCR device for HIV viral load test. Reproduced with permission from ref. 109. Copyright 2011 American Chemical Society. (C) Nuclemeter for HIV viral load testing: (i) An illustration of the nuclemeter's working principle. (ii) Photograph of a nuclemeter chip housing four independent reaction-diffusion columns. (iii) The fluorescence image of reaction-diffusion columns at 32 min after the start of RT-LAMP reaction. The sample chambers connected to columns 1, 2, 3 and 4 contained 104, 103, 102, and 0 copies of HIV RNA. (iv) A photograph of a programmed smartphone for end-point detection of HIV virus with the nuclemeter chip. Reproduced with permission from ref. 90. Copyright 2014 Nature.
Although endpoint nucleic acid lateral flow assays are simple, they cannot quantify the HIV viral genome copies. Yet, it is suitable for early infant diagnosis of HIV infection and use in monitoring treatment responses in settings where determining a simple threshold value is sufficient. Since the majority of participants on therapy in many developing world settings have undetectable viral loads [102], these tests could be used as screens for occult treatment failure. Once detected, however, follow-up tests would be needed to quantify the viral load and/or identify resistance mutations. Additionally, the lateral flow detection poses a high risk of cross contamination due to the transferring of large quantities of amplicons from reactors to lateral flow strip. To this end, Roskos et al. [103] developed a disposable cartridge that carried out isothermal DNA amplification coupled to lateral flow detection in a single sealed device. In addition to lateral flow-based endpoint qualitative detection, various on-chip detection methods, such as electrochemical [104], fluorescence [105], and colorimetric [106–108], have also been reported, avoiding the need for transferring amplicons.
5.3 Digital Nucleic Acid Quantification
Digital PCR provides absolute quantification by means of diluting the sample, dividing it into numerous aliquots, counting the number of positive and negatives partitions, and modeling the total using the Poisson distribution. However, digital PCR is rarely adopted for HIV viral load testing due to the requirement of expensive and complicated instrumentation. Recently, the Ismagilov group developed various digital nucleic acid amplification assays based on their SlipChip platforms for HIV viral load testing. For example, Shen et al. [109] reported rotational microfluidic SlipChips (Fig. 4B) to carry out multivolume digital RT-PCR (MV digital RT-PCR) and quantify HIV and HCV viral loads. The multiplexed SlipChip is capable of achieving a dynamic range of 1.7×102 to 2.0×107 molecules/mL with limit of detection of 40 molecules/mL. To eliminate the need for relatively expensive thermal cyclers, they further combined their SlipChip with the RT-LAMP method and developed a digital RT-LAMP assay for HIV viral load [110]. They found that the performance of a single-step digital RT-LAMP on their SlipChip platform was problematic for HIV viral RNA quantification due to a low efficiency of reverse transcription. To this end, they developed a two-step RT-LAMP protocol for HIV viral load testing on their SlipChip. To minimize the cost, they used a colorimetric dye, instead of an intercalating fluorescence dye, as an indicator. The use of colorimetric dye allows for easy counting of digital output with a cell phone camera [111], thus eliminating the need for a sophisticated fluorescence microscope. However, there are still challenges porting the cell phone technology to real POC molecular diagnostic applications and achieving full regulatory approval due to differences among cell phone models and software versions.
5.4 Nuclemeter Technology
Current HIV viral load testing typically requires either real-time continuous fluorescence monitoring during the polymerization reaction (i.e., qPCR) or droplet preparation and relatively complex data analysis (i.e., dPCR). Minimally-instrumented, inexpensive, endpoint quantitative approaches are desirable for HIV viral load testing. Liu et al. [90] have reported a novel reaction-diffusion-based diagnostic chip (dubbed the “nuclemeter”) for HIV viral load testing (Fig. 4C). The nuclemeter contains one sample chamber and one or more reaction-diffusion columns. Each column contains all the reagents needed for the nucleic acid amplification. When the target nucleic acids are introduced into the sample chamber (column’s inlet), they diffuse into the column and are amplified. After a relatively short time, the column comprises of two regions. In one reacted zone, that including the inlet, the amplification reaction has been carried out. In this region, it appears as a column of light due to the emission of intercalating dye (Fig. 4C(i) and (iii)). The unreacted zone appears dark since target nucleic acids have not yet reached it. The reacted and unreacted zones are separated with a reaction front. The reaction front propagates with a constant velocity and the length of the reacted region increases with time. At any given time, the length of the fluorescence-emitting region correlates with the concentration of nucleic acid targets in the sample (Fig. 4C(iii)). Thus, the number of HIV copies can be read from the position of the reaction-diffusion front, analogous to reading temperature in a “mercury in glass” thermometer. The quantitative detection of HIV viral load can be achieved by a smartphone by taking just single fluorescence image at the end of the enzymatic amplification process (Fig. 4C(iv)). The nuclemeter was able to quantify HIV RNA with a sensitivity of 50 copies per assay, comparable to that of benchtop equipment.
6. Conclusion and Future Perspectives
There is an urgent need for inexpensive, rapid, portable, easy-to-use diagnostic devices that can be used by minimally-trained personnel to quantify HIV nucleic acids at the point of care in the developing world. Remarkable progress has been made towards developing such devices, integrating sample processing, nucleic acid amplification, and result detection. Despite recent technological advances, there are just few examples of fully integrated “sample-to-results” microfluidic devices (Fig. 5) for HIV molecular detection [22,23]. The major challenge is the integration of nucleic acid preparation, amplification and detection steps in a single chip without compromising performance. The fabrication cost is another major consideration for developing POC diagnostic devices that can be used in resource poor settings. Recently emerging paper-based microfluidic devices [112,113] offer attractive alternatives to current plastic-based devices. Mobile phones are now widely available, including in resource poor settings, and POC tests can leverage the mobile phone capabilities for image capturing, data analysis, result display, GPS recording, and communication. In recent years, mobile phones have been widely used for microfluidic-based POC diagnostics [114–116]. The integration of low-cost, microfluidic diagnostic chips with ubiquitous mobile phones to perform HIV diagnostics can greatly improve health monitoring in resource poor settings.
Fig. 5.
Integrated “sample-to-result” microfluidic devices for molecular diagnosis of HIV. (A) Photograph of the dual-path microfluidic device (Rheonix CARD™) capable of simultaneously detecting anti-HIV antibody and HIV RNA. Reproduced with permission from ref. 22. Copyright 2013 hindawi. (B) Photograph of an integrated nucleic acid PCR cassette containing pre-stored reagents. For better visibility, the various food dyes in pouches represent different pre-stored liquid buffers. Reproduced with permission from ref. 23. Copyright 2010 Springer.
Co-infections associated with HIV are a new challenge in HIV/AIDS care [117]. HIV viral load testing and concurrent detection of other pathogens (i.e., HCV, HBV, TB) is critical for patient management. The multiplexing capabilities of single isothermal amplification assays are limited by their requirements for multiple primers (i.e., six for each target in LAMP assay) resulting potential risk of false positives [118,119], or the limitation of high-affinity haptens for lateral flow testing of multiple amplicons [120]. Recently developed two-stage isothermal amplification technology (RAMP) [121] provides an excellent alternative to single isothermal amplification assay for such multiplexed pathogen detection in HIV co-infection. Currently, most microfluidic molecular diagnostic systems consist of a small chip surrounded by a benchtop, electric-powered instrument, perhaps better described as a “chip in a lab” rather than a “lab on a chip”. Minimally or non-instrumented molecular diagnostic devices are of importance in developing simple low-cost diagnostic assay [122]. While HIV molecular diagnostics, especially viral load tests, are perhaps the most prominent and pressing need for microfluidics-based point-of-care tests, integrated microfluidic-based diagnostic devices are valuable for additional diagnostics applications including detecting hepatitis, malaria, and Zika virus.
Table 2.
Isothermal amplification assays for HIV molecular detection
| Amplification methods | Target gene | Limit of detection | Detection time | Reference |
|---|---|---|---|---|
| RT-LAMP | P24, protease | 10–100 copies/reaction | 60 min | 71 |
| RT-LAMP | Reverse transcriptase (RT) gene | 10 copies/ tube (DNA) and 1.7 × 103 copies/ tube (RNA) | 60 min | 49 |
| RT-LAMP | Integrase gene | 102 copies/ mL (DNA) and 104–105 copies/ mL (RNA) | 60 min | 73 |
| RT-LAMP | Integrase gene | 500-5×103 copies/ reaction (RNA) | 60 min | 78 |
| RT-LAMP | gag gene | 196 copies/reaction (RNA) | 60 min | 79 |
| RT-LAMP | Integrase gene | 103–104 copies/mL (DNA) | 60 min | 80 |
| RT-LAMP | pol-integrase gene | 120 copies/mL | 35 min | 81 |
| RPA | Long terminal repeat (LTR) and pol gene | 20 copies/ reaction (DNA) | <20 min | 84 |
| RPA | gag gene | 10 copies of HIV DNA | 15 min | 85 |
| RPA | pol gene | 102 copies /reaction (DNA) | 45 min | 86 |
| RT-HDA | gag gene | 50 copies of HIV RNA | 75 min | 88 |
| HDA | gag gene | ~470 copies/mL of HIV-1 DNA | 75 min | 89 |
Acknowledgments
Dr. Paul H. Edelstein provided his helpful comments and suggestions. The work was supported, in part, by NIH Grants K25AI099160 and Penn Center for AIDS Research (Penn CFAR) Pilot Grant AI045008.
References
- 1. [Accessed 15 September 2016];How aids changed everything. 2015 http://www.unaids.org/sites/default/files/media_asset/MDG6Report_en.pdf.
- 2.Zelin J, Garrett N, Saunders J, Warburton F, Anderson J, Moir K, Symonds M, Estcourt C. International journal of STD & AIDS 19. 2008;10:665–667. doi: 10.1258/ijsa.2008.008132. [DOI] [PubMed] [Google Scholar]
- 3.Granich RM, Gilks CF, Dye C, De Cock KM, Williams BG. Lancet. 2009;373:48–57. doi: 10.1016/S0140-6736(08)61697-9. [DOI] [PubMed] [Google Scholar]
- 4.Malnati MS, Scarlatti G, Gatto F, Salvatori F, Cassina G, Rutigliano T, Volpi R, Lusso P. Nat Protoc. 2008;3:1240–1248. doi: 10.1038/nprot.2008.108. [DOI] [PubMed] [Google Scholar]
- 5.Gibellini D, Vitone F, Gori E, Placa ML, Re MCJ. Virol Methods. 2004;115:183–189. doi: 10.1016/j.jviromet.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 6.Gross R, Bilker WB, Friedman HM, Strom BL. AIDS 532. 2001;15:2109–2117. doi: 10.1097/00002030-200111090-00006. [DOI] [PubMed] [Google Scholar]
- 7. [Accessed 15 September 2016];Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. http://www.who.int/hiv/pub/guidelines/arv2013/en/
- 8.Roberts T, Cohn J, Bonner K, Hargreaves S. Clinical Infectious Diseases. 2016;62:1043–1048. doi: 10.1093/cid/ciw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jani IV, Meggi B, Mabunda N, Vubil A, Sitoe NE, Tobaiwa O, Quevedo JI, Lehe JD, Loquiha O, Vojnov L, Peter TF. JAIDS Journal of Acquired Immune Deficiency Syndromes. 2014;67:e1–e4. doi: 10.1097/QAI.0000000000000250. [DOI] [PubMed] [Google Scholar]
- 10.Tang YW, Ou CY. Emerging microbes & infections. 2012;1:e19. doi: 10.1038/emi.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gale HB, Gitterman SR, Hoffman HJ, Gordin FM, Benator DA, Labriola AM, Kan VL. Clinical Infectious Diseases. 2013;56:1340–1343. doi: 10.1093/cid/cit004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. [Accessed 15 September 2016];Antiretroviral therapy for HIV treatment of adults and adolescents: recommendations for a public health approach 2006 revision. http://www.who.int/hiv/pub/guidelines/artadultguidelines.pdf.
- 13.Bisson GP, Gross R, Bellamy S, Chittams J, Hislop M, Regensberg L, Frank I, Maartens G, Nachega JB. PLoS Med. 2008;5:e109. doi: 10.1371/journal.pmed.0050109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu G, Zaman MH. Bulletin of the World Health Organization. 2012:914–920. doi: 10.2471/BLT.12.102780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. [Accessed 15 September 2016];HIV/AIDS Diagnostics Technology Landscape - 5th edition. http://www.unitaid.eu/images/marketdynamics/publications/UNITAID_HIV_Nov_2015_Dx_Landscape.PDF.
- 16. [Accessed 15 September 2016];HIV diagnostic tests in low- and middle-income countries: forecasts of global demand for 2014–2018. http://apps.who.int/iris/bitstream/10665/179864/1/9789241509169_eng.pdf.
- 17.Chen X, Cui D, Liu C, Li H, Chen J. Analytica chimica acta. 2007;584:237–243. doi: 10.1016/j.aca.2006.11.057. [DOI] [PubMed] [Google Scholar]
- 18.Qiu X, Thompson JA, Chen Z, Liu C, Chen D, Ramprasad S, Mauk MG, Ongagna S, Barber C, Abrams WR, Malamud D. Biomedical microdevices. 2009;11:1175–1186. doi: 10.1007/s10544-009-9334-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C, Mauk MG, Hart R, Bonizzoni M, Yan G, Bau HH. PloS one. 2012;7:e42222. doi: 10.1371/journal.pone.0042222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foudeh AM, Didar TF, Veres T, Tabrizian M. Lab on a Chip. 2012;12:3249–3266. doi: 10.1039/c2lc40630f. [DOI] [PubMed] [Google Scholar]
- 21.Song J, Liu C, Bais S, Mauk MG, Bau HH, Greenberg RM. PLoS Negl Trop Dis. 2015;9:e0004318. doi: 10.1371/journal.pntd.0004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, Abrams WR, Geva E, De Dood CJ, González JM, Tanke HJ, Niedbala RS, Zhou P, Malamud D, Corstjens PL. BioMed research international. 2013 doi: 10.1155/2013/543294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen D, Mauk M, Qiu X, Liu C, Kim J, Ramprasad S, Ongagna S, Abrams WR, Malamud D, Corstjens PL, Bau HH. Biomedical microdevices. 2010;12:705–719. doi: 10.1007/s10544-010-9423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dineva MA, Mahilum-Tapay L, Lee H. Analyst. 2007;132:1193–1199. doi: 10.1039/b705672a. [DOI] [PubMed] [Google Scholar]
- 25.Cui F, Rhee M, Singh A, Tripathi A. Annual review of biomedical engineering. 2015;17:267–286. doi: 10.1146/annurev-bioeng-071114-040538. [DOI] [PubMed] [Google Scholar]
- 26.Kran AMB, Jonassen TØ, Sannes M, Jakobsen K, Lind A, Mæland A, Holberg-Petersen M. Journal of clinical microbiology. 2009;47:2170–2174. doi: 10.1128/JCM.00519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monleau M, Montavon C, Laurent C, Segondy M, Montes B, Delaporte E, Boillot F, Peeters M. Journal of clinical microbiology. 2009;47:1107–1118. doi: 10.1128/JCM.02255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. [Accessed 15 September 2016];Recommendations: Clinical Laboratory Improvement Amendments of 1988 (CLIA) Waiver Applications for Manufacturers of In Vitro Diagnostic Devices. http://www.fda.gov/RegulatoryInformation/Guidances/ucm079632.htm.
- 29.Lee KK, Ahn CH. Lab on a Chip. 2013;13:3261–3267. doi: 10.1039/c3lc50370d. [DOI] [PubMed] [Google Scholar]
- 30.Shim JS, Ahn CH. Lab on a Chip. 2012;12:863–866. doi: 10.1039/c2lc21009f. [DOI] [PubMed] [Google Scholar]
- 31.Zhang XB, Wu ZQ, Wang K, Zhu J, Xu JJ, Xia XH, Chen HY. Analytical chemistry. 2012;84:3780–3786. doi: 10.1021/ac3003616. [DOI] [PubMed] [Google Scholar]
- 32.Dimov IK, Basabe-Desmonts L, Garcia-Cordero JL, Ross BM, Ricco AJ, Lee LP. Lab on a Chip. 2011;11:845–850. doi: 10.1039/c0lc00403k. [DOI] [PubMed] [Google Scholar]
- 33.VanDelinder V, Groisman A. Analytical chemistry. 2006;78:3765–3771. doi: 10.1021/ac060042r. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Liu CC, Li H. Sensors and Actuators B: Chemical. 2008;130:216–221. [Google Scholar]
- 35.Amasia M, Madou M. Bioanalysis. 2010;2:1701–1710. doi: 10.4155/bio.10.140. [DOI] [PubMed] [Google Scholar]
- 36.Wang S, Sarenac D, Chen MH, Huang SH, Giguel FF, Kuritzkes DR, Demirci U. Int J Nanomedicine. 2012;7:5019–5028. doi: 10.2147/IJN.S32579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C, Mauk M, Gross R, Bushman FD, Edelstein PH, Collman RG, Bau HH. Analytical chemistry. 2013;85:10463–10470. doi: 10.1021/ac402459h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, Wang J, Moore H, Rouse R, Umviligihozo G, Karita E. Nature medicine. 2011;17:1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 39.Pavie J, Rachline A, Loze B, Niedbalski L, Delaugerre C, Laforgerie E, Plantier JC, Rozenbaum W, Chevret S, Molina JM, Simon F. PloS one. 2010;5:e11581. doi: 10.1371/journal.pone.0011581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain V, Liegler T, Kabami J, Chamie G, Clark TD, Black D, Geng EH, Kwarisiima D, Wong JK, Abdel-Mohsen M, Sonawane N. Clinical infectious diseases. 2012:cis881. doi: 10.1093/cid/cis881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu C, Liao SC, Song J, Mauk MG, Li X, Wu G, Ge D, Greenberg RM, Yang S, Bau HH. Lab on a Chip. 2016;16:553–560. doi: 10.1039/c5lc01235j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kattenberg JH, Tahita CM, Versteeg IA, Tinto H, Traoré-Coulibaly M, Schallig HD, Mens PF. Trop Med Int Health. 2012;17:550–557. doi: 10.1111/j.1365-3156.2012.02975.x. [DOI] [PubMed] [Google Scholar]
- 43.Alvarez-Muñoz MT, Zaragoza-Rodríguez S, Rojas-Montes O, Palacios-Saucedo G, Vázquez-Rosales G, Gómez-Delgado A, Torres J, Muñoz O. Archives of medical research. 2005;36:382–386. doi: 10.1016/j.arcmed.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Ngo-Giang-Huong N, Khamduang W, Leurent B, Collins I, Nantasen I, Leechanachai P, Sirirungsi W, Limtrakul A, Leusaree T, Comeau AM, Lallemant M. Journal of acquired immune deficiency syndromes (1999) 2008;49:465. doi: 10.1097/QAI.0b013e31818e2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aitken SC, Wallis CL, Stevens W, de Wit TR, Schuurman R. PloS one. 2015;10:e0131541. doi: 10.1371/journal.pone.0131541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smit PW, Elliott I, Peeling RW, Mabey D, Newton PN. The American Journal of Tropical Medicine and Hygiene. 2013;90:195–210. doi: 10.4269/ajtmh.13-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miguel Arredondo, Garrido Carolina, Parkin Neil, Zahonero Natalia, Bertagnolio Silvia, Soriano Vincent, de Mendoza Carmen. Journal of clinical microbiology. 2012;3:569–572. doi: 10.1128/JCM.00418-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Begolo S, Shen F, Ismagilov RF. Lab on a Chip. 2013;13:4331–4342. doi: 10.1039/c3lc50747e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Curtis KA, Rudolph DL, Nejad I, Singleton J, Beddoe A, Weigl B, LaBarre P, Owen SM. PloS one. 2012;7:e31432. doi: 10.1371/journal.pone.0031432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J, Mauk M, Chen D, Qiu X, Kim J, Gale B, Bau HH. Analyst. 2010;135:2408–2414. doi: 10.1039/c0an00288g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Z, Mauk MG, Wang J, Abrams WR, Corstjens PL, Niedbala R, Malamud D, Bau HH. Annals of the New York Academy of Sciences. 2007;1098:429–436. doi: 10.1196/annals.1384.024. [DOI] [PubMed] [Google Scholar]
- 52.Chen X, Cui DF, Liu CC, Li H. Journal of Micromechanics and Microengineering. 2006;17:68. [Google Scholar]
- 53.Wu Q, Bienvenue JM, Hassan BJ, Kwok YC, Giordano BC, Norris PM, Landers JP, Ferrance JP. Analytical chemistry. 2006;78:5704–5710. doi: 10.1021/ac060390t. [DOI] [PubMed] [Google Scholar]
- 54.Hagan KA, Meier WL, Ferrance JP, Landers JP. Analytical chemistry. 2009;81:5249–5256. doi: 10.1021/ac900820z. [DOI] [PubMed] [Google Scholar]
- 55.Liu C, Geva E, Mauk M, Qiu X, Abrams WR, Malamud D, Curtis K, Owen SM, Bau HH. Analyst. 2011;136:2069–2076. doi: 10.1039/c1an00007a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sur K, McFall SM, Yeh ET, Jangam SR, Hayden MA, Stroupe SD, Kelso DM. The Journal of Molecular Diagnostics. 2010;12:620–628. doi: 10.2353/jmoldx.2010.090190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berry SM, LaVanway AJ, Pezzi HM, Guckenberger DJ, Anderson MA, Loeb JM, Beebe DJ. The Journal of Molecular Diagnostics. 2014;16:297–304. doi: 10.1016/j.jmoldx.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berry SM, Pezzi HM, LaVanway AJ, Guckenberger DJ, Anderson M, Beebe DJ. ACS applied materials & interfaces. 2016;8:15040–15045. doi: 10.1021/acsami.6b02555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo Y, Chen J, Chen G. Sensors and Actuators B: Chemical. 2013;184:113–117. [Google Scholar]
- 60.Zhang D, Peng Y, Qi H, Gao Q, Zhang C. Biosensors and Bioelectronics. 2010;25:1088–1094. doi: 10.1016/j.bios.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 61.Kim J, Mauk M, Chen D, Qiu X, Kim J, Gale B, Bau HH. Analyst. 2010;135:2408–2414. doi: 10.1039/c0an00288g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Nucleic acids research. 2000;28:e63–e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu C, Mauk MG, Bau HH. Microfluidics and nanofluidics. 2011;11:209–220. doi: 10.1007/s10404-011-0788-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Gemen B, van Beuningen R, Nabbe A, van Strijp D, Jurriaans S, Lens P, Kievits T. Journal of virological methods. 1994;49:157–167. doi: 10.1016/0166-0934(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 65.Dimov IK, Garcia-Cordero JL, O'Grady J, Poulsen CR, Viguier C, Kent L, Daly P, Lincoln B, Maher M, O'Kennedy R, Smith TJ. Lab on a Chip. 2008;8:2071–2078. doi: 10.1039/b812515e. [DOI] [PubMed] [Google Scholar]
- 66.Lutz S, Weber P, Focke M, Faltin B, Hoffmann J, Müller C, Mark D, Roth G, Munday P, Armes N, Piepenburg O. Lab on a Chip. 2010;10:887–893. doi: 10.1039/b921140c. [DOI] [PubMed] [Google Scholar]
- 67.Shen F, Davydova EK, Du W, Kreutz JE, Piepenburg O, Ismagilov RF. Analytical chemistry. 2011;83:3533–3540. doi: 10.1021/ac200247e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mahalanabis M, Do J, ALMuayad H, Zhang JY, Klapperich CM. Biomedical microdevices. 2010;12:353–359. doi: 10.1007/s10544-009-9391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chander Y, Koelbl J, Puckett J, Moser MJ, Klingele AJ, Liles MR, Carrias A, Mead DA, Schoenfeld TW. Front Microbiol. 2014 doi: 10.3389/fmicb.2014.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Asiello PJ, Baeumner AJ. Lab on a Chip. 2011;11:1420–1430. doi: 10.1039/c0lc00666a. [DOI] [PubMed] [Google Scholar]
- 71.Curtis KA, Rudolph DL, Owen SM. Journal of virological methods. 2008;151:264–270. doi: 10.1016/j.jviromet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 72.Rudolph DL, Sullivan V, Owen SM, Curtis KA. PloS one. 2015;10:e0126609. doi: 10.1371/journal.pone.0126609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuiken C, Yoon H, Abfalterer W, Gaschen B, Lo C, Korber B. Data Mining for Systems Biology. Methods and Protocols. 2013:253–261. doi: 10.1007/978-1-62703-107-3_16. [DOI] [PubMed] [Google Scholar]
- 74.Manak M, Sina S, Anekella B, Hewlett I, Sanders-Buell E, Ragupathy V, Kim J, Vermeulen M, Stramer SL, Sabino E, Grabarczyk P. AIDS research and human retroviruses. 2012;28:594–606. doi: 10.1089/aid.2011.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Curtis KA, Rudolph DL, Owen SM. Journal of medical virology. 2009;81:966–972. doi: 10.1002/jmv.21490. [DOI] [PubMed] [Google Scholar]
- 76.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Aids. 2006;20:W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 77.Chittiprol S, Kumar AM, Shetty KT, Kumar HR, Satishchandra P, Rao RB, Ravi V, Desai A, Subbakrishna DK, Philip M, Satish KS. Clinica Chimica Acta. 2009;409:4–10. doi: 10.1016/j.cca.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ocwieja KE, Sherrill-Mix S, Liu C, Song J, Bau H, Bushman FD. PloS one. 2015;10:e0117852. doi: 10.1371/journal.pone.0117852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zeng Y, Zhang X, Nie K, Ding X, Ring BZ, Xu L, Dai L, Li X, Ren W, Shi L, Ma X. Gene. 2014;541:123–128. doi: 10.1016/j.gene.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 80.Curtis KA, Niedzwiedz PL, Youngpairoj AS, Rudolph DL, Owen SM. Journal of clinical microbiology. 2014;52:2674–2676. doi: 10.1128/JCM.00935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hosaka N, Ndembi N, Ishizaki A, Kageyama S, Numazaki K, Ichimura H. Journal of virological methods. 2009;157:195–199. doi: 10.1016/j.jviromet.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. [Accessed 15 September 2016];TwistAmp® exo, real-time fluorescent detection kit. http://store.twistdx.co.uk/twistdx/twistamp-exo/real-time-fluorescent-dna-detection/prod_4.html.
- 83.Euler M, Wang Y, Nentwich O, Piepenburg O, Hufert FT, Weidmann M. Journal of clinical virology. 2012;54:308–312. doi: 10.1016/j.jcv.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 84.Boyle DS, Lehman DA, Lillis L, Peterson D, Singhal M, Armes N, Parker M, Piepenburg O, Overbaugh J. MBio. 2013;4:e00135–13. doi: 10.1128/mBio.00135-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rohrman BA, Richards-Kortum RR. Lab on a chip. 2012;12:3082–3088. doi: 10.1039/c2lc40423k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crannell ZA, Rohrman B, Richards-Kortum R. PloS one. 2014;9:e112146. doi: 10.1371/journal.pone.0112146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lillis L, Lehman DA, Siverson JB, Weis J, Cantera J, Parker M, Piepenburg O, Overbaugh J, Boyle DS. Journal of virological methods. 2016;230:28–35. doi: 10.1016/j.jviromet.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tang W, Chow WHA, Ying L, Kong H, Tang YW, Lemieux B. Journal of Infectious Diseases. 2010;201:S46–S51. doi: 10.1086/650388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jordan JA, Ibe CO, Moore MS, Host C, Simon GL. Journal of Clinical Virology. 2012;54:11–14. doi: 10.1016/j.jcv.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 90.Liu C, Sadik MM, Mauk MG, Edelstein PH, Bushman FD, Gross R, Bau HH. Scientific reports. 2014;4:7335. doi: 10.1038/srep07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kubota R, Labarre P, Weigl BH, Li Y, Haydock P, Jenkins DM. Chinese Science Bulletin. 2013;58:1162–1168. doi: 10.1007/s11434-012-5634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Singleton J, Osborn JL, Lillis L, Hawkins K, Guelig D, Price W, Johns R, Ebels K, Boyle D, Weigl B, LaBarre P. PloS one. 2014;9:e113693. doi: 10.1371/journal.pone.0113693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lillis L, Lehman D, Singhal MC, Cantera J, Singleton J, Labarre P, Toyama A, Piepenburg O, Parker M, Wood R, Overbaugh J. PLoS One. 2014;9:e108189. doi: 10.1371/journal.pone.0108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Damhorst GL, Duarte-Guevara C, Chen W, Ghonge T, Cunningham BT, Bashir R. Engineering. 2015;1:324–335. doi: 10.15302/J-ENG-2015072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liao SC, Peng J, Mauk MG, Awasthi S, Song J, Friedman H, Bau HH, Liu C. Chemical. 2016;229:232–238. doi: 10.1016/j.snb.2016.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gandelman OA, Church VL, Moore CA, Kiddle G, Carne CA, Parmar S, Jalal H, Tisi LC, Murray JA. PLoS One. 2010;5:e14155. doi: 10.1371/journal.pone.0014155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. [Accessed 5 December 2016];BART reporter system of ERBA Molecular for HIV detection. http://www.erbamdx.co.uk/clinical/hiv.html.
- 98.Toumazou C, Shepherd LM, Reed SC, Chen GI, Patel A, Garner DM, Wang CJA, Ou CP, Amin-Desai K, Athanasiou P, Bai H. Nature methods. 2013;10:641–646. doi: 10.1038/nmeth.2520. [DOI] [PubMed] [Google Scholar]
- 99.Mori Y, Kitao M, Tomita N, Notomi T. Journal of biochemical and biophysical methods. 2004;59:145–157. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 100.Chuang TL, Wei SC, Lee SY, Lin CW. Biosensors and Bioelectronics. 2012;32:89–95. doi: 10.1016/j.bios.2011.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu C, Qiu X, Ongagna S, Chen D, Chen Z, Abrams WR, Malamud D, Corstjens PL, Bau HH. Lab on a Chip. 2009;9:768–776. doi: 10.1039/b814322f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gaolathe T, Wirth KE, Holme MP, Makhema J, Moyo S, Chakalisa U, Yankinda EK, Lei Q, Mmalane M, Novitsky V, Okui L. The Lancet HIV. 2016;3:e221–e230. doi: 10.1016/S2352-3018(16)00037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roskos K, Hickerson AI, Lu HW, Ferguson TM, Shinde DN, Klaue Y, Niemz A. PLoS One. 2013;8:e69355. doi: 10.1371/journal.pone.0069355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nagatani N, Yamanaka K, Saito M, Koketsu R, Sasaki T, Ikuta K, Miyahara T, Tamiya E. Analyst. 2011;136:5143–5150. doi: 10.1039/c1an15638a. [DOI] [PubMed] [Google Scholar]
- 105.Connelly JT, Rolland JP, Whitesides GM. Analytical chemistry. 2015;87:7595–7601. doi: 10.1021/acs.analchem.5b00411. [DOI] [PubMed] [Google Scholar]
- 106.Song J, Mauk MG, Hackett BA, Cherry S, Bau HH, Liu C. Analytical Chemistry. 2016;88:7289–7294. doi: 10.1021/acs.analchem.6b01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Miyamoto S, Sano S, Takahashi K, Jikihara T. Analytical biochemistry. 2015;473:28–33. doi: 10.1016/j.ab.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 108.Tomita N, Mori Y, Kanda H, Notomi T. Nature protocols. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 109.Shen F, Sun B, Kreutz JE, Davydova EK, Du W, Reddy PL, Joseph LJ, Ismagilov RF. Journal of the American Chemical Society. 2011;133:7705–17712. doi: 10.1021/ja2060116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun B, Shen F, McCalla SE, Kreutz JE, Karymov MA, Ismagilov RF. Analytical chemistry. 2013;85:1540–1546. doi: 10.1021/ac3037206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rodriguez-Manzano J, Karymov MA, Begolo S, Selck DA, Zhukov DV, Jue E, Ismagilov RF. ACS nano. 2016;10:3102–3113. doi: 10.1021/acsnano.5b07338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lafleur LK, Bishop JD, Heiniger EK, Gallagher RP, Wheeler MD, Kauffman P, Zhang X, Kline EC, Buser JR, Kumar S, Byrnes SA. Lab on a Chip. 2016 doi: 10.1039/c6lc00677a. [DOI] [PubMed] [Google Scholar]
- 113.Yetisen AK, Akram MS, Lowe CR. Lab on a Chip. 2013;13:2210–2251. doi: 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- 114.Priye A, Wong S, Bi Y, Carpio M, Chang J, Coen M, Cope D, Harris J, Johnson J, Keller A, Lim R. Analytical chemistry. 2016;88:4651–4660. doi: 10.1021/acs.analchem.5b04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Laksanasopin T, Guo TW, Nayak S, Sridhara AA, Xie S, Olowookere OO, Cadinu P, Meng F, Chee NH, Kim J, Chin CD. Science translational medicine. 2015;7:273re1. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- 116.Martinez AW, Phillips ST, Carrilho E, Thomas SW, III, Sindi H, Whitesides GM. Analytical chemistry. 2008;80:3699–3707. doi: 10.1021/ac800112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lacombe K, Bottero J, Lemoine M, Boyd A, Girard PM. Journal of antimicrobial chemotherapy. 2009:dkp414. doi: 10.1093/jac/dkp414. [DOI] [PubMed] [Google Scholar]
- 118.Mahony J, Chong S, Bulir D, Ruyter A, Mwawasi K, Waltho D. Journal of Clinical Virology. 2013;58:127–131. doi: 10.1016/j.jcv.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 119.Liang C, Chu Y, Cheng S, Wu H, Kajiyama T, Kambara H, Zhou G. Analytical chemistry. 2012;84:3758–3763. doi: 10.1021/ac3003825. [DOI] [PubMed] [Google Scholar]
- 120.Crannell Z, Castellanos-Gonzalez A, Nair G, Mejia R, White AC, Richards-Kortum R. Analytical chemistry. 2016;88:1610–1616. doi: 10.1021/acs.analchem.5b03267. [DOI] [PubMed] [Google Scholar]
- 121.Song J, Liu C, Mauk MG, Rankin SC, Lok JB, Greenberg RM, Bau HH. Clinical Chemistry. doi: 10.1373/clinchem.2016.263665. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu C, Mauk MG, Hart R, Qiu X, Bau HH. Lab on a Chip. 2011;11:2686–2692. doi: 10.1039/c1lc20345b. [DOI] [PMC free article] [PubMed] [Google Scholar]